Abstract
Because of their high prevalence in the general population, genetic variants that determine susceptibility to environmental exposures may contribute greatly to the development of occupational diseases in the setting of specific exposures occurring in the workplace. Studies investigating genetic susceptibilities in the workplace may: (1) provide mechanistic insight into the aetiology of disease, in particular the determination of environmentally responsive genes; (2) identify susceptible subpopulations with respect to exposure; and (3) provide valuable input in setting occupational exposure limits by taking genetic susceptibility into account. Polymorphisms in the NAT2 and the HLA-DPB1Glu69 genes provide classic examples of how genetic susceptibility markers have a clear role in identifying disease risk in bladder cancer and chronic beryllium disease, respectively. For diseases with more complex and multifactorial aetiology such as occupational asthma and chronic airways disease, susceptibility studies for selected genetic polymorphisms provide additional insight into the biological mechanisms of disease. Even when polymorphisms for genetic susceptibility have a clear role in identifying disease risk, the value of wide scale genetic screening in occupational settings remains limited due to primarily ethical and social concerns. Thus, large scale genetic screening in the workplace is not currently recommended.
“Nature” versus “nurture” has long presented a dichotomy for scientists and health practitioners when evaluating the importance of individual genetic variation and exogenous and endogenous exposure in disease development. However, with advances in human genetics and risk assessment, current research has shown that the interactions between genetic and environmental factors are critical to disease risk. Consequently, research efforts in this area are directed towards investigating the genetic basis of individual susceptibility to various environmental agents.
All individuals are exposed to a variety of hazardous agents and chemicals in the environment. However, several genetic pathways are thought to have evolved for minimising the adverse effects of these environmental insults. Genes expressed in these pathways, referred to as environmentally responsive genes, exhibit heritable variability that may be associated with altered efficiency of the pathway. Table 1 summarises the different categories of environmentally responsive genes.
Table 1.
Major categories of environmentally responsive genes
| Gene category | Function | Example |
|---|---|---|
| Metabolism | Controls biotransformation (including activation and detoxification) of endogenous and exogenous agents | NAT2 |
| DNA repair | Repairs DNA damage caused by various endogenous and exogenous agents | ERCC2/XPD |
| Cell cycle and cell division | Regulates cell proliferation, growth and differentiation | CDK-2 (cell cycle) |
| Cell signalling | Enables cellular communicatin by converting one kind of signal or stimulus into another | STAT1 |
| Major histocompatibility: class II | Cell-mediated immunity; transplanted tissue rejection; beryllium sensitisation | HLA-DP1 |
Several research programs were launched to promote and facilitate research into environmentally responsive genes. In the 1990s, the National Institute of Environmental Health Sciences (NIEHS), of the U.S. National Institutes of Health, initiated a multiyear project entitled the NIEHS Environmental Genome Project (EGP). The NIEHS EGP focuses on common sequence variations, referred to as genetic polymorphisms, in the environmentally responsive genes. Polymorphisms are defined as mutations existing at a population allele frequency of greater than 1% or 2% of individuals.1
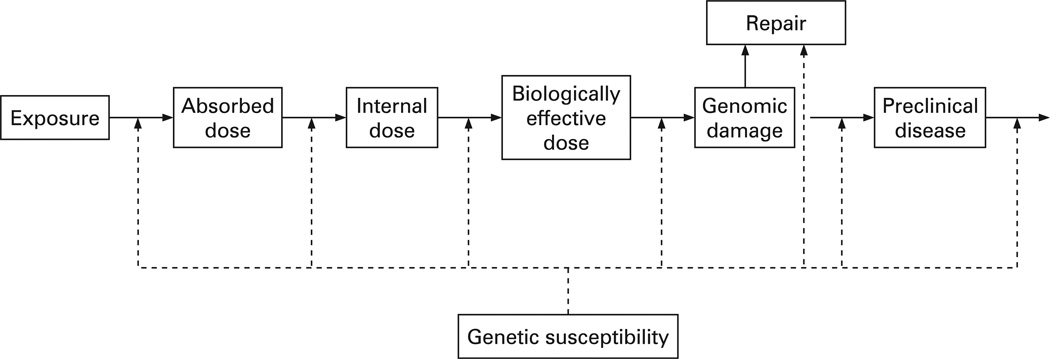
Genetic susceptibility can be viewed as a spectrum encompassing a range of intermediate situations between two extremes. On one side are the rare genetic mutations which give rise to high relative risk conditions (often referred to as disease genes or high-penetrance genes). On the other side of this spectrum are the genetic polymorphisms that are associated with a low-to-moderate risk of the disease (susceptibility genes) and require environmental exposure to result in disease.2,3 Another way to interpret gene penetrance epidemiologically is by considering relative risk. Well-known examples of high-penetrance genes are the inherited mutations in the BRCA1 and BRCA2 genes. Women carrying deleterious mutations in BRCA1 or BRCA2 are estimated to have an 80% lifetime risk of breast cancer in family studies, although this estimate may be overestimated for the general population.4 Despite the high relative risk of breast cancer associated with BRCA1 or BRCA2, mutations in high-penetrance genes are typically rare in the general population and only explain a small proportion of disease aetiology.3,4 It is estimated that only 5–10% of all breast cancers are attributable to mutations in high-penetrance genes such as BRCA1, BRCA2, p53, ATM and PTEN.5,6 In contrast, genetic polymorphisms in low-penetrance genes, because of their higher prevalence in the general population, are expected to contribute more to disease development in combination with various exogenous (eg, hazardous chemicals) and endogenous (eg, hormones) exposures.3,4 For the purpose of this review, we will distinguish between the susceptibility genes with low-to-moderate penetrance and the disease genes with high penetrance.2,7 Figure 1 provides a graphic paradigm of how genetic susceptibility may contribute to the various stages of disease development.8
Figure 1.
Genetic susceptibility in occupational disease development.
Currently there are large scale collaborative efforts between different research institutes, government agencies and private industry to facilitate the discovery and functional analysis of genetic polymorphisms. The most common genetic polymorphism is the single nucleotide polymorphism (SNP), which refers to a single base-pair change in the DNA sequence. Enzymatic and genetic association studies in the literature are a primary source for investigating SNPs with known functionality. However, there are also many publicly available internet SNP databases that serve as useful resources for researchers exploring associations between genetic polymorphisms and disease outcome. Table 2 lists the major internet SNP databases, the principal being dbSNP and HapMap. Some databases (particularly dbSNP and HapMap) catalogue a vast number of human SNPs, and most provide additional information regarding the location of the SNPs, their genetic context, the significance of the changes they introduce, and their reliability. Other databases are more specialised, focusing on genes in a specific cellular response. For instance, the Innate Immunity PGA (IIPGA) website describes genes associated with the innate immunity of humans, while the SeattleSNPs database emphasises genes that are related to inflammation.
Table 2.
Major internet databases (all accessed on 10 March 2008) for single nucleotide polymorphisms (SNPs)
| Organisation/project name and reference | Website |
|---|---|
| Environmental Genome Project56 | http://www.niehs.nih.gov/research/supported/programs/egp/ |
| National Human Genome Research Institute (dbSNP)57 | http://www.ncbi.nlm.nih.gov/projects/SNP/ |
| International HapMap Project58 | http://www.hapmap.org/ |
| National Cancer Institute, Cancer Genome Anatomy Project59 | http://snp500cancer.nci.nih.gov/ |
| SeattleSNPs60 | http://pga.gs.washington.edu/ |
| Innate Immunity in Heart, Lung, and Blood Disease, Programs for Genomic Applications (IIPGA)61 | http://innateimmunity.net/ |
| GeneCards62 | http://www.genecards.org/index.shtml |
The study of genetic susceptibilities, with emphasis on the genetic polymorphisms that alter individual susceptibility to occupational disease, may: (1) provide mechanistic insight into the aetiology of disease; (2) identify susceptible subpopulations with respect to workplace exposure; and (3) provide valuable input in setting exposure limits by taking into account individual susceptibility.3 It should be noted that the process of translating genetics research into practice in environmental and occupational medicine is considered to be at an early phase. Thus, most research findings from genetic susceptibility studies should be communicated with caution to the general public.
This article does not intend to provide a thorough review of the literature on genetic susceptibility in the workplace. Instead, we will summarise two classic examples of genetic susceptibility markers which strongly suggest a biological basis for disease development in combination with a specific occupational exposure. We will also present two additional examples where the biological basis for disease development is less clear. Finally, we will summarise general guidelines for genetic screening in the workplace and offer perspectives on future research and practice needs.
GENETIC SUSCEPTIBILITY TO OCCUPATIONAL CANCER: THE NAT2 POLYMORPHISM AND OCCUPATIONAL BLADDER CANCER
Occupational exposure to arylamines such as 2-naphthylamine, 4-aminobiphenyl and benzidine dyes has been associated with an increased risk of bladder cancer, accounting for 10–20% of cases.9 Arylamines require metabolism to exert their carcinogenic effects. Consequently, genetic polymorphisms in the metabolising genes, which may influence enzyme functionality, are likely to modify individual susceptibility to bladder cancer.
N-acetyltransferase 2 (NAT2) is a key enzyme in the inactivation of arylamines into arylamides. The NAT2 gene is highly polymorphic, with more than 20 different NAT2 alleles identified, including the wild type allele NAT2*4.9–12 The large number of polymorphisms identified in this gene encourages the use of phenotype, rather than genotype, for investigating NAT2 polymorphisms. The phenotype approach summarises individual acetylation status without the necessity of checking the genotype at each identified and unidentified polymorphic site. The NAT2 phenotyping assay is typically measured in vivo using substrates such as isoniazid, sapsone or caffeine. Although results from the NAT2 phenotyping assay provide a continuous measure, most investigators dichotomise results into slow versus rapid acetylator phenotypes.10 The slow acetylation form is prevalent in up to 90% of some Arab populations, in 40–60% of Caucasians of West European origin, and in 5–25% of East Asians.9 Although the phenotype approach may be more useful, recent studies have simultaneously applied phenotyping and genotyping assays in response to the methodological concerns raised by the phenotyping procedure.11
Because the NAT2 enzyme is integral in arylamine detoxification, research has focused on the role of NAT2 polymorphisms in arylamine-associated bladder cancer. As summarised in review papers by Hirvonen,10 Green et al9 and Johns and Houlston,13 studies of bladder cancer and NAT2 acetylation status consistently show an increased bladder cancer risk in individuals with the slow acetylator phenotype and occupational exposure to arylamines. Additionally, a pooled analysis of four case-control studies conducted in four European countries (1530 cases and 731 controls, all Caucasian) suggests that NAT2 acetylation status is not an independent risk factor for bladder cancer but rather modulates the carcinogenic effects of cigarette smoke (probably arylamines) and occupational exposure.14 In contrast, two different cohort studies of benzidine-exposed workers in China do not support an association between the slow acetylator phenotype of NAT2 and elevated bladder cancer risk among individuals who are occupationally exposed,15,16 with a protective association observed between slow acetylation and bladder cancer in benzidine-exposed workers by Carreón et al.16 Findings from the Chinese studies suggest that discrepancies in the metabolism of arylamines may be explained by differences in metabolic enzymes between different ethnic groups11,15 or by variations in the metabolism of different aromatic amines.16
GENETIC SUSCEPTIBILITY FACTORS AND OCCUPATIONAL IMMUNOLOGICAL DISEASE: HLA-DPB1Glu69 AND CHRONIC BERYLLIUM DISEASE
Beryllium is a light-weight metal often alloyed with copper, aluminium and nickel, and is used in the aerospace, ceramic, automotive, electronics, nuclear defence, telecommunications, and tool and die industries, and dental prosthesis manufacturing.17,18 Exposure to beryllium results primarily from inhalation of beryllium particulates during the machining of beryllium-containing products. Since the introduction of beryllium exposure standards, acute beryllium toxicity (mainly pneumonitis) has been virtually eliminated, leaving immune-mediated organ destruction the primary health risk among beryllium-exposed workers.
Exposure to beryllium triggers a type IV, antigen-specific, cell-mediated immune response that results in the proliferation of beryllium-specific T-lymphocytes.18,19 This hypersensitivity to beryllium, or beryllium sensitisation, can be identified through the beryllium-specific lymphocyte proliferation test, an in vitro assay on blood or bronchoalveolar lavage mononuclear cells. Confirmation of chronic beryllium disease (CBD) results if lung pathology reveals sarcoid-like granuloma formations. The cell-mediated response which underlies the pathology of CBD makes it unusual among toxic metal disorders.18,19 Population-based studies show that CBD presents in approximately 2–16% of exposed individuals.17
The low prevalence of CBD among exposed individuals spurred the hypothesis that genes involved in the immunological response to beryllium were significant in the development and progression of CBD. Consequently, the major histocompatibility complex class II (MHC II) molecules were shown to be associated with genetic susceptibility to beryllium sensitisation and CBD, particularly genetic variations in the DP region of the human leukocyte antigen (HLA-DP). Comparing exposed individuals with and without CBD, Richeldi et al showed that the HLA-DPB1*0201 allele was associated with the development of CBD, while the HLA-DPB1*0401 allele was protective.20 Sequence analysis also revealed that the main difference between the two alleles was a glutamic acid residue at position 69 (Glu69).20 The frequency of Glu69-containing HLA-DPB1 alleles (including both *0201 and non-0201 alleles) was also prevalent in 97% of CBD cases compared to 30% of controls, suggesting a potential role of HLA-DPB1Glu69 in the development of CBD.20 Subsequent research has confirmed the association between HLA-DPB1Glu69 and CBD, although the frequency of Glu69 was generally lower among CBD cases than that reported in the original Richeldi study.21–24 Richeldi et al also reported that the presence of aspartic acid and glutamic acid in positions 55 and 56, respectively, occur more frequently among CBD cases than in controls. More recent studies suggest that this latter finding likely results from linkage with Glu69.24 Initially, the presence of Glu69 was considered a marker of disease susceptibility. Despite one report that does not support the association between Glu69 and beryllium sensitisation,23 recent studies have reported comparable frequencies of Glu69 among CBD patients and beryllium-sensitised individuals, suggesting that Glu69 is a risk factor for beryllium sensitisation rather than a susceptibility marker of progression from beryllium sensitisation to CBD.24–27
Other genes (and genetic variations) related to antigen presentation are likely to be involved in predisposing exposed workers to either beryllium sensitisation or CBD. Genes that have been investigated include HLA-DR, HLA-DQ, TNF and ACE.23,25,28,29 For example, studies have suggested that a functional SNP in the promoter region of TNF-α, the -308 guanine to adenine transition, is potentially associated with the progression from beryllium sensitisation to clinical disease.16,23,29
GENETIC SUSCEPTIBILITY FACTORS AND OCCUPATIONAL RESPIRATORY DISEASE: TOLUENE DIISOCYANATE-INDUCED OCCUPATIONAL ASTHMA AND ORGANIC DUST-INDUCED CHRONIC AIRWAY DISEASE
HLA class II genes and toluene diisocyanate-induced occupational asthma
Diisocyanates are widely used in the manufacture of flexible and rigid foams, fibres, coatings such as paints and varnishes, and elastomers. They are a group of low-molecular-weight aromatic and aliphatic compounds characterised by the presence of an –N=C=O group. Exposure to diisocyanates is considered a leading cause of occupational asthma (OA).30 The most important compound is toluene diisocyanate (TDI) and it is generally accepted that 5–15% of exposed subjects develop OA after exposure to TDI.31
Occupational asthma can be characterised “by variable airflow limitation and/or airway hyper-responsiveness due to causes and conditions attributable to a particular occupational environment”,32 and OA has been estimated to account for approximately 15% of asthma in adults.33 Most OA cases induced by high-molecular-weight compounds are often mediated by an immunoglobulin E (IgE) mechanism more frequently observed in atopic subjects. However, for most subjects with TDI-induced OA, it is not possible to demonstrate an IgE-mediated mechanism.34 Although the mechanism for TDI-induced OA is not clear, the pathology for TDI-induced OA is considered a model for non-atopic adult asthma where cell immunity plays an important role in the airway inflammatory process.30 Of particular interest are the HLA class II genes, whose molecules (HLADR, -DQ, and -DP) are involved in binding antigen-derived peptides and presenting them to T-lymphocytes via the T-cell receptor.30
Balboni et al compared the distribution of HLA class II alleles between subjects with TDI-induced OA, TDI-exposed subjects with no OA, and unexposed normal subjects.35 All subjects were European born, unrelated Caucasians working in Northern Italy. The authors identified a significant positive association between disease and HLA-DQB1*0503 and a protective association between disease and HLA-DQB1*0501, which differed in one single position, 57, with aspartic acid in HLA-DQB1*0503 and valine in HLA-DQB1*0501 alleles. When they calculated the phenotypic frequencies of HLA-DQB1-Asp57+, aspartic acid was present in 29 of 30 subjects with TDI-induced OA, compared with 105 of 126 controls (p = 0.05).35 The findings from this study were consistent with previous and later studies in a similar population,30,36 suggesting that HLA class II genes confer susceptibility or protection against TDI-induced OA. In contrast, studies by Rihs et al37 and Bernstein et al38 did not observe an effect of HLA class II alleles on TDI-induced OA. However, Rihs et al exposed subjects to different types of isocyanates, confirmed diagnosis mainly through symptoms, and used airway resistance instead of spirometry to determine airway responsiveness.30 The difference in conclusions between Bernstein et al and the Italian studies collectively may be due to small sample size, the phenotyping methods used and geographical differences between the two study populations.30
Microsomal epoxide hydrolase and organic dust-induced chronic airways disease
Occupational exposure to organic dust has been shown to be associated with chronic airways disease,39–41 and endotoxin, present in organic dust, appears to be a major causative agent contributing to airway inflammation and obstruction.42,43 Among cotton textile and agricultural workers, endotoxin exposure is associated with the development of chronic airways disease.41,42 The presence of endotoxin in the airways triggers a nonspecific immune response including proinflammatory cytokines, chemokines, adhesion molecules and other non-specific inflammatory mediators,44 as well as increased production of oxygen metabolites, such as reactive oxidative species (ROS) that may induce oxidative lung injury.45
Microsomal epoxide hydrolase (mEH) is a phase II biotransformation enzyme which catalyses the hydrolysis of various epoxides and ROS into less reactive and more water-soluble dihydrodiols. Common genetic polymorphisms have been identified in exons 3 (Tyr113His (C113T)) and 4 (His139Arg (G139A)) of the mEH gene. The Tyr113His polymorphism results in a 40–50% decrease in enzyme activity and is considered the “slow allele”. In contrast, the His139Arg polymorphism results in a 25% increase in enzyme activity and is consider the “fast allele”.45 Because mEHs metabolise ROS generated by endotoxin exposure, Hang et al investigated whether polymorphisms in mEH modify the association between endotoxin exposure and faster decline in lung function (a marker for chronic airways disease) in a 20-year prospective cohort study of 265 cotton textile mill workers (endotoxin-exposed) and 234 silk workers (non-exposed) in Shanghai, China.45 Because endotoxin exposure is associated with faster decline in lung function in this study population, investigators hypothesised that decline in lung function would be more pronounced among endotoxin-exposed workers with the “slow allele” for mEH.
Findings from Hang et al indicate that endotoxin exposure was associated with a faster decline in lung function among subjects with “slow” mEH genotypes. Among participants with the His/His allele, endotoxin exposed workers experienced a significantly faster rate of decline in forced expiratory volume in 1 s (FEV1) over 20 years compared to non-endotoxin exposed workers, with an annual difference of −6.7 ml/year.45 When evaluating the joint effect of endotoxin exposure and the Tyr113His polymorphism, the His/His genotype combined with endotoxin exposure was associated with the fastest annual FEV1 decline in comparison with the Tyr/Tyr genotype and no endotoxin exposure.45 Similarly, endotoxin exposure was also significantly associated with a faster rate of FEV1 decline among endotoxin-exposed workers with the His/His genotype of the His139Arg polymorphism.45
GENETIC SCREENING IN THE WORKPLACE
In previous sections, we provided examples of studies demonstrating genetic susceptibility to specific occupational exposures. The first two examples showed how the susceptibility genes under investigation had a clear role in occupational disease risk, while the last two showed how the study of susceptibility genes can provide additional mechanistic insight into occupational diseases that are more complex and multifactorial in aetiology. When susceptibility studies can identify subpopulations at risk with great certainty, their translation into occupational health practice, particularly genetic screening in the workplace, becomes more relevant.
Genetic screening in the workplace aims to identify asymptomatic individuals with certain genetic characteristics that increase the risk of occupational disease.46 Ideally, information from genetic screening allows the employee to choose if they wish to work in a particular environment. The information can also be used by employers for job replacement or relocation to ensure workers most susceptible to a specific risk are placed in the least hazardous environment, with consequent economic advantages through increased safety and reduced health care costs.46,47
In reality, however, genetic screening is used minimally in industrial medical practice; genetic testing to determine susceptibility to workplace hazards has been reported by 16.7% of corporate executives responding to a survey conducted by the American Management Association in 1999.48 The use of genetic screening could increase greatly in the near future, and although it may benefit workers’ health, the risk of harm from its misuse is real and significant and has social and legal ramifications.48
To prevent the misuse of genetic screening that may harm the worker, considerable effort must be made to avoid premature transfer of research knowledge to other settings. Additionally, genetic screening procedures to be applied in the workplace have been suggested to include the following criteria at the minimum:49,50
The goals of screening should be clearly specified, and there should be a definite plan for use of the data.
The test should have an acceptable sensitivity, specificity and positive predictive value.
Workers should have equal access to and/or random participation in the screening.
Protection for worker privacy and confidentially must be guaranteed.
There needs to be clear linkage between the genetic factor being tested and job requirements/duties, demonstrating that the genetic factor is a bona fide prerequisite for adequate job performance.
Even for genetic susceptibility markers that have been shown scientifically to have a clear role in disease risk, the value of wide scale genetic screening in an occupational setting may remain limited. Testing beryllium workers for the HLA-DPB1Glu69 marker provides a real world example of the use of genetic screening to determine genetic susceptibility in the workplace. A recent study that examined voluntary employer-sponsored testing programs with confidential test results found that the longitudinal positive predictive value of the HLA-DPB1Glu69 marker of susceptibility to CBD was only 12%.51 Considering that the positive predictive value of genetic screening is weak given the low prevalence of the disease and high Glu69 frequency among the general population, genetic screening only for Glu69 would not address other susceptibility factors that may be involved in the development of beryllium sensitisation and disease. In addition, difficulties in interpreting test results further limit the utility of the test and may inadvertently result in less attention being paid to industrial hygiene efforts.51
Currently, large scale genetic screening in the workplace is not recommended due to ethical and social concerns.2,7,52–54 Common arguments against the use of genetic screening in the workplace include:
Because of the inherited nature of these genetic characteristics, it is hard to evaluate the psychological and social impacts of genetic testing of these low-to-moderate penetrance genes, especially when no specific interventions are available for carriers of the “risk” alleles.
The genetic screening assays may not have 100% sensitivity and/or specificity. Depending on the allele frequencies, imperfect sensitivity and specificity may have substantial errors at the population level.
Genetic traits can relate differently to different diseases. Consequently, it is difficult to determine whether a particular allele would be associated with an adverse or advantageous health outcome.
The use of genetic screening in the workplace may lead to an increased likelihood of invasion of the privacy and confidentiality of workers; particular attention should be paid to who will have access to the information and for what purpose (eg, employment references, insurance).
Most genetic variations distribute differently in different ethnic and racial groups. Thus, the results of genetic screening must be handled with special care so that potential racial discrimination does not occur.
Identifying workers “at-risk” may erroneously reinforce the trend to focus on this subpopulation while missing many others at risk by failing to recognise that it is the combination of genetic, environmental and behavioural risk factors that leads to the development of occupational disease. Such failure results in lost opportunity to prevent disease.
In summary, the many ethical and social risks outweigh the perceived benefits of implementing genetic screening in the workplace. Although the potential for increased discrimination and invasion of the worker’s privacy is especially troubling, the interpretative challenges of testing may result in less effort being given to exposure reduction to protect all employees at risk. Even when genetic screening is implemented, the information gathered should be viewed as a tool to identify and control harmful exposures rather than simply to exclude the predisposed.55
CONCLUSION
Genetic susceptibility can affect the development of adverse health outcomes, especially in combination with environmental and occupational exposures. Advances in laboratory and computer technologies have facilitated investigation of the susceptibility of environmentally responsive genes in association with occupational disease, which has mainly focused on the genetic polymorphisms of candidate genes in recent years. However, it is generally anticipated that future studies will assess multiple genes within the same pathway simultaneously, which in combination with improved exposure assessment for careful exposure characterisation, will aid full understanding of the role of genetic susceptibility in occupational diseases and in the practice of occupational medicine. Nonetheless, many scientific, social, ethical and legal issues surround the use of genetic screening for susceptibility to workplace exposures. Premature implementation of widespread genetic screening of workers should be avoided at this stage.
Key points.
-
▶
A multitude of publicly available databases, including dbSNP and HapMap, are available for researchers interested in investigating genetic polymorphisms associated with disease outcomes.
-
▶
Polymorphisms in the NAT2 and HLA-DPB1 genes are typical examples of genetic susceptibility markers that demonstrate a strong biological plausibility for disease development in combination with specific occupational exposure.
-
▶
In light of many scientific, ethical, social and legal concerns, implementation of widespread genetic screening of workers should be considered premature at this stage.
Acknowledgements
Supported by NIH grant ES00002 and NIOSH grants T42OH008416 and R01OH2421.
Footnotes
Competing interests: None.
REFERENCES
- 1.Khoury M, Beaty T, Cohen B. Fundamentals of genetic epidemiology. New York: Oxford University Press; 1993. [Google Scholar]
- 2.Christiani DC, Sharp RR, Collman GW, et al. Applying genomic technologies in environmental health research: challenges and opportunities. J Occup Environ Med. 2001;43(6):526–533. doi: 10.1097/00043764-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Vineis P, Malats N, Boffetta P. Why study metabolic susceptibility to cancer? IARC Sci Publ. 1999;(148):1–3. [PubMed] [Google Scholar]
- 4.Dumitrescu RG, Cotarla I. Understanding breast cancer risk -- where do we stand in 2005? J Cell Mol Med. 2005;9(1):208–221. doi: 10.1111/j.1582-4934.2005.tb00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easton D, Ford D, Peto J. Inherited susceptibility to breast cancer. Cancer Surv. 1993;18:95–113. [PubMed] [Google Scholar]
- 6.Hansen RK, Parra I, Lemieux P, et al. Hsp27 overexpression inhibits doxorubicin-induced apoptosis in human breast cancer cells. Breast Cancer Res Treat. 1999;56(2):187–196. doi: 10.1023/a:1006207009260. [DOI] [PubMed] [Google Scholar]
- 7.Vineis P, Christiani DC. Genetic testing for sale. Epidemiology. 2004;15(1):3–5. doi: 10.1097/01.ede.0000101961.86080.f8. [DOI] [PubMed] [Google Scholar]
- 8.Committee on Biological Markers of the National Research Council. Biological markers in environmental health research. Environ Health Perspect. 1987;74:3–9. doi: 10.1289/ehp.74-1474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green J, Banks E, Berrington A, et al. N-acetyltransferase 2 and bladder cancer: an overview and consideration of the evidence for gene-environment interaction. Br J Cancer. 2000;83(3):412–417. doi: 10.1054/bjoc.2000.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirvonen A. Polymorphic NATs and cancer predisposition. IARC Sci Publ. 1999;(148):251–270. [PubMed] [Google Scholar]
- 11.Golka K, Prior V, Blaszkewicz M, et al. The enhanced bladder cancer susceptibility of NAT2 slow acetylators towards aromatic amines: a review considering ethnic differences. Toxicol Lett. 2002;128(1–3):229–241. doi: 10.1016/s0378-4274(01)00544-6. [DOI] [PubMed] [Google Scholar]
- 12.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 13.Johns LE, Houlston RS. N-acetyl transferase-2 and bladder cancer risk: a meta-analysis. Environ Mol Mutagen. 2000;36(3):221–227. doi: 10.1002/1098-2280(2000)36:3<221::aid-em5>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 14.Vineis P, Marinelli D, Autrup H, et al. Current smoking, occupation, N-acetyltransferase-2 and bladder cancer: a pooled analysis of genotype-based studies. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1249–1252. [PubMed] [Google Scholar]
- 15.Ma QW, Lin GF, Chen JG, et al. Polymorphism of N-acetyltransferase 2 (NAT2) gene polymorphism in shanghai population: occupational and non-occupational bladder cancer patient groups. Biomed Environ Sci. 2004;17(3):291–298. [PubMed] [Google Scholar]
- 16.Carreón T, Ruder AM, Schulte PA, et al. NAT2 slow acetylation and bladder cancer in workers exposed to benzidine. Int J Cancer. 2006;118(1):161–168. doi: 10.1002/ijc.21308. [DOI] [PubMed] [Google Scholar]
- 17.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26(10):543–549. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Maier LA. Genetic and exposure risks for chronic beryllium disease. Clin Chest Med. 2002;23(4):827–839. doi: 10.1016/s0272-5231(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 19.McCanlies EC, Kreiss K, Andrew M, et al. HLA-DPB1 and chronic beryllium disease: a HuGE review. Am J Epidemiol. 2003;157(5):388–398. doi: 10.1093/aje/kwg001. [DOI] [PubMed] [Google Scholar]
- 20.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262(5131):242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, White PS, Petrovic M, et al. Differential susceptibilities to chronic beryllium disease contributed by different Glu69 HLA-DPB1 and -DPA1 alleles. J Immunol. 1999;163(3):1647–1653. [PubMed] [Google Scholar]
- 22.Richeldi L, Kreiss K, Mroz MM, et al. Interaction of genetic and exposure factors in the prevalence of berylliosis. Am J Ind Med. 1997;32(4):337–340. doi: 10.1002/(sici)1097-0274(199710)32:4<337::aid-ajim3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 23.Saltini C, Richeldi L, Losi M, et al. Major histocompatibility locus genetic markers of beryllium sensitization and disease. Eur Respir J. 2001;18(4):677–684. doi: 10.1183/09031936.01.00106201. [DOI] [PubMed] [Google Scholar]
- 24.Rossman MD, Stubbs J, Lee CW, et al. Human leukocyte antigen class II amino acid epitopes: susceptibility and progression markers for beryllium hypersensitivity. Am J Respir Crit Care Med. 2002;165(6):788–794. doi: 10.1164/ajrccm.165.6.2104002. [DOI] [PubMed] [Google Scholar]
- 25.Maier LA, McGrath DS, Sato H, et al. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J Immunol. 2003;171(12):6910–6918. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 26.McCanlies EC, Ensey JS, Schuler CR, et al. The association between HLA-DPB1Glu69 and chronic beryllium disease and beryllium sensitization. Am J Ind Med. 2004;46(2):95–103. doi: 10.1002/ajim.20045. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Farris GM, Newman LS, et al. Beryllium sensitivity is linked to HLA-DP genotype. Toxicology. 2001;165(1):27–38. doi: 10.1016/s0300-483x(01)00410-3. [DOI] [PubMed] [Google Scholar]
- 28.Maier LA, Raynolds MV, Young DA, et al. Angiotensin-1 converting enzyme polymorphisms in chronic beryllium disease. Am J Respir Crit Care Med. 1999;159(4 Pt 1):1342–1350. doi: 10.1164/ajrccm.159.4.9806106. [DOI] [PubMed] [Google Scholar]
- 29.Maier LA, Sawyer RT, Bauer RA, et al. High beryllium-stimulated TNF-alpha is associated with the -308 TNF-alpha promoter polymorphism and with clinical severity in chronic beryllium disease. Am J Respir Crit Care Med. 2001;164(7):1192–1199. doi: 10.1164/ajrccm.164.7.2012123. [DOI] [PubMed] [Google Scholar]
- 30.Mapp CE, Beghe B, Balboni A, et al. Association between HLA genes and susceptibility to toluene diisocyanate-induced asthma. Clin Exp Allergy. 2000;30(5):651–656. doi: 10.1046/j.1365-2222.2000.00807.x. [DOI] [PubMed] [Google Scholar]
- 31.Beghe B, Padoan M, Moss CT, et al. Lack of association of HLA class I genes and TNF alpha-308 polymorphism in toluene diisocyanate-induced asthma. Allergy. 2004;59(1):61–64. doi: 10.1046/j.1398-9995.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein IL, Chan-Yeung M, Malo J-L, et al. Definition and classification of asthma. In: Bernstein IL, Chan-Yeung M, Malo J-L, et al., editors. Asthma in the workplace. 2nd edn. New York: Marcel Dekker; 1999. pp. 1–4. [Google Scholar]
- 33.Balmes J, Becklake M, Blanc P, et al. American Thoracic Society statement: occupational contribution to the burden of airway disease. Am J Respir Crit Care Med. 2003;167(5):787–797. doi: 10.1164/rccm.167.5.787. [DOI] [PubMed] [Google Scholar]
- 34.Chan-Yeung M, Malo JL. Occupational asthma. N Engl J Med. 1995;333(2):107–112. doi: 10.1056/NEJM199507133330207. [DOI] [PubMed] [Google Scholar]
- 35.Balboni A, Baricordi OR, Fabbri LM, et al. Association between toluene diisocyanate-induced asthma and DQB1 markers: a possible role for aspartic acid at position 57. Eur Respir J. 1996;9(2):207–210. doi: 10.1183/09031936.96.09020207. [DOI] [PubMed] [Google Scholar]
- 36.Bignon JS, Aron Y, Ju LY, et al. HLA class II alleles in isocyanate-induced asthma. Am J Respir Crit Care Med. 1994;149(1):71–75. doi: 10.1164/ajrccm.149.1.8111601. [DOI] [PubMed] [Google Scholar]
- 37.Rihs HP, Barbalho-Krölls T, Huber H, et al. No evidence for the influence of HLA class II in alleles in isocyanate-induced asthma. Am J Ind Med. 1997;32(5):522–527. doi: 10.1002/(sici)1097-0274(199711)32:5<522::aid-ajim13>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein JA, Munson J, Lummus ZL, et al. T-cell receptor V beta gene segment expression in diisocyanate-induced occupational asthma. J Allergy Clin Immunol. 1997;99(2):245–250. doi: 10.1016/s0091-6749(97)70104-0. [DOI] [PubMed] [Google Scholar]
- 39.Chan-Yeung M, Schulzer M, MacLean L, et al. A follow-up study of the grain elevator workers in the Port of Vancouver. Arch Environ Health. 1981;36:75–81. doi: 10.1080/00039896.1981.10667610. [DOI] [PubMed] [Google Scholar]
- 40.Glindmeyer HW, Lefante JJ, Jones RN, et al. Cotton dust and across-shift change in FEV1 as predictors of annual change in FEV1. Am J Respir Crit Care Med. 1994;149:584–590. doi: 10.1164/ajrccm.149.3.8118622. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz DA, Donham KJ, Olenchock SA, et al. Determinants of longitudinal changes in spirometric functions among swine confinement operators and farmers. Am J Respir Crit Care Med. 1995;151:47–53. doi: 10.1164/ajrccm.151.1.7812571. [DOI] [PubMed] [Google Scholar]
- 42.Christiani DC, Wang XR, Pan LD, et al. Longitudinal changes in pulmonary function and respiratory symptoms in cotton textile workers: a 15-yr follow-up study. Am J Respir Crit Care Med. 2001;163(4):847–853. doi: 10.1164/ajrccm.163.4.2006063. [DOI] [PubMed] [Google Scholar]
- 43.Rylander R, Bake B, Fischer JJ, et al. Pulmonary function and symptoms after inhalation of endotoxin. Am Rev Respir Dis. 1989;140(4):981–986. doi: 10.1164/ajrccm/140.4.981. [DOI] [PubMed] [Google Scholar]
- 44.Milton DK. Endotoxin and other bacterial cell-wall components. In: Macher J, editor. Bioaerosols: assessment and control. Chapter 23. Cincinnati, OH: ACGIH; 1999. [Google Scholar]
- 45.Hang J, Zhou W, Wang X, et al. Microsomal epoxide hydrolase, endotoxin, and lung function decline in cotton textile workers. Am J Respir Crit Care Med. 2005;171(2):165–170. doi: 10.1164/rccm.200407-888OC. [DOI] [PubMed] [Google Scholar]
- 46.Schulte PA, DeBord DG. Public health assessment of genetic information in the occupational setting. In: Khoury MJ, Burke W, Thomson EJ, editors. Genetics and public health in the 21st century: using genetic information to improve health and prevent disease. New York: Oxford University Press; 2000. pp. 203–222. [Google Scholar]
- 47.Vineis P, Ahsan H, Parker M. Genetic screening and occupational and environmental exposures. Occup Environ Med. 2005;62(9):657–662. doi: 10.1136/oem.2004.019190. 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandt-Rauf PW, Brandt-Rauf SI. Genetic testing in the workplace: ethical, legal, and social implications. Annu Rev Public Health. 2004;25:139–153. doi: 10.1146/annurev.publhealth.25.101802.123012. [DOI] [PubMed] [Google Scholar]
- 49.Murray TH. Genetic screening in the workplace: ethical issues. J Occup Med. 1983;25(6):451–454. doi: 10.1097/00043764-198306000-00009. [DOI] [PubMed] [Google Scholar]
- 50.Lappe M. Ethical issues in testing for differential sensitivity to occupational hazards. J Occup Med. 1983;25(11):797–808. [PubMed] [Google Scholar]
- 51.Silver K, Sharp RR. Ethical considerations in testing workers for the -Glu69 marker of genetic susceptibility to chronic beryllium disease. J Occup Environ Med. 2006;48(4):434–443. doi: 10.1097/01.jom.0000200878.16077.3b. [DOI] [PubMed] [Google Scholar]
- 52.Khoury MJ. Will genetics revolutionize medicine? N Engl J Med. 2000;343(20):1497. discussion 1498. [PubMed] [Google Scholar]
- 53.Burke W. Genetic testing. N Engl J Med. 2002;347(23):1867–1875. doi: 10.1056/NEJMoa012113. [DOI] [PubMed] [Google Scholar]
- 54.Vineis P, Schulte P, McMichael AJ. Misconceptions about the use of genetic tests in populations. Lancet. 2001;357(9257):709–712. doi: 10.1016/S0140-6736(00)04136-2. [DOI] [PubMed] [Google Scholar]
- 55.Easton D, Farin F, Omiecinski C, et al. Genetic susceptibility. In: Rom W, editor. Environmental and occupational medicine. Philadelphia: Lippincott-Raven; 1998. pp. 209–221. [Google Scholar]
- 56.Booker SM. Environmental genome project: a positive sequence of events. Environ Health Perspect. 2001;109(1):A22–A23. doi: 10.1289/ehp.109-a22b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 59.Packer BR, Yeager M, Staats B, et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32(Database issue):D528–D532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riva A, Kohane IS. A web-based tool to retrieve human genome polymorphisms from public databases. Proc AMIA Symp. 2001:558–562. [PMC free article] [PubMed] [Google Scholar]
- 62.Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14(8):656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]