Abstract
Persistence and survival under various environmental stresses has been attributed to the capacity of most bacteria to form biofilms. In aquatic environments, the symbiotic bacterium Vibrio fischeri survives variable abiotic conditions during its free-living stage that dictates its ability to colonize the squid host. In the present study, the influence of different abiotic factors such as salt concentration, temperature, static/dynamic conditions, and carbon source availability were tested to determine whether biofilm formation occurred in 26 symbiotic and free-living V. fischeri strains. Statistical analysis indicate that most strains examined were strong biofilm producers under salinity concentrations that ranged between 1–5%, mesophilic temperatures (25–30 °C) and static conditions. Moreover, free-living strains are generally better biofilm formers than the symbiotically competent ones. Geographical location (strain origin) also correlated with biofilm formation. These findings provide evidence that abiotic growth conditions are important for determining whether mutualistic V. fischeri have the capacity to produce complex biofilms, allowing for increased competency and specificity during symbiosis.
Keywords: Biofilm, Environment, Vibrio fischeri
Introduction
Complex communities known as biofilms are increasingly recognized as the predominant form of microbial biomass in the environment [1]. The formation of these dynamic bacterial populations involves attachment and synthesis of a matrix composed primarily of polysaccharides [2] and is one important avenue for host colonization that eventually leads to persistence, antibiotic resistance, and pathogenesis [3]. Various model systems studying bacterial communities have been used to investigate biofilm properties, such as those formed by Pseudomonas aeuruginosa [4] and Staphylococcus spp. [5]. Numerous traits, such as quorum sensing, motility, and exopolysaccharide synthesis have been shown to be essential for correct assembly and maintenance of the biofilm architecture [6–8]. Although many of these characteristics occur in pathogenic Vibrio bacteria, little is known about benign or mutualistic species that have associations with eukaryotic hosts.
Vibrio fischeri is a bioluminescent bacterium that infects the light organs of sepiolid squids and monocentrid fishes establishing mutualistic associations. Free-living Vibrio bacteria colonize aposymbiotic juvenile squids within the first few hours after hatching, and upon colonization form a biofilm in the crypts of the light organ complex [9]. These bacteria produce bioluminescence that is used by the squid to avoid predation in a behavior known as counterillumination [9]. The mutualism is established when the host provides an appropriate niche for the bacteria to reproduce at much higher rates than in their free-living state [10]. Once colonized, symbiotic squids vent 90–95% of their bacteria from their light organ into the environment every morning to repopulate the surrounding bacterio-plankton community [11]. During this cyclical period between symbiotic and free-living states, V. fischeri forms a biolfilm-like aggregate outside the sites of infection prior to colonization [9, 11]. The squid host secretes mucus which is suspended above the sites of colonization [12]. Gram negative bacteria aggregate in this mucus and, by an unknown mechanism, V. fischeri is able to outcompete other bacteria in order to establish the specific mutualism [12]. Although V. fischeri has not been considered a typical model for studying biofilm formation until recently, intercellular signaling or quorum sensing molecules linked to the sociomicrobiology aspect of this particular bacterium make it an attractive organism to examine biofilm regulation. Additionally, recent discovery of hybrid sensor kinases and specific genes such as rpoN (encoding for the σ54) [8], mannose-sensitive hemagglutinin (mshA), uridyl phosphate dehydrogenase (UDPH) [13], and the symbiosis polyssacharide cluster (syp) [14], have demonstrated the importance of biofilm formation with respect to the life history strategy of this marine bacterium prior and during symbiosis. V. fischeri cells that form biofilms exhibit a variety of physiological and molecular characteristics when compared to those in their planktonic state [2]. As a result, the significance of biofilm production has emerged to accommodate a wide variety of sophisticated regulatory procedures allowing survival and adaptation to extreme environmental changes (that is, between a squid light organ and the surrounding seawater).
Symbiotic V. fischeri thrive in many different aquatic environments depending upon which squid host it colonizes; for instance, the genus Eurymna is distributed in the Indo-west Pacific, whereas species in the genus Sepiola are mostly found in Mediterranean waters and along the western margin of the Pacific Ocean [15]. Moreover, some free-living species are symbiotically incompetent or unable to colonize the light organ, even though they are found in the same waters as light organ containing sepiolids [16].
Previous evidence has demonstrated that environmental or abiotic factors influence both growth and physiology of Vibrio bacteria prior to their colonization in a squid light organ [11]. Additionally, significant genetic differences exist among V. fischeri strains from different geographical locations [10]. Therefore, in the present study, we determined whether biofilm formation between free-living and symbiotic V. fischeri strains differed under various environmental conditions (salinity and temperature changes, static-dynamic conditions, and atmosphere) in an effort to describe part of the strategy for V. fischeri survival in aquatic ecosystems.
Materials and methods
Bacterial strains used in the study
V. fischeri strains were isolated from seawater or live squid (Euprymna and Sepiola species; Nishiguchi, 2002) and are listed in Table 1. Isolated strains were stored as glycerol stocks (−80 °C) and re-grown for 24 h in Luria Bertani high salt (LBS; 10 g tryptone, 5 g yeast extract, 20 g sodium chloride, 50 ml 1 M Tris pH 7.5, 3.75 ml 80% glycerol and 950 ml distilled water) medium at 28 °C with moderate shaking (224 rpm). All strains were subsequently sub-cultured again under the same conditions for use in each of the assays. Biofilm quantification under the various conditions of interest was completed as previously described [17], this quantification is achieved by using crystal violet to stain the biofilm and subsequently solubilizing it with 70% ethanol and measuring optical density, which is proportional to the amount of biofilm formed.
Table 1.
Vibrio fischeri strains used in this study.
| Strain | Host | Location |
|---|---|---|
| WH1 | Free-living | USA (Woods Hole, Massachusetts) |
| MDR7 | Free-living | USA (Marina del Rey, California) |
| CB37 | Free-living | Australia (Coogee Bay, Sydney, NSW) |
| ATCC7744 | Free-living | American Type Culture Collection |
| BSM40 | Free-living | France (Banyuls sur mer) |
| BSM46 | Free-living | France (Banyuls sur mer) |
| BSM50 | Free-living | France (Banyuls sur mer) |
| PP3 | Free-living | USA (Kaneohe Bay, O’ahu, Hawaii) |
| PP42 | Free-living | USA (Kaneohe Bay, O’ahu, Hawaii) |
| VLS2 | Free-living | USA (Kaneohe Bay, O’ahu, Hawaii) |
| SR5 | Sepiola robusta | France (Banyuls sur mer) |
| SL518 | Sepiola ligulata | France (Banyuls sur mer) |
| SA1G | Sepiola affinis | France (Banyuls sur mer) |
| SI66 | Sepiola intermedia | Italy (Bari) |
| SI1D | Sepiola intermedia | France (Banyuls sur mer) |
| EM17 | Euprymna morsei | Japan (Tokyo Bay) |
| ET101 | Euprymna tasmanica | Australia (Townsville, QLD) |
| ETWW | Euprymna tasmanica | Australia (Woy Woy, NSW) |
| ETBB20 | Euprymna tasmanica | Australia (Botany Bay, Sydney, NSW) |
| ETSB1 | Euprymna tasmanica | Australia (Shark Bay, WA) |
| EB12 | Euprymna berryi | Japan (Tosa Bay) |
| ES114 | Euprymna scolopes | USA (Kaneohe Bay, O’ahu, Hawaii) |
| ESP915 | Euprymna scolopes | USA (Paiko, O’ahu, Hawaii) |
| ESL5 | Euprymna scolopes | USA (Kaneohe Bay, O’ahu, Hawaii) |
| ESC9 | Euprymna scolopes | USA (Kaneohe Bay, O’ahu, Hawaii) |
Biofilm formation assay
Overnight cultures were washed twice with fresh LBS medium and diluted to a concentration of 1 × 108 Colony Forming Units (CFU)/ml. Aliquots of each V. fischeri isolate were diluted to the same concentration (OD600) and were added to individual wells (5 wells/strain) located on a sterile, flat-bottom, polystyrene 96-well microtitre plate (Corning 96 well plates, Sigma-Aldrich, CLS3628). Three wells were filled with uninoculated sterile LBS as a negative control. Microtiter plates were filled with 100 μl of the medium, covered loosely, and incubated for 24 h at 28 °C. Planktonic (those not forming biofilm) bacteria were removed from each microtiter well by briskly shaking the microtitre plate. The emptied wells were then washed three times with 200 μl of sterile LBS medium, and 125 μl solution of crystal violet (0.2% diluted in sterile water) was then added to each well and incubated for 30 minutes at room temperature (~22 °C) to stain cells adhered to each well. After incubation, crystal violet was removed by shaking the microtiter dish vigorously over a waste tray and washing the plate five times with distilled water and subsequently air-dried. In order to quantify the amount of biofilm, 200 μl of 95% ethanol was added to each stained well (including controls), and incubated for 20 min at room temperature in order to solubilize the dye. The contents of each well were mixed briefly and 125 μl of crystal violet/ethanol solution was transferred to a separate well in an optically clear flat-bottom 96-well plate. Optical density was measured at 562 nm using a plate reader (Biotek FL 800). All samples were repeated in triplicate (3 plates) for a total of 15 replicates per isolate.
Salinity, temperature, carbon source and static/dynamic conditions
The effects of salinity, temperature, carbon source, and aerobic conditions were tested by growing cells under various sodium chloride concentrations, temperatures, carbon sources, or degrees of shaking. Vibrio isolates were grown overnight and sub-cultured the next morning as indicated previously. Washed cells were diluted to equal concentrations (1 × 108 CFU/ml) in modified LBS medium. For salinity measurements, we used sodium chloride concentrations ranging from 0% to 9%, where 1% corresponds to 10 parts per thousand (ppt) [11]. For temperature experiments, cultures were grown at 12, 15, 20, 25, 28, 30 and 32 °C in LBS medium at 32 ppt sodium chloride (or 3.2% sodium chloride). Minimal Ribose Medium (MRM) was also used as a growth medium for the above conditions, since it mimics the nutrient-poor composition of the seawater (composition per liter: NaCl 17.53 g, MgSO4 6.02 g, CaCl2 1.47 g, K2HPO4 0.0575 g, KCl 0.7455 g, NH4Cl 0.5349 g, Ribose 3.0 g, FeSO4·7 H2O 0.00278 g, Tris-HCl 7.88 g) [11]. Isolates were examined by growing cells in different dilutions of MRM (1:2, 1:4, 1:8 and 1:16), using as a diluent a solution with all the components of the MRM except the carbon source (Ribose). Salinity measurements of the different dilutions were calculated using a refractometer.
For static/dynamic conditions, microtiter plates were inoculated and measured after incubation of plates grown at different levels of agitation (0, 60, 120, 180 and 240 rpm) at 28 °C for 24 h on microtitre plates prior to final measurement of biofilm formation.
Data analysis and interpretation
Two different analyses were used to examine the variation of biofilm production from all environmental conditions. Strains were classified according to the amount of biofilm production as described in a previous study [18]: weak [Ac ≤ A ≤ (2 × Ac)], moderate [(2 × Ac) ≤ A ≤ (4 × Ac)] and strong biofilm producers [A > (4 × Ac)]. Ac represents the minimum absorbance for low biofilm production, which is calculated as three standard deviations above A of the control. A two-way ANOVA statistical design was chosen for the analysis of difference between the average mean of free-living and symbiotic Euprymna and Sepiola strains. A randomized complete block design was executed by using the Statistical Analysis Software (SASR) version 5.1.
Results
V. fischeri is a luminous symbiotic bacterium that can also persist solely in the free-living form, flourishes in a wide variety of coastal estuarine and marine waters, and is found as part of microbial biofilms in the environment [11, 13, 19]. Previous studies have focused on growth dynamics and effects of abiotic factors on these bacterial suspensions [4, 5, 11]; however, it is clear that biofilms constitute a distinct and predominantly structurally complex growth phase that is crucial for shaping the majority of aquatic bacterial communities. These biofilm communities are different from their planktonic state due to the physiological cooperation that dictates a high level of organization [20]. The present study attempts to elucidate the role of such environmental conditions on biofilm development of different V. fischeri isolates with various life history strategies.
Table 2 lists an overall view of the capacity to which all Vibrio strains examined aggregate and form biofilms. Most isolates were classified as weak to moderate biofilm formers at temperatures ranging from 12, 15, 20, 25 and 30 °C, whereas strong biofilm production was observed at 25 and 28 °C, corresponding to the optimal temperatures found in waters from the Indo-West Pacific Ocean (where the majority of Euprymna hosts are located) [15]. Strong biofilms were observed at low salinity concentrations (1–5%); conversely higher salinities (6–9%) resulted in significantly less biofilm for all isolates examined.
Table 2.
Summary of biofilm formation of Vibrio fischeri under different environmental conditions.
| Condition | Biofilm formation (n = 26)
|
||||||
|---|---|---|---|---|---|---|---|
| Weak No. Strains | OD + SD | Moderate No. Strains | OD + SD | Strong No. Strains | OD + SD | ||
| Temperature (°C) | 12 | 19* | 0.39 ± 0.21 | 7 | 0.63 ± 0.09 | – | – |
| 15 | 14 | 0.51 ± 0.15 | 12 | 0.79 ± 0.12 | – | – | |
| 20 | – | – | 26 | 0.83 ± 0.25 | – | – | |
| 25 | – | – | 5 | 1.10 ± 0.19 | 21* | 1.69 ± 0.24 | |
| 28 | – | – | – | – | 26 | 1.97 ± 0.29 | |
| 30 | – | – | 26 | 0.78 ± 0.20 | – | – | |
| 32 | 26 | 0.19 ± 0.08 | – | – | – | – | |
| a NaCl (%) | 1 | – | – | – | – | 26 | 1.42 ± 0.44 |
| 2 | – | – | – | – | 26 | 1.57 ± 0.48 | |
| 3 | – | – | – | – | 26 | 1.60 ± 0.53 | |
| 4 | – | – | – | – | 26 | 1.61 ± 0.60 | |
| 5 | – | – | 20* | 1.13 ± 0.09 | 6 | 1.03 ± 0.38 | |
| 6 | 4 | 0.42 ± 0.07 | 19* | 0.95 ± 0.40 | 3 | 1.23 ± 0.06 | |
| 7 | 26 | 0.41 ± 0.38 | – | – | – | – | |
| 8 | 26 | 0.03 ± 0.01 | – | – | – | – | |
| 9 | 26 | 0.01 ± 0.001 | – | – | – | – | |
| Agitation (rpm) | 0 | – | – | – | – | 26 | 1.85 ± 0.39 |
| 60 | – | – | 9 | 1.17 ± 0.12 | 17* | 1.77 ± 0.46 | |
| 130 | 8 | 0.47 ± 0.29 | 10 | 1.20 ± 0.18 | 8 | 1.75 ± 0.32 | |
| 240 | 19 | 0.32 ± 0.11 | 7 | 0.73 ± 0.15 | – | – | |
| MRM (Dilution factor) | Undiluted | 3 | 0.37 ± 0.08 | 21* | 0.84 ± 0.14 | 2 | 1.08 ± 0.07 |
| 1:2 | – | – | 17* | 1.08 ± 0.11 | 9 | 1.73 ± 0.10 | |
| 1:4 | 26 | 0.34 ± 0.28 | – | – | – | – | |
| 1:8 | 26 | 0.15 ± 0.06 | – | – | – | – | |
| 1:16 | 26 | 0.03 ± 0.01 | – | – | – | – | |
1% = 10 parts per thousand
Significant difference within strains (p < 0.05) calculated using the Welsch step-up procedure.
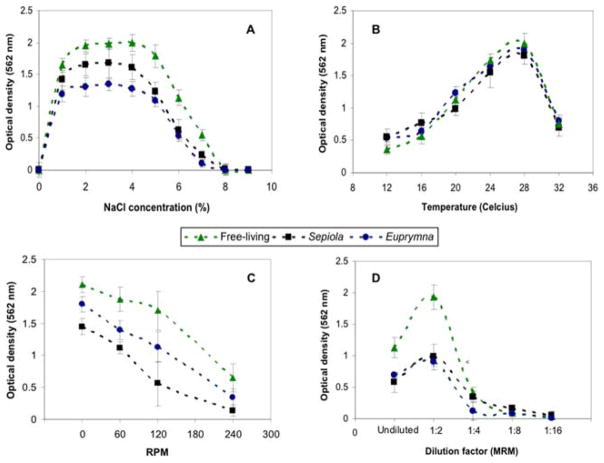
In this study, Sepiola strains were able to form biofilms at higher salinity ranges when compared with strains obtained from Euprymna (Fig. 1A). Previous work also reported the trends for planktonic strains under similar conditions [11]. An interesting finding was observed with free-living V. fischeri strains (WH1, MDR7, CB37) that are symbiotically incompetent (cannot colonize squids). Our results indicate that strains exhibit differences in biofilm formation when the salinity is modified, and free-living strains are generally better biofilm formers than symbiotic isolates at higher salinities (p < 0.05). This provides partial evidence that stress adaptation (including high osmolarity) are dictating bacterial survival and aggregation, and these responses are partially driven by abiotic factors.
Figure 1.
Influence of salinity (A: NaCl concentration, %, where 1% = 10 ppt), temperature (B; °C), carbon source availability (C; Minimal Ribose Media with various concentrations) and dynamic conditions (D; rpm) on biofilm formation when Vibrio fischeri isolates are divided between free-living and symbiotic strains. Data shown represents the mean optical density ± standard deviation (N = 3).
The effect of temperature on biofilm production (Fig. 1B) was not significantly different (p > 0.05) among symbiotic and free-living strains. Biofilm formation was observed in temperatures ranging between 12 °C to 32 °C; temperatures outside of this range reduced bacterial viability by 95% (results not shown). Some V. fischeri strains have been able to adapt and grow at higher (42 °C) and lower (8–10 °C) temperatures (unpublished data); however, biofilms were not observed for these extreme ranges.
Another survival challenge for many bacteria, including V. fischeri, is the degree of dynamism/movement in the environment. These conditions can be related to the effect of water currents on community formation, since it is well known that ocean hydrodynamics differ among habitats. Table 2 indicates that the majority of the strains examined form strong biofilms under static conditions (0 Revolutions Per Minute or RPM) and moderate shaking (60 RPM). Under different levels of dynamic conditions (up to 240 RPM, Fig. 1C), free-living isolates are the best biofilm formers. In a similar manner, Euprymna isolates appear to form greater amounts of biofilm at the same dynamic conditions when compared with Sepiola isolates (Fig. 1C). Finally, aquatic ecosystems differ in their carbon source availability, depending on whether it is in particulate or dissolved carbon form [30]. Carbon is limited in the ocean and is more abundant inside the host’s light organ; however, it is still unclear what preferred metabolic pathway vibrios utilize between their free-living and symbiotic stages [20]. Our study only examined biofilm formation using a nutrient marine medium with compounds that mimic nutrient poor conditions (Minimal Ribose Medium or MRM) [11]. Results demonstrate a significant increase in biofilm formation when the carbon source (Ribose) in the MRM is diluted 1:2 (Fig. 1D). As observed in previous conditions, free-living isolates seem to be the best biofilm formers under oligotrophic environments.
Discussion
It has been shown that abiotic factors such as salinity and temperature are variable among the world’s oceans [www.nodc.noaa.gov] and different luminescent bacteria, including V. fischeri, are able to survive, colonize different host squids, and adapt to such changing conditions [11]. Moreover, microbial dispersal ability and changing water currents can influence bacterial biogeography, radiation, and evolution [21]. For example, the Mediterranean Sea, which has numerous species of Sepiola, has a narrower, yet higher range of salinity (37–40 ppt, or 3.7–4% sodium chloride) when compared with the Indo-West Pacific, where Euprymna hosts are found in lower salinity ranges (30–35 ppt, or 3–3.5% sodium chloride). V. fischeri found in either location are classified as either host generalists (Mediterranean) or host specialists (Indo-West Pacific) due to their association with either sympatric or allopatric host species [11].
It can be observed in Fig. 1A that the maximum quantity of biofilm is formed at the same salinities (2–4% sodium chloride or 20–40 ppt) for every strain; however, our two-way analysis also indicates that there are differences in the quantity of biofilm that is formed under this range of salinity. These differences are related to life history of the strain examined. For example, free-living strains produce more biofilm than either of the symbiotic strains. Additionally, symbiotic strains adapted to high salinity (from Sepiola hosts) produce more biofilm than strains adapted to lower salinity (Euprymna hosts).
Due to the capacity of biofilm formation under different salinity concentrations, mechanisms of osmoadaptation (described in previous reports for free-living Vibrio parahaemolyticus) [22] may be regulated during the biofilm state for V. fischeri. The possibility remains that biofilm regulation is also driven by the squid host, since some conditions inside the squid’s light organ remain undescribed.
Temperature can be a major factor that affects the ability to produce extracellular polymeric substances [23], which are known to enhance adherence capability and biofilm formation in bacteria [24]. Greater quantities of biofilm were observed at 24 and 28 °C, which are optimal growth temperatures for Vibrio and close to the oceanic surface temperatures reported for both Mediterranean (Sepiola) and Indo-West Pacific Oceans (Euprymna) [9]. Since squid hosts are described as “poikilothermic” invertebrates (body temperature is equal to the temperature of its surroundings) [25], strain adaptation to temperature can be attributed solely to environment, and observations of no significant difference in quantity of biofilm between free-living and symbiotic strains supports this preface (according to our two way analysis). Alternatively, earlier studies have reported temperature effects for generation time of planktonic V. fischeri, indicating that the range of temperatures can affect the amount of biofilm produced based on growth of a particular strain [11]. In addition, our findings corroborate earlier data suggesting that temperature is an important factor that dictates specificity of two Vibrio species (V. fischeri and V. logei) within the same squid host [26]. Further experiments need to compare biofilm formation within colonized juvenile squids, to determine if indeed this is true after colonization.
The effect of ocean hydrodynamics can be related to the differences observed in the agitation assays. The strains tested can form biofilms on static conditions and under high degree of dynamism (Fig. 1C), with the general tendency of free-living strains being the best biofilm formers. In some cases, dynamism can be related to oxygen tension or oxygen availability; however, this study is not sufficient to describe the effect of oxygen tension on biofilm formation. This is an important factor that needs to be addressed primarily because the amount of oxygen that is available in the squid light organ is variable [27], and oxygen is an important factor for controlling bioluminescence through expression of the lux operon [28]. In addition, this operon is involved in quorum sensing behavior and colony aggregation or biofilm formation [29]. Future studies are needed to investigate subtle differences attributed to aerobic, microaerophilic, and anaerobic conditions in both free-living and symbiotic states.
Differences observed in various dilutions of MRM may be attributed to bacterial responses associated with coordinated alterations in patterns of gene expression of different metabolic networks that are necessary for efficient carbon utilization [31]. For example, the stringent response has been described as a transcriptional program that mediates prokaryotic adaptation to starvation conditions [32]. Previous work demonstrated that the stringent response activates quorum sensing in Pseudomonas aeruginosa [33], a process that has been proven to control virulence and biofilm formation in Vibrio cholerae [28] and might be an underlying genetic switch that allows V. fischeri to survive and associate only when the medium is diluted 1:2 (Fig. 1D).
Results obtained from the present study show that environmental switches exert an important influence in biofilm formation, and although there is no difference in the patterns observed in Fig. 1, there is a distinction of biofilm quantity between free-living and symbiotic strains. It is unclear at this point if these different responses are a result of a specific genetic change that allows adaptation, or as a simple shift in bacterial physiology related to development of a biofilm.
Microbial biofilms are the predominant form for survival of many species of bacteria in the environment, and are important for determining specificity and persistence in both pathogenic and mutualistic associations. The data presented here represent a quantitative analysis that provides a foundation for determining whether abiotic factors are important for biofilm formation at various life history stages of the bacterium. Additionally, this study examined how biofilm quantity differs significantly depending upon the nature of the strain and the conditions during growth in vitro. These assays can then be extrapolated to variations that occur between environmental or ecological niches; however, additional studies are needed in order to address the importance of conditions inside the squid’s crypts (branched epithelia found inside the squid’s light organ). By examining different environmental parameters, we can better understand bacterial survival and aggregation between free-living and symbiotic V. fischeri isolates to determine what drives specificity and environmental transmission in this mutualistic association.
Acknowledgments
The authors thank S. Soto for advice in the statistical analysis and D. Hogan for help with biofilm studies. The work was in part supported by NSF IOS-0744498, and NIH-NIAID SC1AI081659 to M.K.N. A. Chavez-Dozal was supported by the NASA-New Mexico Space Grant Consortium and NIH-RISE NIGMS R25GM061222 at NMSU.
References
- 1.Kokare CR, Chakraborty S, Khopade AN, Mahadik KR. Biofilm: importance and applications. Indian J Biotechnol. 2007;8:159–168. [Google Scholar]
- 2.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis K. Riddle of biofilm resistance. Antimicrobial Agents and Chemotherapy. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 5.Beenken KB, Dunman PM, McAleese F, Macapagal D, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadeli CD, Xavier JB, Levin SA, Foster KR. The evolution of quorum sensing in bacterial biofilms. PLoS Biology. 2008;6:171–179. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell CL, Hussa EA, Visick KL. The putative sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J Bacteriol. 2008;190:4941–4950. doi: 10.1128/JB.00197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfe AJ, Milikan DS, Campbell JM, Visick KL. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence and colonization. Appl Environ Microbiol. 2004;70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones BW, Nishiguchi MK. Counterillumination in the bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda) Marine Biol. 2004;144:1151–1155. [Google Scholar]
- 10.Jones BW, Maruyama A, Ouverney CC, Nishiguchi MK. Spatial and temporal distribution on the Vibrionaceae in coastal waters of Hawaii, Australia and France. Microbial Ecol. 2007;54:314–323. doi: 10.1007/s00248-006-9204-z. [DOI] [PubMed] [Google Scholar]
- 11.Soto W, Gutierrez J, Remmega MR, Nishiguchi MK. Salinity and temperature effects on physiological responses of Vibrio fischeri from diverse ecological niches. Microbial Ecol. 2009;57:140–150. doi: 10.1007/s00248-008-9412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Role of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbionts colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;70:2520–2524. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariyakumar DS, Nishiguchi MK. Characterization of two specific host genes, mannose sensitive hemagglutinin (mshA) and uridyl phosphate dehydrogenase (UDPH) that are involved in the Vibrio fischeri-Euprymna tasmanica mutualism. FEMS Microbiol. 2009;299:65–73. doi: 10.1111/j.1574-6968.2009.01732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and σ54- dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 15.Nesis KN. Distribution of recent Cephalopoda and implications for plio-pleistoscene events. Berliner Palaeo-biologische Abhandlungen. 2003;3:199–224. [Google Scholar]
- 16.Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrit JH, Kadouri DE, O’Toole GA. Growing and analyzing static biofilms. Curr Protocols Microbiol. 2005;Chapter 1(Unit 1B):1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Bonaventura G, Stepanovic S, Picciani C, Pompilio A, Piccolomini R. Effect of environmental factors on biofilm formation by clinical Streptomonas maltophilia isolates. Folia Microbiologica. 2007;52:86–90. doi: 10.1007/BF02932144. [DOI] [PubMed] [Google Scholar]
- 19.Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- 20.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyholm SV, Nishiguchi MK. The evolutionary ecology of a sepiloid squid-Vibrio association: from cell to environment. Vie et Milleu. 2008;58:175–184. [PMC free article] [PubMed] [Google Scholar]
- 22.Naughton LM, Blumerman SL, Carlberg M, Boyd EF. Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus. Appl Environ Microbiol. 2009;75:2802–2810. doi: 10.1128/AEM.01698-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 2006;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 25.Zuur AF, Pierce GJ. Common trends in northeast Atlantic squid time series. J Sea Res. 2004;52:57–72. [Google Scholar]
- 26.Nishiguchi MK. Temperature affects species distribution in symbiotic populations of Vibrio. Appl Environ Microbiol. 2000;66:3550–3555. doi: 10.1128/aem.66.8.3550-3555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- 28.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 29.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 30.Hader DP, Kumar HD, Smith RC, Worrest RC. Effects on aquatic ecosystems. J Photochem Photobiol B: Biology. 1998;46:53–68. [Google Scholar]
- 31.Bruckner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. 2006;209:141–148. doi: 10.1111/j.1574-6968.2002.tb11123.x. [DOI] [PubMed] [Google Scholar]
- 32.Jain V, Kumar M, Chatterji D. ppGpp: Stringent response and survival. J Microbiol. 2005;44:1–10. [PubMed] [Google Scholar]
- 33.Van Delden C, Comte R, Bally AM. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]