Abstract
The kinetics of thymine-thymine cyclobutane pyrimidine dimer (TT-CPD) formation was studied at 23 thymine-thymine base steps in two 247-base pair DNA sequences irradiated at 254 nm. Damage was assayed site-specifically and simultaneously on both the forward and reverse strands by detecting emission from distinguishable fluorescent labels at the 5’-termini of fragments cleaved at CPD sites by T4 pyrimidine dimer glycosylase and separated by gel electrophoresis. The total DNA strand length of nearly 1000 bases made it possible to monitor damage at all 9 tetrads of the type XTTY, where X and Y are non-thymine bases. TT-CPD yields for different tetrads were found to vary by as much as an order of magnitude, but similar yields were observed at all instances of a given tetrad. Kinetic analysis of CPD formation at 23 distinct sites reveals that both the formation and reversal photoreactions depend sensitively on the identity of the nearest-neighbour bases on the 5’ and 3’ side of a photoreactive TT base step. The lowest formation and reversal rates occur when two purine bases flank a TT step, while the highest formation and reversal rates are observed for tetrads with at least one flanking C. Overall, the results show that the probabilities of CPD formation and photoreversal depend principally on interactions with nearest-neighbour bases.
Introduction
The most prevalent photoproduct formed in DNA by UV irradiation is the cyclobutane pyrimidine dimer (CPD).1–3 CPDs form in a 2+2 cycloaddition reaction that covalently bonds two pyrimidine bases that in duplex DNA are usually adjacent in the same DNA strand. This photoproduct interferes with subsequent DNA processing and accounts for much of the UV-induced mutation spectrum.4, 5
The rates of CPD formation throughout the genome determine in part the overall genotoxic burden. Several variables influence CPD yields. First, the yield of CPDs under UVC irradiation at a given site in isolated DNA decrease in the order TT > TC > CT > CC.6 Second, the yield of a given CPD such as the TT-CPD is influenced by the sequence of nearby bases.1, 7–13 Third, yields can depend on the irradiation wavelength and absorbed dose.
Despite decades of study and the apparent simplicity of the underlying photochemistry, it is still difficult to rationalize observed CPD yields along a particular DNA strand, even under well-defined in vitro conditions. Disentangling steric from electronic effects on the photochemistry has proven particularly challenging.14, 15 Previous studies have shown that CPD yields depend strongly on the conformation of a pyrimidine-pyrimidine step.9, 11, 16–18 Using femtosecond time-resolved infrared spectroscopy, Schreier et al.19 demonstrated that the TT-CPD is formed in (dT)18 in less than one picosecond. Because backbone motions that affect the conformation of stacked pyrimidine bases occur on longer time scales, ultrafast photoproduct formation indicates that conformation at the instant of photoexcitation is a key factor governing the probability of reaction.20
Ground-state conformational distributions obtained from classical molecular dynamics simulations have been used to explain CPD yields in some simple model systems,14, 21–23 but this approach has not yet been applied to the wider set of sequence contexts found in longer DNAs. Despite the success of the steric models, there are indications that electronic factors can play a role in CPD formation rates. Recently, Cannistraro et al.24 and Pan et al.14, 15 suggested that the rate of dimer formation could be reduced by deactivation of the reactive excited state via the formation of charge-transfer exciplexes between a bipyrimidine base steps and flanking bases.
Importantly, UV radiation can both form CPDs and cleave them to the starting bases. Direct excitation of a CPD results in photoreversal with a quantum yield that approaches unity.25 Because CPDs absorb at shorter wavelengths than undamaged nucleobases, photoreversal is thought to be most important at shorter (i.e. UVC) wavelengths, but this may not be the case for C-containing CPDs (i.e. TC, CT, and CC), which have larger absorption cross sections at UVB wavelengths than the TT-CPD.26, 27 Direct photosplitting warrants further study for this reason and because of the large number of model studies performed with UVC irradiation.
A second possibility for photoinduced CPD splitting that has been the focus of recent attention is electron transfer from a photoexcited base that is proximal to the CPD.12 This photolyase-like repair mechanism is thought to require a good electron-donor base such as guanine12, 28 or 8-oxoguanine.29 Of course, CPD reversal, whether it occurs by this proposed electron transfer mechanism or by sensitized electron transfer, requires absorption of a UV photon by the photoproduct. Its study therefore presupposes high doses that maximize the probability of exciting a CPD or a flanking base proximal to a dimer created by an earlier photon. Many studies have focused on low dose conditions that provide insight into formation rates,1, 5, 7, 10, 11 but are insensitive to photoreversal. Others30, 31,30, 31 have measured photostationary yields under high dose conditions,30, 31 but this approach cannot determine the individual rates. Consequently, little is known about the sequence-dependence of photoreversal.
Recently, Rokita and co-workers studied TT-CPD formation kinetics over a wide range of doses in double-stranded oligonucleotides for a limited set of sequences.12 Inspired by this approach, we have studied the kinetics of TT-CPD formation in two 247 base pair DNA sequences (Fig. S1, Electronic Supplementary Information). By design, each sequence has an identical 50 base pair sequence at either end that serves to monitor the number of photoproducts formed. By using different fluorescent labels on the 5’-end of both the forward and reverse strands of the DNA sequences, we were able to determine dimer formation kinetics simultaneously on both strands. Although fluorescence has been used previously to detect UV damage site-specifically in DNA,32 this is the first time to our knowledge that both strands of a duplex have been labelled by fluorescent dyes, allowing simultaneous detection of lesions on both strands in a single irradiation experiment.
The results show that TT-CPD formation and photoreversal are highly predictable based on the identity of the two immediately flanking bases. This suggests that the steric and/or electronic effects that govern CPD production in duplex DNA reflect local interactions with nearest-neighbour bases.
Experimental Methods
Preparation of DNA Samples
The method described in Manohar et al.33 was used to prepare a 247 base pair (bp) dsDNA molecule by PCR from plasmids containing either the MP2+50 nucleosome positioning sequence34 or the 5S+50 nucleosome positioning sequence.35 The resulting dsDNA molecule has a Cy5 label at the 5' terminus of the forward strand, and a Cy3 label at the 5' terminus of the reverse strand. Bipyrimidine steps are designated by listing the DNA sequence and the strand followed by a number that indicates the distance of the first (5’) pyrimidine base from the 5’ strand end, i.e. MP2+50 ATTC Cy5–227.
1,3-dimethyluracil (DMU) was purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. Aqueous solutions were prepared using H2O from a commercial water ultrapurifier. All solutions and enzymes used in the enzymatic digestion of DNA samples were used as received from New England Biolabs, Inc.
Photoirradiation and Actinometry
DNA samples were irradiated in 0.5X TE buffer consisting of 5 mM Tris buffer, pH 7.5 and 0.5 mM ethylenediaminetetraacetic acid (EDTA). 10 mM potassium fluoride was added to inhibit the formation of UV-induced single strand breaks.36 The concentration of DNA used in all experiments was 10 nM, assayed from its absorbance at 260 nm using a NanoDrop 2000 spectrometer (Thermo Scientific, Inc, Wilmington, DE). The molar mass of a base pair of DNA was assumed to be 600 g/mol, while a 50 ng/µL DNA solution was assumed to have an absorbance of 1 at 260 nm in a 1 cm path length cell.
110 µL DNA samples were placed inside a NMR tube (Wilmad LabGlass Vineland, NJ, part no. 507-PP-7 (4.2 mm ID)) and irradiated at room temperature using a G8T5 germicidal UVC low-pressure mercury lamp (8W output power, from Ushio America, Cypress, CA, with peak emission at 254 nm). Although the NMR tubes used are made from Class A borosilicate glass, which absorbs UVC radiation strongly, the very thin wall thickness of 0.38 mm nonetheless transmits approximately 10% of the incident UVC photons.
Actinometry measurements confirmed that a DMU solution absorbs approximately ten times as many quanta when held in a fused silica optical cuvette placed in an identical position in the photoreactor compared to when a DMU solution is held in a borosilicate glass NMR tube. Absorption by the borosilicate glass (see, for example, the spectrum in Fig. S2 of ref. 37) attenuates the weaker UVB lines of the low-pressure mercury lamp much less than the strongest line at 254 nm. Consequently, the radiation absorbed by DNA in the NMR tube contains a stronger UVB component than would be the case with a fused silica vessel. Although there is a UVB component present, we do not expect it to be photochemically active because the total photon flux of the UVB lines is still less than that from the 254 nm line.
Samples were not deaerated as oxygen has a negligible effect on CPD formation in base multimers. DNA damage was followed in time by removing 7 – 20 µL of DNA solution by microsyringe every few minutes and analysing this solution for damage site-specifically by enzymatic digestion and gel electrophoresis as described below. Damage was quantitated at approximately 10 time points in each irradiation run.
Irradiation conditions were maintained as constantly as possible from run to run and the incident photon flux was estimated by DMU actinometry before each trial. DMU was dissolved in H2O and held in a standard 1 cm path length fused silica cuvette the centre of which was located at the same position as the centre of the NMR tube using in DNA experiments. Absorption spectra of the DMU solution before and after irradiation were measured using a PerkinElmer Lambda 5 spectrometer (PerkinElmer, Inc., Waltham, MA), and the incident photon flux was estimated following the procedure described in Law et al.22
Enzymatic Digestion and Gel Electrophoresis
5.0 µL of DNA solution sampled during an irradiation run was combined with 1 µL of 10 ng/µL T4 pyrimidine dimer glycosylase (T4 PDG) dissolved in 10 mM tris buffer (pH 7.5), 0.1 mM EDTA, 0.1 mM dithiothreitol (DTT) and 50% (v/v) glycerol. The resulting solution was diluted to a final volume of 20 µL with ddH2O and a reaction buffer made from 1 M NaCl, 10 mM DTT, 10 mM EDTA and 250 mM phosphate buffer (pH 7.2). Bovine serum albumin (BSA) was added to this solution to prevent sticking of the sample to the container walls. The final 20 µL solution contained 100 mM NaCl, 1mM DTT, 1 mM EDTA, 25 mM phosphate buffer, and 0.1 mg/mL BSA and was incubated at 37°C for 30 minutes.
After incubation, 1 µL of a 1:1 phenol:chloroform mixture was added to terminate enzymatic digestion and separate T4 PDG from the DNA. Each DNA sample was treated with 21 µL of formamide and heated at 95°C for at least 20 minutes to ensure denaturation of the DNA.
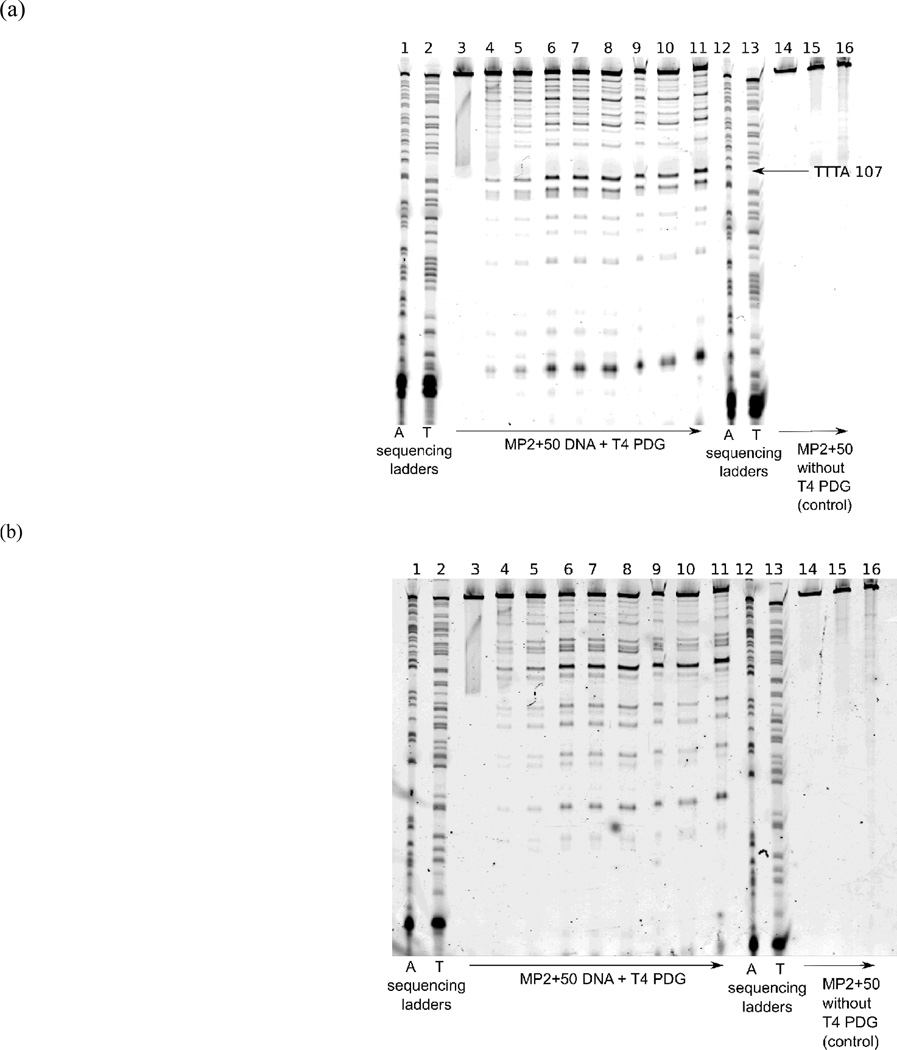
Next, the samples were analysed using a 10% (w/w) polyacrylamide electrophoresis gel in 9 M urea and TBE (90 mM tris buffer (pH 8.0), 90 mM boric acid and 2 mM EDTA), following the procedure described in Manohar et al.33 In addition to control and digested aliquots, ddATP and ddTTP sequencing ladders specific to the DNA sequence were loaded onto the gel. The position of TT, TC and CT dimer formation sites on gels were determined by comparison with the ddATP and ddTTP sequencing ladders. The effects of photobleaching of the fluorescent dye labels are discussed in the ESI. Strand breaks in the DNA gel were visualized using a Typhoon imager (GE HealthCare, Piscataway, NJ) to detect the fluorescent end-labels. Typical gel images for the MP2+50 sequence are reproduced in figure 1. 5S+50 samples were analyzed similarly.
Fig. 1.
A typical denaturing polyacrylamide gel used for determination of CPD formation kinetics in the MP2+50 DNA sequence imaged with the (a) Cy5 and (b) Cy3 dye. The arrows at bottom indicate the direction of increasing UVC irradiation time. The lane assignments are as follows: (lanes 1 and 12) ddATP sequencing ladder, (2 and 13) ddTTP sequencing ladder, (3–11) MP2+50 DNA after digestion with T4 PDG before irradiation (3), and after exposing the DNA to a photon fluence in units of Einsteins cm−2 of (4) 1.5 × 10−7, (5) 3.0 × 10−7, (6) 1.2 × 10−6, (7) 1.8 × 10−6, (8) 2.7 × 10−6, (9) 3.6 × 10−6, (10) 4.8 × 10−6, and (11) 6.3 × 10−6. Control lanes 14–16 depict MP2+50 DNA without T4 PDG digestion before irradiation (14) and after a photon fluence of (15) 2.7 × 10−6 and (16) 6.3 × 10−6 Einsteins cm−2.
Control experiments were carried out on samples collected after various irradiation times to ensure that strand breaks are the result of T4 PDG digestion. In this case, the same protocols were followed, except that purified H2O was used in place of the T4 PDG solution.
Quantification of CPDs from Gel Images
The integrated fluorescence intensity was measured in a bounding box drawn about each band using the ImageQuant 5.2 software (GE HealthCare, Pistacaway, NJ). The size of the bounding box was held constant for a given band across all lanes of a given gel. The background fluorescence intensity was equated to the median intensity of the pixels on the boundary of the bounding box and subtracted from the intensity of each pixel within the box prior to integration.
Gel Band Normalization
Because the fluorescent label is present at the 5’ strand end, only the strand break nearest the 5’ end is directly detected. In order to account for the possibility of multiple strand breaks per strand, the concentration of CPDs at a particular site, [CPD], was corrected using the equation due to Brash,38
| (1) |
In eq. 1, [DNA]0 is the initial concentration of DNA in the sample, Iband is the fluorescence intensity of the bands corresponding to the site of interest, and ∑Iband is the sum of the fluorescence yield of all bands representing strand breaks 3’ to the band under consideration and including the fluorescence intensity associated with undamaged DNA. The concentration of dimers detected at time zero was subtracted from all concentration values at later times for each band in order to account for strand breaks and incompletely formed DNA in the initial sample.
CPD concentration, [CPD], vs. the incident photon flux was fit using the equation,12
| (2) |
In equation 2, [DNA]0 is the initial concentration of DNA strands, kf is the rate of dimer formation, kr is the rate of dimer photoreversal, and x is the incident photon flux measured by DMU actinometry. In this work, kf and kr values for both DNA strands are reported relative to the measured kf value for the CTTA Cy5–34 site found in both DNA sequences.
Results and Discussion
The reaction kinetics of CPDs formed at TT steps in the MP2+50 and 5S+50 sequences were analysed in detail. The TT CPD is formed in the highest yield of any bipyrimidine lesion at both UVC and UVB wavelengths,6 making it easy to characterize sequence-dependent reactivity trends. Comparing the DNA sequences used (Fig. S1 in ESI) with sequencing ladders like those in Fig. 1, we find that our sequencing gels have 2 – 3 bp resolution. Consequently, dimers formed at TT steps and separated by at least one nonreactive (i.e. purine) base are fully resolved. Gel bands at positions with two or more immediately adjacent TT dimer formation sites, such as TTTA MP2+50, Cy5–107, could not be resolved. Sites containing three or more consecutive thymine bases were therefore excluded from analysis, although they were used in the intensity normalization procedure described above.
Dimer Formation and Photoreversal Kinetics Depend Primarily on the Identity of Nearest Neighbours
CPD yields were measured as a function of UVC dose at a total of 23 sites on the two DNA sequences. Each site is one of 9 possible base tetrads of the form XTTY, where X and Y are any two bases other than T. The photodimer yield for the GTTG site at the Cy3–28 positions of both the MP2+50 and 5S+50 sequences was lower at all doses than for any other tetrad. Due to the low yield, it was not possible to determine kf and kr at this site. CPD yields were measured, however, as a function of dose at each of the 8 other tetrads at a minimum of two sites with the exception of ATTA, which only occurs at Cy3–154 on the MP2+50 strand. Tetrads located very near a strand terminus were excluded, as were sites with potentially significant levels of CPD formation from adjacent CPD-forming sites (e.g. GTTCTC at 5S+50 Cy3–156).
Figs. 2 and 3 show representative kinetic data for two tetrads found at various sites in the MP2+50 and 5S+50 sequences. The response is linear at low dose, but reaches a plateau as the dose is increased. The plateau is the photostationary or equilibrium dimer yield obtained when the rates of dimer formation and splitting are in balance.8, 30, 39 Of course, this is an apparent and not a rigorous photostationary yield because at very high levels of irradiation the concentration of CPDs will decrease due to competition from (6-4) photoadduct and photohydrate formation, both of which are not photoreversible. In this study, no significant decrease in [CPD] was observed at the fluence levels used, and such kinetics were therefore neglected.
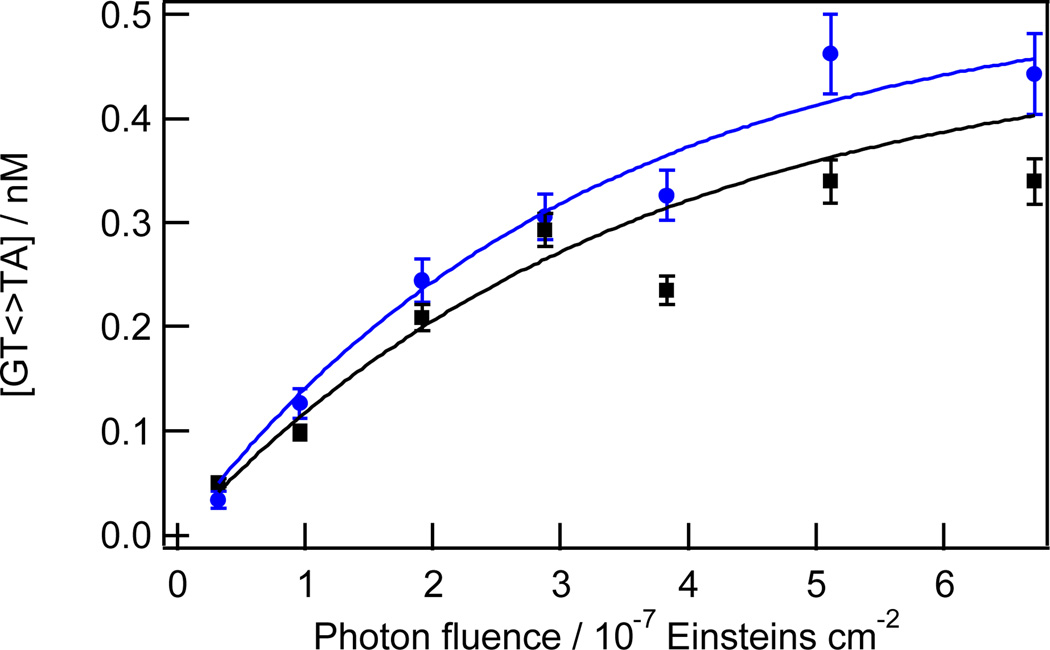
Fig. 2.
Kinetics of TT CPD formation for the GTTA tetrads at Cy5–166 (blue circles) and Cy5–140 (black squares) in the 5S+50 sequence. Solid curves are fits to points of the same colour using equation 2.
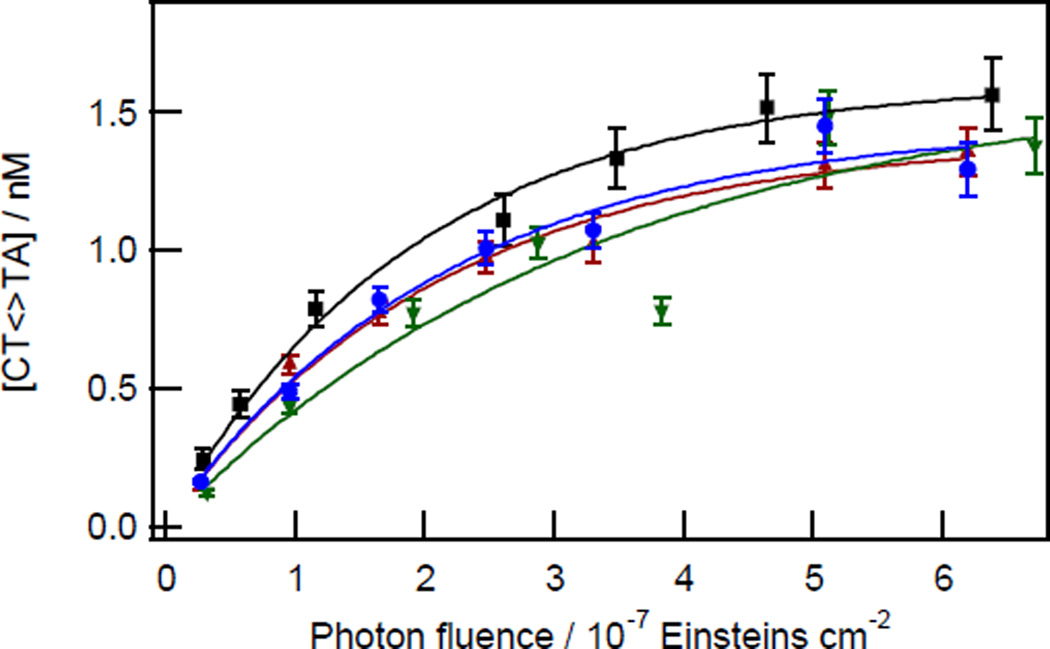
Fig. 3.
Kinetics of TT CPD formation for CTTA tetrads at Cy5–34 (black squares) of the MP2+50 duplex and at the following sites on the 5S+50 duplex: Cy5–34 (blue circles), Cy5–182 (green downward triangles), Cy3–105 (red upward triangles). Solid curves are fits to points of the same colour using equation 2.
The equilibrium dimer yield for CTTA is reached at a dose of ~7 × 10−7 Einsteins cm−2 or ~3 kJ m−2. A comparable saturating photon fluence was reported by Haseltine et al.,30 while slightly higher39 and lower12, 31 saturating fluences have been reported by others for UVC-irradiated DNA. The fact that photostationary yields are attained at similar doses as in past UVC work is evidence that the weak UVB component experienced by our DNA samples (see Experimental Methods) does not affect the kinetics. Douki et al. report that TT-CPD formation is linear with UVB dose up to 100 kJ m−2,39 a value that is perhaps 100 times greater than the maximum UVB levels in our experiments.
For each of the 23 tetrad sites studied, CPD vs. dose curves like the ones shown in Figs. 2 and 3 were fitted to eq. 2 in order to determine kf and kr. These rates are listed in Table 1 relative to the value of kf observed for the CTTA site at Cy5–34 in each DNA sequence. The rates kf and kr are very similar for all instances of a given tetrad even though the lowest and highest rates for different tetrads vary by a factor of approximately five. The good agreement between rates measured at different occurrences of each tetrad corroborates earlier reports that flanking bases strongly influence CPD yields under both low dose and high dose conditions.10, 30, 40
Table 1.
Rates of dimer formation (kf) and reversal (kr) relative to kf for CTTA at Cy5–34.
| Tetrad | Sequence | Position | kf | kr |
|---|---|---|---|---|
| ATTA | MP2+50 | Cy3−154 | 0.35 ± 0.05 | 1.9 ± 0.4 |
| ATTC | MP2+50 | Cy5−227 | 0.65 ± 0.13 | 8.9 ± 2.1 |
| 5S+50 | Cy5−227 | 0.44 ± 0.16 | 6.0 ± 1.6 | |
| 5S+50 | Cy5−101 | 0.58 ± 0.12 | 5.0 ± 1.2 | |
| 5S+50 | Cy3−51 | 0.54 ± 0.12 | 6.3 ± 1.6 | |
| Average | 0.56 ± 0.08 | 6.5 ± 1.7 | ||
| ATTG | MP2+50 | Cy5−168 | 0.21 ± 0.02 | 2.4 ± 0.3 |
| MP2+50 | Cy3−81 | 0.19 ± 0.02 | 2.9 ± 0.4 | |
| Average | 0.20 ± 0.01 | 2.6 ± 0.3 | ||
| CTTA | MP2+50 | Cy5−34 | 1 (defined) | 4.2 ± 0.5 |
| 5S+50 | Cy5−34 | 1 (defined) | 5.5 ± 1.8 | |
| 5S+50 | Cy5−182 | 0.43 ± 0.11 | 2.7 ± 1.1 | |
| 5S+50 | Cy3−105 | 0.65 ± 0.13 | 3.8 ± 0.9 | |
| Average | 0.83 ± 0.09 | 4.0 ± 0.6 | ||
| CTTC | MP2+50 | Cy5−190 | 0.70 ± 0.07 | 3.9 ± 0.5 |
| 5S+50 | Cy5−74 | 0.54 ± 0.11 | 6.1 ± 1.3 | |
| Average | 0.62 ± 0.12 | 5.0 ± 1.6 | ||
| CTTG | MP2+50 | Cy5−155 | 0.43 ± 0.05 | 3.4 ± 0.5 |
| MP2+50 | Cy5−100 | 0.52 ± 0.06 | 5.4 ± 0.8 | |
| MP2+50 | Cy3−107 | 0.33 ± 0.04 | 2.8 ± 0.4 | |
| MP2+50 | Cy3−61 | 0.33 ± 0.03 | 3.6 ± 0.4 | |
| 5S+50 | Cy5−130 | 0.28 ± 0.05 | 3.5 ± 1.0 | |
| 5S+50 | Cy5−69 | 0.31 ± 0.05 | 4.1 ± 0.8 | |
| Average | 0.37 ± 0.02 | 3.8 ± 0.3 | ||
| GTTA | 5S+50 | Cy5−166 | 0.18 ± 0.04 | 3.2 ± 1.0 |
| 5S+50 | Cy5−140 | 0.15 ± 0.03 | 2.9 ± 0.9 | |
| Average | 0.16 ± 0.03 | 3.1 ± 0.7 | ||
| GTTC | MP2+50 | Cy5−133 | 0.50 ± 0.10 | 15.1 ± 3.3 |
| 5S+50 | Cy3−196 | 0.28 ± 0.12 | 12.5 ± 5.4 | |
| Average | 0.38 ± 0.15 | 13.8 ± 1.8 |
A potential complication occurs at sites where C flanks a TT step on either the 5’ or 3’ side. In such cases, the CPD yield at TT could contain contributions from TC- and/or CT-CPDs because these overlapping sites are not resolved at our gel resolution. However, CPDs at isolated TC and CT sites form with very different kinetics than at isolated TT sites. This is illustrated for two TC sites in Figs. S4 and S5 in the ESI. The photostationary yields at these sites are 5 to 10 times lower than at TT sites. In addition, the photostationary state is established at much lower fluences. Both observations are consistent with high reversal rates for C-containing dimers.41 This is a consequence of their greater absorption at UVC wavelengths compared to the TT-CPD.27 Earlier, Gordon and Haseltine found that the equilibrium yield of the CC-CPD is reached at a UVC dose four times lower than the one needed to establish equilibrium amounts of the TT CPD.8
The relative rates of dimer formation and photoreversal at TC or CT sites are difficult to measure precisely on account of the low photostationary yields. Nevertheless, comparison of TT vs. TC dimerization kinetics at sites having the same flanking bases (compare Table 1 with Table S1 in the ESI) indicates that TC formation rates are approximately 30% to 100% as large as TT formation rates, in agreement with previous low-fluence studies,42 while dimer reversal rates are much higher for the TC lesions.
The low photostationary yields of TC-CPDs and the rapid attainment of the photostationary state mean that these lesions negligibly affect the yield of TT-CPDs when both types of lesions contribute to a given gel band. Consequently, when reactivity trends are discussed below, we shall assume that the effect of a flanking C in a tetrad like CTTA is to modulate the photoreactivity at the central TT step, and not to contribute significant numbers of unresolved C-containing CPDs. There is also no evidence that the photostationary state is approached on two separate timescales and this justifies our assumption that the photodimerization kinetics at C-flanked TT sites are dominated by the formation and reversal of the central TT-CPD.
CPD Formation and Photoreversal Trends for Different Tetrads
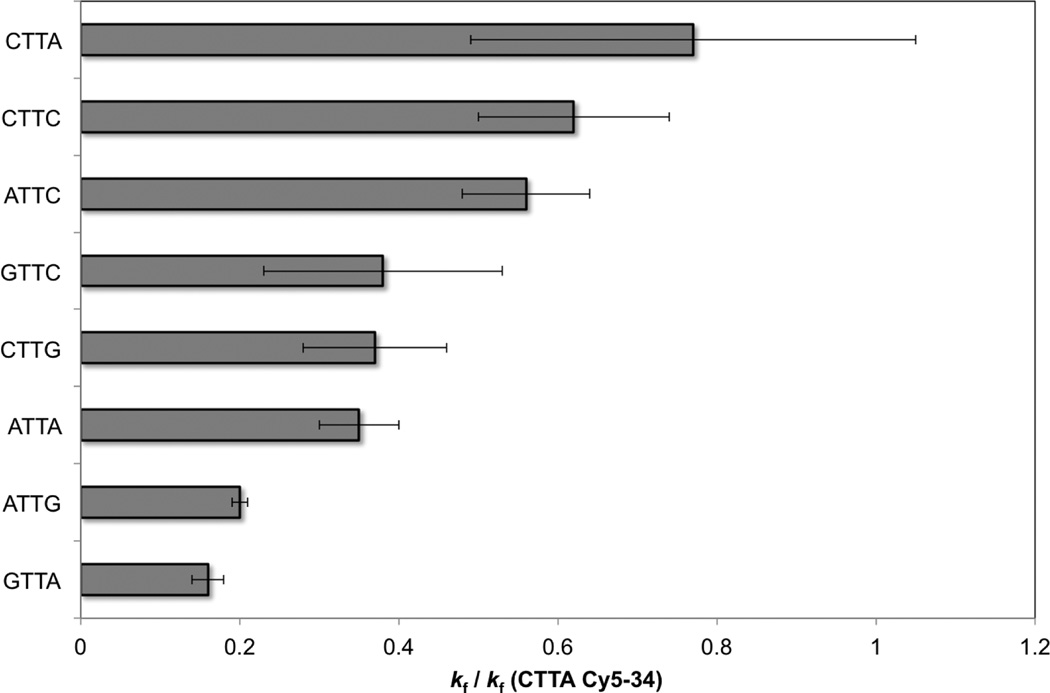
For low irradiation doses, the CPD yield at a given tetrad is just kf multiplied by the dose. The average kf values are ordered from largest to smallest in the bar graph in Fig. 4. As mentioned earlier, we were unable to obtain rates for GTTG due to its very low equilibrium yield. Others have noted the strong suppression of the TT-CPD by flanking Gs.10, 11, 32, 40
Fig. 4.
Average dimer formation rates and error bars for the indicated tetrads relative to kf for the Cy5–34 site of the MP2+50 and 5S+50 sequences.
The average formation rates (kf, Table 1 and Fig. 4) for ATTG and GTTA are approximately equal and roughly half as large as the ATTA rate. This is in excellent agreement with results from Holman et al.12 for these three tetrads in 18-bp double-stranded oligonucleotides containing a single TT base step. Fig. 4 reveals that the formation rates seen in tetrads with two flanking purines are lower than for any of the other tetrads. On the other hand, replacing one or both of the purines by C dramatically enhances kf.
Average kf values in Table 1 vary by a factor of 5 for the 8 tetrads studies (Table 1 and Fig. 4). Variation by a factor of ~2 was observed by Haseltine et al.30 for the smaller set of six tetrads in their investigation. If kf for GTTG is half as large as for ATTG, as reported by Holman et al.,12 then the kf values for the 9 XTTY tetrads span approximately one order of magnitude. Supporting this spread in reactivity, Yoon et al.1 found that the frequency of the TT-CPD varies by a factor of 10 for the different sequence contexts in their study. We note that in vivo CPD yields may differ due to effects of nucleosome binding.17, 43
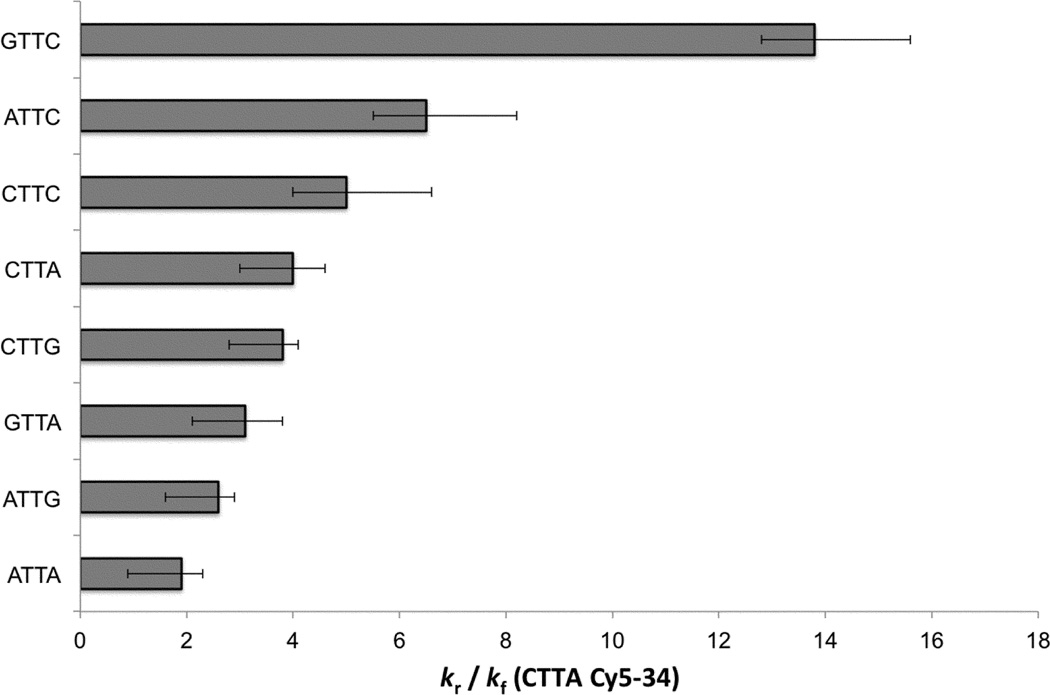
Having established that the trends in TT formation rates in our study are in excellent agreement with past studies where comparisons are possible, we now turn to the measured photoreversal rates, which have been studied much less often. Reversal rates (kr in Table 1 and Fig. 5 bar graph) also depend strongly on the flanking bases. As with the formation rates, the reversal rates are smallest for tetrads containing two purine bases, while replacement of one or both purines by C leads causes kr to increase. The largest reversal rates are observed with a purine that is 5’ to the lesion and a 3’ pyrimidine. Overall, the reversal rates increase in roughly the same order as seen for the formation rates. The exception is the GTTC tetrad, which has one of the lowest kf values, but the largest value of kr.
Fig. 5.
Average dimer reversal rates and error bars for the indicated tetrads relative to kf for the Cy5–34 site of the MP2+50 and 5S+50 sequences.
The study by Holman et al.12 is the only one to our knowledge to discuss photoreversal rates for a bipyrimidine lesion at specific sites along a DNA strand. Even so, these authors studied only sequences in which a TT step is flanked by two purines. They observed high kr rates for GTTG and GTTA, but kr was found to vary less than kf for the four possible purine-purine tetrads.12
We find that when the full group of XTTY tetrads is considered that GTTA, ATTG, and ATTA have the smallest kr values (Fig. 5). Tetrads with one or more Cs have even larger rates of photoreversal. While GTTC has the highest rate of photoreversal, the pattern seen in Fig. 5 suggests that this has more to do with a 3’ C than with a 5’ G.
We note that GTTG, the sole non-T-flanked TT tetrad whose rate constants could not be measured, is not expected to have the lowest kr value based on the measurements by Holman et al.12 Consequently, the total variation of ~6 in photoreversal rates is somewhat smaller than the range of kf values.
The highest kr values are observed for C-flanked TT CPDs. We do not believe that these rates are influenced by photoreversal of TC-CPDs occurring alongside reversal of TT-CPDs. TC- and CT-CPDs are formed in low yields under our conditions, as discussed above. Furthermore, we observe the highest photoreversal rate for CTTA, which should have only a minor contribution from the CT-CPD, which forms in the lowest overall yield of the four CPDs.6
Equilibrium TT-CPD yields are obtained in the high dose limit and are given by kf/(kf+kr) (eq. 2). These yields, which depend solely, on the ratio of kf to kr, vary as follows: GTTG < GTTC < GTTA < ATTG ≈ ATTC ≈ CTTG < CTTC < CTTA ≈ ATTA. These results agree reasonably well with equilibrium TT-CPD yields determined by Haseltine et al. for isolated, UVC-irradiated DNA: GTTA ≈ ATTG < ATTC < CTTA < CTTC < ATTA.
According to these results, the three tetrads with a 5’G have the lowest equilibrium yields. Photosensitized reversal by electron donation from G, the mechanism stressed by Holman et al.12 may be one contributing factor, but the equilibrium yields are already suppressed by the low TT-CPD formation rates for tetrads with a 5’G. Although we were unable to measure rates for GTTG, Holman et al.12 report that kr for this tetrad is 50% larger than for ATTA, putting it roughly in the middle of measured kr values. We conclude that low yields of TT CPD at TT steps flanked by G have more to do with the very low formation rates observed for these tetrads than with photoreversal rates, which are not particularly exceptional. Importantly, the latter effect is the sole cause of low yields in the low dose regime.
The fact that similar kr values are measured for different sites of a given tetrad indicates that CPD photosplitting, whether it occurs directly via photoreversal,25 or indirectly through charge-transfer mediated self-repair,12 is primarily a local phenomenon that does not depend on long-range electronic or conformational effects.
As discussed above, the measured photoreversal rates (Fig. 5) do not appear to provide strong support for electron donation from a 5’ G. While the three highest photoreversal rates do decrease with the 5’ base in the order G > A > C, the same order expected for electron donor ability,44 a much lower rate of reversal is observed for GTTA compared to GTTC even though both have a 5’ G. This would appear to rule out a major role for electron transfer from a flanking purine to the TT lesion. Pan et al.45 recently found no evidence that excitation of a purine base flanking a cis-syn TT-CPD leads to scission within experimental uncertainty. Of course, this could still be attributable to poor stacking between the putative electron donor base and the CPD in the trinucleotides studied.45 A further possibility is that electron transfer-repair by a neighbouring base is highly inefficient. That this is so is established by the results of this study. In particular, the much larger absorption cross section for G compared to T<>T means that G would be excited at a nearly one hundred-fold higher rate at 254 nm, yet a proximal G changes the photoreversal rate by a much smaller amount. More work is needed into the intriguing possibility of DNA self-repair.12
Overall these results, which encompass a more diverse array of sequence contexts, provide strong support for the hypothesis that flanking bases exert the greatest influence on CPD formation.8, 12 Long-range conformational or electronic effects such as energy transfer17 do not appear to play a role based on the fact that rates are determined mainly by the identity of the flanking bases.
The finding that interactions with nearest-neighbour bases dominate the photochemistry of a TT step that is not flanked by thymines is insufficient to decide between steric vs. electronic effects. Conformational properties relevant to CPD formation (base pair flexibility, stacking energy, etc.) are not expected to extend more than one to two base pairs away from the dimer formation site, while the notion that electronic states are restricted to a pair of stacked bases is consistent with the most recent understanding of DNA excitations.46, 47 Pan et al.14 have suggested that ground-state conformation and not electronic effects play the biggest role in determining dimer yields in model trinucleotides, but it is uncertain whether the trends in ground-state conformation identified in molecular dynamics simulations will carry over to double-stranded structures.
It is somewhat surprising that photoreversal rates depend on local sequence as strongly as seen here. After all, direct photoreversal involves electronic excitation of a CPD, the electronic structure of which is unlikely to be significantly perturbed by neighbouring bases on account of their very different transition energies. In addition, photoexcitation is the rate-limiting step due to the small absorption cross-section of the TT-CPD at 254 nm.
It is difficult to envision how the photoexcitation rate could depend on conformation, but the flanking base or bases could modulate the excited-state dynamics near the conical intersection that is thought to mediate splitting.48 It is also possible that a photon absorbed by the TT-CPD creates an excited state that is preferentially quenched by neighbouring bases, possibly in dependence on their transition energies as well as on geometric factors such as base-base overlap that determine the electronic coupling. Quantum chemical calculations can usefully explore these possibilities.
Conclusions
DNA strands with a total length of nearly 1000 bases were irradiated at 254 nm and then analysed at a sequencing resolution sufficient to detect thymine-thymine CPDs in a variety of sequence contexts. The results strengthen the conclusion that CPDs at TT steps are formed in yields that depend foremost on the identity of flanking bases and extend this finding to a larger class of tetrads containing a central TT step. A full kinetic analysis of yield vs. dose measurements reveals that photoreversal rates for the TT-CPD also depend sensitively on the nature of the flanking bases.
Supplementary Material
Acknowledgments
Funding from the US National Science Foundation (CHE-1112560, awarded to BK) and the US National Institutes of Health (GM083055, awarded to MGP) is gratefully acknowledged.
Footnotes
Electronic Supplementary Information (ESI) available: Base sequences of the MP2+50 and 5S+50 constructs, supplementary figures and tables, and discussion of photobleaching of Cy5 and Cy3 dyes.
Notes and references
- 1.Yoon J-H, Lee C-S, O'Connor TR, Yasui A, Pfeifer GP. The DNA damage spectrum produced by simulated sunlight. J. Mol. Biol. 2000;299:681–693. doi: 10.1006/jmbi.2000.3771. [DOI] [PubMed] [Google Scholar]
- 2.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 3.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You Y-H, Szabo PE, Pfeifer GP. Cyclobutane pyrimidine dimers form preferentially at the major p53 mutational hotspot in UVB-induced mouse skin tumors. Carcinogenesis. 2000;21:2113–2117. doi: 10.1093/carcin/21.11.2113. [DOI] [PubMed] [Google Scholar]
- 5.Rochette PJ, Therrien J-P, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. Uva-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douki T, Cadet J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of cc photolesions. Biochemistry. 2001;40:2495–2501. doi: 10.1021/bi0022543. [DOI] [PubMed] [Google Scholar]
- 7.Brash DE, Haseltine WA. UV-induced mutation hotspots occur at DNA damage hotspots. Nature. 1982;298:189–192. doi: 10.1038/298189a0. [DOI] [PubMed] [Google Scholar]
- 8.Gordon LK, Haseltine WA. Quantitation of cyclobutane pyrimidine dimer formation in double- and single-stranded DNA fragments of defined sequence. Radiat. Res. 1982;89:99–112. [PubMed] [Google Scholar]
- 9.Becker MM, Wang Z. Origin of ultraviolet damage in DNA. J. Mol. Bio. 1989;210:429–438. doi: 10.1016/0022-2836(89)90120-4. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell DL, Jen J, Cleaver JE. Sequence specificity of cyclobutane pyrimidine dimers in DNA treated with solar (ultraviolet B) radiation. Nucleic Acids Res. 1992;20:225–229. doi: 10.1093/nar/20.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kundu LM, Linne U, Marahiel M, Carell T. RNA is more uv resistant than DNA: The formation of UV-induced DNA lesions is strongly sequence and conformation dependent. Chem. Eur. J. 2004;10:5697–5705. doi: 10.1002/chem.200305731. [DOI] [PubMed] [Google Scholar]
- 12.Holman MR, Ito T, Rokita SE. Self-repair of thymine dimer in duplex DNA. J. Am. Chem. Soc. 2007;129:6–7. doi: 10.1021/ja0668365. [DOI] [PubMed] [Google Scholar]
- 13.Hariharan M, Lewis FD. Context-dependent photodimerization in isolated thymine-thymine steps in DNA. J. Am. Chem. Soc. 2008;130:11870–11871. doi: 10.1021/ja804950j. [DOI] [PubMed] [Google Scholar]
- 14.Pan ZZ, McCullagh M, Schatz GC, Lewis FD. Conformational control of thymine photodimerization in purine-containing trinucleotides. J. Phys. Chem. Lett. 2011;2:1432–1438. [Google Scholar]
- 15.Pan ZZ, Hariharan M, Arkin JD, Jalilov AS, McCullagh M, Schatz GC, Lewis FD. Electron donor-acceptor interactions with flanking purines influence the efficiency of thymine photodimerization. J. Am. Chem. Soc. 2011;133:20793–20798. doi: 10.1021/ja205460f. [DOI] [PubMed] [Google Scholar]
- 16.Becker MM, Wang JC. Use of light for footprinting DNA in vivo . Nature. 1984;309:682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- 17.Gale JM, Nissen KA, Smerdon MJ. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc. Natl. Acad. Sci. USA. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyamichev V. Unusual conformation of (dA)n·(dT)n-tracts as revealed by cyclobutane thymine-thymine dimer formation. Nucleic Acids Res. 1991;19:4491–4496. doi: 10.1093/nar/19.16.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernández CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Thymine dimerization in DNA is an ultrafast photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner PJ. Conformational flexibility and photochemistry. Acc. Chem. Res. 1983;16:461–467. [Google Scholar]
- 21.Johnson AT, Wiest O. Structure and dynamics of poly(T) single-strand DNA: Implications toward CPD formation. J. Phys. Chem. B. 2007;111:14398–14404. doi: 10.1021/jp076371k. [DOI] [PubMed] [Google Scholar]
- 22.Law YK, Azadi J, Crespo-Hernández CE, Olmon E, Kohler B. Predicting thymine dimerization yields from molecular dynamics simulations. Biophys. J. 2008;94:3590–3600. doi: 10.1529/biophysj.107.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCullagh M, Hariharan M, Lewis FD, Markovitsi D, Douki T, Schatz GC. Conformational control of TT dimerization in DNA conjugates. A molecular dynamics study. J. Phys. Chem. B. 2010;114:5215–5221. doi: 10.1021/jp100983t. [DOI] [PubMed] [Google Scholar]
- 24.Cannistraro VJ, Taylor JS. Acceleration of 5-methylcytosine deamination in cyclobutane dimers by G and its implications for UV-induced C-to-T mutation hotspots. J. Mol. Biol. 2009;392:1145–1157. doi: 10.1016/j.jmb.2009.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher GJ, Johns HE. In: Photochemistry and photobiology of nucleic acids. Wang SY, editor. New York: Academic Press; 1976. pp. 225–294. [Google Scholar]
- 26.Varghese AJ. Photochemical reactions of cytosine nucleosides in frozen aqueous solution and in deoxyribonucleic acid. Biochemistry. 1971;10:2194–2199. doi: 10.1021/bi00788a003. [DOI] [PubMed] [Google Scholar]
- 27.Fenick DJ, Carr HS, Falvey DE. Synthesis and photochemical cleavage of cis-syn pyrimidine cyclobutane dimer analogs. J. Org. Chem. 1995;60:624–631. [Google Scholar]
- 28.Chinnapen DJF, Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. USA. 2004;101:65–69. doi: 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen KV, Burrows CJ. A prebiotic role for 8-oxoguanosine as a flavin mimic in pyrimidine dimer photorepair. J. Am. Chem. Soc. 2011;133:14586–14589. doi: 10.1021/ja2072252. [DOI] [PubMed] [Google Scholar]
- 30.Haseltine WA, Gordon LK, Lindan CP, Grafstrom RH, Shaper NL, Grossman L. Cleavage of pyrimidine dimers in specific DNA sequences by a pyrimidine dimer DNA-glycosylase of m. Luteus . Nature. 1980;285:634–641. doi: 10.1038/285634a0. [DOI] [PubMed] [Google Scholar]
- 31.Lippke JA, Gordon LK, Brash DE, Haseltine WA. Distribution of UV light-induced damage in a defined sequence of human DNA: Detection of alkaline-sensitive lesions at pyrimidine nucleoside-cytidine sequences. Proc. Natl. Acad. Sci. USA. 1981;78:3388–3392. doi: 10.1073/pnas.78.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sage E, Cramb E, Glickman BW. The distribution of UV damage in the lacI gene of Escherichia coli: Correlation with mutation spectrum. Mutation Res. 1992;269:285–299. doi: 10.1016/0027-5107(92)90211-j. [DOI] [PubMed] [Google Scholar]
- 33.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong F, Hansen JC, Van Holde KE. DNA and protein determinants of nucleosome positioning on sea-urchin 5S ribosomal-RNA gene-sequences in vitro . Proc. Natl. Acad. Sci. USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Görner H. Photochemistry of DNA and related biomolecules: quantum yields and consequences of photoionization. J. Photochem. Photobiol B. 1994;26:117–139. doi: 10.1016/1011-1344(94)07068-7. [DOI] [PubMed] [Google Scholar]
- 37.Hong HZ, Wang YS. Formation of intrastrand cross-link products between cytosine and adenine from UV irradiation of d(BrCA) and duplex DNA containing a 5-bromocytosine. J. Am. Chem. Soc. 2005;127:13969–13977. doi: 10.1021/ja0531677. [DOI] [PubMed] [Google Scholar]
- 38.Brash DE. In: DNA repair: A laboratory manual of research procedures. Friedberg EC, Hanawalt PC, editors. New York: Marcel Dekker; 1983. pp. 327–345. [Google Scholar]
- 39.Douki T, Court M, Sauvaigo S, Odin F, Cadet J. Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J. Biol. Chem. 2000;275:11678–11685. doi: 10.1074/jbc.275.16.11678. [DOI] [PubMed] [Google Scholar]
- 40.Bourre F, Renault G, Seawell PC, Sarasin A. Distribution of ultraviolet-induced lesions in simian virus 40 DNA. Biochimie. 1985;67:293–299. doi: 10.1016/s0300-9084(85)80071-7. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire DGE, Ruzsicska BP. Quantum yields and secondary photoreactions of the photoproducts of dTpdT, dTpdC and dTpdU. Photochem. Photobiol. 1993;57:755–769. doi: 10.1111/j.1751-1097.1993.tb09207.x. [DOI] [PubMed] [Google Scholar]
- 42.Douki T. Low ionic strength reduces cytosine photoreactivity in UVC-irradiated isolated DNA. Photochem. Photobiol. Sci. 2006;5:1045–1051. doi: 10.1039/b604517k. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Mann DB, Suquet C, Springer DL, Smerdon MJ. Ultraviolet damage and nucleosome folding of the 5S ribosomal RNA gene. Biochemistry. 2000;39:557–566. doi: 10.1021/bi991771m. [DOI] [PubMed] [Google Scholar]
- 44.Seidel CAM, Schulz A, Sauer MHM. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- 45.Pan ZZ, Chen JQ, Schreier WJ, Kohler B, Lewis FD. Thymine dimer photoreversal in purine-containing trinucleotides. J. Phys. Chem. B. 2012;116:698–704. doi: 10.1021/jp210575g. [DOI] [PubMed] [Google Scholar]
- 46.Middleton CT, de La Harpe K, Su C, Law YK, Crespo-Hernández CE, Kohler B. DNA excited-state dynamics: From single bases to the double helix. Annu. Rev. Phys. Chem. 2009;60:217–239. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- 47.Su C, Middleton CT, Kohler B. Base-stacking disorder and excited-state dynamics in single-stranded adenine homo-oligonucleotides. J. Phys. Chem. B. 2012;116:10266–10274. doi: 10.1021/jp305350t. [DOI] [PubMed] [Google Scholar]
- 48.Boggio-Pasqua M, Groenhof G, Schäfer LV, Grubmüller H, Robb MA. Ultrafast deactivation channel for thymine dimerization. J. Am. Chem. Soc. 2007;129:10996–10997. doi: 10.1021/ja073628j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.