Abstract
The left ventricle (LV) responds to a myocardial infarction (MI) with an orchestrated sequence of events that results in fundamental changes to both the structure and function of the myocardium. This collection of responses is termed LV remodeling. Myocardial ischemia resulting in necrosis is the initiating event that culminates in the formation of an extracellular matrix (ECM)-rich infarct scar that replaces necrotic myocytes. While the cardiomyocyte is the major cell type that responds to ischemia, infiltrating leukocytes and cardiac fibroblasts coordinate the subsequent wound healing response. The matrix metalloproteinase (MMP) family of enzymes regulates the inflammatory and ECM responses that modulate scar formation. Matridomics is the proteomic evaluation focused on ECM, while degradomics is the proteomic evaluation of proteases as well as their inhibitors and substrates. This review will summarize the use of proteomics to better understand MMP roles in post-MI LV remodeling.
Keywords: matridomics, degradomics, matrix metalloproteinases, myocardial infarction, proteomics
Introduction
Following a myocardial infarction (MI), the left ventricle (LV) undergoes a series of events that substantially alters LV structure and function. This process is termed LV remodeling and occurs in three primary, but overlapping, phases.
The first phase starts immediately after MI and lasts for approximately three days. During this time, the infarct tissue expands resulting in LV chamber dilation, and the inflammatory response is initiated [1]. In the absence of reperfusion, neutrophils are the first inflammatory cells to infiltrate the necrotic myocardium and release reactive oxygen species and proteases. With reperfusion, all leukocyte types enter simultaneously [2].
During the second phase that occurs at 3–7 days post-MI, the LV continues to dilate and becomes spherical, and there is a reduction in ejection fraction and an increase in myocardial strain (Figure 1). Necrotic cardiomyocytes in the infarct region are removed while viable myocytes in the peri-infarct region undergo compensatory hypertrophy. Macrophage infiltration peaks to remove necrotic myocytes and apoptotic neutrophils, as well as activate cardiac fibroblasts that secrete extracellular matrix (ECM) for infarct scar formation [3]. The formation of the infarct scar results from a balance between ECM degradation and synthesis. Excessive ECM degradation by matrix metalloproteinases (MMPs) can lead to excessive thinning of the LV free wall with resultant aneurysm or rupture [4]. As a result, the LV is most vulnerable to rupture during this time period in both animal models of permanent artery occlusion and humans who are not successfully reperfused. Excessive ECM degradation can also disrupt cardiomyocyte alignment and impair contraction or electrical signaling [5]. Conversely, excessive ECM synthesis by fibroblasts can lead to a stiff and non-compliant LV, the development of diastolic dysfunction, and ultimately progression to heart failure. Therefore, successful wound healing post-MI relies on a balance between sufficient ECM degradation and synthesis.
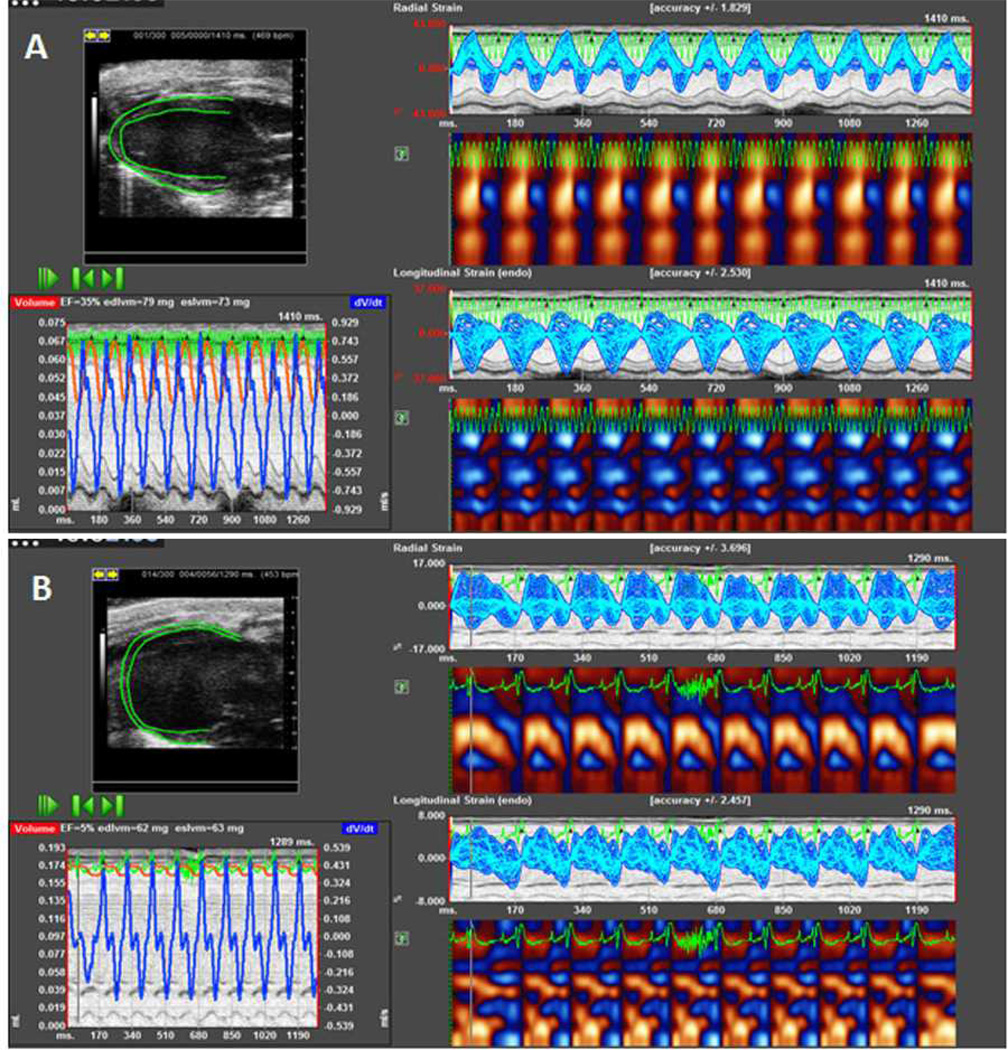
Figure 1.
A representative speckle tracking-based strain echocardiographic analysis of the left ventricle (LV) pre- and post-myocardial infarction (MI). A: Baseline and B: Day 7 post-MI. The post-MI image illustrates LV dilation, reduced ejection faction, and decreased radial and longitudinal strains. Images were acquired with a Vevo 2100 (Visualsonics; our own unpublished data). Analysis was conducted using the VevoStrain™ software.
The third phase begins around day 7 post-MI and continues indefinitely. This phase involves the chronic LV remodeling response that occurs at a highly variable rate in both animal models and patients. The possible outcomes of this phase ranges from formation of minor scar tissue with no further progression of fibrosis and no residual symptoms to extensive adverse remodeling with resultant congestive heart failure [6].
Currently, over 90% of acute MI patients that present to the emergency department survive beyond 30 days, which primarily reflects the benefit of reperfusion strategies [7–9]. However, 20–45% of MI survivors will subsequently develop adverse LV remodeling and heart failure despite currently available therapies (i.e., angiotensin converting enzyme inhibitors, angiotensin receptor inhibitors, statins, and beta-adrenergic blockers) [10–12]. Because of this limitation in current therapeutic options, novel strategies are needed to diagnose and treat patients who are at risk for progression to heart failure [8, 13–15].
Identifying ECM changes that regulate the physiological response to MI is essential to understanding LV remodeling [16–18]. Matridomics provides a global, integrated view of the ECM network at the protein level, and degradomics is the proteomic evaluation of MMPs, their inhibitors, and substrates. These approaches provide promising avenues to elucidate mechanisms and identify therapies to limit adverse post-MI LV remodeling [19].
Strategies to focus on ECM
Matridomics is defined as the proteomic evaluation of all the components of the ECM present in a tissue at the time of evaluation. This approach examines multiple ECM proteins in a high throughput way, which has several advantages over examining ECM at the transcriptional level or the individual ECM protein level. For one, mRNA levels do not always correlate with protein levels. Further, mRNA levels do not provide information on protein quality, and this is especially true for the highly post-translationally modified ECM. In addition, a matridomics approach provides direct information about ECM proteins, including quantity and quality (e.g., presence of post-translational modifications) [19]. Post-translational modifications can dramatically alter the signaling transduction networks that link intercellular and extracellular communication [20, 21]. Post-translational modifications relevant to the ECM include glycosylation, citrullination, and proteolytic processing [20]. The latter is of especial importance for MMP regulation of ECM.
Strategies to study ECM proteins include evaluation of ECM secreted from isolated cells (secretome) or within a tissue (matridome; Figure 2). Using reverse phase liquid chromatography coupled to mass spectrometry, Stastna et al. identified 83 unique proteins present in media obtained from cultured rat cardiac stem cells compared to cultured neonatal rat ventricular myocytes. Atrial natriuretic protein and connective tissue growth factor were found to be derived from myocytes, while interleukin-1 receptor-like 1 protein (ST2) was found to be derived from cardiac stem cells [22]. Stable isotope labeled amino acids in cell culture (SILAC) labeling has also been used to quantify the secretome of transforming growth factor- β (TGFβ) signaling-deficient mammary fibroblasts. Over 1000 proteins were identified in the conditioned media as being differentially expressed between fibroblasts with or without an intact TGFβ receptor II, including colony stimulating factor-1, TIMP-2, and TIMP-3 [23].
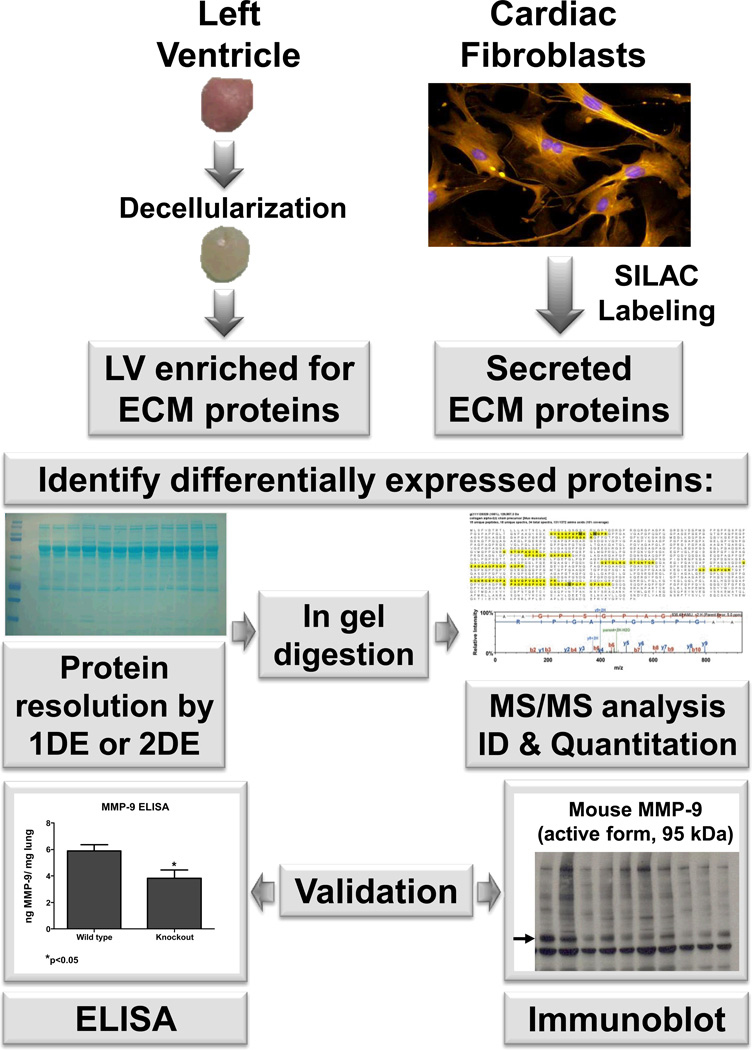
Figure 2.
A representative experimental design for a matridomics study. Two strategies typically used to identify ECM proteins differentially expressed are: 1) decellularization of the tissue to focus in on the extracellular matrix environment; or 2) SILAC labeling to examine the fibroblast secretome. SILAC- stable isotope labeling by amino acids in cell culture; ECM- extracellular matrix; ELISA- enzyme-linked immunosorbent assay; LV- left ventricle; and TIMP- tissue inhibitor of metalloproteinases.
Compared to isolated cell studies, analyzing ECM proteins in complex tissue is at least a magnitude more difficult. Secreted proteins can be collected from serum-free conditioned cell media to separate out from cellular proteins. However, ECM proteins within tissue surround cells in a highly organized and cross-linked scaffold that complicates their analysis at several levels. Many ECM proteins are large, difficult to solubilize, and undergo extensive post-translational modifications. In addition, multiple cell types within the myocardium contribute a large number of intracellular proteins that interfere with the analysis of ECM proteins. These cell types include myocytes, endothelial cells, vascular smooth muscle cells, fibroblasts, infiltrating leukocytes, and myofibroblasts. Because of the high cellular content, intracellular proteins should be removed to enrich the ECM fraction. One approach is to decellularize the tissue sample to enrich for ECM [24, 25]. Several versions of this technique have been developed, with the main difference being that sodium dodecyl sulfate (SDS) with or without Triton X-100 was used to fragment cell and organelle membranes. Once the cellular constituents are solubilized, they can be removed from the sample.
The Mayr laboratory has utilized the decellularization approach to examine both vascular and cardiac ECM [26–28]. They used a multi-step extraction approach that sequentially enriched for ECM proteins. The first extraction step used 0.5 M NaCl to extract highly soluble proteins. The second extraction step used 0.08% SDS to remove cellular components. The final extraction used 4 M guanidine HCl to solubilize the decellularized ECM. Using this approach, they identified 103 ECM proteins in human aortas and 125 ECM proteins in human abdominal aortic aneurysms. The Mayr laboratory also examined the left ventricles of pigs and humans that had been reperfused [19, 27]. A total of 139 ECM proteins were identified in decellularized porcine LVs that had been exposed to 2 h of ischemia and 15 or 60 days of reperfusion. For 15 of the proteins, this was the first report linking them to cardiac ECM. In addition, several of the newly identified cardiac ECM proteins have been previously linked to cartilage homeostasis. A major strength of this study was that both the border and infarct regions were analyzed, which provided spatial and temporal information on ECM scar composition changes in response to reperfusion. Combining the protein signatures of acute and chronic remodeling stages with an analysis of protein network interactions, the investigators identified transforming growth factor β1 as a pivotal regulator of ECM remodeling in the setting of ischemia and reperfusion. The study also identified ECM proteins which are known to play a role in cardiac remodeling such as cartilage intermediate layer protein 1, matrilin-4, extracellular adipocyte enhancer binding protein 1, collagen α-1 (XIV). As evidenced from the results described above, using a matridomic approach provides an unbiased method to focus in on ECM changes that occur during LV remodeling.
Degradomics
Degradomics is broadly defined as the characterization of all proteases, inhibitors, and substrates in a tissue at the time of evaluation [29]. In humans, there are more than 500 proteases, 150 protease inhibitors, and hundreds of identified substrates and interactors [30]. Time and tissue specificity limits the number of proteases present at evaluation, as not every protease is present in all tissues at any one given time Degradomics is a shotgun approach that can be used with both label and label-free mass spectrometry. By design, shotgun proteomic approaches are unbiased; our means to analyze and interpret the results, however, are still confined by reductionist concepts. Because of the high complexity of results obtained, coupling degradomics datasets with sophisticated bioinformatics is necessary to fully appreciate the rich supply of proteomic information.
Some techniques that have been used to screen for MMP substrates include substrate phage display, proteomic identification of protease cleavage sites, and combinatorial peptide libraries [31]. Protein topography and migration analysis platform (PROTOMAP) is a technique that couples one dimensional electrophoresis with mass spectrometry to directly map cleavage sites and identify substrates. This approach has been used to identify caspase substrates and would likely be applicable for MMP substrates. More recently, N-terminomics has been used to search for MMP substrates [29]. This approach isolates proteolytically generated N-termini to simultaneously identify substrates and cleavage sites in a single experiment. Four specific approaches that use N-terminomics include terminal amine isotopic labeling of substrates (TAILS), combined fractional diagonal chromatography (COFRADIC), acetylation of N-termini, and selective biotinylation of unblocked N-terminal α-amines chemically or by subtiligase [32–37].
Using the above approaches, different comparison groups have been used to identify protease substrates. In vivo, most experimental designs use wild type vs. null or transgenic mice. In vitro, isolating cells from these mice or using inactive catalytic domains to capture substrates has frequently been used [31]. Because MMP cleavage of the substrate is a temporary event, several groups have used exosite scanning techniques to determine MMP binding partners. Exosites are domains ancillary to the catalytic domain that mediate interactions and facilitate substrate binding to modulate affinity, efficiency, and sequence specificity [30]. MMP exosites include the collagen binding domain and the hemopexin domain [38]. Because protease-substrate interactions are transient and difficult to analyze kinetically, using an exosite approach takes advantage of the fact that the binding is more stable, particularly when used in the absence of a catalytic domain or with a mutated catalytic domain [31]. Recombinant exosites can also be used as competitive inhibitors to find substrates. Exosite scanning has been used with a quantitative proteomic approach to identify monocyte chemotactic protein-3 as a MMP-2 substrate [39, 40].
The identification of a broad substrate list has allowed the development of predicted cleavage site consensus sequences, and bioinformatics can be used to search for candidate substrates. Figure 3 shows confirmed and candidate substrates of MMP-9 as an example of the coupling of these approaches to obtain a molecular network for a particular MMP. Candidate substrates that are derived from in silico approaches need to be biochemically confirmed using in vitro and in vivo assays. Further, identifying whether a protein is a MMP substrate is much easier than identifying where the substrate is cleaved. While knowing that a protein is a substrate provides mechanistic insight, knowledge of the exact cleavage site(s) is necessary to understand the functional consequences of the cleavage. For some proteins, cleavage results in activation while for other proteins it results in inactivation. Only a small proportion of candidate substrates have been validated in vitro, fewer have been validated in vivo, and fewer still have been mapped to identify the cleavage site(s). The MEROPS database (merops.sanger.uk) has been developed, which provides integrated information on proteolytic enzymes, their substrates, and inhibitors along with a collection of known cleavage sites.
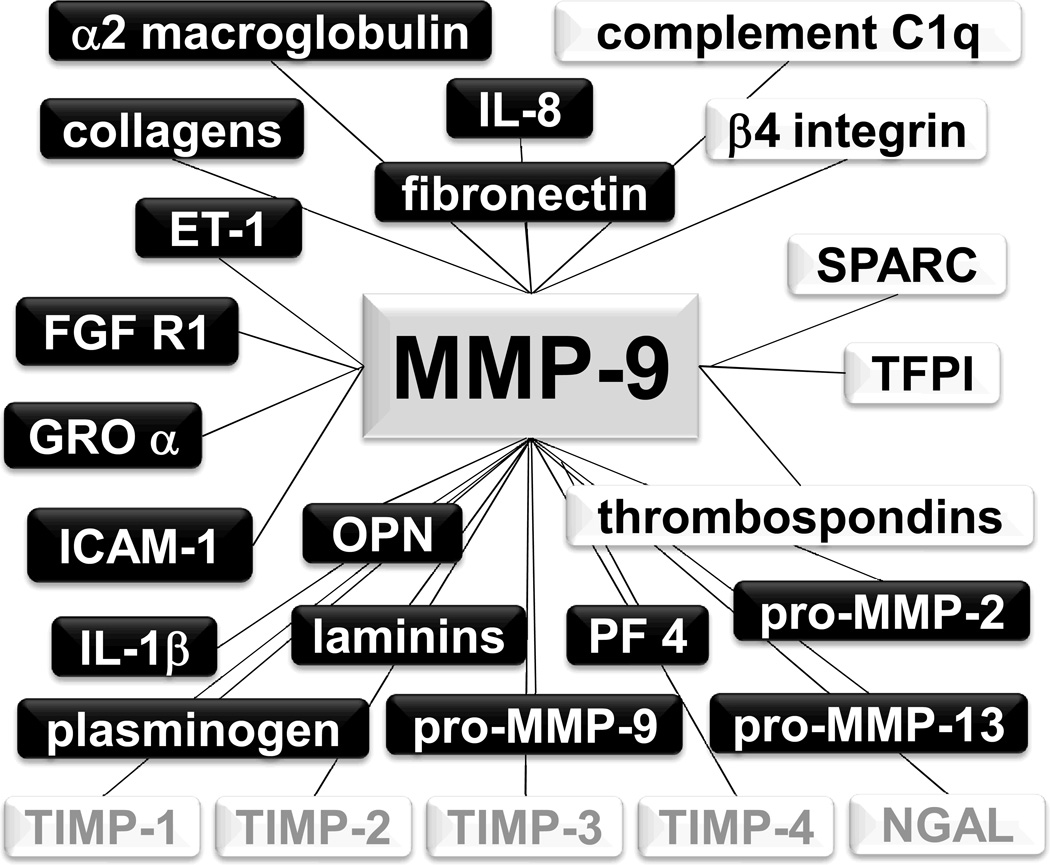
Figure 3.
A selection of the MMP-9 molecular interaction network. Known substrates are shown in the black boxes, while candidate substrates are shown in the white boxes. Factors that bind to MMP-9, but are not substrates, are shown in gray. IL-8- interleukin-8; SPARC- secreted protein acidic and rich in cysteine; TFPI- tissue factor pathway inhibitor; PF 4- platelet factor 4; IL-1β- interleukin-1β; ICAM-1- intercellular adhesion molecule-1; OPN- osteopontin; GRO α- growth related oncogene alpha; FGF R1- fibroblast growth factor receptor 1; ET-1- endothelin-1; and NGAL- neutrophil gelatinase-associated lipocalin.
The net function of an MMP is defined by its substrate repertoire. The main factors that determine if an MMP will have a beneficial or detrimental consequence are the MMP source, location, and time of induction. Therefore, once the substrate list is developed, it will be important to rank substrates based on both preference and importance. In vitro cleavage of a substrate by MMP does not mean that the substrate is preferred in vivo, and this is a major difference between traditional biochemical approaches and proteomics techniques. When unbiased screens are used to identify novel MMP substrates, fewer than 20% of substrates identified are ECM. The low percentage suggests that signaling regulation is a key MMP function and many MMP substrates are non-ECM proteins, or that difficulties in resolving ECM are responsible for the low percentage of ECM substrates observed (or maybe both) [41]. The former indicates an important role for MMPs in chemokine processing; and more than 35 chemokines are known to be regulated by MMPs [42]. Quantitative proteomics can be used with competitive assays to see which substrate(s) in a complex mixture are preferred. Developing a hierarchy of preference will be essential to understanding the net biological consequence of MMP activity. In the setting of pathology, the background proteolysis (which occurs to maintain system homeostasis) may need to be subtracted if it contributes noise that makes interpretation difficult. Finally, studies to determine which MMPs cleave a particular substrate and if there is a hierarchical preference are needed. If several MMPs process the same substrate, but at different cleavage sites, the differences in the cleavage fragments produced may result in diverse downstream effects. This highlights the strong need to identify MMP substrates from in vivo samples, which are the most biologically relevant.
Using Degradomics to Develop Better MMP Inhibitors
In clinical cardiovascular studies, MMP inhibitors have not proven efficacious for multiple reasons, including trial design, patient selection, inadequate (or nonspecific) dosing issues, and an incomplete picture of MMP biology [43]. One important concept brought out from degradomic studies is that MMPs proteolyze substrates that have a deleterious role in remodeling as well as those that have a beneficial role. Global, nonspecific inhibition strategies have not worked; and strategies that only focus on inhibiting one MMP may not work either. Rather, strategies that inhibit the upstream activators or the downstream substrates may prove more useful. Inhibiting MMPs by targeting upstream pathways will only work if this is a “leaky strategy,” since both the positive and negative effects will be blocked if all upstream signaling is inhibited. A more fruitful approach may be to target the downstream substrates. In order for this to be viable, the most biologically important substrates need to be identified, as the most obvious ones may not be the most important.
Pharmacoproteomics is the global analysis of the effects of a drug on the system assessed, using proteomic techniques to map effects [38]. This approach can be used as a high throughput method to screen for candidate drugs or to refine inhibition strategies if the critical positive and negative substrates are known. Degradomic approaches highlight the complexity of the protease network, which has been described as a web, with interconnections among the protease families [29]. Understanding the interconnectivity and dependence of components and mapping these effects will help to increase the efficiency of pre-clinical inhibitor evaluations and may help to limit severe side effects that have been observed during early clinical development. For example, factors that exacerbate adverse remodeling need to be identified and compared to those that are protective, such as factors that resolve the inflammatory response. Using the protease web to identify critical intersections where protease pathways cross to affect these factors will provide mechanistic insight and help to identify therapeutic targets. The complexity of the interclass connections also highlight that interpretations for MMP null studies often do not consider both the direct and indirect effects of the gene deletion.
ECM Proteins Involved in LV Remodeling
It is estimated that approximately 140 different protein components make up the ECM [29], and several ECM components are known to be involved in post-MI LV remodeling. ECM proteins include those that provide structure (e.g., collagens, fibronectin, and laminin) as well as those that provide support roles. The latter includes matricellular proteins (e.g. secreted protein acidic and rich-in-cysteine (SPARC) and thrombospondin-1 (TSP-1)), as well as the MMPs. Matricellular proteins are a group of ECM proteins that do not play a direct role in formation of structural elements but indirectly regulate cell-matrix interactions [44]. We will briefly summarize below the known roles for each of these components in the post-MI remodeling response, to describe how ECM proteomics can be used to understand remodeling.
Collagens
Collagen I is the most abundant collagen type in the normal adult myocardium, and collagen degradation is robust during the first and second phases of post-MI remodeling. In a rat MI model, Weber et al showed that collagenolytic activity increased on the second day post-MI and remained elevated through day 7 [45]. Cannon and colleagues reported significant collagen degradation in the rat on the first day after coronary artery ligation [46], and Villareal and colleagues reported collagen I degradation beginning 15–30 minutes post occlusion and continuing for up to 48 hours post-MI [47]. There are multiple collagen subtypes, and the major collagens synthesized in the post-MI infarct region are collagens I and III. Collagen I and III mRNA levels increase at day 2 and remain elevated for at least 21 days post-MI [48]. Other collagens known to increase post-MI include types IV, V, and VI. Interestingly, collagen VI alpha 1 null mice show reduced LV dilation and collagen deposition compared to wild type at 8 weeks post-MI [49]. Collagen VI has also been shown to induce myofibroblast differentiation post-MI [50].
There are several additional collagen types in the heart, including collagen type 15 (Col XV or Col15a1) and collagen type 18 (Col XVIII or Col18a1). Mice lacking Col XV demonstrated irregularly organized ECM matrix [51]. Of interest, both Col XV and Col XVIII can be proteolytically processed to generate fragments. Due to high homology, two Col XV fragments (restin and the C terminal fragment NC1) form similar fragments from Col XVIII (termed endostatin and NC1, respectively) [52, 53]. These fragments are generally anti-angiogenic, although their roles can vary depending on whether the fragment is soluble or immobilized.
Fibronectin and Laminin
The glycoproteins fibronectin and laminin are normally expressed at high levels during development, at low levels in the adult, and are robustly re-expressed post-MI [54, 55]. Fibronectin is present in two forms: a soluble circulating form and an insoluble extracellular matrix form [56]. In the normal myocardium, fibronectin is localized to the basement membrane that surrounds endothelial and smooth muscle cells [57]. Fibronectin coordinates multiple cellular processes, including adhesion and migration as well as growth and differentiation [58]. For example, the adherence of fibrillar collagen to cardiac myocytes is mediated through adhesion to fibronectin [59]. George and colleagues demonstrated the importance of fibronectin for early embryonic development, as fibronectin null mice are embryonic lethal and show notable cardiac defects [60].
Fibronectin levels robustly increase early post-MI. Knowlton and colleagues demonstrated a 13-fold increase in fibronectin mRNA in the rabbit heart at day 1 post-MI [61]. Mice lacking the extra domain-A (EDA domain) of fibronectin showed decreased mortality, better systolic function, and less LV dilatation at 7 days post-MI compared to wild-type mice [62]. Fibronectin has also been shown to influence monocyte migration into the infarcted myocardium, by binding to the VLA-5 receptor [63]. The 120 kDa fibronectin fragment stimulates tissue-infiltrating macrophages into the damaged myocardium, which in turn prevents apoptotic death of viable cardiac myocytes [64].
Similar to fibronectin, laminin also coordinates cell adhesion, migration, growth, and differentiation [65]. Laminins are a family of basement membrane proteins that naturally exist as heterotrimeric polypeptides. Currently, 16 laminin trimer groups have been identified [66]. Several groups report increased laminin levels post-MI, including increases in laminins α2, α4, α5, β1, β2, and γ1 [67, 68]. A study in rat ventricles showed the presence of laminin in the basal membranes of cardiac myocytes using confocal microscopy [69]. Another study in cat showed that freshly isolated adult cardiac myocytes readily attach to laminin via β1 integrin receptors [70]. The role of laminin post-MI, however, has not been evaluated by null or overexpression strategies.
In the post-MI heart, degradation of both fibronectin and laminin occur in wild type control mice, and this was attenuated in MMP-2 null mice to indicate a direct or indirect role for MMP-2 in the cleavage of these particular ECM proteins. Cleavage of fibronectin and laminin was associated with increased macrophage infiltration into the infarcted area [71]. Although the exact sequences have not been mapped, the cleavage of both proteins plays an integral role in the post-MI inflammatory response. Interestingly, the proteolytic processing of fibronectin and laminin is not likely to be MMP-2 selective, as MMP-9 has also been shown to cleave these proteins in vitro [72, 73]. The degradomic approaches described above may provide useful information on the processing of these ECM proteins in the post-MI setting.
SPARC and TSP-1
Several matricellular proteins are also present in the post-MI heart. Matricellular proteins are ECM proteins that modulate cell function by serving as accessory ECM proteins. However, unlike fibrillar ECM proteins, matricellular proteins do not contribute directly to fibril and basil laminae organization in the heart [44].
Two noteworthy matricellular proteins in the post-MI LV are SPARC and TSP-1, both of which are secreted by fibroblasts. SPARC, also known as osteonectin, is an extracellular Ca2+ binding protein expressed at high levels in post-MI LV [74]. SPARC mRNA and protein levels are significantly increased in the LV at days 2–14 post-MI [75], [76]. Reed and colleagues showed a positive correlation between SPARC expression and the level of inflammatory response, particularly an increase in leukocyte infiltration that began at day 2 post-MI [77].
The absence of SPARC correlates with improved early LV function at day 3 post-MI but increased mortality by day 14 post-MI [78], due to a significant increase in the incidence of cardiac rupture in SPARC null mice [27].Consistent with this finding, fibroblast activation was blunted in the nulls at day 3 post-MI. Mice with adenoviral overexpression of SPARC showed improved collagen maturation and decreased cardiac dilatation and dysfunction post-MI when compared to wild type [27].
SPARC effects on LV structure and function in the post-MI setting are likely due in part to its interactions with collagen. Collagens I, II, and III contain major binding sites for SPARC [79]. In addition, collagen levels in the conditioned media of SPARC null fibroblasts are decreased compared to WT fibroblasts [80]. Similarly, decreased collagen I secretion has been seen in SPARC-null mesangial cells [81]. Therefore, SPARC may play an important role in formation of scar tissue post-MI by regulating the inflammatory response, fibroblast activation, and collagen assembly into the scar.
The thrombospondins are a group of five secreted Ca++-binding glycoproteins. TSPs -1 and -2 exist in a trimeric form, while TSPs -3, -4, and -5 exist as pentamers [82]. One of the major roles of TSP is to interact with membrane proteins, such as integrins and proteoglycans, to regulate cell-ECM signaling and alter cell migration and adhesion [83, 84]. In the post-MI rat myocardium, TSP-1 increases and is expressed by fibroblasts and macrophages [85]. In TSP-1 null mice, macrophage and myofibroblast infiltration increases in the infarct area at day 3 post-MI compared to wild type controls. In addition, TSP-1 null mice demonstrate increased LV end diastolic volume post-MI, indicating that TSP-1 deletion can affect global LV function [86]. TSP-1 has also been shown to activate TGF-β1, further implicating TSP-1 in the post-MI inflammatory and fibrotic responses [87].
Both SPARC and TSP-1 interact with MMPs. SPARC upregulates membrane type MMP-1, as well as MMP-2 [88]. TSP-1 upregulates MMP-9 expression in breast cancer and gastric cancer tissue, as well as in endothelial cells [89, 90]. In vascular smooth muscle cells, TSP-1 increased MMP-2 activity [89]. However, the role of TSP-1 in directly regulating MMPs has not been resolved and likely involves biphasic effects. In addition, both SPARC and TSP-1 have been shown to be MMP substrates by in vitro or in silico approaches [91].
Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases (TIMPS)
Matrix metalloproteinases (MMPs) are proteolytic, zinc-dependent enzymes responsible for turnover of ECM and non-ECM substrates [6]. The MMP family is currently composed of 25 proteinases loosely categorized into five groups based on in vitro substrate preferences or localization: collagenases, gelatinases, stromelysins, matrilysins, and membrane-type MMPs [6]. The majority of MMPs are secreted as an inactive pro-MMP and are later activated by a cysteine switch mechanism that releases the pro-domain from the catalytic site [6]. The membrane type MMPs (MT-MMPs), MMP-11, and MMP-28 are exceptions as each are intracellularly activated by furin [92]. Four TIMPs have been identified to date.
MMPs and TIMPs are involved in both the inflammatory and reparative responses to MI [93]. Every inflammatory cell type expresses at least one MMP and TIMP, and MMP activation can be observed in the LV within 15 minutes of reperfusion [93, 94]. The inflammatory phase not only involves the degradation of existing ECM by MMPs, but also involves MMP processing of cytokines and chemokines along with growth factors, all of which coordinate the wound healing response [93]. In addition to inhibiting MMP, TIMPs also effect cell proliferation and apoptosis [95, 96]. Of the 25 MMPs and 4 TIMPs presently identified, MMPs -1, -2, -3, -7, -8, -9, -12, -13, -14 and TIMPs-1-4 have been evaluated post-MI (Table 1 and Table 2) [97]. With the exception of MMP-2, the expression levels of the other MMPs are low in the normal myocardium and are robustly increased post-MI in both temporal and spatial specific ways [6, 97]. Endogenous MMP inhibitors are listed in Table 3. MMP and TIMP roles, post-MI, have been reviewed previously [97, 98].
Table 1.
Matrix metalloproteinase (MMP) levels post-MI.
| MMP | Post-MI Levels |
|---|---|
| MMP-1 | ↑ from days 3–7 post-MI [6, 50, 100] |
| MMP-2 | ↑ from day 4, peaks at day 7 and ↓ to pre-MI levels by day 14 post-MI [6] |
| MMP-3 | ↑ 48 hours post-MI, peaks by day 4 [6, 46, 49] |
| MMP-7 | ↑ in 1st week post-MI and ↓ to pre-MI levels by 8 weeks post-MI [6] |
| MMP-8 | ↑ at 2 weeks post-MI and stays elevated [6, 61, 77] |
| MMP-9 | ↑ in first week post-MI, reduces to control levels by 14 days post-MI [6] |
| MMP-13 | ↑ in 72 hours and declines by 14 days post-MI [6, 76] |
| MMP-14 | ↑ in 3 days post-MI and further elevates by 16 days post-MI [6] |
Table 2.
Tissue inhibitor of metalloproteinase (TIMP) cardiac cell expression and levels post-MI
| TIMP | Cardiac Cell Expression | Post-MI levels |
|---|---|---|
| TIMP-1 | cardiac myocytes, leukocytes, and fibroblasts [97] | Protein levels ↑ 3 d post-MI in the infarct region of mice; mRNA ↑ 6 h post-MI and ↓ after 2 d in the infarct region of rats [97, 101] |
| TIMP-2 | cardiac fibroblasts [97] | Significant ↓ in protein levels at 3 d and 1 w post-MI in mice; no change observed in first week post MI but ↑ 2 and 16 w post-MI in rats [97, 102] |
| TIMP-3 | cardiac fibroblasts [97] | ↓ protein levels at 3 d and 1 w post-MI in mice; significantly low levels in the infarct regions of sheep at 8 weeks post-MI [97, 101] |
| TIMP-4 | cardiac myocytes and cardiac fibroblasts [97] [103] | ↓ protein levels at 1 w post-MI in mice; ↓protein at 1 and 8 w post-MI in rats [97, 101] |
Table 3.
Endogenous MMP Inhibitors.
Post-MI LV remodeling involves the dynamic interaction between the ECM and the MMPs that break down ECM components; the relationship between MMPs and the endogenous TIMPs that block MMP activity; and the interconnection between cytokines and growth factors, ECM components, and MMPs [99]. Therefore, consideration of MMP effects is crucial for successful therapeutic approaches for MI. Table 1 summarizes the current literature with regard to MMP and TIMP levels post-MI
Conclusions
Matridomics and degradomics are emerging proteomic techniques that hold promise to drive the ECM remodeling field forward. However, before that success is achieved, several issues need to be resolved. Current ECM enrichment protocols do not likely solubilize the entire ECM, and an incomplete analysis of the ECM composition limits the full potential of this approach. More effective homogenizing buffers that can completely dissolve the ECM need to be evaluated for compatibility with downstream mass spectrometry approaches, whether used as a gel-free method or coupled to 1-DE analysis. As with all other proteomic approaches, consistent and highly reproducible sample preparation is a key step to both matridomic and degradomic strategies. Finally, systems biology approaches and close collaboration with bioinformaticians are needed for adequate data interpretation. Harnessing the complexity of the ECM environment will likely provide critical information that will allow us to develop novel therapeutic strategies to limit the progression of adverse LV remodeling, improve quality of life, decrease morbidity, and improve survival.
Acknowledgements
We acknowledge support from the Rapoport Foundation for Cardiovascular Research to RAL; to UTHSCSA CTSA to GA; and from NIH/NHLBI HHSN 268201000036C (N01-HV-00244) for the San Antonio Cardiovascular Proteomics Center and R01 HL075360, the Max and Minnie Tomerlin Voelcker Fund, and the Veteran’s Administration (Merit) to MLL.
Disclosures
M. L. Lindsey has received grant funding from Novartis, Amylin Pharmaceuticals, and Canopus Pharmaceuticals. All projects are unrelated to this article.
Abbreviations
- ECM
extracellular matrix
- LV
left ventricle
- MMP
matrix metalloproteinase
- MI
myocardial infarction
- SPARC
secreted protein, acidic and rich-in-cysteine
- TSP-1
thrombospondin-1
- TIMP
tissue inhibitor of metalloproteinases
- TGFβ
transforming growth factor- β
References
- 1.Tao Z-Y, Cavasin MA, Yang F, Liu Y-H, Yang X-P. Temporal changes in matrix metalloproteinase expression and inflammatory response associated with cardiac rupture after myocardial infarction in mice. Life sciences. 2004;74:1561–1572. doi: 10.1016/j.lfs.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 2.Bowers SL, Baudino TA. Laying the groundwork for growth: Cell-cell and cell-ECM interactions in cardiovascular development. Birth Defects Res C Embryo Today. 2010;90:1–7. doi: 10.1002/bdrc.20168. [DOI] [PubMed] [Google Scholar]
- 3.Baicu CF, Stroud JD, Livesay VA, Hapke E, et al. Changes in extracellular collagen matrix alter myocardial systolic performance. American journal of physiology. 2003;284:H122–H132. doi: 10.1152/ajpheart.00233.2002. [DOI] [PubMed] [Google Scholar]
- 4.Sam F, Sawyer DB, Chang L-F, Eberli FR, et al. Progressive left ventricular remodeling and apoptosis late after myocardial infarction in mouse heart. American journal of physiology. 2000;279:H422–H428. doi: 10.1152/ajpheart.2000.279.1.H422. [DOI] [PubMed] [Google Scholar]
- 5.Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 6.Vanhoutte D, Schellings M, Pinto Y, Heymans S. Relevance of matrix metalloproteinases and their inhibitors after myocardial infarction: A temporal and spatial window. Cardiovascular research. 2006;69:604–613. doi: 10.1016/j.cardiores.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Takemura G, Nakagawa M, Kanamori H, Minatoguchi S, Fujiwara H. Benefits of reperfusion beyond infarct size limitation. Cardiovascular research. 2009;83:269–276. doi: 10.1093/cvr/cvp032. [DOI] [PubMed] [Google Scholar]
- 8.Opie LH, Commerford PJ, Gersh BJ, Pfeffer MA. Controversies in ventricular remodelling. Lancet. 2006;367:356–367. doi: 10.1016/S0140-6736(06)68074-4. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa K, Kimura A, Taniwa T, Takenaka T, et al. Modification of treatment strategies over a period of 14 years has markedly reduced cardiac events among post-myocardial infarction patients. Circ J. 2002;66:881–885. doi: 10.1253/circj.66.881. [DOI] [PubMed] [Google Scholar]
- 10.Lewis EF, Velazquez EJ, Solomon SD, Hellkamp AS, et al. Predictors of the first heart failure hospitalization in patients who are stable survivors of myocardial infarction complicated by pulmonary congestion and/or left ventricular dysfunction: a VALIANT study. European heart journal. 2008;29:748–756. doi: 10.1093/eurheartj/ehn062. [DOI] [PubMed] [Google Scholar]
- 11.Hellermann JP, Goraya TY, Jacobsen SJ, Weston SA, et al. Incidence of heart failure after myocardial infarction: is it changing over time? Am J Epidemiol. 2003;157:1101–1107. doi: 10.1093/aje/kwg078. [DOI] [PubMed] [Google Scholar]
- 12.Lewis EF, Moye LA, Rouleau JL, Sacks FM, et al. Predictors of late development of heart failure in stable survivors of myocardial infarction: the CARE study. Journal of the American College of Cardiology. 2003;42:1446–1453. doi: 10.1016/s0735-1097(03)01057-x. [DOI] [PubMed] [Google Scholar]
- 13.Anavekar NS, McMurray JJV, Velazquez EJ, Solomon SD, et al. Relation between Renal Dysfunction and Cardiovascular Outcomes after Myocardial Infarction. The New England journal of medicine. 2004;351:1285–1295. doi: 10.1056/NEJMoa041365. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer MA. ACE Inhibitors in Acute Myocardial Infarction. Circulation. 1998;97:2192–2194. doi: 10.1161/01.cir.97.22.2192. [DOI] [PubMed] [Google Scholar]
- 15.Hu K, Gaudron P, Ertl G. Long-Term Effects of Beta-Adrenergic Blocking Agent Treatment on Hemodynamic Function and Left Ventricular Remodeling in Rats With Experimental Myocardial Infarction: Importance of Timing of Treatment and Infarct Size. J of American College of Cardiology. 1998;31:692–700. doi: 10.1016/s0735-1097(97)00527-5. [DOI] [PubMed] [Google Scholar]
- 16.Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circulation research. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindsey ML, Mann DL, Entman ML, Spinale FG. Extracellular matrix remodeling following myocardial injury. Annals of medicine. 2003;35:316–326. doi: 10.1080/07853890310001285. [DOI] [PubMed] [Google Scholar]
- 18.Spinale FG. Myocardial Matrix Remodeling and the Matrix Metalloproteinases: Influence on Cardiac Form and Function. Physiological reviews. 2007;87:1285–1342. doi: 10.1152/physrev.00012.2007. [DOI] [PubMed] [Google Scholar]
- 19.Deleon KY, de Castro Bras LE, Lange RA, Lindsey ML. Extracellular matrix proteomics in cardiac ischemia/reperfusion: the search is on. Circulation. 2012;125:746–748. doi: 10.1161/CIRCULATIONAHA.111.086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leeming DJ, Bay-Jensen AC, Vassiliadis E, Larsen MR, et al. Post-translational modifications of the extracellular matrix are key events in cancer progression: opportunities for biochemical marker development. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2011;16:193–205. doi: 10.3109/1354750X.2011.557440. [DOI] [PubMed] [Google Scholar]
- 21.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 22.Stastna M, Chimenti I, Marban E, Van Eyk JE. Identification and functionality of proteomes secreted by rat cardiac stem cells and neonatal cardiomyocytes. Proteomics. 2010;10:245–253. doi: 10.1002/pmic.200900515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu BJ, Yan W, Jovanovic B, An AQ, et al. Quantitative analysis of the secretome of TGF-beta signaling-deficient mammary fibroblasts. Proteomics. 2010;10:2458–2470. doi: 10.1002/pmic.200900701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeQuach JA, Mezzano V, Miglani A, Lange S, et al. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PloS one. 2010;5:e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singelyn JM, DeQuach JA, Seif-Naraghi SB, Littlefield RB, et al. Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials. 2009;30:5409–5416. doi: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Didangelos A, Yin X, Mandal K, Baumert M, et al. Proteomics characterization of extracellular space components in the human aorta. Molecular & cellular proteomics : MCP. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 28.Didangelos A, Yin X, Mandal K, Saje A, et al. Extracellular matrix composition and remodeling in human abdominal aortic aneurysms: a proteomics approach. Molecular & cellular proteomics : MCP. 2011;10:M111. doi: 10.1074/mcp.M111.008128. 008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler GS, Overall CM. Updated biological roles for matrix metalloproteinases and new "intracellular" substrates revealed by degradomics. Biochemistry. 2009;48:10830–10845. doi: 10.1021/bi901656f. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 31.Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Molecular & cellular proteomics : MCP. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Gevaert K, Impens F, Van Damme P, Ghesquiere B, et al. Applications of diagonal chromatography for proteome-wide characterization of protein modifications and activity-based analyses. The FEBS journal. 2007;274:6277–6289. doi: 10.1111/j.1742-4658.2007.06149.x. [DOI] [PubMed] [Google Scholar]
- 33.Staes A, Van Damme P, Helsens K, Demol H, et al. Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC) Proteomics. 2008;8:1362–1370. doi: 10.1002/pmic.200700950. [DOI] [PubMed] [Google Scholar]
- 34.McDonald L, Beynon RJ. Positional proteomics: preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nature protocols. 2006;1:1790–1798. doi: 10.1038/nprot.2006.317. [DOI] [PubMed] [Google Scholar]
- 35.McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nature methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- 36.Timmer JC, Enoksson M, Wildfang E, Zhu W, et al. Profiling constitutive proteolytic events in vivo. The Biochemical journal. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahrus S, Trinidad JC, Barkan DT, Sali A, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison CJ, Butler GS, Rodriguez D, Overall CM. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21:645–653. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 39.McQuibban GA, Gong JH, Tam EM, McCulloch CA, et al. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 40.Overall CM, McQuibban GA, Clark-Lewis I. Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biological chemistry. 2002;383:1059–1066. doi: 10.1515/BC.2002.114. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez D, Morrison CJ, Overall CM. Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochimica et biophysica acta. 2010;1803:39–54. doi: 10.1016/j.bbamcr.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 42.Cox JH, O C. Cytokine Substrates: MMP Regulation of Inflammatory Signaling Molecules. 2008:519–539. [Google Scholar]
- 43.Peterson JT. The importance of estimating the therapeutic index in the development of matrix metalloproteinase inhibitors. Cardiovascular research. 2006;69:677–687. doi: 10.1016/j.cardiores.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 44.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Current Opinion in Cell Biology. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 45.Cleutjens JPM, Kandala JC, Guarda E, Guntaka RV, Weber KT. Regulation of Collagen Degradation in the Rat Myocardium after Infarction. Journal of molecular and cellular cardiology. 1995;27:1281–1292. doi: 10.1016/s0022-2828(05)82390-9. [DOI] [PubMed] [Google Scholar]
- 46.Cannon RO, III, Butany JW, McManus BM, Speir E, et al. Early Degradation of Collagen After Actue Myocardial Infarction in the Rat. The American journal of cardiology. 1983;52:390–395. doi: 10.1016/0002-9149(83)90145-5. [DOI] [PubMed] [Google Scholar]
- 47.Villarreal F, Omens J, Dillmann W, Risteli J, et al. Early degradation and serum appearance of type I collagen fragments after myocardial infarction. Journal of molecular and cellular cardiology. 2004;36:597–601. doi: 10.1016/j.yjmcc.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Cleutjens J, Verluyten M, Smiths J, Daemen M. Collagen remodeling after myocardial infarction in the rat heart. The American journal of pathology. 1995;147:325–338. [PMC free article] [PubMed] [Google Scholar]
- 49.Luther DJ, Thodeti CK, Shamhart PE, Adapala RK, et al. Absence of Type VI Collagen Paradoxically Improves Cardiac Function, Structure, and Remodeling After Myocardial Infarction. Circulation research. 2012 doi: 10.1161/CIRCRESAHA.111.252734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naugle JE. Type VI collagen induces cardiac myofibroblast differentiation: implications for postinfarction remodeling. American Journal of Physiology: Heart and Circulatory Physiology. 2006;290:H323–H330. doi: 10.1152/ajpheart.00321.2005. [DOI] [PubMed] [Google Scholar]
- 51.Rasi K, Piuhola J, Czabanka M, Sormunen R, et al. Collagen XV is necessary for modeling of the extracellular matrix and its deficiency predisposes to cardiomyopathy. Circulation research. 2010;107:1241–1252. doi: 10.1161/CIRCRESAHA.110.222133. [DOI] [PubMed] [Google Scholar]
- 52.Nikolova A, Ablasser K, Wyler von Ballmoos MC, Poutias D, et al. Endogenous angiogenesis inhibitors prevent adaptive capillary growth in left ventricular pressure overload hypertrophy. The Annals of thoracic surgery. 2012;94:1509–1517. doi: 10.1016/j.athoracsur.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishihara T, Ochi M, Sugimoto K, Takahashi H, et al. Subcutaneous injection containing IL-3 and GM-CSF ameliorates stab wound-induced brain injury in rats. Experimental neurology. 2011;229:507–516. doi: 10.1016/j.expneurol.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Lal A, Veinot JP, Ganten D, Leenen FH. Prevention of cardiac remodeling after myocardial infarction in transgenic rats deficient in brain angiotensinogen. Journal of molecular and cellular cardiology. 2005;39:521–529. doi: 10.1016/j.yjmcc.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Oliviero P, Chassagne C, Salichon N, Corbier A, et al. Expression of laminin alpha2 chain during normal and pathological growth of myocardium in rat and human. Cardiovascular research. 2000;46:346–355. doi: 10.1016/s0008-6363(00)00034-1. [DOI] [PubMed] [Google Scholar]
- 56.Amrani DL, Falk MJ, Mosesson MW. Studies of fibronectin synthesized by cultured chick hepatocytes. Experimental cell research. 1985;160:171–183. doi: 10.1016/0014-4827(85)90246-0. [DOI] [PubMed] [Google Scholar]
- 57.Hein S, Schaper J. The extracellular matrix in normal and diseased myocardium. J Nucl Cardiol. 2001;8:188–196. doi: 10.1067/mnc.2001.113331. [DOI] [PubMed] [Google Scholar]
- 58.Hocking DC. Fibronectin matrix deposition and cell contractility: implications for airway remodeling in asthma. Chest. 2002;122:275S–278S. doi: 10.1378/chest.122.6_suppl.275s. [DOI] [PubMed] [Google Scholar]
- 59.Villarreal FJ, Dillmann WH. Cardiac hypertrophy-induced changes in mRNA levels for TGF-beta 1, fibronectin, and collagen. The American journal of physiology. 1992;262:H1861–H1866. doi: 10.1152/ajpheart.1992.262.6.H1861. [DOI] [PubMed] [Google Scholar]
- 60.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 61.Knowlton AA, Connelly CC, Romo GM, Mamuya W, et al. Rapid Expression of Fibronectin in the Rabbit Heart after Myocardial Infarction with and without Reperfusion. The Journal of clinical investigation. 1992;89:1060–1068. doi: 10.1172/JCI115685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arslan F, Smeets MB, Riem Vis PW, Karper JC, et al. Lack of fibronectin-EDA promotes survival and prevents adverse remodeling and heart function deterioration after myocardial infarction. Circulation research. 2011;108:582–592. doi: 10.1161/CIRCRESAHA.110.224428. [DOI] [PubMed] [Google Scholar]
- 63.Trial J, Baughn RE, Wygant JN, McIntyre BW, et al. Fibronectin fragments modulate monocyte VLA-5 expression and monocyte migration. The Journal of clinical investigation. 1999;104:419–430. doi: 10.1172/JCI4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trial J, Rossen RD, Rubio J, Knowlton AA. Inflammation and Ischemia: Macrophages Activated by Fibronectin Fragments Enhance the Survival of Injured Cardiac Myocytes. Experimental Biology and Medicine. 2004;229:538–545. doi: 10.1177/153537020422900612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tashiro K, Sephel GC, Weeks B, Sasaki M, et al. A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. J. Biol. Chem. 1989;264:16174–16182. [PubMed] [Google Scholar]
- 66.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, et al. A simplified laminin nomenclature. Matrix Biology. 2005;24:326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 67.Research, I. o. L. A., Sciences, C. o. L., Council, N. R. Guide for the Care and Use of Laboratory Animals. The National Academies Press; 1996. [PubMed] [Google Scholar]
- 68.Morishita N, Kusachi S, Yamasaki S, Kondo J, Tsuji T. Sequential changes in laminin and type IV collagen in the infarct zone--immunohistochemical study in rat myocardial infarction. Jpn Circ J. 1996;60:108–114. doi: 10.1253/jcj.60.108. [DOI] [PubMed] [Google Scholar]
- 69.S~etersdal T, Rotevatn TTLlS, Dalen H, Scheie P. Fibronectin and laminin in transverse tubules of cardiac myocytes studied by laser confocal microscopy and immunocytochemistry. Histochemistry. 1992;98:73–80. doi: 10.1007/BF00716997. [DOI] [PubMed] [Google Scholar]
- 70.Hill DR, Kessler SP, Rho HK, Cowman MK, de la Motte CA. Specific-sized hyaluronan fragments promote expression of human beta-defensin 2 in intestinal Epithelium. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.356238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsumura S-i, Iwanaga S, Mochizuki S, Okamoto H, et al. Targeted deletion or pharmacological inhibition of MMP-2 prevents cardiac rupture after myocardial infarction in mice. J. Clin. Invest. 2005;115:599–609. doi: 10.1172/JCI22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miles AJ, Skubitz AP, Furcht LT, Fields GB. Promotion of cell adhesion by single-stranded and triple-helical peptide models of basement membrane collagen alpha 1(IV)531–543. Evidence for conformationally dependent and conformationally independent type IV collagen cell adhesion sites. The Journal of biological chemistry. 1994;269:30939–30945. [PubMed] [Google Scholar]
- 73.Xu D, Suenaga N, Edelmann MJ, Fridman R, et al. Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol Cell Proteomics. 2008;7:2215–2228. doi: 10.1074/mcp.M800095-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sage H, Vernon RB, Decker J, Funk S, Iruela-Arispe ML. Distribution of the calcium-binding protein SPARC in tissues of embryonic and adult mice. J Histochem Cytochem. 1989;37:819–829. doi: 10.1177/37.6.2723400. [DOI] [PubMed] [Google Scholar]
- 75.Schellings MW, Vanhoutte D, Swinnen M, Cleutjens JP, et al. Absence of SPARC results in increased cardiac rupture and dysfunction after acute myocardial infarction. The Journal of experimental medicine. 2009;206:113–123. doi: 10.1084/jem.20081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Komatsubara I, Murakami T, Kusachi S, Nakamura K, et al. Spatially and temporally different expression of osteonectin and osteopontin in the infarct zone of experimentally induced myocardial infarction in rats. Cardiovascular Pathology. 2003;12:186–194. doi: 10.1016/s1054-8807(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 77.Reed MJ, Puolakkainen P, Lane TF, Dickerson D, et al. Differential expression of SPARC and thrombospondin 1 in wound repair: immunolocalization and in situ hybridization. J Histochem Cytochem. 1993;41:1467–1477. doi: 10.1177/41.10.8245406. [DOI] [PubMed] [Google Scholar]
- 78.McCurdy SM, Dai Q, Zhang J, Zamilpa R, et al. SPARC Mediates Early Extracellular Matrix Remodeling Following Myocardial Infarction. American journal of physiology. Heart and circulatory physiology. 2011 doi: 10.1152/ajpheart.01070.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giudici C, Raynal N, Wiedemann H, Cabral WA, et al. Mapping of SPARC/BM-40/Osteonectin-binding Sites on Fibrillar Collagens. The Journal of biological chemistry. 2008;283:19551–19560. doi: 10.1074/jbc.M710001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rentz TJ, Poobalarahi F, Bornstein P, Sage EH, Bradshaw AD. SPARC Regulates Processing of Procollagen I and Collagen Fibrillogenesis in Dermal Fibroblasts. J. Biol. Chem. 2007;282:22062–22071. doi: 10.1074/jbc.M700167200. [DOI] [PubMed] [Google Scholar]
- 81.Francki A, Bradshaw AD, Bassuk JA, Howe CC, et al. SPARC regulates the expression of collagen type I and transforming growth factor-beta1 in mesangial cells. The Journal of biological chemistry. 1999;274:32145–32152. doi: 10.1074/jbc.274.45.32145. [DOI] [PubMed] [Google Scholar]
- 82.Risher WC, Eroglu C. Thrombospondins as key regulators of synaptogenesis in the central nervous system. Matrix biology : journal of the International Society for Matrix Biology. 2012;31:170–177. doi: 10.1016/j.matbio.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chatila K, Ren G, Xia Y, Huebener P, et al. The role of the thrombospondins in healing myocardial infarcts. Cardiovascular & hematological agents in medicinal chemistry. 2007;5:21–27. doi: 10.2174/187152507779315813. [DOI] [PubMed] [Google Scholar]
- 84.Lawler J. The functions of thrombospondin-1 and-2. Current Opinion in Cell Biology. 2000;12:634–640. doi: 10.1016/s0955-0674(00)00143-5. [DOI] [PubMed] [Google Scholar]
- 85.Sezaki S, Hirohata S, Iwabu A, Nakamura K, et al. Thrombospondin-1 is induced in rat myocardial infarction and its induction is accelerated by ischemia/reperfusion. Exp Biol Med (Maywood) 2005;230:621–630. doi: 10.1177/153537020523000904. [DOI] [PubMed] [Google Scholar]
- 86.Frangogiannis NG. Critical role of endogenous thrombospondin-1 in preventing expansion of healing myocardial infarcts. Circulation. 2005;111:2935–2942. doi: 10.1161/CIRCULATIONAHA.104.510354. [DOI] [PubMed] [Google Scholar]
- 87.Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 88.McClung HM, Thomas SL, Osenkowski P, Toth M, et al. SPARC upregulates MT1-MMP expression, MMP-2 activation, and the secretion and cleavage of galectin-3 in U87MG glioma cells. Neurosci Lett. 2007;419:172–177. doi: 10.1016/j.neulet.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schellings MW, van Almen GC, Sage EH, Heymans S. Thrombospondins in the heart: potential functions in cardiac remodeling. J Cell Commun Signal. 2009;3:201–213. doi: 10.1007/s12079-009-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang TN, Albo D, Tuszynski GP. Fibroblasts promote breast cancer cell invasion by upregulating tumor matrix metalloproteinase-9 production. Surgery. 2002;132:220–225. doi: 10.1067/msy.2002.125353. [DOI] [PubMed] [Google Scholar]
- 91.Sage EH, Reed M, Funk SE, Truong T, et al. Cleavage of the Matricellular Protein SPARC by Matrix Metalloproteinase 3 Produces Polypeptides That Influence Angiogenesis. J. Biol. Chem. 2003;278:37849–37857. doi: 10.1074/jbc.M302946200. [DOI] [PubMed] [Google Scholar]
- 92.Phatharajaree W, Phrommintikul A, Chattipakorn N. Matrix metalloproteinases and myocardial infarction. The Canadian journal of cardiology. 2007;23:727–733. doi: 10.1016/s0828-282x(07)70818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nahrendorf M, Pittet MJ, Swirski FK. Monocytes: Protagonists of Infarct Inflammation and Repair After Myocardial Infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobaczewski M, Gonzalez-Quesada C, Frangogiannis NG. The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. Journal of molecular and cellular cardiology. 2010;48:504–511. doi: 10.1016/j.yjmcc.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo Y-H, Gao W, Li Q, Li P-F, et al. Tissue inhibitor of metalloproteinases-4 suppresses vascular smooth muscle cell migration and induces cell apoptosis. Life sciences. 2004;75:2483–2493. doi: 10.1016/j.lfs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 96.Egea V, Zahler S, Rieth N, Neth P, et al. Tissue inhibitor of metalloproteinase-1 (TIMP-1) regulates mesenchymal stem cells through let-7f microRNA and Wnt/beta-catenin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E309–E316. doi: 10.1073/pnas.1115083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lindsey ML, Zamilpa R. Temporal and spatial expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases following myocardial infarction. Cardiovascular therapeutics. 2012;30:31–41. doi: 10.1111/j.1755-5922.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parker BL, Palmisano G, Edwards AV, White MY, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Molecular & cellular proteomics : MCP. 2011;10:M110. doi: 10.1074/mcp.M110.006833. 06833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindsey ML, Borg TK. Understanding the role of the extracellular matrix in cardiovascular development and disease: where do we go from here? Journal of molecular and cellular cardiology. 2010;48:431–432. doi: 10.1016/j.yjmcc.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu L, Zhang JQ, Ramires FJ, Sun Y. Molecular and cellular events at the site of myocardial infarction: from the perspective of rebuilding myocardial tissue. Biochemical and biophysical research communications. 2004;320:907–913. doi: 10.1016/j.bbrc.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 101.Komori H, Watanabe H, Shuto T, Kodama A, et al. alpha1-acid glycoprotein up-regulates CD163 via TLR4/CD14 pathway: possible protection against hemolysis-induced oxidative stress. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.353771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kandalam V, Basu R, Abraham T, Wang X, et al. TIMP2 Deficiency Accelerates Adverse Post-Myocardial Infarction Remodeling Because of Enhanced MT1-MMP Activity Despite Lack of MMP2 Activation. Circulation research. 2010;106:796–808. doi: 10.1161/CIRCRESAHA.109.209189. [DOI] [PubMed] [Google Scholar]
- 103.Tummalapalli CM, Heath BJ, Tyagi SC. Tissue inhibitor of metalloproteinase-4 instigates apoptosis in transformed cardiac fibroblasts. J Cell Biochem. 2001;80:512–521. doi: 10.1002/1097-4644(20010315)80:4<512::aid-jcb1005>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 104.Joseph D, Raffetto aRAKb. Matrix metalloproteinases and their inhibitors in vascular remodeling and vascular disease. doi: 10.1016/j.bcp.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J Cell Sci. 2002;115:3719–3727. doi: 10.1242/jcs.00063. [DOI] [PubMed] [Google Scholar]