Abstract
Nuclear receptor coactivator 6 (NCOA6) also known as PRIP/RAP250/ASC-2 anchors a steady state complex of cofactors and function as a transcriptional coactivator for certain nuclear receptors. This is the first study to identify NCOA6 as an HNF4α interacting protein. CYP2C9 is an important enzyme which metabolizes both commonly used therapeutic drugs and important endogenous compounds. We have previously shown that CAR (a xenobiotic sensing receptor) upregulates the CYP2C9 promoter through binding to a distal site, while HNF4α trancriptionally upregulates CYP2C9 via proximal sites. We demonstrate ligand enhanced synergistic cross talk between CAR and HNF4α. We now identify NCOA6 as crucial to the underlying mechanism of this crosstalk. NCOA6 was identified as an HNF4α interacting protein in this study using a yeast two hybrid screen and GST-pulldown assays. Furthermore, we identified NCOA6, CAR, and other coactivators as part of a mega complex of cofactors associated with HNF4α in HepG2 cells. While the interaction of NCOA6 with CAR is specifically through the first LXXLL motif of NCOA6, both LXXLL motifs are involved in its interaction with HNF4α. Silencing of NCOA6 abrogated the synergistic activation of the CYP2C9 promoter and the synergistic induction of the CYP2C9 gene by CAR-HNF4α. ChIP analysis revealed that NCOA6 can pull down both the proximal HNF4α and distal CAR binding sites of the CYP2C9 promoter and provides the basis for the recruitment of other cofactors. We conclude that the coactivator NCOA6 mediates the mechanism of the synergistic activation of the CYP2C9 gene by CAR and HNF4α.
Cytochrome P450 2C9 (CYP2C9) is a major member of the cytochrome P450 superfamily in human liver, metabolizing numerous therapeutically used drugs and physiologically important endogenous compounds (Goldstein, 2001). Hepatic expression of CYP2C9 exhibits considerable interindividual variability in humans. Some of this interindividual variability is due to upregulation of CYP2C9 levels by prior exposure to drugs and xenobiotics such as rifampicin, hyperforin, phenobarbital and taxol (Komoroski et al., 2004; Madan et al., 2003; Raucy et al., 2002). Studies in primary hepatocytes and clinical studies in vivo in humans have confirmed that CYP2C9 levels and the clearances of CYP2C9 substrates are increased after the administration of drugs (Henderson et al., 2002; Williamson et al., 1998).
Recent studies have shown that the constitutive androstane receptor (CAR) and the pregnane X receptor (PXR) both bind to responsive elements (RE’s) in the CYP2C9 promoter and are responsible for the transcriptional upregulation of CYP2C9 by various drugs (Chen et al., 2004; Ferguson et al., 2002; Gerbal-Chaloin et al., 2002). There is considerable over lap between the two receptors for similar sets of responsive elements in the promoters of various genes (Goodwin et al., 2001; Smirlis et al., 2001; Xie et al., 2000). Each of these receptors is preferentially activated by a wide range of structurally unrelated compounds (Moore et al., 2000; Sueyoshi et al., 1999). Human CAR is activated preferentially by compounds such as phenobarbital, chlorpromazine, clotrimazole, methoxyclor, and CITCO (Timsit and Negishi, 2007), while PXR is activated preferentially by ligands such as rifampicin (rifampin) (Timsit and Negishi, 2007), taxol (paclitaxel), and hyperforin (Xie et al., 2000). The critical feature for activation of CAR by xenobiotics is its translocation from the cytoplasm to the nucleus, where it heterodimerizes with RXR (retinoid X receptor) which facilitates its binding to CAR-RE’s within the DNA of various promoters (Honkakoski et al., 1998; Kawamoto et al., 1999; Moore, 2005; Sueyoshi et al., 1999; Suino et al., 2004; Xu et al., 2004). Activation of CAR elicits a pleiotropic response regulating diverse pathways including various CYP enzymes, liver growth, and liver tumor promotion by phenobarbital (Yamamoto et al., 2004).
Regulation of various promoters by CAR is substantially influenced by other nuclear receptors and transcription factors. We have shown that the hepatic enriched transcriptional factor HNF4α (hepatic nuclear factor 4α) transcriptionally upregulates the CYP2C9 promoter after binding to at least two proximal direct repeats; furthermore, HNF4α and CAR synergistically activate the CYP2C9 promoter in HepG2 cells and mutation of the HNF4α sites reduces or abolishes CAR-mediated induction of CYP2C9 (Chen et al., 2005). This suggests a potential cross talk between a CAR site at −1839 bp and one of the two proximal HNF4α binding sites in the CYP2C9 promoter. The present study addresses the mechanism of this cross-talk.
Cofactors interact with nuclear receptors in the presence of ligands to bring about successful completion of gene transcription (McKenna and O'Malley, 2002; Rosenfeld and Glass, 2001). These cofactors have been found to be associated as complexes; several such complexes have been purified: DRIP (Rachez et al., 1999), TRAP (Fondell et al., 1996) and PRIC (Surapureddi et al., 2002). Cofactors identified in such complexes include activators of the p160 family (McKenna and O'Malley, 2002; Rosenfeld and Glass, 2001), CREB binding protein/p300 (Chrivia et al., 1993; Eckner et al., 1994), and Mediator proteins such as PBP (Zhu et al., 1997), PRIP (Zhu et al., 2000) and PGC-1α (Puigserver et al., 1999). These cofactors all contain one or more conserved LXXLL motifs which have been found to be necessary for ligand-dependent interaction with the AF-2 domain (Heery et al., 1997). PGC-1α has been extensively studied, also as a coactivator of HNF4α (Lin et al., 2005). In the present study, we identify NCOA6 as a new interacting partner of HNF4α. NCOA6 is reported to belong to a novel steady state complex called as ASCOM (ASC-2/PRIP complex) that contains a subset of trithorax group of proteins (Goo et al., 2003). The current model of NR-mediated transcription proposes that subsets of coactivator complexes contribute sequentially to the multiple subreactions of the transcription process (Lonard and O'Malley, 2006; McKenna et al., 1999).
The present study first identifies the cofactor NCOA6 as a new HNF4α-interacting partner using yeast 2-hybrid screens, and protein-protein interaction studies. NCOA6, HNF4α and CAR were identified as part of a mega complex in HepG2 cells. ChIP assays show that antibody to NCOA6 precipitated both the CAR and HNF4α binding sites of the CYP2C9 promoter. Finally, silencing NCOA6 abolished the synergistic activation of the CYP2C9 gene by CAR and HNF4α and reduced the recruitment of coactivators and methyltransferases to the HNF4α sites. These results strongly indicate that NCOA6 is crucial for the formation of a bridge between the CAR and HNF4α receptor sites in the CYP2C9 promoter and for the cross-talk between these two receptors.
Material and Methods
Yeast two-hybrid screening
ProNet technologies automated two-hybrid screening was performed by Myriad Genetics, Salt Lake City, UT, as previously described (Garrus et al., 2001). Full length HNF4α or partial domains were used as bait. Human liver was used to prepare “prey” libraries. Baits were mated with Prey and selection was based on the dropout media (-Trp, -Leu, -His, -Ade) plates. Interactions between bait and prey molecules were identified using His and Ade selections. Plasmids isolated were re-transformed into yeast, and interactions were confirmed by liquid β-galactosidase assays. The identities of the prey were determined by DNA sequencing.
Plasmids and adenovirus mediated expression and RNA interference
CAR, HNF4α, NCOA6, PBP and PGC-1α were cloned in pcDNA3.1 by PCR amplification. GST-CAR and GST-HNF4α were cloned in pGEX-4T1. NCOA6 domains, NCOA6 I (1–353 aa), NCOA6 II (338 −673 aa), NCOA6 III (648–998 aa containing 1st LXXLL motif at 851 aa), NCOA6 IV (986–1327 aa), NCOA6 V(1292–1641 aa coding for 2nd LXXLL motif at 1491 aa) NCOA6 VI (1625–2065aa) and NCOA6 VII (648–1641 aa) were cloned in pGEX-4T1 and pcDNA3.1. CYP2C9-1.9Kb/pGL3 construct has been previously described (Chen et al., 2005), used for transient transfection assay of CYP2C9 promoter expression. Adeno virus expressing full-length CAR and HNF4α were made with AdEasy™ XL Adenoviral Vector system (Stratagene, CA). Virus particles were purified on continuous cesium chloride, dialyzed and stored in Tris buffered sucrose. siRNA targets for NCOA6 were identified using Genscript’s target finder, construct builder and sequence scrambler for construction of negative control siRNA. The following siRNA were designed to silence NCOA6 mRNA coding sequence, NC-I (@146 bp): 5’-TTGTGGCCTTCAAAGGAAATA-3’; NC-II (@500 bp):5’-TGGCAAGTGGTCCAGGAATAA-3’; NC-III (@ 2241 bp): 5’-CTCCGAACATGCAA GGAAATA-3’; NC-IV (@ 2451 bp): 5’-GATGCCTGATGTTAGCATTCA-3’ and NC-V (@ 3262 bp): 5’-GTGCCACCATCA\CCTGATAAA-3’. Using the construct builder, double stranded shRNA oligos were designed for pRNAT-H1.1/Adeno (SD-1219) with H1 promoter and cGFP as the marker. NC-III siRNA target sequence was used to design the scrambled siRNA 5’-CACTGAAGTATACCAAGAGCA-3’. Adeno viruses expressing each shRNA were prepared and purified. HepG2 cells were routinely infected with 2.5× 109 virus particles (VP).
Cell culture, transient transfections and ligands
HepG2 cells were maintained in the Eagle’s minimal essential medium supplemented with 10% fetal bovine serum and penicillin-streptomycin at 37°C under 5% CO2. All transient transfections were carried out as described in Lipofectamine 2000 protocol (Invitrogen, CA). Briefly, 0.2 µg of CYP2C9 luciferase construct, 0.1 µg of each receptor construct(s), and 0.1 µg of coactivator construct (2:1:1) with 0.02 µg of pRL-TK vector as internal control, pcDNA 3.1 as the empty vector to make the total amount of DNA transfected to 0.8 µg were combined in 50 µL OPTI-MEM, mixed with transfection reagent as suggested and overlayed on 80–90% confluent HepG2 cells in serum containing media. Twenty-four hours later, medium was replaced, and ligands were added at the appropriate concentrations (0.1% of DMSO and 0.1 µM CITCO). Ligands were incubated with the HepG2 cells for 24 h and assayed for promoter activity using Promega’s dual luciferase assay kit (Promega, Madison, WI). Firefly luciferase readings were normalized with renilla readings to calculate promoter activity.
CAR, HNF4α expression HepG2 nuclear extracts, isolation of interacting proteins and immunoblotting
Ten plates (15 cm) of 90% confluent HepG2 cells were infected with 2.5 × 1011 VP for 48 h with either AdCAR or AdHNF4α. The cells were harvested after a quick wash in cold PBS; nuclear extracts were prepared according Dignam method ((Dignam et al., 1983). GST and GST-HNF4α fusion proteins immobilized on GSH-sepharose beads were incubated with 5–10 mg of AdCAR overexpressing HepG2 nuclear extracts (AdCAR-NE’s) overnight. The beads were extensively (100 volumes) washed after incubation with TEGN buffer (20 mM Tris, pH 8.0, 0.2 mM EDTA, 10% glycerol, 0.1% NP-40) containing 180 mM of NaCl and once with TEGN buffer containing no salt. The bound proteins were eluted in SDS-PAGE sample buffer and heat denatured. The proteins were separated on a 4–20% gradient gels and silver stained for visualization. GST and GST-HNF4α bound proteins after separation on 4–20% gradient gels were transferred onto nitrocellulose membrane, blocked with 5% milk in TBS-T (Tris buffered saline with Tween-20) and immunoblotted with antibodies for CAR (sc-13065, lot # I2404) (sc= Santa Cruz Biotechnology, Inc., Santa Cruz CA and reconfirmed with a monoclonal CAR antibody from Research and Diagnostics Systems, Inc., Minneapolis MN (Cat# PP-N4111-00 and clone # N4111), HNF4α (sc8987, lot# A2304), CBP (sc369x, lot# L0706), PIMT (sc23114, lot# B2406), PGC-1α (sc13067, lot# I2904) and NCOA6 (Bethyl laboratories, A 310–411A).
The Protein Microcharacterization Facility at NIEHS provided protein sequencing-Mass spectrometry
Protein bands of GST and GST-HNF4α lanes were sliced in 2mm thickness and tryptic digested. Coomassie blue stained SDS-PAGE gels were cut and proteins digested with trypsin essentially as described in Choi, et al. Resulting peptide digests were then analyzed by nano-LC ESI-MS and MS/MS on and Agilent XCT Ultra ion trap mass spectrometer and data processed and searched against the NCBI non-redundant database as previously described (Choi et al., 2007).
Expression and purification of recombinant proteins, GST-Pull Down assay
Full-length recombinant proteins of CAR and HNF4α were expressed as GST fusion proteins in E.coli BL21 (DE3). GST pull-down assays were performed by incubating 5 µL of 35S methionine labeled proteins in a 500 µL of NETN buffer containing 1mg/ml fatty acid free BSA in the presence and absence of respective ligands. The bound proteins were washed with 3X with NETN buffer and were heat denatured in SDS sample buffer. These proteins were separated on 4–20% gradient gels and after fixing and amplifying the signals, the gels were dried and autoradiographed.
qPCR
Total RNA was extracted using RNeasy mini prep system (QIAGEN, Valencia, CA). RT-PCR analysis was performed in two steps by initial reaction with Superscript II (Invitrogen, Carlsburg, CA) reverse transcriptase. PCR with Taqman® Universal PCR Master Mix (Applied Biosystems (ABI), Foster City CA) was then performed with gene-specific primers using relative quantification methods (2X−ΔΔCT) and measured on an Applied Biosystems Geneamp PCR System 9700 using taqman probes (ABI) for CYP2C9 (Cat# HS00426397_m1. lot# 465955), NCOA6 (HS00204160_m1, lot# 426728), PGC-1α (HS00173304_m1, lot# 440759), PBP (HS00191130_m1, lot# 426852), SRC-1(HS00186661_m1, lot# 442260), GRIP-1(HS00896112_m1, lot# 587691) and HNF4α (HS00230853_m1, lot# 465073) with TBP as the internal control (4333769F, lot# 0706051).
Chromatin immunoprecipitation (ChIP) Analysis
Five plates (15 cm) of 90% confluent HepG2 cells were infected with 2.5 × 1011 VP for 48 h each with adenovirus expressing, lacZ, CAR, HNF4α, CAR-HNF4α, CAR-HNF4α with siRNA for NCOA6 (NC-III) and CAR-HNF4α with siRNA for NCOA6 (NC-IV), individually. After 48h, the cells were cross linked with 1% formaldehyde directly in the media for 10 min and the chromatins were prepared (Qi et al., 2003). Chromatin extracts were precleared by incubating with rabbit serum for 3h and the IgG bound proteins were pulled down by incubating with pre-swollen Protein-A beads. The supernatant was stored or used for immunoprecipitations. All the chromatins used in the IPs were checked to contain equal amounts of the target gene by PCR amplification and adjusted to 100 µl, which were further diluted to 500 µL to be used in the immunoprecipitation with 2µg of each antibody. CAR, CBP, NCOA6, PIMT immunoprecipitates were washed 2X with buffer C and 4X with buffer D. HNF4α antibody precipiates were washed 3X with buffer C and 6X with buffer D (buffers described previously)(Qi et al., 2003). The DNA binding proteins bound cognate cis acting elements and DNA fragments from the chromatin extracts (inputs) were purified and used as control for PCR reactions. The primers used for amplification of the human CYP2C9 promoter are 5’-TAAAGACAGCAACCGAGC-3’ and 5’-TACAATGATTCAGGATTTCG-3’ spanning for the CAR response element and 5’-ATATACAAGGCATAGAATATGGCC-3’ and 5’-GACCAATCACCTAGGTCCAC-3’ spanning the HNF4α response elements. Negative control primers are 5’-ATGGTTGCCACTGGGGATCT-3’ and 5’-TGCCAAAGCCTAGGGGAAGA-3’.
Statistical Comparisons
Results were analyzed by two-way or one way analysis of variance and pairwise comparisons were made using Bonferroni t-test.
Results
Isolation and cloning of NCOA6 interacting protein by yeast two hybrid screening
In the initial studies, yeast two hybrid screens were performed using two human liver cDNA libraries with HNF4α as the bait. Two out of eleven HNF4α baits (93–352 aa and 165–355 aa) identified several strong interacting proteins that could pass two rounds of screening. One of the prey proteins identified by both the bait molecules was NCOA6 (1993–2726 bp and 2322–2724 bp) coding for the first LXXLL motif. This is the first time NCOA6 has been identified as an HNF4α interacting protein. Other known coactivators of HNF4α identified by the screen include PGC-1α. The complete coding sequence of human NCOA6 protein was assembled in pcDNA3.1 (+) using IMAGE and EST clones (Invitrogen).
Identification of nuclear proteins, which bind selectively to, immobilized GST-HNF4α
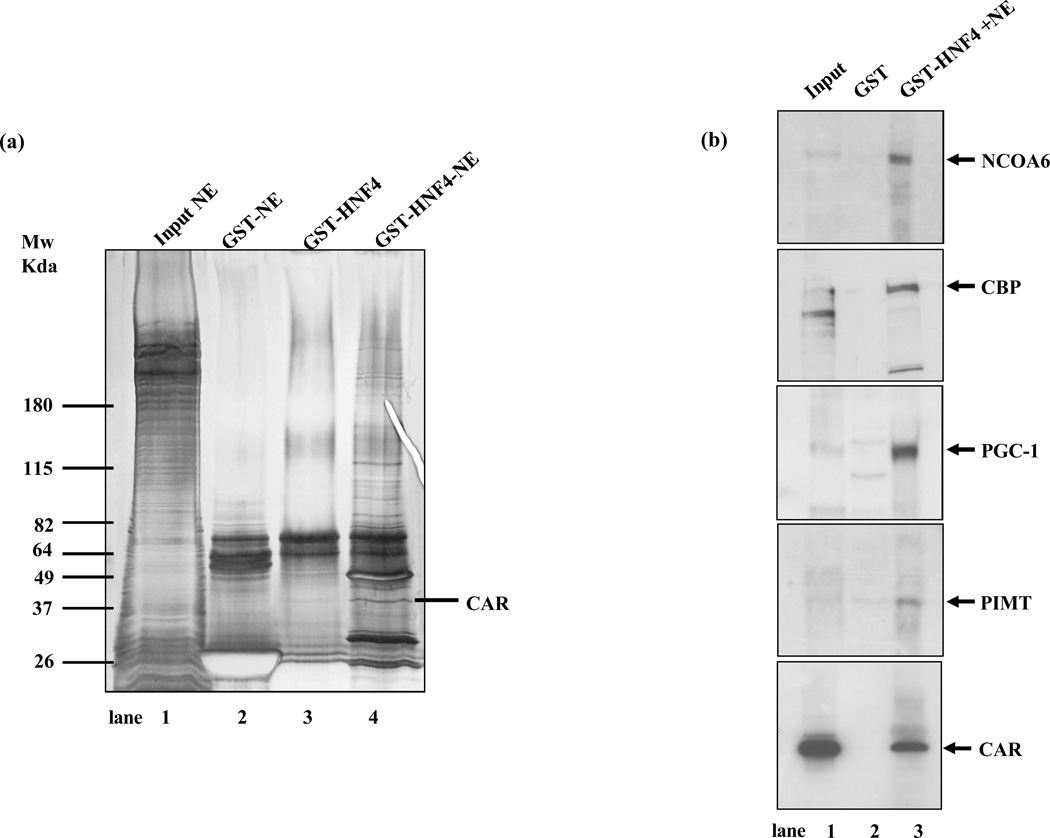
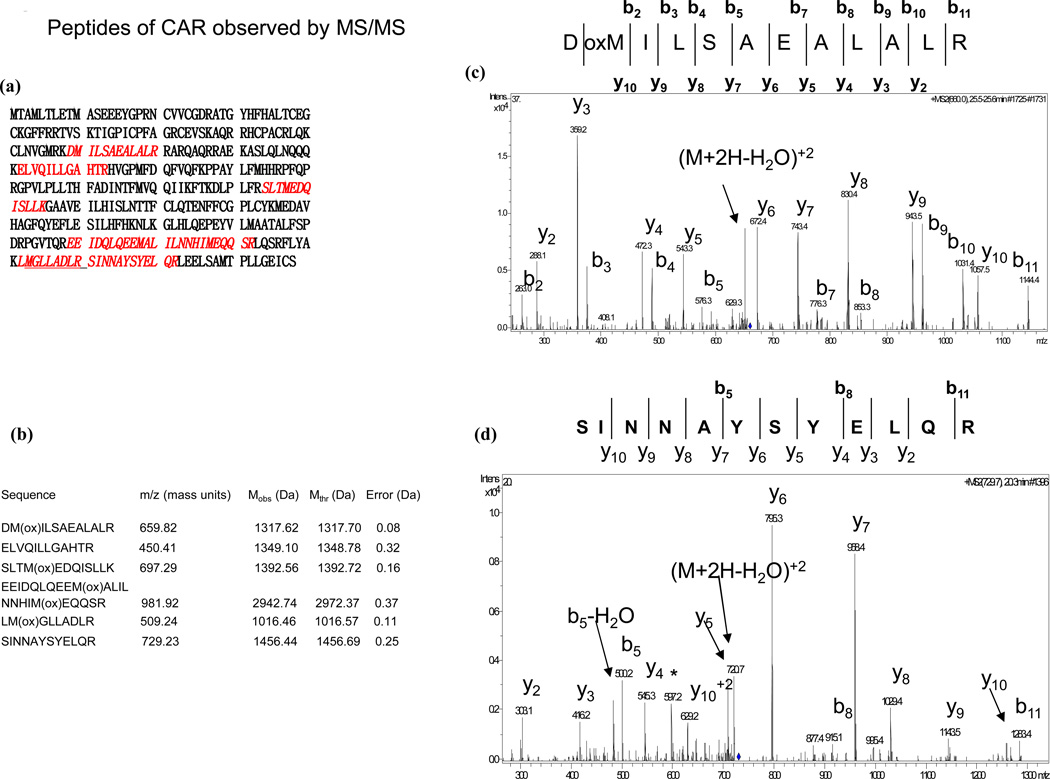
Nuclei were isolated from HepG2 cells infected with AdCAR for 48 h to ensure the translocation of CAR and its binding proteins to induce the transcriptional activity of the target gene of interest viz.CYP2C9. To maximize the capture of proteins, we used full length HNF4α. HepG2 nuclear extracts enriched with CAR were incubated with immobilized GST and GST-HNF4α. After extensively washing, the bound proteins were denatured and subjected to SDS-PAGE. Very few nuclear proteins were bound to GST alone (Fig.1a, lane 2). In contrast, more than 25 nuclear proteins with electrophoretic motilities above 40kda were detected which bound to GST-HNF4α (lane 4). An equivalent amount of GST-HNF4α purified from bacteria is shown as a control (lane 3). Protein sequencing data identified a polypeptide band with a molecular weight of ~40kd as the constitutive androstane receptor (Fig. 2). Constitutive androstane receptor, CAR (gi number 83921568), was positively identified in the GST-HNF4α bound proteins and was not observed in the GST only bound proteins. CAR was identified with a SpectrumMill distinct summed MS/MS search score of 94.23 with 22% sequence coverage (Fig. 2a). Specifically, 6 different tryptic peptides (Fig. 2B) that are unique to CAR were observed via MS and MSMS. Each of the 6 peptides was unambiguously identified by extensive b- and y-series ions in MSMS experiments as can be seen from the representative MS/MS spectra shown in Fig. 2 (panels C and D).
Fig. 1. Purification and identification of nuclear proteins interacting with HNF4α and CAR.
(a) Isolation of HNF4α binding complex from nuclear extracts of HepG2 cells in which Ad-CAR was over expressed (AdCAR-NEs). SDS/gradient gel analysis (4–20%) and silver staining of HNF4α binding complexes from 7.5 mg of AdCAR-NEs bound to GST (lane 2) and GST-HNF4α (lane 4). GST-HNF4α without the addition of NE is shown as a control (lane 3) and 50µg of AdCAR-NE is shown as input (lane 1). (b) Immunoblots identifying CAR and various coactivators in the GST-HNF4α bound complex. Samples from Input: AdCAR expressing NEs, GST: GST bound proteins from AdCAR-NEs and GST-HNF4α bound proteins from AdCAR-NEs were immunoblotted with specific antibodies for CBP, PGC-1α, NCOA6, PIMT and CAR. Lane 1: NE input, lane 2: NE bound to GST as a control and lane 3: GST-HNF4α complex from Ad-CAR-NEs.
Fig. 2.
Mass spectrometric identification of CAR. Six peptides of CAR (gi 83921568) shown in red, resulting in 22% sequence coverage, were observed by MS and MSMS (A). Generally, the precursor ions show good agreement between observed masses and theoretical masses for the predicted peptides (B). Extensive b- and y-series ions were observed for all six peptides. The MS/MS spectra of the ion (m/z 659.82) corresponding to the most N-terminal peptide observed (residues 89–100) and of the ion (m/z 729.23) corresponding to the most C-terminal peptide observed (residues 331–342) are shown as examples (C and D).
Although current identification of additional large proteins by protein microsequencing is in progress, we used immunoblotting to identify some of the expected cofactors in the GST-HNF4α binding complex. We also detected CAR by immunoblotting in both the input and in the proteins bound to GST-HNF4α but not in proteins eluting from GST confirming the MS/MS results (Fig 1b). Since we identify CAR and HNF4α in nuclear complexes in HepG2 cells but were unable to show evidence of a direct interaction between CAR and HNF4α using GST-HNF4α and radiolabeled CAR or GST-CAR and radiolabeled HNF4α (not shown), we hypothesized that cofactors might be responsible for bringing these two nuclear receptors together as a nuclear mega-complex. We have identified several cofactors including NCOA6, CBP, PGC-1α and PIMT by immunoblotting the Ad-CAR expressing nuclear extracts (inputs) and in proteins bound to GST-HNF4α but not in proteins retained by GST alone, suggesting that the HNF4α-CAR complex represents a functional transcriptional complex (Fig 1a and Fig 1b). The identification of NCOA6 and PGC-1α as HNF4α interacting proteins by immunoblotting was consistent with the results of our two hybrid screen. PIMT is also a known NCOA6 interacting protein with a methyltransferase domain.
NCOA6 interacts with nuclear receptors CAR and HNF4α
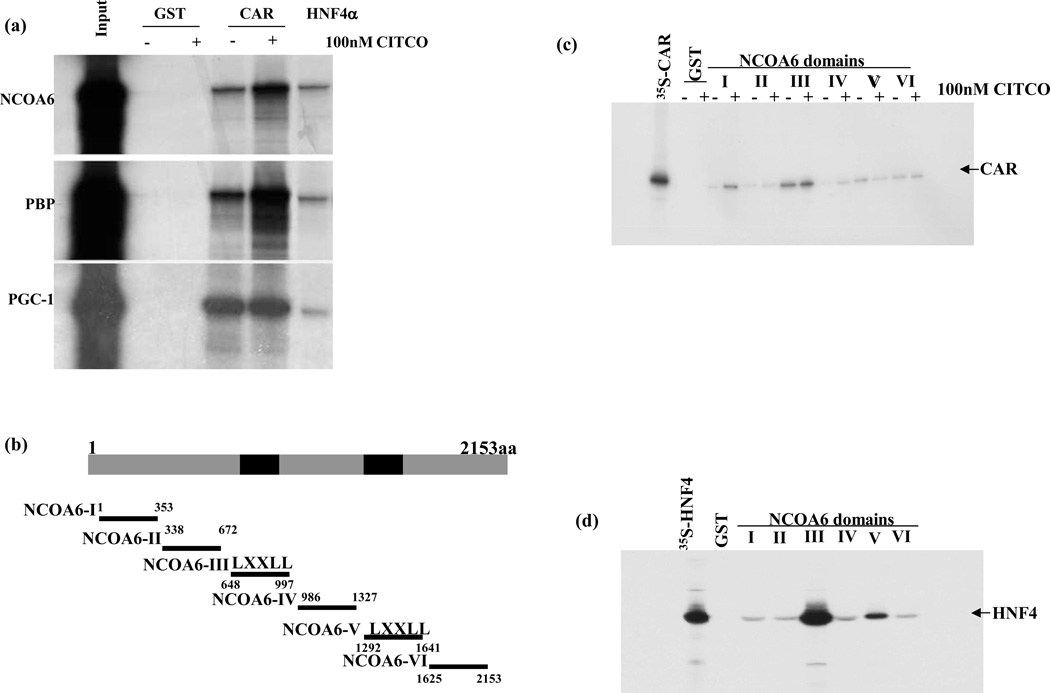
To identify NCOA6 interactions, we first used a GST fusion protein of a truncated NCOA6 (648 to 900 aa; coding for the first LXXLL motif). GST-NCOA6 was used for binding assays with 35S methionine labeled HNF4α and CAR. Both the nuclear receptors bound to GST-NCOA6 strongly in the absence and presence of CITCO which is known to be a high affinity ligand for human CAR (Maglich et al., 2003). To verify the reciprocal relationships, GST-CAR and GST-HNF4α were incubated with in vitro translated full length NCOA6 and the known coactivators PBP and PGC-1α were used as a positive control. GST-CAR and GST-HNF4α fusion proteins retained the radio-labeled NCOA6, PBP and PGC-1α in presence and absence of 100 nM CITCO: GST alone did not retain NCOA6 (Fig. 3a). The CAR ligand CITCO augmented the retention of NCOA6 and PBP by GST-CAR. As expected, GST-CAR and GST-HNF4α fusion proteins also retained other in vitro translated known coactivators such as SRC-1 and GRIP (data not shown). To define the interacting domain(s) of NCOA6, GST-NCOA6 domains were allowed to interact with in vitro 35S-methionine translated CAR, or HNF4α. NCOA6 domains expressed as GST- fusion proteins are shown schematically in Fig. 3b. GST-NCOA6-III domain (648–998aa; the first LXXLL at 851aa) interacted significantly with CAR (Fig. 3c) in a ligand independent manner. The other NCOA6 domains did not interact appreciably with CAR suggesting that only the first LXXLL motif was involved in the interaction. The first LXXLL motif of NCOA6 has been previously shown to interact with other nuclear receptors such as PPAR (α and γ) TRα and ERα (Zhu et al., 2000). The NCOA6-CAR interaction differs from PBP-CAR interactions in that PBP requires both LXXLL motifs to bind with CAR (Jia et al., 2005). The interacting domains between NCOA6 and HNF4α were similarly mapped with 35S-methionine-HNF4α (Fig.3d), the GST-NCOA6-III domain strongly retained the radiolabeled HNF4α, suggesting a robust interaction. Surprisingly, there was also a moderate interaction between the GST-NCOA6-V domain (containing the second LXXLL motif) and HNF4α, suggesting a new role for this LXXLL motif. The second LXXLL motif of NCOA6 was also recently shown to be involved in the interaction with LXR (another liver specific receptor) in regulating lipogenesis and cholesterol/bile acid homeostasis in the liver and interaction with ERα (Li et al., 2007a).
Fig. 3. In vitro protein-protein interactions between coactivators NCOA6, and the nuclear receptors CAR, HNF4α and domain mapping.
The first LXXLL motif of NCOA6 interacts with HNF4α and CAR. (a) GST-CAR and HNF4α were immobilized on GSH-sepharose beads. NCOA6 and PBP as a positive control were translated in vitro in the presence of 35S methionine using Promega’s in vitro system. Full length radiolabeled proteins were allowed to interact with GST, GST-CAR, and GST-HNF4α (+ 100 nM CITCO or DMSO as the control). Bound proteins were heat denatured, separated by electrophoresis on a 4–20% gradient gels, fixed, signal was amplified and dried for autoradiography. 1/10th of the transcription/translation mixes were loaded as input. (b) Schematic representation of NCOA6 domains. GST-NCOA6 domains or GST control were incubated with (c) 35S methionine labeled-CAR (in the presence and absence of 100 nM CITCO) or (d) 35S methionine labeled-HNF4α (no ligand) and subjected to SDS-gel electrophoresis, gels were dried and autoradiographed as described in Methods.
CAR and HNF4α synergistically activate the CYP2C9 promoter in a ligand enhanced manner and NCOA6 modestly augments this effect
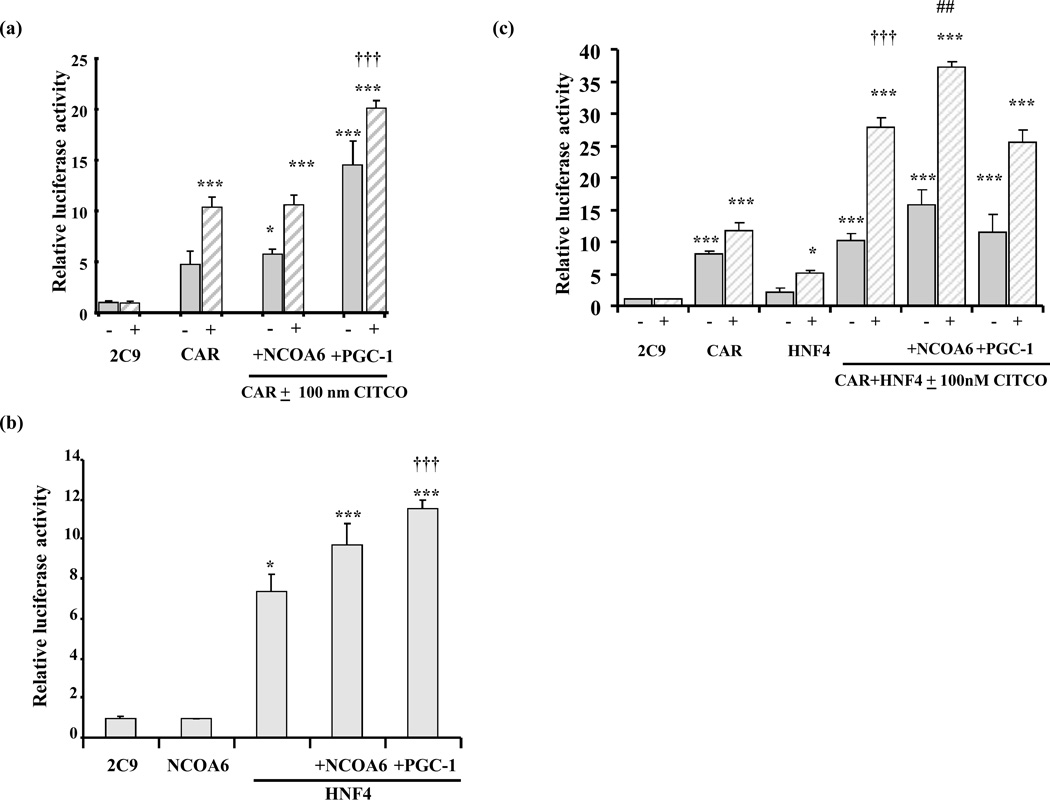
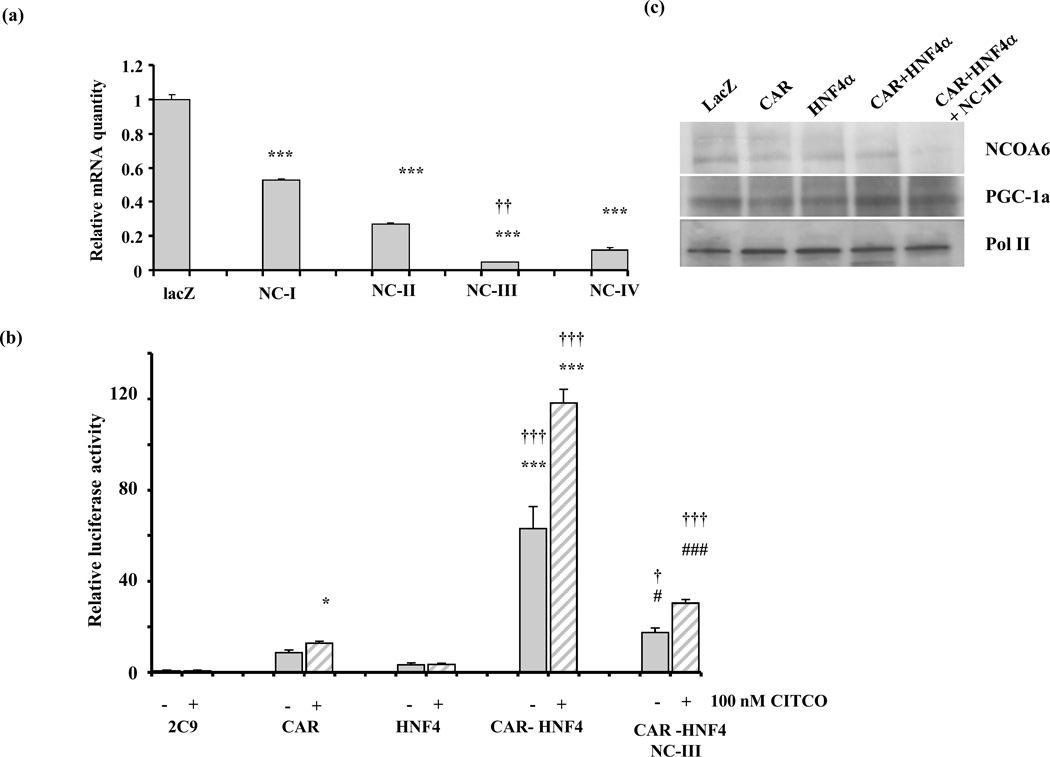
CAR transactivates the CYP2C9 promoter, and the CAR ligand CITCO enhances this effect (p<0.01). The activation is ~5-fold in the absence of ligand, and 11-fold in the presence of 100 nM CITCO (Fig 4a). This concentration of ligand was used because initial experiments verified that it is specific for CAR, while higher doses are not specific since they activate another receptor, PXR. Coexpression of NCOA6 did not significantly enhance CAR mediated promoter activation, while PGC-1α, a known CAR coactivator significantly enhanced activation (p<0.001). HNF4α transactivated CYP2C9 promoter activity (~7-fold) (p<0.05) (Fig 4b) and coexpression of NCOA6 with HNF4α modestly enhanced this activation to ~9-fold (p<0.001), although the enhancement by NCOA6 was not quite significant (p=0.09). NCOA6 by itself does not transactivate CYP2C9 promoter. As a positive control, expression of PGC-1α, a known coactivator of HNF4α (which is known to be expressed at low levels in HepG2 cells), significantly increased HNF4α activation (p<0.01) to ~12-fold. PGC-1α had no effect on the 2C9 promoter activity alone (not shown). When both CAR and HNF4α are coexpressed with the CYP2C9 promoter in the presence of the CAR ligand CITCO, there is a synergistic 26-fold transactivation of CYP2C9 promoter activity when compared to 11-fold activation by CAR alone or 4.8-fold by HNF4α alone (Fig 4c). Cotransfection with NCOA6 slightly increased this activation by CAR and HNF4α to 35-fold, while PGC-1α produced no significant increase in this experiment. In summary, there is a synergistic activation of CYP2C9 promoter expression by the nuclear receptors CAR and HNF4α in the presence of the CAR ligand, CITCO. Exogenous NCOA6 had no effect on CAR activation, but modestly enhanced the activation of the CYP2C9 promoter by HNF4α, and the synergistic activation by CAR and HNF4α showing it is a coactivator of HNF4α.
Fig. 4. Effects of coactivators on the synergistic activation of CYP2C9 promoter expression by HNF4α and CAR in HepG2 cells.
(a) NCOA6 does not effect CAR mediated transactivation. HepG2 cells were transfected with CAR, NCOA6 and PGC-1α along with CYP2C9 reporter construct, in the presence (hatched bars) and absence (solid bars) of 100nM CITCO as the ligand with pRL as the internal control. All transfections were performed in triplicate. Values are means of triplicates + SE. CAR significantly upregulates the CYP2C9 promoter in the absence and presence of ligand at *, p<0.05, **,p<0.01, ***, p<0.001. Transfection with PGC-1α and CAR significantly transactivates the CYP2C9 promoter more than CAR alone at †††, p<0.001, while NCOA6 had no effect.
(b) Effect of NCOA6 and PGC-1α on HNF4α mediated transactivation. HNF4α and NCOA6 were expressed along with the CYP2C9 promoter in HepG2. *, p<0.05 **, p<0.01and ***, p<0.001.values are significantly greater than CYP2C9 promoter alone. NCOA6 alone does not transactivate CYP2C9 promoter. Cotransfection with PGC-1α and HNF4 α activates the CYP2C9 promoter significantly greater than HNF4α alone at ††† , p<0.001. (c) CAR and HNF4α synergistically activate CYP2C9 promoter expression in the presence of CITCO and the effect of coactivatosr NCOA6 and PGC-1α . HepG2 cells were transfected with CAR and HNF4α individually and in combination, coactivators NCOA6 and PGC-1α were added to CAR-HNF4 combination in the presence (hatched bars) and absence of 100 nM CITCO as the ligand with CYP2C9 reporter constructs. All transfections were performed in triplicate and values represent the means + SE. The values represent fold activation over CYP2C9 promoter alone. CAR, HNF4α or CAR+ HNF4α significantly upregulate CYP2C9 promoter activity compared to CYP2C9 promoter alone, at *, p<0.05, **, p<0.01, ***, p<0.001. †††, designates synergistic rather than additive response to cotransfection with HNF4α and CAR at p<0.001 (ANOVA with interaction). Transfection with NCOA6 significantly enhances the activation of CAR and HNF4α at p<0.01, ##, while transfection with PGC-1α had no significant effect.
NCOA6 as the bridging coactivator for nuclear receptors CAR and HNF4α
To more definitely test the transcriptional role of NCOA6 in the synergistic activation of CYP2C9 by CAR and HNF4α, we then expressed small interfering RNA (siRNA) directed against NCOA6 in HepG2 cells using adenoviral system to silence endogenous NCOA6 expression. Among five targets tested, NC-V was not effective. Of the remaining four, NC-III and NC IV reduced the expression of NCOA6 mRNA levels by 95% and 88% respectively as quantified by qPCR (p<0.001)(Fig. 5a). NC-III produced a significantly lower expression than any of the other targets. Western blot analysis confirmed that NCOA6 protein was markedly decreased in cells infected with NC-III (Fig. 5c), while PGC-1α and Pol II remain unchanged. To test whether silencing of NCOA6 effects the ligand dependent synergistic activation of the CYP2C9 promoter by CAR and HNF4α, the CYP2C9-luc promoter construct and CAR, HNF4α, or CAR-HNF4α were transiently transfected into HepG2 cells, and 24 h later the cells were infected with adenovirus expressing scrambled or siRNAs for NCOA6. It should be noted that the magnitude of the synergistic activation is larger than in Fig 4c, probably because transfection efficiencies are different during culture involving adenoviral infection (serum absent from the medium but is added later). Serum is absent from the medium during adenoviral treatment of the cells, but is added later which would shock the cells altering transfection efficiency. The synergistic activation of the CYP2C9 promoter by CAR-HNF4α was dramatically suppressed by the expression of siRNA (NC-III) for NCOA6 in the presence and absence of ligand (Fig 5b).
Fig. 5. Silencing of NCOA6 abrogates the synergistic effects of HNF4α with CAR on CYP2C9 gene expression.
(a) Screening of siRNAs for silencing NCOA6 mRNA in HepG2 cells. Five potential targets for silencing NCOA6 were identified in silico. HepG2 cells were infected with 2.5×109 VP/ml for each of the siRNAs for NCOA6 (NC-I to NC-V) individually with scrambled siRNA as control. After 48h, total RNA was prepared to estimate NCOA6 mRNA levels using qPCR. One target, NC-V was not effective (not shown). All the remaining targets significantly suppressed NCOA6 mRNA, ***, p<0.001, although NC-III was the most effective p<0.001, ††. Values represent means ± SE of triplicate analyses.
(b) siNCOA6 (NC-III) dramatically decreases the synergistic effects of CAR and HNF4α on CYP2C9 promoter activation. HepG2 cells were cotransfected with the CYP2C9 promoter and CAR alone or in combination with HNF4α as in Fig. 3c, and the cells were infected with scrambled or siNCOA6 (NC-III) 2.5×109 VP/ml in the presence(+) and absence (−) of 100 nM CITCO. Values represent means ± SE of triplicate transfections. Transfection with CAR or CAR+HNF4α significantly increased CYP2C9 promoter activity, at *, p<0.05, **, p<0.01, *** p<0.001. †, ††, and †††, indicate synergistic rather than additive response to HNF4α and CAR at p<0.05, p<0.01, and p<0.001 respectively (ANOVA with interaction). NC-III significantly decreases the synergistic action of CAR-HNF4α in both the absence and presence of CITCO at #, p<0.05, and ##, p<0.001 respectively.
(c) Immunoblots of endogenous NCOA6 and PGC-1a in HepG2 cells infected with nuclear receptors. 100 µg of NEs from HepG2 cells infected with LacZ, CAR, HNF4α and CAR-HNF4α, CAR-HNF4α+ siNCOA6 (NC-III) were immunoblotted for NCOA6 and PGC-1α. NCOA6 is clearly down regulated in HepG2 cells infected with siNCOA6 (NC-III), while PGC-1α expression levels remain unchanged. 10 µg of NE’s were blotted for Pol II as a control and its expression level remain unchanged.
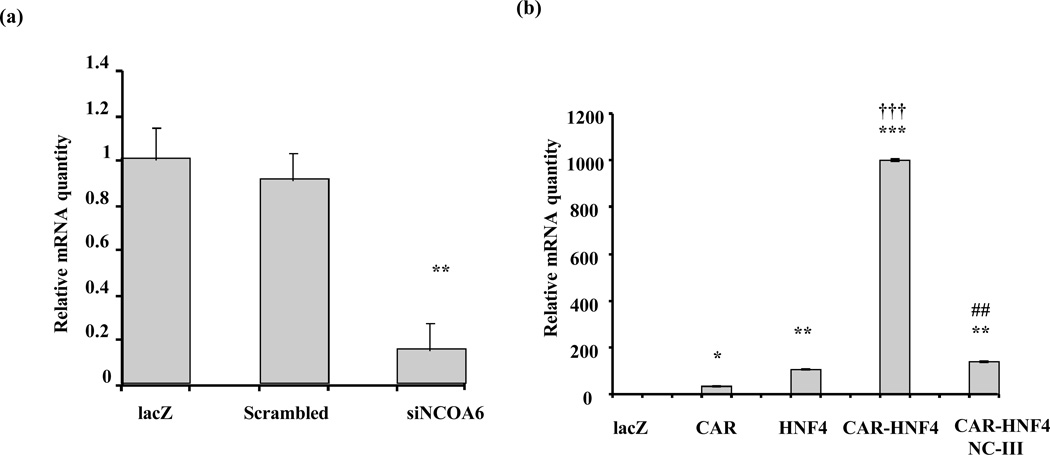
Silencing of NCOA6 down regulates constitutive and the synergistic induction of CYP2C9 mRNA in HepG2 cells
We examined the effect of NCOA6 siRNA on CYP2C9 mRNA in HepG2 cells. Cells were infected with either adenovirus expressing scrambled or NCOA6 siRNAs for 48 h with lacZ infected cells as control. Scrambled siRNA did not change CYP2C9 mRNA expression significantly in normal HepG2 cells. siNCOA6 (NC-III) on the other hand down regulated endogenous CYP2C9 mRNA expression more than 75% (Fig. 6a). Since we have earlier shown that CAR and HNF4α synergistically activate the CYP2C9 promoter and that the down regulation of NCOA6 levels greatly inhibits this synergistic activation, we examined whether there was any synergistic induction of CYP2C9 mRNA by CAR and HNF4α, and effect of siNCOA6 (NC-III) on the synergistic increase in CYP2C9 mRNA in vivo. We infected AdCAR and AdHNF4α individually and in combination in HepG2 cells and simultaneously down regulated NCOA6 levels with siRNA for NCOA6 (NC-III). Adenoviral expression of CAR and HNF4α produces a synergistic rather than additive 1000-fold induction of CYP2C9 mRNA in HepG2 cells compared to a 100-fold induction by HNF4α alone and a 35-fold induction by CAR alone. The synergistic effects of HNF4α and CAR on CYP2C9 mRNA in HepG2 cells were abolished by down regulating NCOA6 with adenoviral siRNA (p<0.01) (Fig. 6b). In contrast, silencing NCOA6 did not affect the induction of CYP2C9 mRNA by CAR alone or HNF4α alone (data not shown). Silencing of NCOA6 also did not affect the expression of mRNA of other known coactivators such as SRC-1, PBP, or GRIP-1 although siRNA for NCOA6 modestly decreased mRNA for PGC-1α (~40%) (data not shown). Taken together, the effects of NCOA6 siRNA on the synergistic transactivation by CAR and HNF4α and their mRNAs strongly indicate that NCOA6 acts as a bridging partner between CAR and HNF4α responsible for the synergistic upregulation of CYP2C9 gene expression.
Fig. 6. NCOA6 siRNA (III) prevents the synergistic effects of CAR and HNF4α on CYP2C9 mRNA in vivo in HepG2 cells.
(a) SiNCOA6 decreases the endogenous CYP2C9 expression. HepG2 cell were infected with adenovirus containing scrambled or siNCOA6(NC-III) for 48h. The cells were harvested, and total RNA was prepared to estimate constitutive CYP2C9 mRNA levels using qPCR. NC-III significantly decreases endogenous CYP2C9 mRNA at *(p<0.001). (b) Silencing of NCOA6 abolishes the synergistic effects of adenoviral expression of CAR and HNF4α on CYP2C9 mRNA in HepG2 cells. HepG2 cells were infected with adenovirus expressing lacZ, CAR, HNF4α, CAR-HNF4α and CAR-HNF4α with siNCOA6(NC-III) for 48h. The cells were harvested and total RNA was used to measure CYP2C9 mRNA by qPCR. CAR or HNF4α significantly increased CYP2C9 mRNA at *, p<0.05, **p<0.01, ***p<0.001. †††, synergistic rather than additive response to transfection with HNF4α and CAR at p<0.001. ##, NCIII significantly reduced the CYP2C9 mRNA response to HNF4α and CAR. Values represent means ± SE of triplicates.
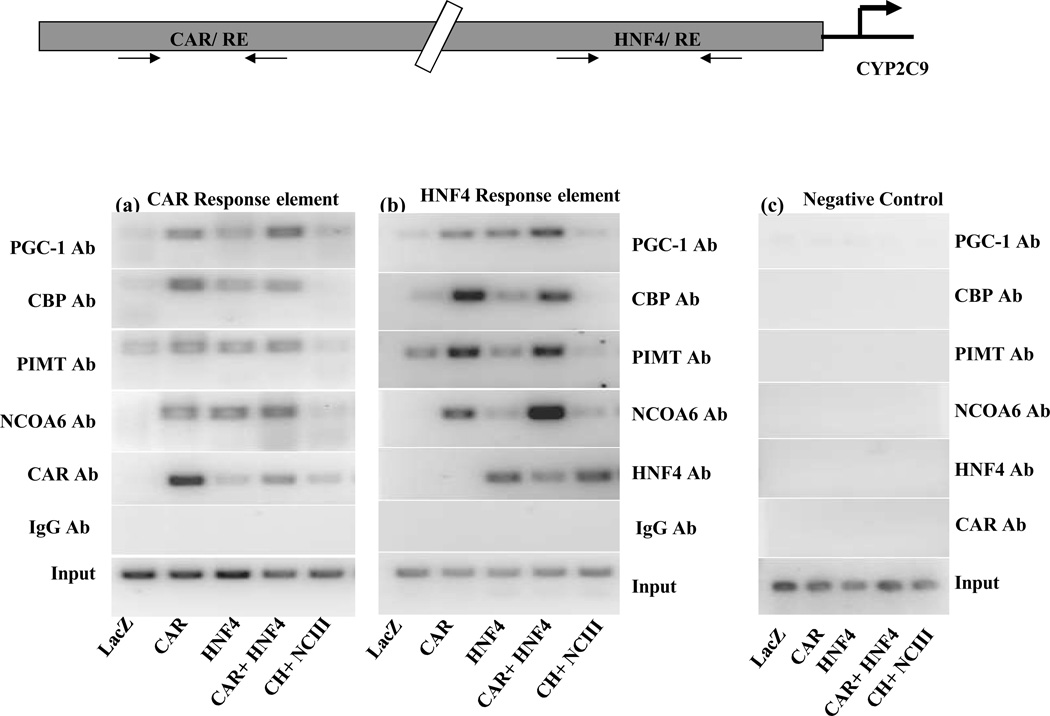
Recruitment analysis of CAR, HNFα and cofactors on the CAR response element and HNF4α response element of CYP2C9 promoter
ChIP assays were performed on the chromatin extracts from the HepG2 cells described above in Fig 6b to demonstrate the recruitment of nuclear receptors CAR and HNF4α to their respective binding sites on CYP2C9 promoter and the effect of silencing of NCOA6 on the association of coactivators to these nuclear receptors (Fig 7). The following PCR products were generated by primer pairs for the CAR-RE (Fig. 7a), HNF4α -RE (Fig. 7b) or for a negative control primers from GAPDH-CNAP-1 gene (Fig 7c). Input chromatin and non-immune IgG was used as a negative control are shown at the bottom of each panel. These ChIP assays demonstrate that CAR is recruited robustly to the CAR binding site (Fig 7a) on the promoter region of CYP2C9 in chromatin extracts prepared from HepG2 cells, particularly in cells overexpressing CAR and moderately in cells overexpressing CAR-HNF4α with or without the siRNA for NCOA6 (NC-III). As expected, HNF4α was recruited to its binding site (Fig.7b) on the CYP2C9 promoter in all the chromatin extracts from cells overexpressing HNF4α, CAR-HNF4α and siRNA for NCOA6 (NC-III) did not affect this recruitment. Antibodies for the coactivator NCOA6 robustly precipitated the HNF4α binding sites in chromatin extracts from cells overexpressing CAR, HNF4α and CAR-HNF4α, particularly in cells overexpressing both CAR and HNF4α, and also precipitated the CAR binding site although less robustly. This association of NCOA6 to both the HNF4α binding sites and CAR binding site was negligible in chromatin extracts from cells expressing NCOA6 siRNA. The coactivator PGC-1α was robustly associated with both the HNF4α binding sites and CAR binding sites in chromatin extracts from cells overexpressing CAR, HNF4α and CAR-HNF4α, while PIMT was associated with primarily with the HNF4α site. Silencing of NCOA6 with NC-III greatly reduced or abolished the association of CBP, PGC-1α and PIMT to the CYP2C9 promoter, suggesting that NCOA6 is required for the recruitment of cofactors from the mediator complex. Panel 7c shows no amplification of a non target gene from immunoprecipitated chromatins. Thus ChIP assays show that CAR and HNF4α interact with their respective binding sites on the CYP2C9 promoter in vivo in its native context, NCOA6 interacts with both the nuclear receptors and brings down both sites by its interaction with the nuclear receptors CAR and HNF4α, and silencing NCOA6 prevents not only the recruitment of NCOA6 but also other coactivators to the HNF4α -RE.
Fig. 7. Analysis of recruitment of the nuclear receptors CAR, HNF4α and coactivators to the CYP2C9 promoter by Chromatin Immunoprecipitation analysis.
ChIP analysis for CAR-HNF4α mediated recruitment of nuclear receptor cofactors to the CYP2C9 gene promoter. Chromatin extracts isolated from HepG2 cells infected individually with adenovirus expressing, lacZ, CAR, HNF4α, CAR-HNF4α and CAR-HNF4α with siNCOA6 (NC-III) were pre-cleared as described in Material and Methods and immunoprecipitated with antibodies for CAR, HNF4α, NCOA6, PIMT, CBP and PGC-1α . PCR was used to analyze the CYP2C9 promoter at the CAR binding sites (a) and HNF4α binding sites (b) control negative primers (c). Expression of CAR and HNF4α increased their recruitment to their respective sites. Immunoprecipitation with NCOA6, PIMT, CBP and PGC-1α showed their association with both CAR and HNF4α binding sites on CYP2C9 promoter. Silencing of NCOA6 essentially abolishes the recruitment of NCOA6, PIMT, CBP and PGC-1α to the HNF4α sites as well as many of the cofactors to the CAR and HNF4α binding sites.
Discussion
In the present study we address the mechanism of the synergistic activation of the human CYP2C9 promoter by CAR and HNF4α. Importantly, we identify NCOA6 as a new HNF4α-interacting protein using the HNF4α as the bait in yeast two hybrid screening. To identify the proteins that interact with full length HNF4α, we used a GST-pull down approach using nuclear extracts from HepG2 cells expressing CAR. Both CAR and CYP2C9 are expressed at very low levels in HepG2 cells (Ferguson et al., 2002), but overexpression of CAR (by infection with AdCAR) induces CYP2C9 mRNA expression in these cells. Therefore, we used nuclear extracts from HepG2 cells infected with AdCAR to identify binding protein partners in the CAR-HNF4α complex. Mass spectral analysis identified CAR, and immunoblotting identified NCOA6 and CAR as well as, CBP, PGC-1α and PIMT in this mega complex. As discussed earlier, nuclear receptor coactivators exist in sub-complexes (McKenna et al., 1999). Although there are two schools of thought whether these sub complexes form sequentially or combinatorially, recent evidence points to proteins that facilitate such linkage of these sub complexes, even for a transient period (Misra et al., 2002). Examples of such proteins include NCOA6, linking CBP to a PBP complex and PIMT, directly linking CBP with PBP. Consistent with this bridging function, we found proteins belonging to the known CBP sub-complex and Mediator sub-complex in the isolated CAR-HNF4α complexe from HepG2 cells. The synergistic activation of HNF4α and CAR on CYP2C9 promoter activity, effects of silencing NCOA6 on this synergism and the results of ChIP assays suggest that after recruitment of CAR and HNF4α to their respective binding sites, a set of coactivators are recruited to form a bridge between the receptors initiating the observed surge in transcriptional activity. siRNA studies suggest that NCOA6 is necessary for the formation of this bridge between HNF4α and CAR and for recruitment of other coactivators to the proximal HNF4α sites.
PBP (Zhu et al., 1997) and PRIP/NCOA6 (Zhu et al., 2000) are known to individually regulate the expression of the ap2 gene which is involved in adipogenesis (Qi et al., 2003). GRIP-1 and PBP have also been shown to function as coactivators for a CAR mediated increase in Cyp2b10 gene expression (Jia et al., 2005; Min et al., 2002). Although NCOA6 has shown to be a coactivator for CAR (Choi et al., 2005), CAR mediated gene expression of CYP2b10 was not affected in NCOA6 null mice (Guo et al., 2006; Sarkar et al., 2007). GRIP-1 belongs to the p160 family of coactivators (Leo and Chen, 2000). Although GRIP-1 could interact with CAR and HNF4α individually, GRIP-1 is not a major player in bridging function. Deletion of the SRC-1, SRC-2/GRIP-1, SRC-3/pCIP genes in knockout mice have no effect on CAR regulated gene transcription (Xia et al., 2007). PBP on the other hand, acts as a coactivator for CAR and studies in PBP-null mice indicate that PBP is necessary for the translocation of CAR to the nucleus and PBP regulates the hepatic expression of CAR (Jia et al., 2005). In the present study, we show that NCOA6 acts as an interacting partner for HNF4α (Fig.2). Since in vitro data shows that PBP can bind to CAR, and HNF4α binds to NCOA6 (Fig.2), we considered the possibility of a bridge formation either directly or through PIMT (Zhu et al., 2001). PIMT has been reported to from a bridge between NCOA6 and PBP (Misra et al., 2002).
Although there is also a possibility of an indirect interaction/mechanism between CAR and HNF4α with PIMT or PIMT like methyl transferases, one likely scenario for the CAR-HNF4α bridge formation is by the direct interaction of NCOA6 with both the receptors simultaneously. The NR-1 box containing the first LXXLL motif is found to interact with CAR alone (Fig.5b), whereas HNF4α interacted with the NR boxes coding for both the first LXXLL and the second LXXLL motifs (Fig. 5c). This opens the possibility that the NR-1 box binds to CAR and the NR-2 box binds to HNF4α, thereby directly bridging CAR and HNF4α to bring about the synergistic activation of the CYP2C9 promoter and the induction of CYP2C9 mRNA. Non-redundant coactivators like CBP and its binding proteins (HATs; histone acetylating proteins) are known to interact with both CAR and HNF4α, similarly the redundant coactivator PGC-1α (a member of the PGC-1 family known to be involved in splicing) interacts with both CAR and HNF4α. Although these coactivators are probably part of the CAR-HNF4α transcription complex, our data suggests it is unlikely that they are required for the bridge between CAR and HNF4α. For example, the redundant coactivator PGC-1 could rescue not the silencing effect of NCOA6 in CYP2C9 transactivation assays.
Promoter regions of genes have binding sites for numerous nuclear proteins and nuclear receptors. Therefore, the possibility of cross talk between binding partners is not only feasible but essential to transduce complex and sometimes contradictory signals for the regulation of a gene. Such a cross talk between GR and PPARγ has been documented on ap2 gene expression and between the coactivators PBP and NCOA6 (Qi et al., 2003). CAR also cross-talks with the forkhead transcription factor FOXO1 to repress its activation of an insulin response sequence in the glucose-6-phosphatase gene (Kodama et al., 2004). PXR also cross-talks with the insulin response transcription factor FoxA2 to repress transcription of genes involved in lipid metabolism in the mouse (Nakamura et al., 2007) and PXR represses glucagon activation of the glucose-6-phosphatase gene by binding to CREB, the cAMP response element binding protein, inhibiting CREB interaction with its DNA binding elements (Kodama et al., 2007). Several examples of either inhibitory cross-talk or potentiation between HNF4α and PXR or CAR have been reported. PXR appears to interfere with HNF4α-upregulation of CYP7A1 and the regulation of cholesterol metabolism (Bhalla et al., 2004; Li et al., 2007b). HNF4α has long been suggested to have a positive role in the PXR-mediated induction of CYP3A4. Possible potentiation between HNF4α and CAR has also been recently reported in the regulation of CYP3A4 (Li and Chiang, 2006) and steroid and bile acid sulfotransferase (SULT2A1) (Echchgadda et al., 2007).
Our study provides evidence for a new mechanism for the synergistic effect of HNF4α with CAR on expression of the human CYP2C9 gene, wherein cofactors are proposed to bridge distant receptor sites in the promoter, resulting in a synergistic effect on gene expression. In particular, NCOA6 appears to be an essential factor possibly providing a platform for recruitment of cofactors to this bridge.
Acknowledgments
The authors thank Drs. Georges Frech and Karen Heichman for their oversight of the two-hybrid project at Myriad Genetics, Salt Lake City. The authors acknowledge and thank Dr Jason Williams, Protein Microcharacterization facility NIEHS, for mass spectrometric analysis and Dr.Grace Kissling, Biostatistics branch, NIEHS for statistical analysis. The authors also thank Joyce Blaisdell and Sherry Coulter, NIEHS, for their assistance in this work.
This research was supported by the Intramural Research Program of the NIH and NIEHS (S.S., R.R. and J.A.G.)
Abbreviations used
- CAR
Constitutive Activate/Androstane Receptor
- HNF4α
Hepatocyte Nuclear Factor 4 alpha
- RE
Response element
- CYP
Cytochrome P-450
- NCOA6
Nuclear receptor coactivator 6 (PRIP/RAP250/ASC-2)
- PBP
PPAR binding protein (TRAP220/MED-1/DRIP205)
- CBP
CREB binding protein
- SRC-1
Steroid receptor coactivator-1
- PGC-1α
PPAR gamma coactivator-1
- PIMT
PRIP-interacting protein with methyltransferase domain
- NR-1
Nuclear receptor interacting box
- GRIP-1/SRC-2
glucocorticoid receptor interacting protein/steroid receptor coactivator-2
- ChIP
Chromatin immunoprecipitation
- Q-PCR
Quantitative polymerase chain reaction
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4- dichlorobenzyl)oxime
References
- Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279(43):45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, Goldstein JA. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308(2):495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kissling G, Negishi M, Goldstein JA. The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther. 2005;314(3):1125–1133. doi: 10.1124/jpet.105.087072. [DOI] [PubMed] [Google Scholar]
- Choi E, Lee S, Yeom SY, Kim GH, Lee JW, Kim SW. Characterization of activating signal cointegrator-2 as a novel transcriptional coactivator of the xenobiotic nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19(7):1711–1719. doi: 10.1210/me.2005-0066. [DOI] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282(42):30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. The Xenobiotic-Sensing Nuclear Receptors Pregnane X Receptor, Constitutive Androstane Receptor, and Orphan Nuclear Receptor Hepatocyte Nuclear Factor 4{alpha} in the Regulation of Human Steroid-/Bile Acid-Sulfotransferase. Mol Endocrinol. 2007;21(9):2099–2111. doi: 10.1210/me.2007-0002. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8(8):869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, LeCluyse EL, Negishi M, Goldstein JA. Regulation of human CYP2C9 by the constitutive androstane receptor: discovery of a new distal binding site. Mol Pharmacol. 2002;62(3):737–746. doi: 10.1124/mol.62.3.737. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A. 1996;93(16):8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem. 2002;277(1):209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52(4):349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, Roeder RG, Azorsa DO, Meltzer PS, Suh PG, Song EJ, Lee KJ, Lee YC, Lee JW. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23(1):140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Moore LB, Stoltz CM, McKee DD, Kliewer SA. Regulation of the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol. 2001;60(3):427–431. [PubMed] [Google Scholar]
- Guo D, Sarkar J, Ahmed MR, Viswakarma N, Jia Y, Yu S, Sambasiva Rao M, Reddy JK. Peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP) but not PPAR-interacting protein (PRIP) is required for nuclear translocation of constitutive androstane receptor in mouse liver. Biochem Biophys Res Commun. 2006;347(2):485–495. doi: 10.1016/j.bbrc.2006.06.129. [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Henderson L, Yue QY, Bergquist C, Gerden B, Arlett P. St John's wort (Hypericum perforatum): drug interactions and clinical outcomes. Br J Clin Pharmacol. 2002;54(4):349–356. doi: 10.1046/j.1365-2125.2002.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Moore R, Washburn KA, Negishi M. Activation by diverse xenochemicals of the 51-base pair phenobarbital- responsive enhancer module in the CYP2B10 gene. Mol Pharmacol. 1998;53(4):597–601. doi: 10.1124/mol.53.4.597. [DOI] [PubMed] [Google Scholar]
- Jia Y, Guo GL, Surapureddi S, Sarkar J, Qi C, Guo D, Xia J, Kashireddi P, Yu S, Cho YW, Rao MS, Kemper B, Ge K, Gonzalez FJ, Reddy JK. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc Natl Acad Sci U S A. 2005;102(35):12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19(9):6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol. 2004;24(18):7931–7940. doi: 10.1128/MCB.24.18.7931-7940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama S, Moore R, Yamamoto Y, Negishi M. Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene. Biochem J. 2007 doi: 10.1042/BJ20070481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoroski BJ, Zhang S, Cai H, Hutzler JM, Frye R, Tracy TS, Strom SC, Lehmann T, Ang CY, Cui YY, Venkataramanan R. Induction and inhibition of cytochromes p450 by the St. John's wort constituent hyperforin in human hepatocyte cultures. Drug Metab Dispos. 2004;32(5):512–518. doi: 10.1124/dmd.32.5.512. [DOI] [PubMed] [Google Scholar]
- Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245(1):1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Li Q, Chu MJ, Xu J. Tissue- and Nuclear Receptor-specific Function of the C-terminal LXXLL Motif of Coactivator NCoA6/AIB3 in Mice. Mol Cell Biol. 2007a doi: 10.1128/MCB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen W, Chiang JY. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J Lipid Res. 2007b;48(2):373–384. doi: 10.1194/jlr.M600282-JLR200. [DOI] [PubMed] [Google Scholar]
- Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos. 2006;34(5):756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125(3):411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Madan A, Graham RA, Carroll KM, Mudra DR, Burton LA, Krueger LA, Downey AD, Czerwinski M, Forster J, Ribadeneira MD, Gan LS, LeCluyse EL, Zech K, Robertson P, Jr., Koch P, Antonian L, Wagner G, Yu L, Parkinson A. Effects of prototypical microsomal enzyme inducers on cytochrome P450 expression in cultured human hepatocytes. Drug Metab Dispos. 2003;31(4):421–431. doi: 10.1124/dmd.31.4.421. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, Moore JT. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278(19):17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108(4):465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O'Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol. 1999;69(1–6):3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- Min G, Kemper JK, Kemper B. Glucocorticoid receptor-interacting protein 1 mediates ligand-independent nuclear translocation and activation of constitutive androstane receptor in vivo. J Biol Chem. 2002;277(29):26356–26363. doi: 10.1074/jbc.M200051200. [DOI] [PubMed] [Google Scholar]
- Misra P, Qi C, Yu S, Shah SH, Cao WQ, Rao MS, Thimmapaya B, Zhu Y, Reddy JK. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. J Biol Chem. 2002;277(22):20011–20019. doi: 10.1074/jbc.M201739200. [DOI] [PubMed] [Google Scholar]
- Moore DD. CAR: three new models for a problem child. Cell Metab. 2005;1(1):6–8. doi: 10.1016/j.cmet.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, Collins JL, Kliewer SA. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem. 2000;275(20):15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Moore R, Negishi M, Sueyoshi T. Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver. J Biol Chem. 2007;282(13):9768–9776. doi: 10.1074/jbc.M610072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Qi C, Surapureddi S, Zhu YJ, Yu S, Kashireddy P, Rao MS, Reddy JK. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem. 2003;278(28):25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398(6730):824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Raucy J, Warfe L, Yueh MF, Allen SW. A cell-based reporter gene assay for determining induction of CYP3A4 in a high-volume system. J Pharmacol Exp Ther. 2002;303(1):412–423. doi: 10.1124/jpet.102.038653. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276(40):36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- Sarkar J, Qi C, Guo D, Ahmed MR, Jia Y, Usuda N, Viswakarma N, Rao MS, Reddy JK. Transcription coactivator PRIP, the peroxisome proliferator-activated receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PParalpha and CAR, the constitutive androstane receptor, in mouse liver. Gene Expr. 2007;13(4–5):255–269. doi: 10.3727/000000006780666948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirlis D, Muangmoonchai R, Edwards M, Phillips IR, Shephard EA. Orphan receptor promiscuity in the induction of cytochromes p450 by xenobiotics. J Biol Chem. 2001;276(16):12822–12826. doi: 10.1074/jbc.M005930200. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274(10):6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16(6):893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Surapureddi S, Yu S, Bu H, Hashimoto T, Yeldandi AV, Kashireddy P, Cherkaoui-Malki M, Qi C, Zhu YJ, Rao MS, Reddy JK. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U S A. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72(3):231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson KM, Patterson JH, McQueen RH, Adams KF, Jr., Pieper JA. Effects of erythromycin or rifampin on losartan pharmacokinetics in healthy volunteers. Clin Pharmacol Ther. 1998;63(3):316–323. doi: 10.1016/S0009-9236(98)90163-1. [DOI] [PubMed] [Google Scholar]
- Xia J, Liao L, Sarkar J, Matsumoto K, Reddy JK, Xu J, Kemper B. Redundant enhancement of mouse constitutive androstane receptor transactivation by p160 coactivator family members. Arch Biochem Biophys. 2007;468(1):49–57. doi: 10.1016/j.abb.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, Guzelian PS, Evans RM. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14(23):3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell. 2004;16(6):919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res. 2004;64(20):7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Kan L, Qi C, Kanwar YS, Yeldandi AV, Rao MS, Reddy JK. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Cao WQ, Yeldandi AV, Rao MS, Reddy JK. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc Natl Acad Sci U S A. 2001;98(18):10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272(41):25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]