Abstract
The transcription factor E2F1 is a key regulator of proliferation and apoptosis but the molecular mechanisms that mediate these cell fate decisions remain unclear. Here, we identify FOXO transcription factors as E2F1 target genes that act in a feed-forward regulatory loop to reinforce gene induction of multiple apoptotic genes. We found that E2F1 forms a complex with FOXO1 and FOXO3. RNAi-mediated silencing of FOXO impaired E2F1 binding to the promoters of cooperative targets genes. A FOXO3 mutant insensitive to inactivation by survival kinases rescued the inhibitory effect of growth factor signaling on E2F1-mediated transcription and apoptosis. The E2F1/FOXO axis is frequently blocked in cancer, as evidenced by the specific down regulation of the FOXO-dependent E2F1 transcriptional program in multiple cancer types and by the association of a reduced E2F1/FOXO transcriptional program with poor prognosis. HDAC and PI3K inhibitors were identified as specific activators of E2F1/FOXO transcription, acting to enhance E2F1-induced apoptosis in a FOXO3-dependent manner. Notably, combining the HDAC inhibitor vorinostat with a PI3K inhibitor led to enhanced FOXO-dependent apoptosis. Collectively, our results identify E2F1/FOXO cooperation as a regulatory mechanism that places E2F1 apoptotic activity under the control of survival signaling. Therapeutic reactivation of this tumor suppressive mechanism may offer a novel broad-acting therapy for cancer.
Introduction
The role of the retinoblastoma tumor suppressor (Rb) in the control of E2F transcription factors function is now recognized as the key step in the regulation of cell cycle entry. In response to growth factor signaling Rb is inhibited by cyclin-dependent kinases (CDKs) leading to E2F activation and G0 to G1/S transition. Disruption of various components of this control pathway leads to deregulated proliferation and is central to the development of many forms of human cancer (1).
Previous studies have found that among the E2F family E2F1 protein is unique in its ability to induce apoptosis in addition to its more conventional role in the control of cellular proliferation (2). For example, E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis (3) and thymocytes derived from E2F1−/− mice are resistant to apoptotic stimuli (4). Following DNA damage E2F1 is stabilized by ATM and Chk2 phosphorylation leading to apoptosis induction (5, 6). The E2F1-dependent apoptosis is mediated through transcriptional induction of numerous proapoptotic genes and repression of survival genes (6–10). Collectively, these and other studies suggest that frequently deregulated E2F1 activity in cancer cells represents a potential Achilles heel that might be exploited in cancer therapy. However, in order to effectively harness this therapeutic potential, we must better understand the mechanisms that inactivate the apoptotic potential of E2F1 in cancer.
Previous studies from our group have demonstrated that the decision to proliferate or undergo a cell death response following E2F1 activation was regulated by PI3K/Akt function, coinciding with a specific repression of only a subset of E2F1 target genes (11, 12). Left unclear from these observations is the mechanism by which PI3K signaling can specifically prevent the induction of apoptotic but not the proliferative E2F1 target genes. Given the evidence for combinatorial mechanisms of transcription control involving other E2F family members, we hypothesized that the outcome of E2F1 activation might also be affected by the status of its different transcriptional partners.
Similar to E2F1, the FOXO family of transcription factors plays an important role in various cellular processes. Activation of FOXO activity can lead to growth arrest, apoptosis, increased stress resistance, differentiation, and metabolic responses in a system-specific manner (13). Phosphorylation of FOXO proteins by kinases such as AKT and SGK, downstream of PI3K activation by growth factor signaling, leads to their nuclear exclusion and subsequent degradation (14, 15). FOXO are also regulated by CK1, DYRK1A kinases and SIRT1 deacetylase. Different posttranslational modifications not only control FOXO localization but also might affect their transcriptional specificity (16). Thus FOXO proteins integrate the information on the cell state from multiple signaling pathways and translate it into transcriptional responses. Here we identify FOXO family of transcription factors as E2F1 transcriptional partners that control E2F1 transcriptional specificity and apoptosis providing a mechanistic link between PI3K signaling and E2F1.
Materials and Methods
Detailed Materials and Methods are available in Supplementary Information. Catalog numbers and oligonucleotide sequences used in this study can be found in Supplementary Table S7.
Cell Culture and Drugs
U2OS human osteosarcoma cells stably expressing ER-HA-E2F1 were obtained from Dr. Rotter. IMR90, 293T and U2OS cells were grown in DMEM with 10% FCS. Cell line identity was authenticated by DNA STR profiling assay. 4-hydroxy tamoxifen (OHT), LY294002, and G418 were from Sigma. Vorinostat (SAHA) was from ChemieTek.
Microarray analysis
For microarray analysis of U2OS ER-E2F1 cells RNA was prepared using RNeasy kit (Qiagen). and analyzed on Affymetrix U133A 2.0 microarrays. Microarray expression data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE39136.
Cell Viability and Apoptosis Assays
Relative cell numbers were quantified using MTS or CellTiter-Glo® assays (Promega). Caspase 3/7 activity assays were performed using Caspase-GLO 3/7 luminogenic substrate (Promega).
Adenovirus, Lentivirus Infection and siRNA
Adenoviruses expressing FOXO3 AAA and LacZ (Vector Biolabs) were used at MOI 10. For construction of pLEX-FOXO3, HindIII+XbaI fragment from pCDNA3-FLAG-FOXOs (Addgene) was cloned into BamHI site of pLEX-puro (Open Biosystems).
Reporter Assays
APAF1 promoter reporter construct (−396/+208) was kindly provided by Dr. Helin and described in (17). FOXO binding site was mutated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Reporter activities were measured using Dual-Glo® Luciferase Assay System (Promega).
Chromatin Immunoprecipitation
Chromatin immunoprecipitations of ER-HA-E2F1 were performed using EZ-ChIP kit (Millipore) according to manufacturer’s instructions.
Co-immunoprecipitation and Western blotting
FLAG-FOXO1 or FLAG-FOXO3 (Addgene) were precipitated from 293 cell lysates with anti-FLAG (M2) beads (Sigma) and cotransfected E2F1 was detected by Western blotting. The following antibodies were used: anti-OctA-probe (Flag, sc-807), anti-E2F1 (sc-193), anti-β-tubulin (sc-8035) from Santa-Cruz; anti-APAF1 (ALX-804-348) from Enzo; anti-cleaved PARP (9546), anti-FOXO1 (2880) from Cell Signaling); anti-FOXO3 (07-702) from Millipore.
Statistical analysis
All pooled results are presented as mean and SEM of triplicate experiments. Kaplan–Meier survival curves and their significance levels were generated using GraphPad software and compared using and the logrank test. p-values for the significance of non-zero linear regression trend in the drug sensitivity analysis was generated using GraphPad software.
Results
A feed-forward gene regulatory circuit involving E2F1 and FOXO transcription factors
Previous studies have suggested that specificity and consequently the outcome of transcription factor (TF) activation might be determined by the status of its TF partners (18–20). Accordingly, in considering possible mechanisms that underlie the specificity of E2F1 function, we have focused on the identification of TFs that might function together with E2F1. In this context, we made use of findings from the system biology field demonstrating that feed-forward regulation is the most highly overrepresented network motif in transcriptional networks (21). In this motif, one transcription factor activates a second transcription factor that then cooperates with the initial inducer to regulate target genes (Fig. 1A). Using the feed-forward mode of regulation as a working hypothesis, we decided to look for potential E2F1 cooperating transcription factors among the genes that are induced by E2F1. To identify TFs that are induced by E2F1, we performed expression microarray experiment in an inducible system of U2OS cells that express an ER-E2F1 fusion protein. Addition of 4-hydroxytamoxifen (OHT) leads to nuclear translocation of the chimeric protein and activation of E2F1-mediated transcription. This system was characterized previously and microarray analysis demonstrated the lack of OHT effect on transcription in the parental U2OS cells (22).
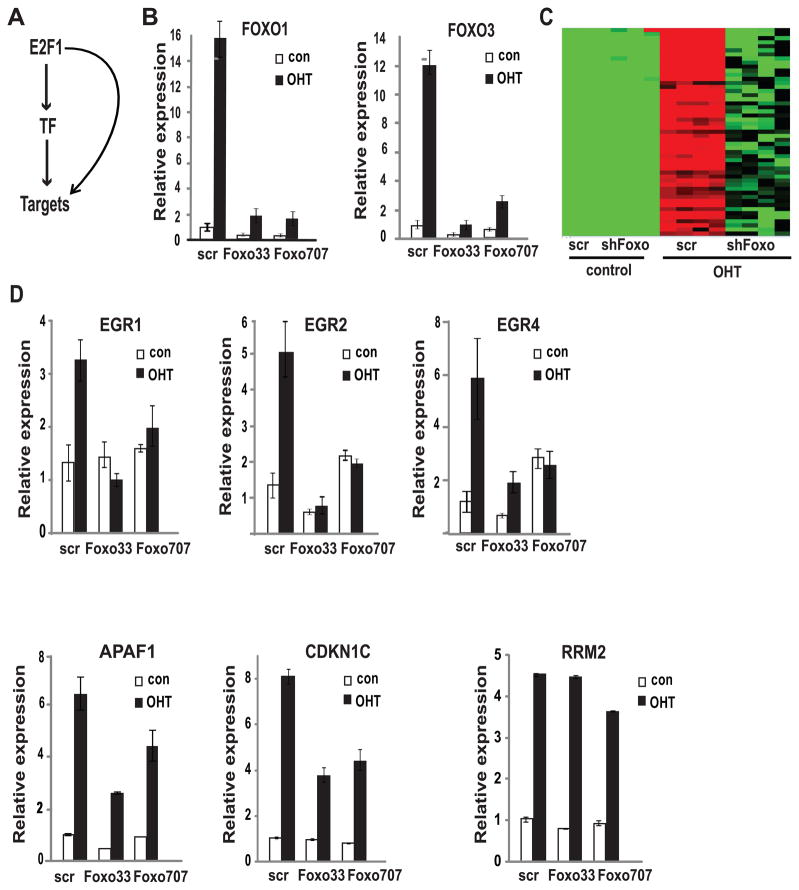
Figure 1. FOXO-dependent E2F1 transcriptional targets.
A. Schematic of a coherent feed-forward loop.
B. Real time RT-PCR analysis of FOXO1 and FOXO3 levels in U2OS ER-E2F1 cells infected with lentiviruses encoding shRNA constructs targeting FOXO1 and FOXO3 (FOXO33, FOXO707) or scrambled shRNA (scr). Twenty four hours post infection cells were serum starved for twenty four hours and then medium was replaced to serum-free medium with (OHT) or without (con) 20 nM OHT for six hours.
C. Microarray analysis of the effect of FOXO knockdown on E2F1-mediated transcription in the experiment described in (B). The heatmap represents the expression levels of top 52 FOXO-dependent E2F1 targets (red-high expression, green-low expression, black-intermediate expression).
D. Real-time PCR validation of the microarray analysis in (C) for EGR1, EGR2, EGR4, APAF1, CDKN1C and RRM2. See also Fig. S1 and Fig. S2
Amongst 1098 genes activated more than two fold in response to E2F1, 113 genes encoded transcriptional regulators (Table S1), suggesting that regulation of other TFs represents an important part of E2F1 transcriptional program. Upregulation of two members of the FOXO family of transcription factors, FOXO1 and FOXO3, was of particular interest to us given their established role in apoptosis (23). In addition, the well established inactivation of FOXO by PI3K signaling coupled with our previous findings on the role of this survival signaling pathway in the specific repression of the apoptotic E2F1 transcriptional program suggested that FOXO TFs might function as E2F1 apoptotic transcriptional cofactors (11, 14, 15). Notably, the induction of FOXO1 and FOXO3 mRNA by E2F1 is direct as it occurred in the presence of protein synthesis inhibitor and E2F1 binding to FOXO1 and FOXO3 promoters was demonstrated by chromatin immunoprecipitation experiments (24).
To explore a role of the FOXO proteins in E2F1-mediated transcription activation, we knocked down their expression and performed a microarray analysis of gene expression before and after induction of E2F1 activity by 4-OHT. Given previous work that has suggested overlapping and potentially compensatory function for FOXO1 and FOXO3 (25), at this stage we have not sought to distinguish individual roles for these proteins in the function of E2F1 and used two different shRNA constructs (Foxo33 and Foxo707) targeting both FOXO1 and FOXO3. Real time PCR analysis validated the strong induction of both FOXO1 and FOXO3 by OHT in cells infected with control shRNA and demonstrated the efficient knockdown of both FOXO family members by each of the shRNA constructs (Fig. 1B). Comparison of expression levels after E2F1 activation in cells infected with scrambled shRNA control to those in cells with FOXO knockdown demonstrated that among 1140 genes induced by E2F1 at least two fold, 278 (25%) were induced to significantly lower levels following FOXO knockdown (Fig. 1C and Table S2). The fact that FOXO knockdown did not affect E2F1-mediated induction of the majority of E2F1 target genes rules out the possibility of shFOXO effects on ER-E2F1 levels or nuclear translocation.
Examination of the list of FOXO-dependent E2F1 targets identified several genes that had been previously linked with apoptosis, including EGR genes (26–29), APAF1 (30), PTCH1 (31), EPHA7 (32), NLRP3 (33), and CTSB (34). Notably, the list also contained NR4A3, identified as serum-repressed apoptotic E2F1 target in a previous study (12). In addition, a CDK inhibitor CDKN1C (p57), was also induced by E2F1 in a FOXO-dependent manner. Finally, we also note the E2F1-mediated induction of numerous developmental genes, including several components of the Wnt pathway that were attenuated by FOXO knockdown.
To further explore the role of E2F1 and FOXO in the control of these genes, RT-PCR analysis was performed. As shown in Fig. 1D, the E2F1-mediated induction of the EGR1, EGR2, and EGR4 genes was abrogated following FOXO knockdown. In addition to the EGR genes, E2F1-mediated induction of APAF1, a key component of apoptosis pathway was also attenuated by FOXO knockdown (30). RT-PCR analysis also confirmed a role for FOXO in E2F1-mediated induction of additional genes, including CDKN1C (Fig. 1D) and several components of Wnt pathway (Fig. S1). In contrast, the induction of a cell cycle-promoting gene encoding ribonucleotide reductase subunit M2 (RRM2) was unaffected by FOXO knockdown (Fig. 1D).
To further establish a role for FOXO in the E2F1-mediated induction of selected genes we used a complementary approach in which FOXO3 was ectopically expressed in the U2OS ER-E2F1 cells. RT-PCR analysis following induction of E2F1 activity by OHT demonstrated that E2F1-mediated activation of the EGR4, APAF1, and CDKN1C genes by was enhanced by FOXO3. This cooperation was gene-specific since RRM2 was also induced by E2F1 but this induction was unaffected by FOXO3 (Fig. S2).
E2F1 and FOXO transcriptional cooperation
In considering the mechanism by which E2F1 and FOXO proteins might synergize in the activation of target genes, we found that E2F1 physically interacts with both FOXO1 and FOXO3 in co-immunoprecipitation experiments using tagged proteins (Fig. 2A). Similar experiments using endogenous E2F1 and FOXO proteins were inconclusive, likely due to technical limitations of the antibodies used. To address a role for FOXO in the function of E2F1, we carried out chromatin immunoprecipitation assays to measure the interaction of E2F1 with target promoters. As shown in Fig. 2B, the induction of E2F1 with OHT led to a recruitment of E2F1 to the APAF1 promoter. Importantly, this binding was dependent on FOXO since combined knockdown of FOXO1 and FOXO3 eliminated the binding of E2F1 to the APAF1 promoter. A similar result was seen for E2F1 recruitment to the EGR1 promoter. In contrast, binding of E2F1 to the promoter of a proliferative gene MCM4 was only marginally affected by FOXO knockdown demonstrating promoter specificity of FOXO effect.
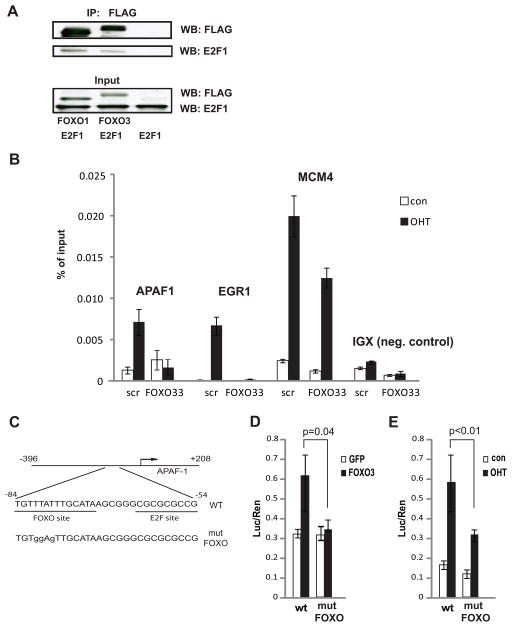
Figure 2. E2F1 and FOXO transcriptional cooperation.
A. 293T cells were cotransfected with the indicated combinations of E2F1 and either FLAG-tagged FOXO1, Flag-tagged FOXO3, or empty vector expression plasmids. Lysates were immunoprecipitated using anti-FLAG antibody and precipitates as well as inputs were analyzed by western blot for the presence of FOXO (using anti-FLAG antibody) and of E2F1 (using anti-E2F1 antibody).
B. U2OS ER-E2F1 cells were infected with lentiviruses encoding an shRNA construct targeting FOXO1 and FOXO3 (FOXO33) or scrambled shRNA (scr). Twenty four hours post infection cells were serum starved for twenty four hours and then medium was replaced to serum-free medium with (OHT) or without (con) 30 nM OHT for four hours.
Chromatin immunoprecipitation was performed with anti-HA antibody to detect binding of ER-HA-E2F1 to the indicated promoters.
C. Schematic representation of APAF1 promoter reporter constructs used in luciferase assays.
D. U2OS cells were co-transfected with APAF1 promoter reporter constructs depicted in (C) as indicated below the graphs together with FOXO3 or GFP control expression constructs. Luciferase assays were performed 48 hours later.
E. U2OS ER-E2F1 cells were transfected with equal amounts of APAF1 promoter reporter constructs depicted in (C) as indicated below the graphs. Twenty four hours post infection cells were serum starved for twenty four hours and then medium was replaced to serum-free medium with (OHT) or without (con) 60 nM OHT. Luciferase assays were performed five hours later.
To test whether direct binding of FOXO proteins to the APAF1 promoter is necessary for modulation of the E2F1-mediated APAF1 induction, we utilized a reporter gene under the control of APAF1 promoter sequence (Fig. 2C). The activity of the reporter was induced by cotransfected FOXO3 (Fig. 2D). We identified a potential FOXO recognition sequence located just upstream of an E2F binding site (Fig. 2C). Mutation of this site abolished the activation of the reporter by FOXO3 demonstrating that this site is, indeed, a functional FOXO site responsible for the induction of APAF1 by FOXO (Fig. 2D). Importantly, the E2F1-mediated induction of APAF1 promoter reporter was also attenuated by the mutation of the FOXO site (Fig. 2E). Taken together, these results establish APAF1 as novel direct FOXO target, cooperatively regulated by E2F1 and FOXO.
Survival signaling regulates the capacity of E2F1 to activate transcription and apoptosis through FOXO
A role for FOXO in the activation of a subset of E2F1 target genes, coupled with previous work demonstrating that FOXO activity is inhibited by growth factor-induced survival signaling, suggest a mechanism by which the apoptotic function of E2F1 could be controlled independent of the role of E2F1 in proliferation. To determine the extent to which FOXO-dependent E2F1 transcription is regulated by growth factor signaling, we examined the induction of two representative cooperative target genes, APAF1 and CDKN1C. As shown in Fig. 3A, the induction of these genes by E2F1 was inhibited in the presence of serum (FCS). To assess the contribution of FOXO inactivation to the inhibition of E2F1-mediated transcription by serum, we made use of a FOXO3 mutant that is immune to phosphorylation by survival kinases AKT and SGK (FOXO3-AAA)(14, 15). As demonstrated in Fig. 3A, this constitutively active FOXO3 mutant rescued the induction of APAF1 and CDKN1C by E2F1 in the presence of serum. In contrast, induction of a FOXO-independent proliferative E2F1 target, RRM2, was unaffected by serum and inhibited by FOXO3 AAA.
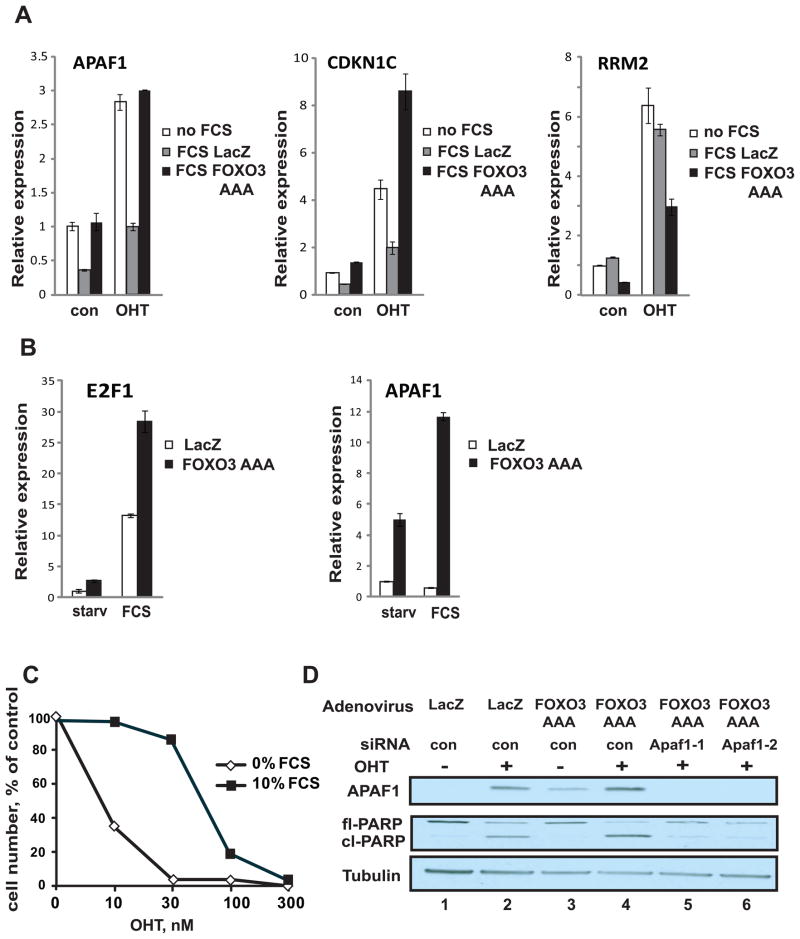
Figure 3. Survival signaling regulates the capacity of E2F1 to activate transcription and apoptosis through FOXO.
A. U2OS ER-E2F1 cells were either serum-starved (no FCS) or infected in the presence of serum with adenovirus encoding FOXO 3 AAA constitutively active mutant (FCS FOXO3 AAA) or with control LacZ adenovirus (FCS LacZ). Twenty four hours later medium was replaced with (OHT) or without (con) 30 nM OHT for 5 hours. Expression levels of the indicated genes were determined by real time RT-PCR.
B. IMR90 were infected with adenovirus encoding FOXO 3 AAA constitutively active mutant (FOXO3 AAA) or with control LacZ adenovirus (LacZ). in serum-free medium. After 40 hr cells medium was replaced to either serum-free mediun (starv) or medium containing 10% serum (FCS) for 20 hours. Expression levels of the indicated genes were determined by real time RT-PCR.
C. Dose-response curves in U2OS cells treated with a range of OHT concentrations in the presence (10% FCS) or the absence (0% FCS) of serum. Relative cell numbers were determined using MTS assay forty eight hours after OHT addition. See also Fig. S3. Measurements were done in triplicate wells with standard deviations smaller than the symbols representing the averages.
D. U2OS ER-E2F1 cells were transfected with the indicated siRNA duplexes in the presence of 10% FCS. Twenty four hours later cells were infected with adenoviruses encoding FOXO 3 AAA constitutively active mutant (FOXO3) or with control LacZ adenovirus. Twenty four hours later medium was replaced with (+) or without (−) 20 nM OHT for 48 hours. Expression of APAF1 and PARP was analyzed by Western blotting. Tubulin served as a loading control.
To extend the observation of cooperation between E2F1 and FOXO to normal cells and endogenous E2F1, we evaluated the effects of FOXO3 AAA on serum induced transcription in IMR90 fibroblasts. Whereas E2F1 was robustly upregulated following addition of serum to starved cells infected with a control LacZ virus, APAF1 was not induced. Infection of cells with FOXO3 AAA adenovirus resulted in significant induction of APAF1 mRNA in starved cells and enabled its further upregulation by serum (Fig. 3B).
Previous work has shown that E2F1-induced apoptosis can be suppressed by survival signals provided by the addition of serum (12). This result is recapitulated by the experiment in Fig. 3C, demonstrating that the induction of E2F1 activity in U2OS ER-E2F1cells following OHT addition resulted in a dose-dependent cell death response that was inhibited by serum (FCS). This apoptosis was E2F1-dependent since similar treatment of parental U2OS cells with OHT did not affect their growth (Fig. S3). Making use of low OHT concentrations where apoptosis is inhibited by serum, we have further explored the role of FOXO and APAF1 in E2F1-mediated apoptosis.
As shown by the Western blot analysis in Fig. 3D, activation of E2F1 by OHT led to a moderate induction of APAF1 in the presence of serum (lane 1, 2) as did overexpression of a phosphorylation-resistant FOXO3-AAA mutant (compare lanes 1 and3). The combined action of E2F1 and FOXO led to a further increase in APAF1 levels (lane 4). Coincident with the induction of APAF1 was the cleavage of Poly (ADP-ribose) polymerase (PARP) as seen by the reduction in full length PARP and the appearance of a cleaved form of the protein. Importantly, knockdown of APAF1 using two independent siRNAs strongly attenuated this apoptotic response (lane 5, 6) establishing a central role for APAF1 in the E2F1/FOXO apoptotic axis.
The E2F1/FOXO transcriptional program is reduced in cancer
Amongst the various changes accompanying the development of an oncogenic phenotype is the loss of capacity to initiate cell death (35). Given the apparent role of FOXO transcription factors in defining the transcriptional specificity of E2F1 to include the induction of apoptosis, we evaluated the pattern of expression of FOXO-dependent E2F1 target genes as cells transition from a normal state to an oncogenic state. To develop a measure of the E2F1/FOXO transcription program, we first identified those probe sets on the microarray that were induced at least two fold by E2F1 in the presence of FOXO activity. Then, we identified the subset from this group whose induction by E2F1 was reduced at least 1.5 fold by FOXO knockdown. This yielded a total of 52 probe sets representing 51 genes (Table S3). We then compared the average expression of this group of genes in normal and tumor samples making use of several publicly available expression datasets.
We started our analysis with a sarcoma dataset, given the fact that our initial microarray experiments were performed in the osteosarcoma cell line U2OS. As seen in Fig. 4A, the expression of the FOXO-dependent E2F1 transcriptional signature was significantly reduced in sarcomas as compared to normal fat tissue. To evaluate the extent to which this distinction between normal and tumor samples was a reflection of the FOXO-dependent component of the E2F1 transcriptional program, we generated a second signature that included genes whose induction by E2F1 was not affected by FOXO in our microarray analysis (Table S4). As seen in Fig. 4A, this signature failed to exhibit a significant distinction between tumor and normal samples.
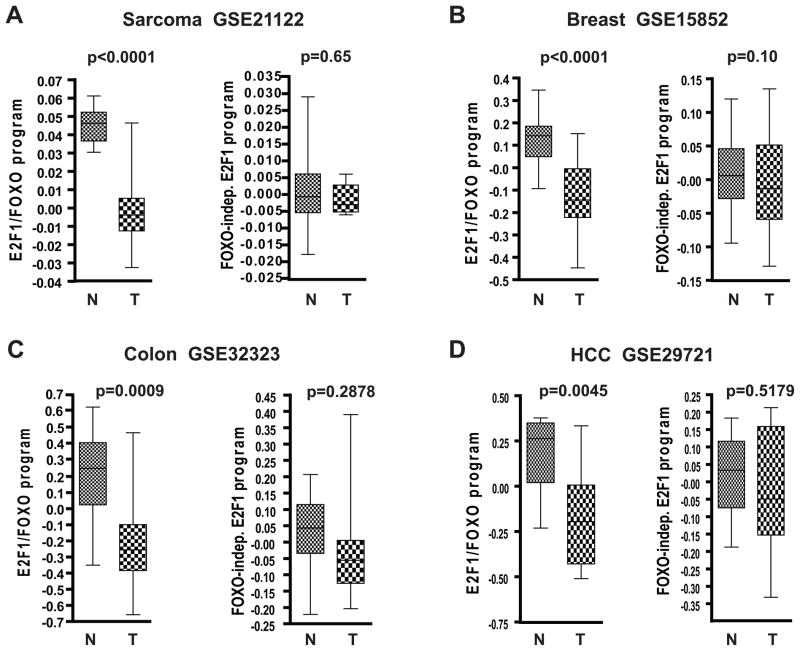
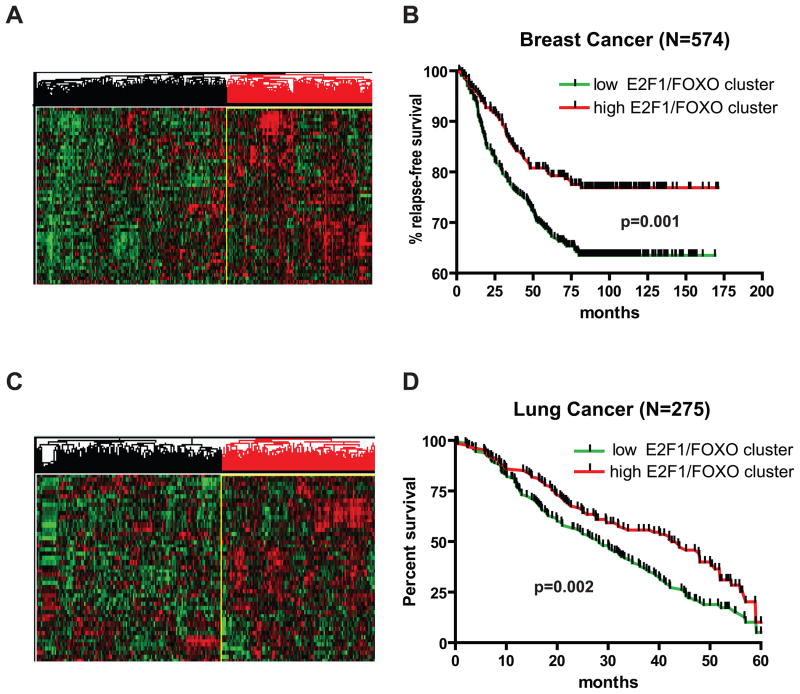
Figure 4. Inactivation of E2F1/FOXO transcriptional program is observed in multiple cancer types and associates with poor prognosis.
E2F1/FOXO targets in the ER-E2F1 microarray analysis were defined as probes induced at least two fold by OHT in sh-scr samples, whose fold induction by OHT was attenuated at least 1.5-fold by shFOXO (Table S3). FOXO-independent E2F1 targets were defined as probes induced at least two fold by OHT in sh-scr samples, whose induced levels did not differ by more than 10% between sh-scr and sh-FOXO samples (Table S4).
The box shows 25th percentile to 75th percentile with a line at the median. The whiskers indicate the highest and the lowest values. In the analysis of matched tumor and normal samples from the same individual we used paired t-test to calculate the significance of the differences in expression. For unmatched samples Mann-Whitney test was used for p-value calculations. N-normal, T-tumor.
A. Analysis of 149 sarcomas and 9 normal fat samples
B. Analysis of 43 matched pairs of breast tumors and normal samples
C. Analysis of 17 matched pairs of colorectal tumors and normal samples
D. Analysis of 11 matched pairs of hepatocellular carcinomas (HCC) and normal liver samples
To extend our observations to epithelial cancer types we analyzed five additional datasets representing breast, colorectal and liver cancers. The characteristics of all datasets and summary of the results are shown in Table S5. In all datasets we found a significant reduction of the FOXO-dependent E2F1 transcriptional signature in tumor samples (Fig. 4 and Table S5). In contrast, the expression of the FOXO-independent E2F1 transcriptional signature did not significantly differ between tumor and normal samples in four datasets (Fig. 4 and Table S5) and was downregulated to a smaller extent than the FOXO-dependent E2F1 program in two datasets (Table S5). The difference in the extent of downregulation in tumors between FOXO-dependent and FOXO-independent E2F1 signatures was statistically significant in a combined analysis of all six datasets (p=0.03, Wilcoxon paired signed-rank test). Taken together, these results demonstrate that specific inactivation of the FOXO-dependent arm of E2F1 transcriptional program occurs in multiple cancer types.
A reduced FOXO-dependent E2F1 signature is associated with poor prognosis in breast and lung cancer
To evaluate the potential role of the FOXO-dependent E2F1 transcriptional program in cancer outcomes, we performed survival analysis studies in breast and lung cancer. We extracted the expression values of the 52 FOXO-dependent E2F1 target probes sets described above (Table S3) from a large breast cancer dataset (36). Unsupervised clustering of samples resulted in identification of two main sample clusters, stratifying the tumors into high and low expressors of the FOXO-dependent E2F1 transcriptional program (Fig. 5A). Survival analysis demonstrated that patients whose tumors were characterized by a high expression of this program had significantly better prognosis than those from the low expression cluster. Similar results were obtained in an analysis of large lung adenocarcinoma dataset (37) (Fig. 5B).
Figure 5. Inactivation of E2F1/FOXO transcriptional associates with poor prognosis in breast and lung cancer.
A. Expression data for the E2F1/FOXO signature described above (Table S3) was extracted from the previously compiled combined BC1143 breast cancer dataset of 1143 tumors (36). This dataset consists of ten independent publicly available datasets representing various subtypes and stages of breast cancer. Genes and samples were clustered using unsupervised hierarchical clustering as described in Methods. Red color indicates high expression and green color indicates low expression.
B. 574 patients from the BC1143 dataset, for which survival data was available, were stratified into two groups according to the clustering results in (A) and survival curves were constructed for each group.
C, D Five year survival analysis of 275 lung cancer patients was performed as in A, B using a publicly available dataset (37).
In summary, lower expression of the FOXO-dependent arm of E2F1 transcriptional program associates with poor prognosis in breast and lung cancer. Taken together with our results on a widespread downregulation of this program in multiple cancer types, and the experimental results on the cooperation between E2F1 and FOXO3 in apoptosis induction, these results suggest that FOXO-dependent part of E2F1 transcriptional program is an important novel tumor suppressive mechanism, frequently targeted in cancer.
HDAC and PI3K inhibitors activate E2F1/FOXO transcriptional program and apoptosis
The potential tumor suppressive function of FOXO/E2F1 cooperation and its loss in various cancer types, prompted us to identify opportunities to reactivate the FOXO-dependent E2F1 transcriptional program in tumors as a therapeutic strategy. To this end, we made use of the Connectivity Map, a reference collection of gene-expression profiles from three human cancer cell lines treated with bioactive small molecules that enables the identification of compounds that induce the expression of a particular gene signature (38). A query with the gene expression signature consisting of FOXO-dependent E2F1 targets identified multiple HDAC inhibitors (HDACi) including trichostatin A (TSA), vorinostat (SAHA) and scriptaid as the top ranking inducers of the E2F1/FOXO transcriptional program. Notably, highly significant upregulation of E2F1/FOXO signature was observed in all three cell lines used for the Connectivity Map analysis representing prostate, breast and blood cancers (Table S6 and Fig. 6A). The induction of the FOXO-dependent E2F1 signature by HDACi was highly specific to this signature as demonstrated by low specificity values.
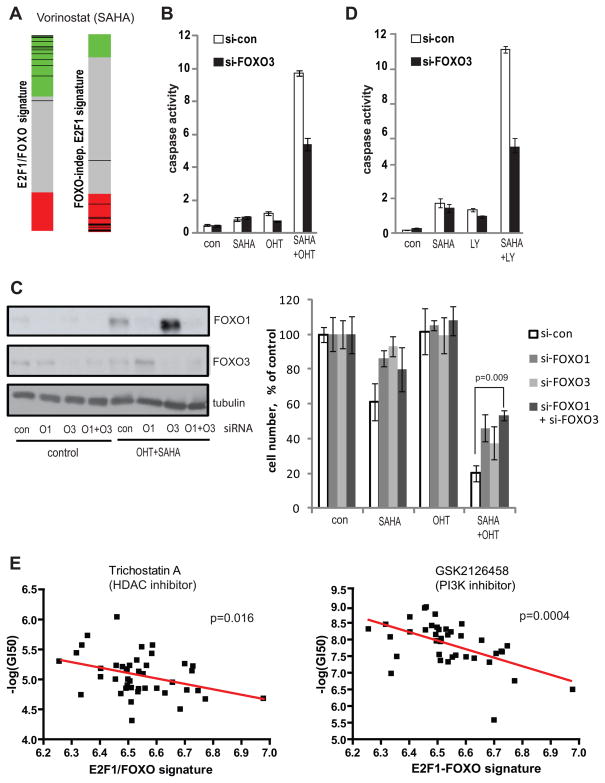
Figure 6. HDAC and PI3K inhibitors activate E2F1/FOXO transcriptional program and apoptosis.
A. Connectivity Map analysis of vorinostat (SAHA) effect on the expression of FOXO-dependent and FOXO-independent E2F1 signatures. Each black line represents a connectivity score from an individual vorinostat experiment. Experiments where vorinostat induced the corresponding signature are located in green areas (positive score), whereas experiments where vorinostat downregulated the corresponding signature are located in red areas (negative score).
B. U2OS ER-E2F1 cells were transfected with siRNA targeting FOXO3 (si-FOXO3) or with a non-silencing control (si-con). Twenty four hours later cells were treated with 1 uM SAHA, 20 nM OHT or their combination for 20 hours in the presence of 10% serum. Combined activity of caspase 3 and caspase 7 was measured using luminogenic substrate and normalized to cell numbers.
C. U2OS ER-E2F1 cells were transfected with siRNA targeting FOXO1 (O1) and FOXO3 (O3) separately or in combination, or with a non-silencing control (si-con). Twenty four hours later cells were treated with 1 uM SAHA, 10 nM OHT or their combination for 72 hours in the presence of 10% serum.
Left panel: Western blot for FOXO1, FOXO3 and tubulin (loading control) demonstrating knockdown efficiency. Right panel: Cell Titer Glo assay measurements of relative cell numbers as percentages of untreated controls.
D. U2OS cells were transfected with siRNA targeting FOXO3 (si-FOXO3) or with a non-silencing control (si-con). Twenty four hours later cells were treated with 1 uM SAHA, 50 uM LY294002 (LY) or their combination for 20 hours in the presence of 10% serum. Combined activity of caspase 3 and caspase 7 was measured using luminogenic substrate and normalized to cell numbers.
E. Publicly available gene expression and drug sensitivity data for a panel of fifty breast cancer cell line was from Heiser et. al. (39). Expression of the E2F1/FOXO transcriptional program was quantified as described in Fig. 4 and correlated with the sensitivity to indicated drugs. p-values represent the significance of non-zero linear regression trend.
In stark contrast to the activation of the FOXO-dependent E2F1 signature by HDACi, the signature consisting of FOXO-independent E2F1 targets was repressed by HDAC inhibitors as demonstrated in Fig. 6A for vorinostat. This result suggests that HDAC inhibitors specifically induce FOXO-specific subset of E2F1 targets while repressing FOXO-independent E2F1 targets. Similar specific activation of the FOXO-dependent E2F1 signature was observed for a PI3K inhibitor, LY294002, in agreement with the established inhibitory role of PI3K signaling on FOXO activity (Table S6) (14, 15).
To examine the role of HDAC inhibitors in E2F1-induced apoptosis we treated U2OS ER-E2F1 cells with SAHA alone, OHT alone or with their combination. While treatment of cells with SAHA alone resulted in only mild apoptotic response, it potently enhanced the apoptotic activity of E2F1 (Fig. 6B). Further, this apoptotic caspase activation was attenuated by FOXO3 knockdown, suggesting important role of FOXO3 in mediating the apoptotic synergy between E2F1 and SAHA. We then studied the effect of individual and combined knockdown of FOXO1 and FOXO3 on cell viability following separate and combined treatment of cells with SAHA and OHT (Fig. 6C). Western blot analysis confirmed efficient knockdown of FOXO1 and FOXO3 by corresponding siRNAs. Whereas the low concentration of OHT did not affect cell proliferation in the presence of serum, SAHA strongly synergyzed with OHT in decreasing cell numbers. Importantly, single knockdowns of FOXO1 or FOXO3 significantly attenuated this toxicity with combined knockdown resulting in the greatest degree of protection.
Next, we combined SAHA with a PI3K inhibitor (LY294002) to further activate FOXO activity. As demonstrated in Fig. 6D, treatment of U2OS with this combination led to a synergistic apoptosis induction, which was again attenuated by FOXO3 knockdown. Thus, the synergy between HDAC and PI3K inhibitors during apoptosis induction is mediated at least in part through FOXO-dependent pathway.
Our results so far suggested that HDAC inhibitors and PI3K inhibitors specifically activate FOXO-dependent E2F1 transcription and that activation of this transcriptional program has tumor suppressive properties. Accordingly, we would expect that these drugs will be more active in tumors where this program is inactivated, reflected by a lower basal E2F1/FOXO signature. To test this hypothesis, we made use of publicly available data from a recent study that profiled 50 breast cancer cell lines for drug sensitivity and basal gene expression (39). As demonstrated in Fig. 6E, we indeed found a statistically significant inverse correlation between the average expression of FOXO-dependent E2F1 signature and sensitivity to an HDAC inhibitor, trichostatin A. Similar result was observed for a PI3K inhibitor GSK2126458.
In summary, we identify HDAC inhibitors and PI3K inhibitors as specific activators of FOXO-dependent E2F1 transcriptional program and demonstrate FOXO-dependent apoptotic synergy of HDAC inhibitor with E2F1 and with PI3K inhibitor. Growth inhibition by PI3K and HDAC inhibitors correlates with the extent of E2F1/FOXO signature downregulation, suggesting potential use of this signature as a mechanism-based biomarker for patient selection in future clinical trials for these drugs.
Discussion
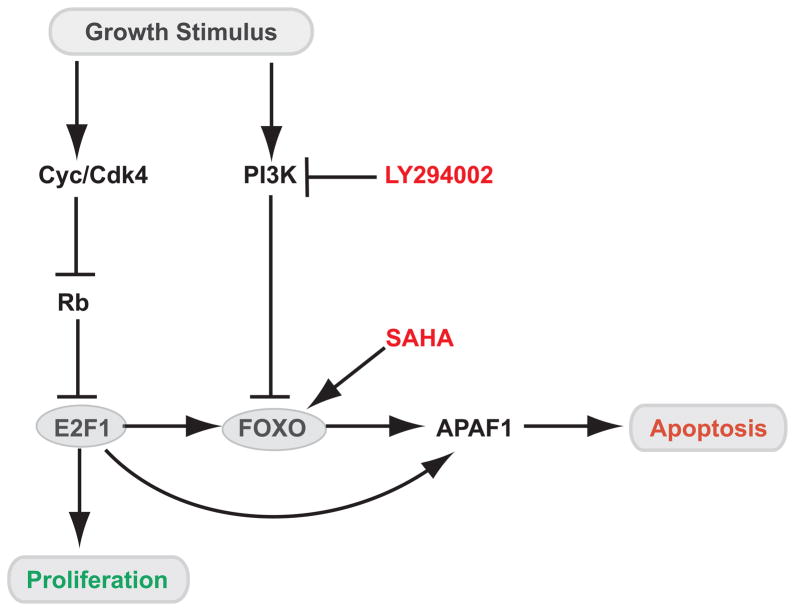
The role of the Rb/E2F pathway in the control of critical cell fate decisions including proliferation, quiescence, development, differentiation, metabolism and cell death, has been well established in previous work (40). Particularly intriguing is the apparent dual role of the E2F1 transcription factor in the activation of gene expression programs leading to proliferation and apoptosis (41). The identification of FOXO as component of the E2F1-induced apoptotic program, coupled with the established role of survival signaling events regulating FOXO activity, provides a molecular basis by which survival signals can control the outcome of E2F1 activation and block the death component (Fig. 7).
Figure 7. Model for a role of FOXO in E2F1-dependent transcription and apoptosis.
Growth stimuli result in activation of cyclin-dependent kinase activity with subsequent inactivation of the retinoblastoma protein (Rb) and accumulation of free E2F1. E2F1 induces expression of multiple proliferative genes, as well as FOXO genes, and then cooperates with FOXO proteins to induce a subset of target genes, including apoptotic genes such as APAF1. PI3K signaling initiated by growth factors specifically represses the induction of this subset of E2F1 targets through phosphorylation and subsequent nuclear exclusion of FOXO. Since the induction of proliferative E2F1 targets is FOXO-independent, it is not inhibited by growth factors. FOXO-dependent E2F1 transcription and apoptosis could be activated in the presence of growth stimulus through inhibition of PI3K (i.e by LY294002) and/or using HDAC inhibitors (i.e SAHA).
The role for E2F1 and FOXO acting in combination highlights an example of the concept of combinatorial transcription control by which individual transcription factors mediate their function through cooperative interactions. The significance of combinatorial transcription control is at least two-fold. First, a mechanism whereby two transcription factors act in combination generates a much greater specificity of promoter recognition than can be achieved with either factor alone since it is the recognition of the combined target sequences that defines function. Second, the opportunity for the individual transcription factors to form multiple combinations provides the potential for a much greater repertoire of specificity than what could be achieved through the action of single proteins. Further, the coherent feed-forward loop (FFL) established by E2F1 and FOXO may enhance the cell’s ability to distinguish normal and abnormal E2F1 signals by generating a delay in the activation of targets such as APAF1 due to the time required to accumulate FOXO. Upon E2F1 removal, however, there is no delay in shutting off APAF1 even though FOXO remains present. Such an asymmetric delay device can thus work as a noise filter, which responds only to sustained activity of the upstream activator and rejects response to transient input.
We also believe the spectrum of control involving the combination of E2F1 and FOXO goes beyond the activation of an apoptotic program, given the identification of multiple genes as FOXO-dependent E2F1 targets. For instance, the identification of multiple developmental genes, including several Wnt signaling pathway components, as cooperative targets suggests a potential role for E2F1/FOXO crosstalk in development and differentiation. In addition, we identify CDKN1C (p57), a negative regulator of the cell cycle through CDK inhibition, as FOXO-dependent E2F1 target (Fig. 1C). Thus, the feed forward loop between E2F1 and FOXO family might contribute not only to apoptosis but also to growth arrest. The repression of proliferative E2F1 targets such as RRM2 by FOXO3 AAA (Fig. 3A) is consistent with this notion and might be either direct (as described previously for Cyclin D) or indirect through the induction of CDK inhibitors (42). Although additional studies will be required to better understand the role of E2F1/FOXO cooperation in different cellular contexts, the overall effect seems to be tumor suppressive based on the observation that the E2F1/FOXO transcriptional program is reduced as normal cells transition to an oncogenic state. The implication of this finding is that while E2F1 likely participates in various cellular processes, it is the FOXO-dependent subset of E2F1 targets that identifies a tumor suppressing transcriptional program. Given the frequent functional inactivation of FOXO in human tumors (43), this underscores the significance of recognizing the complexity of gene control mediated by individual transcription factors and the importance of identifying the combinations that define unique transcriptional regulatory programs.
Intriguingly, in addition to joint regulation by E2F1 and FOXO, APAF1, as well as other apoptotic genes, are regulated by a tumor suppressor, p53. Since p53 itself is activated by E2F1, this suggest an even more complex regulatory scheme where activities of multiple checkpoint pathways interconnected through an array of FFLs and controlled by upstream signaling pathway need to be activated in order to trigger apoptosis (44).
Finally, the identification of a regulatory system involving E2F1 and FOXO, and the importance of control of this pathway in determining oncogenic outcomes, provides an opportunity to develop new and novel therapeutic strategies that could reactivate the E2F1/FOXO program. The identification of HDAC and PI3K as targets, and the potential of a SAHA/PI3K inhibitor combination to have therapeutic benefit in tumors lacking the E2F1/FOXO program, represents one such opportunity. SAHA is already approved for the treatment of cutaneous T-cell lymphoma and currently is tested in clinical trials for additional cancer types. Likewise, many PI3K inhibitors are currently in early phase clinical studies. The strong apoptotic synergy between SAHA and a PI3K inhibitor suggests that a combination of these two drugs, coupled with the selective use in a population of patients exhibiting reduced E2F1/FOXO program, could represent a novel therapeutic strategy to test in future clinical studies.
Supplementary Material
Acknowledgments
We would like to thank Dr. Rotter for U2OS ER-E2F1 cells, Dr. Helin for the APAF1 reporter construct, Drs. Mathey-Prevot and Milyavsky for the critical reading of the manuscript, Dr. Lucas for statistical advice and all members of the Nevins lab for helpful discussions. This work was supported by grants from the NIH (CA104663 and CA106520).
Footnotes
Potential conflict of interest: None
References
- 1.Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: where cancer meets the cell cycle. Exp Biol Med. 2006;231:1271–81. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg D. E2F1 pathways to apoptosis. FEBS Lett. 2002;529:122–5. doi: 10.1016/s0014-5793(02)03270-2. [DOI] [PubMed] [Google Scholar]
- 3.Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, et al. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–61. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 5.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 6.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 7.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–8. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 8.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;21:21. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 9.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–34. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 10.Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene. 2002;21:1359–69. doi: 10.1038/sj.onc.1205157. [DOI] [PubMed] [Google Scholar]
- 11.Hallstrom TC, Nevins JR. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100:10848–53. doi: 10.1073/pnas.1831408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–50. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–65. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 16.Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moroni MC, Hickman ES, Denchi EL, Caprara G, Colli E, Cecconi F, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–8. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 18.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci U S A. 2003;100:5136–41. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–80. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 22.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011;1813:1978–86. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Nowak K, Killmer K, Gessner C, Lutz W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta. 2007;1769:244–52. doi: 10.1016/j.bbaexp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed MM, Sells SF, Venkatasubbarao K, Fruitwala SM, Muthukkumar S, Harp C, et al. Ionizing radiation-inducible apoptosis in the absence of p53 linked to transcription factor EGR-1. J Biol Chem. 1997;272:33056–61. doi: 10.1074/jbc.272.52.33056. [DOI] [PubMed] [Google Scholar]
- 27.Lim JH, Park JW, Min DS, Chang JS, Lee YH, Park YB, et al. NAG-1 up-regulation mediated by EGR-1 and p53 is critical for quercetin-induced apoptosis in HCT116 colon carcinoma cells. Apoptosis. 2007;12:411–21. doi: 10.1007/s10495-006-0576-9. [DOI] [PubMed] [Google Scholar]
- 28.Pignatelli M, Luna-Medina R, Perez-Rendon A, Santos A, Perez-Castillo A. The transcription factor early growth response factor-1 (EGR-1) promotes apoptosis of neuroblastoma cells. Biochem J. 2003;373:739–46. doi: 10.1042/BJ20021918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie B, Wang C, Zheng Z, Song B, Ma C, Thiel G, et al. Egr-1 transactivates Bim gene expression to promote neuronal apoptosis. J Neurosci. 2011;31:5032–44. doi: 10.1523/JNEUROSCI.5504-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein, homologous to C. elegans Ced-4, participates in cytochome c-dependent activation of caspase-3. Cell. 1997;90:405–13. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 31.Thibert C, Teillet MA, Lapointe F, Mazelin L, Le Douarin NM, Mehlen P. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science. 2003;301:843–6. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 32.Depaepe V, Suarez-Gonzalez N, Dufour A, Passante L, Gorski JA, Jones KR, et al. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–50. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- 33.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–6. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 34.Deiss LP, Galinka H, Berissi H, Cohen O, Kimchi A. Cathepsin D protease mediates programmed cell death induced by interferon-gamma, Fas/APO-1 and TNF-alpha. EMBO J. 1996;15:3861–70. [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 36.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 39.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–9. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–7. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 41.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cel Biol. 2002;22:7842–52. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 44.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–48. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.