Abstract
Insect viruses have evolved strategies to control the host RNAi antiviral defense mechanism. In nature Drosophila C Virus (DCV) infection causes low mortality and persistent infection, whereas the closely related Cricket Paralysis Virus (CrPV) causes a lethal infection. We show these viruses use different strategies to modulate the host RNAi defense machinery. The DCV RNAi suppressor (DCV-1A) binds to long double-stranded RNA (dsRNA) and prevents processing by Dicer2. In contrast, the CrPV suppressor (CrPV-1A) interacts with the endonuclease Ago2 and inhibits its activity, without affecting the miRNA-Ago1 mediated silencing. The link between viral RNAi suppressors and the outcome of infection was examined using recombinant Sindbis viruses encoding either CrPV-1A or DCV-1A. Flies infected with Sindbis virus expressing CrPV-1A showed a dramatic increase in virus production, spread and mortality. In contrast, Sindbis pathogenesis was only modestly increased by expression of DCV- 1A. We conclude that RNAi suppressors function as virulence factors.
Keywords: viral defense, Drosophila C virus, Cricket Paralysis virus, RNAi suppressor, acquired immunity, viral persistence
Introduction
RNA interference (RNAi) is a highly conserved, post-transcriptional gene expression control mechanism involved in a number of critical cellular processes1. In insects and plants, RNAi functions as a major antiviral defense mechanism2. In Drosophila, Dicer2, a class III dsRNA endonuclease, processes viral dsRNA structures into small interfering RNAs (siRNAs). siRNAs assemble into RNA induced silencing complex (RISC) to serve as guides for the endonuclease Ago2, which targets the viral RNA for degradation. As a countermeasure to RNAi antiviral defense, viruses encode suppressors of RNA silencing (VSR) to modulate host antiviral response.
The study of viral RNAi suppressors has provided insights into the mechanistic details of siRNA and miRNA (microRNA) production, as well as RISC assembly. A number of factors including helicases, double-stranded RNA binding proteins, and endonucleases are required for the proper assembly of functional RISC3,4. Accordingly, viral suppressors can potentially target any of these factors to circumvent the host RNAi defense. Intriguingly, most RNAi suppressors are RNA binding proteins that interact with dsRNA precursors and/or siRNAs to inhibit dsRNA processing by Dicer and assembly of RISC5. However, some plant viral RNAi suppressors also target components of the RNAi machinery itself. For instance, P0 suppressor protein of the plant polerovirus inhibits downstream events in RISC assembly by promoting Ago1 degradation6–8. Another plant virus RNAi suppressor, Cucumber mosaic virus (CMV) 2b, inhibits both siRNA and miRNA pathway by blocking Ago1 cleavage activity9. Furthermore, inhibition of RNA silencing by most plant viral suppressors also affects miRNA function, causing developmental abnormalities that may contribute to the pathogenic consequences of infection9,10.
Several insect viruses also encode RNAi suppressors2,11. Drosophila C virus (DCV) is a positive strand RNA virus within the Dicistroviridae family that infects many strains of D. melanogaster and in nature establishes a non-lethal persistent infection12–17. A closely related dicistrovirus, Cricket paralysis virus (CrPV), however, produces lethal infections in field crickets18 and in fruit fly19. The viral RNA genomes are comprised of two distinct open reading frames, termed ORF1 and ORF2, and the expression of both is determined by internal ribosome entry site (IRES)-mediated translation initiation20. ORF1 encodes the non-structural replication proteins and ORF2 encodes the structural proteins that form the viral capsid. Experimental infection by DCV and CrPV is dramatically exacerbated in flies with genetic defects lacking the RNAi effector proteins Dicer2 and Ago2, indicating that RNAi is a bona fide anti-viral defense mechanism in insects21–23. The differential outcomes of DCV and CrPV infections in nature, led us to examine whether these closely related viruses employ distinct strategies to control the host antiviral response. We previously reported that DCV encodes a dsRNA binding protein, DCV-1A, that suppresses RNA silencing by specifically blocking Dicer2 processing of virus dsRNA into siRNAs22. On the other hand, the mechanism employed by CrPV to counteract the Drosophila immune system has not been established.
Here we show that CrPV encodes a potent VSR, CrPV-1A that interacts with Ago2 and inhibits RISC activity. Furthermore, unlike plant virus RNAi suppressors, this hitherto undescribed RNAi inhibitor neither affects the miRNA pathway nor alters the normal development and physiology of the animal. Given the distinct pathogenic outcomes observed in DCV and CrPV infections in nature, we hypothesized that RNAi suppressors may function as virulence factors. Indeed, we found that CrPV and DCV RNAi suppressor can modulate the outcome of Sindbis virus infection in flies. Recombinant Sindbis virus expressing CrPV-1A increases virus production resulting in high mortality, whereas DCV-1A expression resulted in only a modest enhancement of infection. We propose that insect virus RNAi suppressors are key modulators of the host immune response that fine-tune the outcome of infection in line with the evolutionary viral strategy for successful host transmission.
Results
CrPV infection blocks RNAi in S2 cells
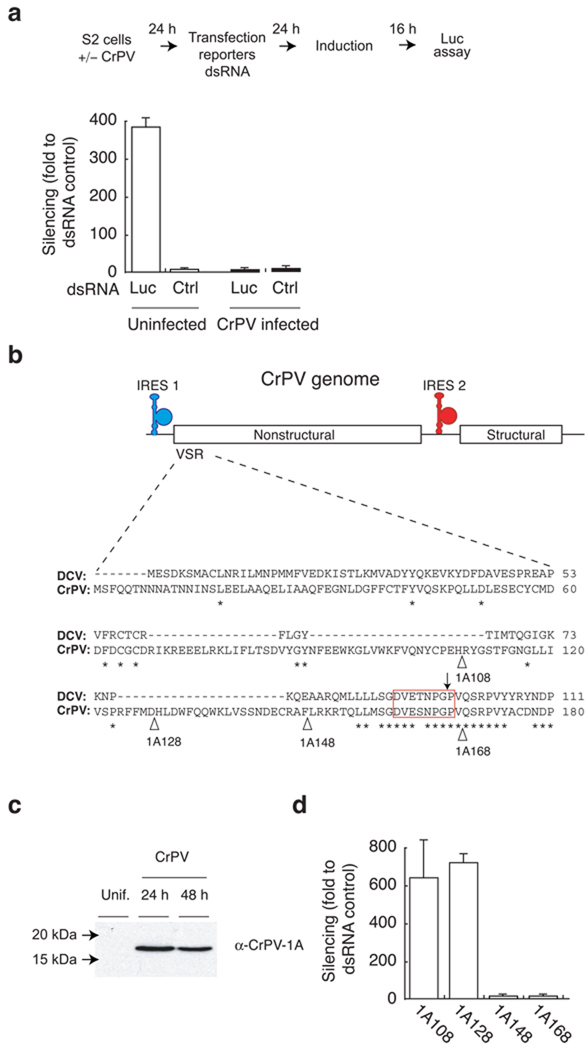
Drosophila melanogaster deficient in key RNAi endonucleases, Dicer2 and Ago2, are highly susceptible to CrPV infection suggesting that the fly RNAi machinery plays an essential role in antiviral defense21–23. Therefore, we examined the possibility that CrPV may encode a suppressor of RNAi to control the RNA silencing machinery. RNAi suppression was analyzed in S2 cells using a dual luciferase reporter system consisting of a firefly luciferase (FLuc) expressing plasmid and a specific 200 nt dsRNA targeting the firefly luciferase mRNA or a eGFP dsRNA control (Ctrl) (Fig. 1a)22. Drosophila S2 cells were either CrPV or mock infected for 24 hour prior to co-transfection with the reporter system for RNAi silencing activity. An internal control using renilla luciferase (RLuc) was also included in each experiment. In uninfected control cells, we observed efficient silencing of firefly luciferase (suppression by a factor of 380) as compared to a control dsRNA. In contrast, silencing was completely suppressed in CrPV infected cells (Fig. 1a), indicating that CrPV encodes a potent suppressor of RNAi.
Figure 1.
Cricket paralysis virus (CrPV) antagonizes RNAi in S2 cells. (a) CrPV infected S2 cells or uninfected S2 cells were co-transfected with firefly, renila luciferase plasmid and either double stranded RNA (dsRNA) targeting the firefly luciferase (Luc) or eGFP dsRNA control (Ctrl). Silencing efficiency in S2 cells were monitored by comparing the ratio of firefly to renila luciferase expression. (b) CrPV genome encodes both non-structural and structural proteins whose translation is regulated by internal ribosome entry site (IRES) 1 and 2 respectively. The extended broken line (−) represents alignment of the viral suppressor of RNA silencing (VSR) domains that include N-terminal 180 amino acid of the CrPV ORF1 and 111 amino acid of the DCV ORF1. - indicates gap, * indicates complete homology, red open box represents the conserved octameric sequence, ↓ indicates non-proteolytic cleavage site, and Δ indicates sites of deletion in the VSR region (c) S2 cells were either mock (Unif) or CrPV infected (at MOI 1) for 24hrs and 48 hrs. Harvested samples were analyzed for 1A expression by western blot analysis using rabbit polyclonal antibody raised against the suppressor protein (α-CrPV-1A). (d) Plasmids encoding C-terminal amino acid deletion of putative CrPV suppressor domain (168,148,128,108) were cotransfected with dual luciferase reporter system along with FLuc dsRNA targeting firefly luciferase or eGFP dsRNA control as described in figure a. Silencing was represented as fold silencing compared to the control dsRNA. Data in (a) and (d) indicate averages and standard deviations of four independent experiments.
The N-terminal region of CrPV ORF1 encodes an RNAi suppressor protein
DCV and CrPV are closely related species within the cripavirus genus of Dicistroviridae family. Amino acid sequence alignment between DCV and CrPV indicated a high degree (ca. 55%) of identity within open reading frame 1 (ORF1). A dsRNA binding motif (DSRM) was previously identified within the N-terminal 99 amino acid region of DCV ORF1, spanning residues 25–88, and shown to encode a potent suppressor of RNAi (DCV-1A)22. Based on these observations, we set out to determine whether the suppression of RNAi by CrPV could be attributed to a DCV-1A-like RNAi suppressor that mapped to the N-terminus of CrPV ORF1.
Alignment of DCV and CrPV in this region revealed no substantial homology as compared to homologous downstream amino acid sequences, which includes a highly conserved octamer sequence DVExNPGP (Fig. 1b). This octamer sequence serves as a signal for co-translational protein cleavage between the glycine and proline residues in several picornaviruses24. We hypothesized that this DVExNPGP sequence may serve a similar function during DCV and CrPV polyprotein processing. We previously reported a DSRM within sequences upstream of the DVExNPGP site that functions as a potent RNAi suppressor protein in DCV22. We detected a protein with a molecular weight consistent with cleavage at the DVExNPGP site in CrPV infected cells by western blot analysis using a polyclonal antibody raised against the first 148 N-terminal amino acids of CrPV ORF1 (Fig. 1c).
Next, we evaluated the ability of the CrPV sequence upstream of the DVExNPGP motif to suppress RNAi in S2 cells. Using the dual reporter assay, luciferase (Fluc, Rluc) expression plasmids were co-transfected with dsRNA that specifically targets firefly luciferase and plasmids encoding C-terminal deletions of the putative CrPV RNAi suppressor protein (N-terminal 168, 148, 128, or 108 amino acids). Both CrPV-1A168 and CrPV-1A148 efficiently blocked RNA silencing (Fig.1d). In contrast, CrPV-1A128 and CrPV-1A108 expression were unable to suppress RNAi (Fig.1d). Hence, the N-terminal 148 amino acids of CrPV ORF1 are sufficient to block RNAi in S2 cells. To examine the effects of CrPV-1A suppressor in vivo, we generated transgenic flies expressing the CrPV-1A148 protein, the inactive CrPV-1A108 fragment, or GFP. In order to monitor the effects of the CrPV-1A148 suppressor in vivo, these transgenic flies were crossed with flies expressing a hairpin dsRNA (inverted repeat) targeting the white gene (IR[white]). The expression of two mini white genes would be suppressed by expression of this hairpin dsRNA, thus providing a colorimetric readout for RNA silencing. The white gene was efficiently silenced in flies expressing GFP and the IR[white] hairpin causing an orange eye color due to the loss of red eye pigmentation (Fig. 2a). In contrast, RNA silencing was suppressed in flies expressing CrPV-1A148, resulting in expression of the white gene and, thus, a red eye color. Flies expressing the inactive CrPV-1A108 were able to suppress the white gene similar to the GFP flies (Fig. 2a). Therefore, our observations demonstrate that CrPV-1A148 is a potent RNAi suppressor in Drosophila.
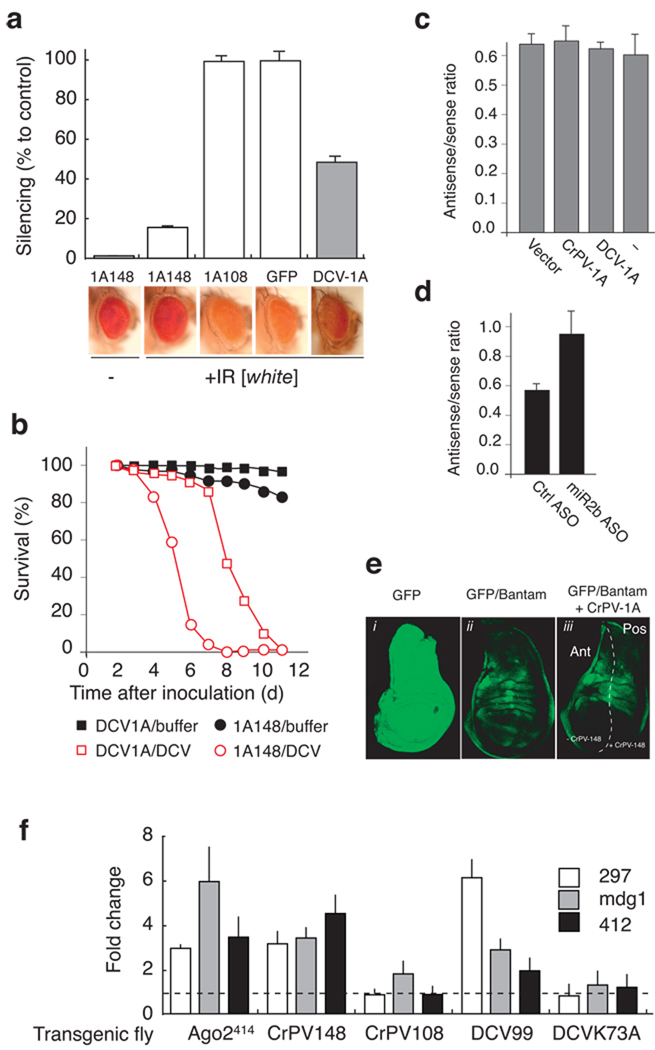
Figure 2.
CrPV-1A is a potent RNAi suppressor and does not interfere in the microRNA pathway. (a) Transgenic flies expressing CrPV-1A148 or CrPV-1A108, or DCV-1A99 or GFP and an inverted repeat [IR] directed against the white gene were analyzed for their ability to carry out efficient RNAi. Loss of red eye pigments in presence of indicated suppressor proteins is expressed as % silencing compared to GFP control fly. (b) Transgenic flies expressing CrPV-1A and DCV-1A were injected intra-thoracically with 100 TCID50 DCV or Tris buffer control and survival rate was monitored daily. (c) Luciferase reporter plasmid carrying miR2b target sequences in sense and antisense orientation (Supplementary Fig. 1b) in the 3’ untranslated region (UTR) was transfected into S2 cells. Luciferase expression in presence of CrPV-1A, DCV-1A was expressed as ratio of luciferase counts produced by luciferase miR2b antisense over luciferase miR2b sense reporter transfection. (d) S2 cells were transfected with miR-2b ASO followed by trasnsfection of reporter system (miR2b antisense). miR-2b ASO restored luciferease expression up to 50% compared to Ctrl ASO (e) The effect of CrPV-1A on bantam mediated translational repression in the imaginal disc was analyzed in flies expressing GFP mRNA containing three complementary bantam miRNA target sites. (f) Fold changes in retrotransposons RNA levels were calculated relative to 297, mdg1 and 412 RNA levels measured in control flies. Fold changes in homozygous Ago2414 mutants were calculated relative to heterozygous ago2414 flies. Data in (a), (c), (d) and (f) indicate standard deviations from three independent experiments.
To further characterize CrPV-1A148 (herein named CrPV-1A) suppressor, we compared its RNAi suppressing efficiency with that of the RNAi suppressor of DCV, DCV-1A99 (herein named DCV-1A). Suppression by CrPV-1A and DCV-1A was assessed in transgenic flies by measuring the accumulation of red eye pigments in flies expressing each of the RNAi suppressor proteins under the control of identical promoters (Fig. 2a). The red eye pigment absorbance (OD480) from control flies expressing GFP and IR[white] was defined as 100% silencing, and the OD480 value from flies not expressing IR[white] (Fig. 2a, Fig. 1A148 -) was defined as the maximum inhibition of silencing. Flies expressing CrPV-1A and IR[white] strongly inhibited RNA silencing (14% silencing) thus restored the red eye phenotype, whereas DCV-1A expression resulted a modest effect on RNA silencing (50% silencing) (Fig. 2a, compare 1A148 with DCV-1A). Control flies expressing CrPV-1A108 and GFP exhibited maximum silencing induced by the dsRNA hairpin targeting white gene (Fig. 2a). Western blot analysis demonstrated that each suppressor protein accumulated to similar levels (Supplementary Fig. 1a). Furthermore, the pathogenic effect of DCV infection was exacerbated by the expression of CrPV-1A resulting in higher mortality as compared to transgenic flies expressing DCV-1A (Fig. 2b). Together, these data indicate that CrPV-1A is a more potent RNAi suppressor than DCV-1A.
CrPV-1A inhibits siRNA function but not the miRNA pathway
In plants, many VSR also interfere with the miRNA pathway and are often associated with disease symptoms resembling plant developmental defects25. We, thus, examined whether CrPV-1A also inhibits Drosophila the miRNA pathway. We carried out experiments in S2 cells using a firefly luciferase reporter containing two perfect complementary miR2b sequences (antisense) in its 3' untranslated region (UTR)22(Supplementary Fig. 1b). 4A firefly luciferase mRNA containing two copies of the miR2b sequence in the opposite orientation (sense) was used as a control. S2 cells were co-transfected with firefly luciferase miR2b reporter constructs (sense or antisense), a plasmid encoding Renilla luciferase (transfection control), and either the CrPV-1A or DCV-1A expressing construct. The ratio of luciferase (miR2b antisense/miR2b sense) expression was identical in cells transfected with DCV-1A, CrPV-1A, empty vector, and no plasmid control (Fig. 2c). The dynamic range of the miR2b reporter assay was determined using anti-sense oligos (ASO) targeting endogenous miR2b miRNA. ASO reduced miR2b silencing effect up to 50% compared to non-specific ASO control (Fig. 2d). We concluded that CrPV-1A does not affect the miRNA gene silencing pathway in S2 cells.
The effect of CrPV-1A on the miRNA pathway was further examined in third instar larvae. Transgenic flies expressing CrPV-1A were crossed with transgenic flies containing a bantam miRNA sensor construct that allows in vivo imaging of bantam miRNA activity26. A GFP reporter containing three copies of a perfect bantam target sequence in its 3’UTR was expressed under the control of the tubulin promoter (Supplementary Fig. 1c). Since bantam miRNA expression is temporally and spatially regulated in response to patterning cues, the expression of GFP will now be under similar control and a characteristic GFP expression pattern was observed in wing imaginal discs from transgenic larvae (Fig. 2e, GFP/bantam). In contrast, a GFP transgene construct lacking the bantam target sequences showed ubiquitous GFP expression in wing imaginal discs (Fig. 2e, GFP). To monitor the effects of CrPV-1A on bantam, we used an engrailed-GAL4 driver to express CrPV-1A in the posterior compartment of the imaginal discs27. No effect was observed on bantam miRNA-mediated GFP repression in the presence of CrPV-1A (Fig. 2e, GFP/bantam + CrPV-1A, and Supplementary Serial Optical Sections 1 and 2). Additionally, ubiquitous expression of CrPV-1A throughout development does not generate any defect in adult flies, further suggesting CrPV-1A does not affect the miRNA pathway in the intact animal (not shown). Thus, unlike plant virus RNAi suppressors, CrPV-1A does not interfere with the host miRNA pathway.
Recently a new class of small RNA called endogenous small interfering RNA (esiRNAs) has been described in Drosophila28–30. The esiRNAs derive from repetitive sequence elements like retrotransposons and control their replication in somatic cells. To examine whether CrPV-1A perturbs the esiRNA pathway, steady state level of three distinct retrotransposons 297, mdg1, 41228,29 were analyzed in female adult head of transgenic flies expressing functional RNAi suppressor CrPV-1A148, DCV-1A99 and control flies expressing non-functional suppressor CrPV-1A108, dsRNA binding mutant DCV-1A-K73A22. Homozygous Ago2414 mutant flies were used as positive control51 (Fig. 2f). Expression of suppressor DCV-1A and CrPV-1A, but not suppressor mutant controls significantly increased retrotransposon expression levels, indicating that the virus suppressors also inhibit the esiRNA pathway.
The CrPV-1A blocks Ago2 cleavage activity
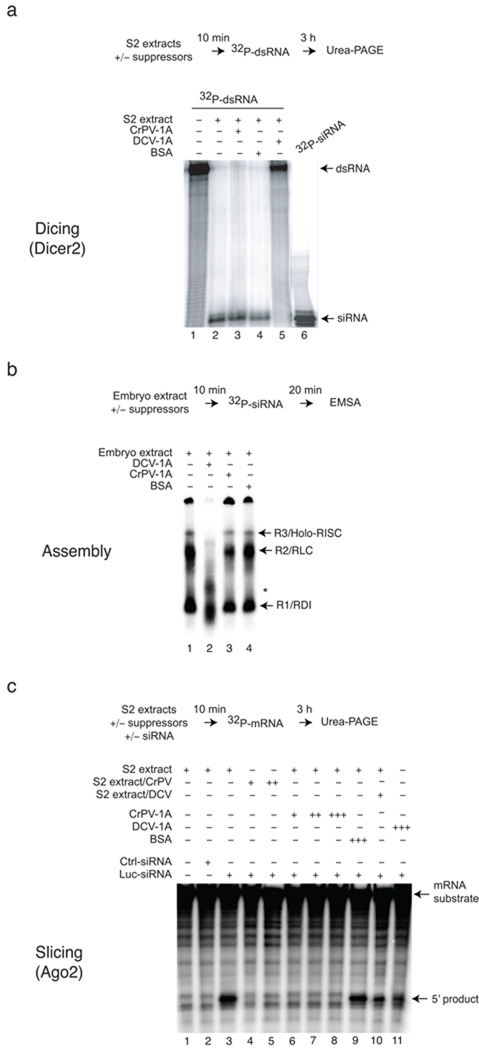
We next examined the mechanism by which CrPV-1A inhibits RNAi. Since most RNAi suppressors bind dsRNA to inhibit silencing5, we first determined whether CrPV-1A binds dsRNA. Electrophoretic mobility shift experiments were performed using radio-labeled dsRNA (200 bp) or siRNA (21 bp) probes and purified recombinant CrPV-1A or DCV-1A (Supplementary Fig. 2a). Consistent with our previous findings22 DCV-1A efficiently bound to long dsRNA (Supplementary Fig. 2a, lane 1, left panel) and to a lesser extent to siRNA (lane 1, right panel). In contrast, CrPV-1A did not bind dsRNA or siRNA (Supplementary Fig. 2a, lanes 2–7 and 9–14). Next, we examined the effect of CrPV-1A on Dicer2 activity using an in vitro assay. In S2 cell extracts, dsRNA was efficiently processed to 21 bp siRNAs (Fig. 3a, lane 2). Production of siRNA was inhibited by the addition of recombinant DCV-1A (Fig. 3a, lane 5), but not by recombinant CrPV-1A or BSA control (Fig. 3a, lane 3 and 4). These experiments indicated that CrPV-1A inhibits RNAi at a different step than DCV-1A, functioning downstream of siRNA production. Indeed, CrPV-1A, unlike DCV-1A, effectively blocked silencing induced by transfection of both long dsRNA in S2 cells as well as 21bp siRNA (Supplementary Fig. 2b), supporting a role downstream of siRNA production.
Figure 3.
CrPV-1A inhibits RISC activity downstream of Dicer processing. (a) Radio-labeled dsRNA substrates (32P-dsRNA) were incubated in S2 buffer control (lane 1) or in S2 extracts (lane 2). Processing of long dsRNA to siRNA by Dicer2 in presence of recombinant CrPV-1A (0.35 µM, lane 3), DCV-1A (0.35 µM, lane 5) or bovine serum albumin (BSA) (0.35 µM, lane 4) was monitored using 12% denaturing gel with an end-labeled 21bp synthetic siRNA marker. (b) Recombinant suppressor proteins DCV-1A (0.35 µM, lane 2), CrPV-1A (0.35 µM, lane 3) or BSA protein (0.35 µM, lane 4) were incubated in Drosophila embryo extract for 10 min. 32P-labeled siRNAs duplex were subsequently added to the reaction for another 20 min. siRNP complexes (R1, R2 and R3) were analyzed by electrophoretic mobility shift assay (EMSA) using 4% native polyacrylamide gel electrophoresis. (c) Target mRNA cleavage assay was analyzed in absence of siRNA (lane 1), in presence of GAPDH (Ctrl) siRNA (lane 2) or Luc siRNA (lane 3-lane 11). To analyze the effects of CrPV-1A, assays were carried out using CrPV infected S2 extracts (MOI 1 and MOI 2) in lane 4, lane 5 or by incubating recombinant CrPV-1A with increasing concentration (0.15 µM, lane 6), (0.25 µM, lane 7), (0.35 µM, lane 8) in uninfected S2 extracts. The activity of the DCV-1A was analyzed using infected DCV S2 extracts (MOI 1, lane 10) or by incubating recombinant DCV-1A (0.35 µM, lane 11) in uninfected S2 extracts. The 5’ cleaved products were analyzed by denaturing polyacrylamide gel.
siRNAs produced by Dicer2 are incorporated into the RISC to guide mRNA cleavage. Three siRNA nucleoprotein (siRNP) complexes occurring during this assembly process can be resolved by native gel electrophoresis, including an initiator R1 (R2D2-Dcr2 initiator complex/RDI), an intermediate R2 (RISC loading complex/RLC), and an effector R3 (holo-RISC) complex31–33. We thus employed native gel analysis to examine whether CrPV-1A affects RISC assembly. Incubation of radio-labeled siRNA duplex in Drosophila embryo extract resulted in formation of R1, R2 and R3 complexes (Fig. 3b, lane 1). The presence of CrPV-1A did not affect the assembly of these three complexes (Fig. 3b, lane 3). In contrast, addition of DCV-1A inhibited formation of holo-RISC (Fig. 3b, lane 2), yielding two intermediate complexes (Fig. 3b, denoted by asterisks). These experiments indicated that CrPV-1A does not affect siRNA loading and holo-RISC assembly. Furthermore, we find that DCV-1A has a hitherto undescribed function affecting RISC assembly, in addition to its previously described function to bind long dsRNA, suggesting that DCV-1A acts at several levels to inhibit RNAi function.
Next we tested whether CrPV-1A affects RISC-mediated mRNA cleavage using an in vitro target cleavage assay (Fig. 3c)34. S2 extracts, programmed with a FLuc siRNA, or a non-specific control siRNA, were incubated with a 592 nt radio-labeled cap FLuc mRNA substrate. In control reactions, the FLuc mRNA target was efficiently cleaved (5´ product) in the presence of a FLuc specific siRNA, as expected (Fig. 3c, compare lane 3 with lanes 1 and 2). While purified DCV-1A addition had little effect on target cleavage, FLuc mRNA cleavage was strongly inhibited upon addition of purified recombinant CrPV-1A (Fig. 3c, lane 6, 7 and 8 for CrPV and lanes 10, 11 for DCV). Similarly, cleavage was also inhibited in extracts from CrPV-infected S2 cells (Fig. 3c, lane 4 and 5). These experiments demonstrate that CrPV-1A suppresses RNAi by inhibiting RISC activity without affecting Dicer processing or RISC assembly.
CrPV-1A interacts with Ago2
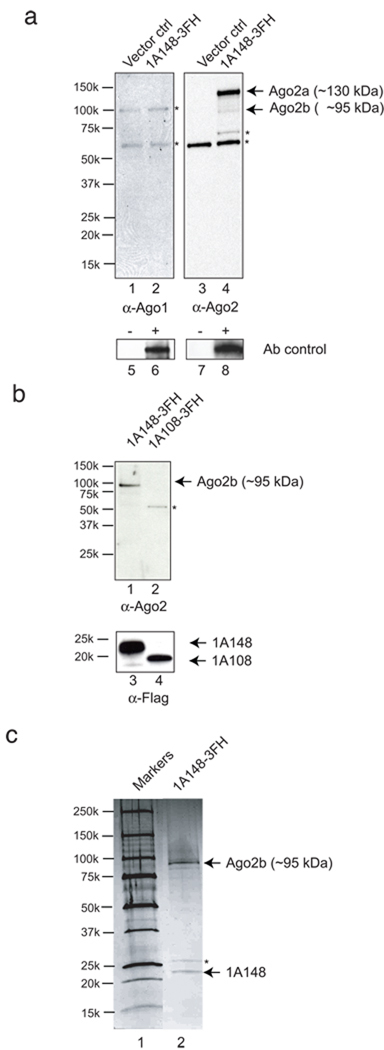
We hypothesized that CrPV-1A inhibits RISC activity by associating with one or more components of holo-RISC. To examine this possibility, CrPV-1A carrying a C-terminal tandem affinity (Flag and His) purification tag (CrPV-1A-3FH) was immunoisolated from S2 cells. Western blot analysis using Ago2 specific antibody revealed that CrPV-1A co-immunoprecipitated with Ago2 (Fig. 4a, lane 4, right panel). In contrast, an antibody directed against Ago1 did not detect any Ago1 association with CrPV1A (Fig. 4a. lane 2, left panel), consistent with our results showing that CrPV-1A does not affect the Ago1-dependent miRNA pathway. The Ago2 antibody used detected two Ago2 forms of 130 kDa and 95 kDa (Obbard et al., 2006). It is possible that the smaller form is the product of partial proteolysis during affinity tag purification, as we often detect 95 kDa isoform (Fig 4b and Fig 4c). To further analyze the interaction between CrPV-1A and Ago2, we transiently expressed CrPV-1A148-3FH and CrPV-1A108-3FH in S2 cells. While full length CrPV-1A148-3HF co-precipitated with Ago2, the truncated CrPV-1A108 was unable to interact with Ago2 (Fig 4b, lane 2). The interaction of CrPV-1A with Ago2 was further analyzed by two-step affinity purification from S2 extracts stably expressing CrPV-1A148-3FH. Commassie staining of the purified materials revealed the presence of only two distinct bands not present in the control sample (Supplementary Fig. 3). Mass-spectroscopy analysis identified the two bands as CrPV-1A and Ago2. Furthermore, silver staining of CrPV-1A associated proteins uncovered no additional proteins (Fig. 4c, lane 2). These experiments suggested that CrPV-1A blocks catalytic activity of the RISC by direct interaction with endonuclease Ago2.
Figure 4.
CrPV-1A interacts with Argonaute 2 in S2 cells. (a) S2 cells were transfected with plasmids transiently expressing CrPV-1A-3FH (lane 2, 4) or empty vector control (lane 1, 3). Flag-His tagged CrPV-1A was immunopurified with anti-Flag antibody and western blotted with Drosophila Argonaute 2 (α-Ago2, right panel) or Argonaute 1 (α-Ago1, left panel) polyclonal antibodies. Two isoforms of Ago2 (Ago2a and Ago2b) were detected with Ago2 specific antibodies. Asterisks (*) indicate cross-reacting host-protein band. Lane 6 and 8 show the positive detection of E.coli expressed Drosophila Ago1 and endogenous Drosophila Ago2 with Ago1 and Ago2 antibody respectively (Ab control). (b) Flag-His tagged CrPV-1A148, CrPV-1A108 were transfected in to S2 cells, immunopurified using anti-Flag antibody and probed with Ago2 antibody (α-Ago2). To detect the expression of 1A protein, the blot was stripped and further probed with anti-Flag antibody (α-Flag) (lane 3 and lane 4, bottom panel). Asterisks (*) indicates a cross-reacting host-protein band (c) Stable S2 cell lines either expressing CrPV-1A-3FH (induced with CuSO4) or uninduced control was lysed, immunopurified with anti-Flag antibody followed by Talon magnetic bead purification. The silver stained gel shows the co-purification of the CrPV-1A and Argonaute 2 (Ago2b isoform) from S2 cells (right lane). Left lane represents the molecular weight marker. Asterisk (*) indicates non-specific contaminant band (see Supplementary figure 3).
CrPV-1A acts on preassembled holo-RISC
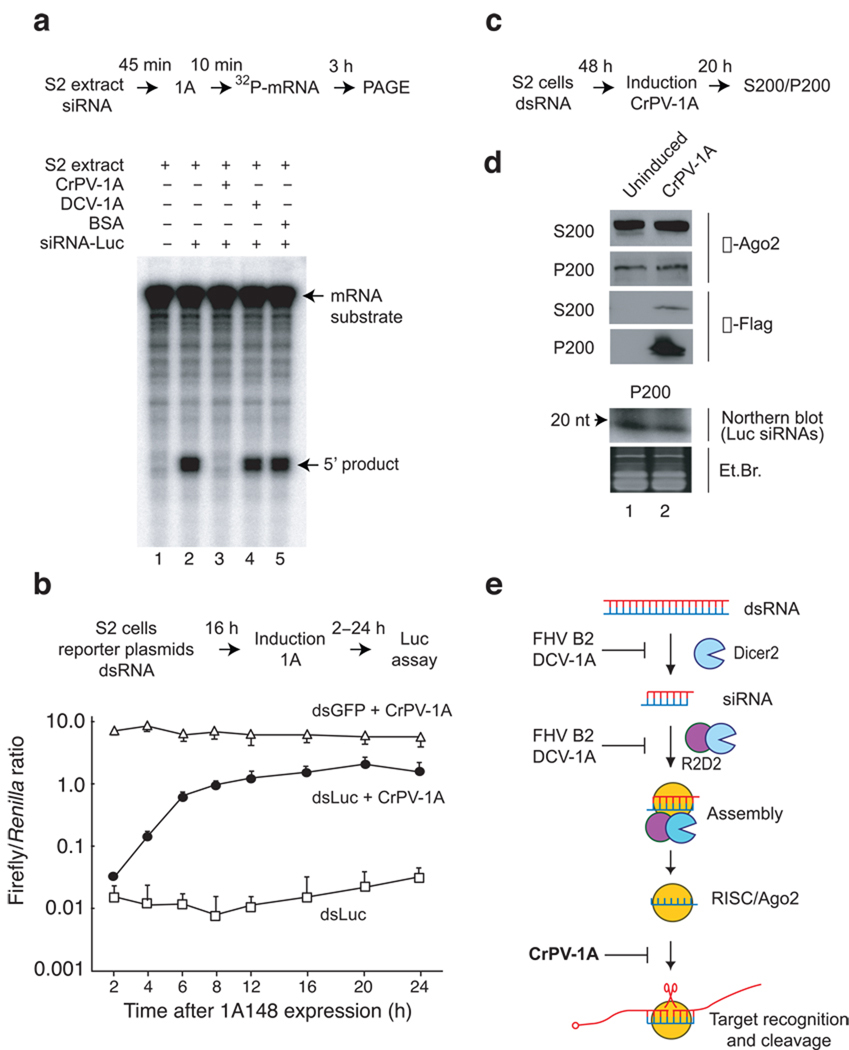
The observation that CrPV-1A does not affect siRNA-RISC assembly in vitro (Fig. 3b), but nonetheless inhibits RISC cleavage activity (Fig. 3c), suggests that CrPV-1A interferes with the function of assembled holo-RISC. To directly test this possibility luciferase mRNA target cleavage assay was performed. S2 cell extracts were pre-incubated with duplex siRNA to allow holo-RISC assembly31–33 followed by recombinant CrPV-1A addition. We observed that CrPV-1A efficiently inhibited holo-RISC enzymatic activity even after assembly of holo-RISC (Fig. 5a, lane 3). As expected, DCV-1A and BSA control had no effect on pre-assembled RISC cleavage (Fig. 5a, lanes 4 and 5).
Figure 5.
CrPV-1A interferes with the function of pre-programmed holo-RISC in vitro and in vivo. (a) S2 extracts were programmed with siRNA for 45 min followed by addition of 1A [(0.35 µM CrPV-1A, lane 3), (0.35 µM DCV-1A, lane 4) or (BSA, lane 5)] for 10 min. RISC assay was initiated by incubating 32P-mRNA substrate for additional 3hrs and 5′ cleaved product was analyzed using denaturing polyacrylamide gel electrophorsis (PAGE). (b) Silencing of firefly luciferase in stable S2 cells was programmed by co-transfecting firefly and renila reporter plasmid with specific FLuc dsRNA (dsLuc) or control eGFP dsRNA (dsGFP). 16 hrs post-transfection, luciferase expression was monitored either in presence (dsLuc+CrPV-1A, dsGFP+ CrPV-1A) or absence (dsLuc) of CrPV-1A for a period of 24 hrs. (c) Stable S2 cells were transfected with FLuc dsRNA for 48 hrs followed by induction of suppressor for additional 20 hrs. The post-nuclear cytoplasmic extract was centrifuged at 200,000×g to separate the ribosomal pellet (P200) from the supernatant (S200). (d) P200 and S200 fractions were western blotted using Drosophila Argonaute 2 (α-Ago2) polyclonal and anti-Flag (α-Flag) monoclonal antibodies. RNA extracted from P200 fraction was analyzed for Luc siRNA by northern blot. P200 ethidium bromide (Et. Br) represents RNA isolated from soluble high salt P200 extraction. (e) DCV-1A and FHV B2 either inhibits Dicer 2 processing of long dsRNA or siRNA incorporation in to RISC by virtue of their dsRNA or siRNA binding activity. Conversely, CrPV-1A acts at the level of holo-RISC by protein-protein interaction with Drosophila Argonaute 2.
The ability of CrPV-1A to inhibit holo-RISC activity in vivo was also examined using the dual-luciferase reporter system in S2 cells expressing CrPV-1A under the control of an inducible metallothionein promoter (Fig 5b). Cells were co-transfected with plasmids expressing Renilla and firefly luciferase and dsRNA targeting firefly luciferase (dsLuc), or GFP (dsGFP) for 16 hrs to allow for maximal silencing of luciferase activity (Fig. 5b, dsLuc). Induction of CrPV-1A expression at 16 hrs resulted in near-complete restoration of firefly luciferase expression (Fig. 5b, dsLuc + CrPV-1A) compared to no CrPV-1A expression control (dsLuc). Together, these results indicate that CrPV-1A can efficiently inhibit pre-assembled holo-RISC function.
Previous findings indicated that, in Drosophila, Ago2 is a component of active holo-RISC, which sediments with a ribosomal fraction following high-speed centrifugation (200,000×g)35. To examine whether CrPV-1A associates with active holo-RISC, cytoplasmic extracts from CrPV-1A expressing S2 cells were subjected to high-speed centrifugation to separate a high molecular weight fraction (P200) from supernatant (S200). Fractions were analyzed by immunoblot using antibodies that specifically recognize Ago2 and CrPV-1A. Expression of CrPV-1A did not affect the distribution of Ago2 between the S200 and P200 fractions (Fig. 5d, α-Ago2). Furthermore, CrPV-1A expression did not affect siRNA association with the P200 fraction (Fig. 5d, northern blot, compare lanes 1, 2). Interestingly, CrPV-1A fractionates preferentially with the high molecular weight P200 fraction, despite the fact that purified recombinant CrPV-1A migrates as a smaller soluble protein in gel filtration analysis (data not shown). We concluded that CrPV-1A neither induces Ago2 degradation nor causes disassembly of the holo-RISC, instead it is specifically recruited to the holo-RISC containing P200 fraction, where it exerts an inhibitory effect through binding to Ago2.
RNAi suppressors are virulence determinants
It is puzzling that two viruses with such overall homology as CrPV and DCV exhibit dramatically different virulence levels in natural infection12,13,18,19. Both DCV and CrPV induce high mortality in fruit flies by intra-thoracic injection in experimental laboratory condition21,22. In contrast, DCV causes mild and persistent infection while CrPV infection is highly lethal by oral inoculation 13,14,16,19,36,37. Furthermore in S2 cells, DCV establishes a persistent infection following an initial acute phase, while CrPV is highly lytic with no establishment of persistent infection (data not shown).
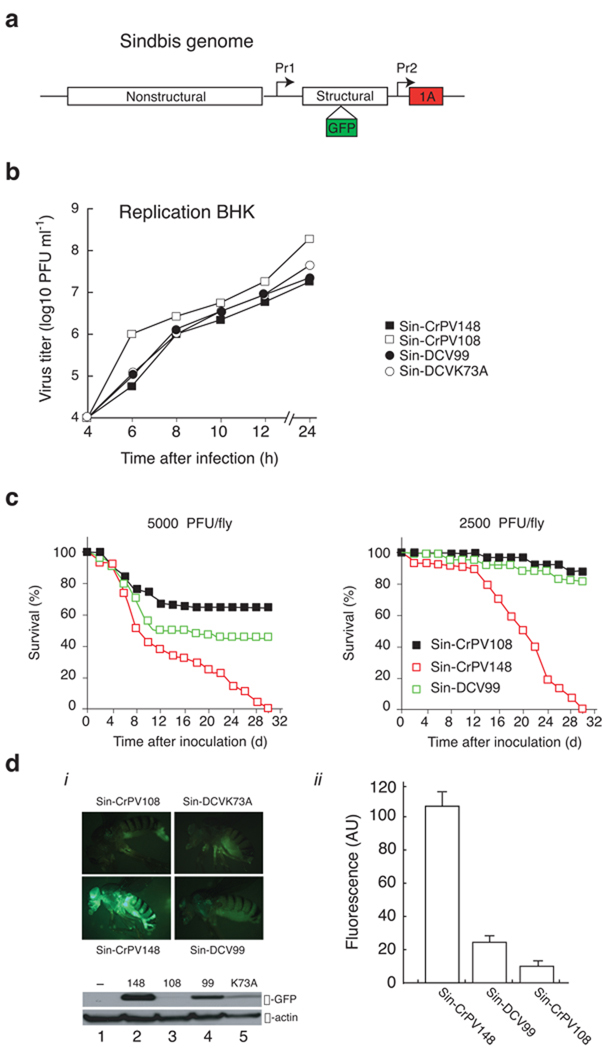
Virus pathogenesis is determined by many factors, including the ability of the virus to evade host immune response. Given the different properties of each suppressor, we considered whether they might determine the outcome of infection. Ideally, this hypothesis should be tested by engineering a virus carrying either of the different RNAi suppressors in an otherwise isogenic backgrounds. Given that no infectious clones are available for either DCV or CrPV, we examined the effect of CrPV-1A or DCV-1A in the context of a recombinant Sindbis virus infection. We engineered recombinant Sindbis viruses to express a green fluorescent protein (GFP) and RNAi suppressors CrPV-1A (Sin-CrPV148) or DCV-1A (Sin-DCV99) under the control of duplicated sub-genomic promoter (Fig. 6a)15. Control Sindbis viruses expressed inactive suppressors -- a truncated version of CrPV-1A (Sin-CrPV108) or DCV-1A carrying a mutation in the DSRM (Sin-DCVK73A)22. In addition to infecting insect cells, Sindbis also infects mammalian cells, which do not appear to use RNAi to control the virus, affording an unbiased control system for viral replication efficiency. No appreciable differences in virus replication kinetics were observed for these recombinant viruses in mammalian BHK-21 cells (Fig. 6b), indicating that expression of RNAi suppressors does not affect virus fitness in cells where the RNAi machinery does not play a major role in antiviral defense38.
Figure 6.
RNAi suppressors determine spread and pathogenesis of Sindbis virus in fruit fly. (a) Expression of the Sindbis virus structural proteins is regulated by a sub-genomic promoter (Pr1). CrPV-1A and DCV-1A were cloned under the control of a duplicated subgenomic promoter (Pr2) in the 3’ untranslated region (UTR) of the sindbis genome. These recombinant viruses also express green fluorescence protein (GFP) that cleaves at both N and C- terminus during structural polyprotein processing15. (b) BHK cells were infected with recombinant Sindbis viruses at MOI 1, virus sample were collected over a period of 24 hrs (h). Titer of the each sample [plaque forming unit (PFU) per ml] was measured by plaque assay and replication kinetics of each virus was determined. (c) Intrathoracic injections of flies with engineered Sindbis virus expressing CrPV-1A (Sin-CrPV148) induced higher mortality and tissue spread at similar dose of infection (2500–5000 PFU) compared to Sindbis virus expressing DCV-1A (Sin-DCV99). Survival rate of flies (%) was monitored daily (d) for thirty days. (d) (i) Flies injected with recombinant Sindbis viruses (Sin-CrPV148, Sin-CrPV108, Sin-DCV99 and Sin-DCVK73A) were imaged by GFP microscopy. For western blot analysis 4–5 flies from each injection at 11 days post infection were homogenized and blotted with anti-GFP antibodies (α-GFP). The same blot was stripped and probed with anti-actin antibody (α-actin). (ii) GFP fluorescence (AU) for recombinant Sindbis viruses (Sin-CrPV148, Sin-CrPV108, Sin-DCV99) was measured and data represents the means and standard error of the mean for 4–5 individual flies.
Sindbis virus does not appear to encode an RNAi suppressor (data not shown) and establishes a persistent infection in Drosophila39. We thus determined the effect of CrPV-1A and DCV-1A on Sindbis infection following intra-thorax inoculations in Drosophila. Both Sin-CrPV148 and Sin-DCV99 increased fly mortality as compared to the control virus expressing an inactive suppressor, Sin-CrPV108 (Fig. 6c). Consistent with the observation that CrPV-1A is a more effective RNAi suppressor than DCV-1A (Fig. 2a and 2b), Sin-CrPV148 induced higher mortality in flies than Sin-DCV99 (Fig. 6c). This enhanced pathogenicity correlated with a dramatic increase in virus replication (Fig. 6d, α–GFP and fluorescence AU). Strikingly, Sin-CrPV148 infection generated a broader GFP tissue distribution as seen in the thorax, abdomen, head, and legs of injected flies (Fig. 6d, i). Infection with control Sin-CrPV108 caused a localized GFP expression at the site of injection, suggesting that the absence of a functional suppressor limits both infectivity (Fig. 6d, ii) and virus spread (Fig. 6d, i). We measured a modest increase in GFP expression during Sin-DCV99 infection that was localized predominately in the thorax, and to a lesser extent in the head of injected flies (Fig. 6d, ii). Quantification of GFP expression by western blot and fluorescence microscopy demonstrated that GFP expression was approximately 5 fold higher in flies infected with Sin-CrPV148 as compared to Sin-DCV99 (Fig.6d, α–GFP and Fluorescence AU). GFP expression steadily increased during the first 10 days post Sin-CrPV148 infection, at which point GFP levels remained constant (data not shown). Importantly, infected flies exhibited maximum GFP expression 10 days post Sin-DCV99 infection followed by a gradual reduction in fluorescence, suggesting that flies were able to control the infection. We thus observed a striking correlation between the enhancement of Sindbis infection severity by CrPV-1A, the potency of this suppressor, and the virulence of natural CrPV infections. In contrast, DCV, which produces a moderate natural infection, carries a suppressor that causes a modest increase in Sindbis virulence. Taken together, these results indicated that these distinctly acting RNAi suppressors are virulence factors that determine the outcome of infection.
Discussion
RNA interference (RNAi) can provide sequence specific antiviral immunity in plants and insects40–42. To counteract this antiviral mechanism, viruses have evolved suppressor proteins with RNAi-modulating activity. We here identified a unique mode of action employed by CrPV for suppressing RNAi-based antiviral immunity. This suppression activity mapped to the N-terminal region of CrPV ORF1, encoding the protein CrPV-1A. We find that CrPV-1A specifically interacts with Ago2 in the context of preassembled holo-RISC and can potently suppress RNAi in Drosophila.
The mechanism of CrPV-1A is different from RNAi suppressors from FHV (B2) and DCV (1A), which appear to prevent viral dsRNA processing by Dicer 2 and siRNAs to assemble into RISC 2,11. This widely used strategy presumably relies on physical shielding of viral dsRNAs from the RNAi machinery. In contrast, CrPV-1A, shows no detectable RNA binding affinity (Supplementary Fig. 2) and does not prevent the loading of siRNAs (Fig. 3b, Fig. 5d) or the formation of holo-RISC (Fig. 3b, Fig. 5d). We find that CrPV-1A interacts with Ago2 (Fig 4c) and inhibits RISC activity in vivo and in vitro without affecting Dicer processing or RISC assembly (Fig. 3–5). CrPV-1A, in the absence of other viral proteins, is preferentially recruited to high molecular weight pre-assembled holo-RISC fractions containing Ago2 and siRNAs (Fig. 5d), and inhibits holo-RISC catalytic activity following RISC pre-formation and assembly (Fig. 5a). In principle, additional CrPV-1A functions within the CrPV replication cycle cannot be excluded, given the multifunctional nature of many viral proteins. However, the fact that CrPV-1A alone both inhibits RNAi and suffices to strongly enhance pathogenicity of the evolutionarily unrelated Sindbis virus demonstrates that CrPV-1A is itself an RNAi suppressor with an autonomous pathogenesis modulating function, which is transferable to other viruses.
While the existence of virus-encoded RNAi suppressors in animals is now firmly documented, relatively little is known about how these factors intersect with host functions and how they contribute to the overall balance between viral replication and transmission. Comparison of RNAi suppression strategies of two closely related viruses, DCV and CrPV, yield insights into both issues. Notably, we find that both viruses are specific for the host siRNA pathway, as neither CrPV-1A nor DCV-1A affected the miRNA gene-silencing pathway (Fig. 2). Indeed, CrPV-1A suppresses the siRNA-mediated gene-silencing pathway by selectively interacting with Ago2, and not related Ago1 homolog (Fig 4a) involved in miRNA-mediated RISC function. Both DCV and CrPV affect the esiRNA mediated retrotransposons silencing by virtue of their abilities to counteracts the functions of Dicer2 and Ago2 respectively (Fig 2f). These observations contrast with known plant virus RNAi suppressors, which do not discriminate between the siRNA and miRNA pathways9,25,43. Due to aberrations in the miRNA pathway during infection, plant viruses often cause physiological and developmental abnormalities within the host9,25,43. The difference between plant and animal viruses may be explained by the interdependent nature of the plant siRNA and miRNA pathways as opposed to the distinctly separate pathways observed in Drosophila melanogaster3. Since miRNAs are important for host physiology and development, the selectivity in RNAi suppression in insect viruses may afford them an evolutionary advantage. By controlling the host RNAi antiviral response without perturbing normal host physiology, viruses may replicate efficiently and establish a persistent infection, as well as facilitating vertical transmission of the virus to the host progeny. These strategies would ensure virus transmission and survival within a host population.
DCV and CrPV are closely related viruses within the Dicistroviridae family. While both viruses encode RNAi suppressor at N-terminus of their genomes, these suppressors exhibit very different mechanisms of action and RNAi suppressor potency. Our data suggests that their potency for inhibition of the host antiviral defense contributes to their differential pathogenicity. Indeed, engineering Sindbis virus to carry DCV-1A and CrPV-1A altered the pathogenicity of an otherwise benign Sindbis infection in flies to an acute one of varying severity. Since both RNAi suppressors were expressed within the same site of the Sindbis virus, using the same promoter and in the absence of other CrPV or DCV proteins, these experiments provide a direct readout of the link between their autonomous RNAi suppressor function and pathogenicity. The expression of the more potent RNAi suppressor, CrPV-1A, significantly enhanced Sindbis virus pathogenicity and correlated with a dramatic increase in virus replication and fly mortality. On the other hand Sin-DCV99 was only partially more pathogenic. It is tempting to speculate that the differential potency of the CrPV and DCV RNAi suppressors stems from their very different modes of action. DCV-1A relies on nonspecific RNA binding activity to protect long dsRNA from Dicer2 processing. Complete RNAi inhibition likely requires high levels of DCV-1A production to saturate all Dicer2 processing sites. In contrast, CrPV-1A acts by directly inhibiting the catalytic component of RISC (Fig. 5e), a multi-turn over enzyme44. Thus, inhibition of the siRISC by CrPV-1A may provide a more efficient strategy to prevent viral genome degradation, which may lead to high viral titer production.
The differential potency of the RNAi suppressors of DCV and CrPV, and their differential effects on Sindbis pathogenicity resonate with the very different types of infection caused by these otherwise very similar viruses. CrPV is a highly lytic virus that induces substantial mortality in flies19. In contrast, DCV, in nature, establishes a symbiotic, chronic infection in Drosophila without causing appreciable mortality12,13,16,45. Indeed, DCV was first found in field populations by random screening of the flies and not as a result of any particular syndrome13. Given that the pathogenic characteristics of DCV and CrPV infection are replicated by simply transferring the respective RNAi suppressors to Sindbis, we argue that these proteins are attuned with the natural virus infection strategy. For instance, the chronic, persistent infection of Drosophila characterized by DCV may be attributed to the modest RNAi suppressing activity exhibited by DCV-1A. Our results point to an exquisite equilibrium between the host antiviral defense machinery and the viral RNAi suppressors that serves as an evolutionary fine tune of the host-virus interactions and determines the pathogenic outcome of infection, virus survival, and virus spread.
Methods
RNAi reporter assay
We cultured Drosophila S2 cells (Supplementary Fig. 4), on 96 well plates. We transfected S2 cells with plasmids encoding CrPV-1A or infected with CrPV (MOI 1) and carried out RNAi reporter assay as described22. We tested the effect of CrPV-1A on microRNA (miRNA) miR2b function as described22. We designed antisense oligonucleotides (ASO) against endogenous miR2b46and transfected in to S2 cells using Dharmafect 4 reagent (Dharmacon) for 48 hrs followed by co-transfection of the reporter systems. We synthesized 2’-O-Me 3’ cholesterol modified miR-2b ASO (C.A.A.A.U.G.C.U.C.C.U.C.A.A.A.G.C.U.G.G.C.U.G.U.G.A.U.A.A.U.U.C.U.3'-Chl) from Dharmacon.
Dicer assay
We performed Dicer assay in S2 cell extracts as described22,34. We incubated recombinant DCV-1A, CrPV-1A or BSA in the reaction for 10 min followed by addition of uniformly labeled 200 bp GL3 dsRNA (105 cpm) for 3 hrs at 25°C.
Slicer assay
We performed RISC cleavage assays as described33 using a 5’ capped FLuc mRNA target (592 nt) with a specific siRNA that generates a 100nt 5’ cleaved product. We either used CrPV or DCV infected S2 extract or supplemented recombinant CrPV-1A and DCV-1A in uninfected S2 extract. We performed a 35 µl reaction (40% v/v S2 extracts) for 3hr at 25°C.
Assembly assay
We performed RISC assembly experiments as described31,32. We incubated recombinant suppressor protein in embryo extract for 10 min, subsequently we added 32P-radiolabeled siRNA duplex to the reaction for another 20 min. The 5µl reactions contained 40% v/v embryo lysates.
Generation of stable S2 cell lines
We cloned CrPV-1A in frame with C-terminal 3xFLAG and 6xHis tags in vector pMT/V5-HisA (Invitrogen). We generated stable cell lines according to manufacturer’s instruction (Invitrogen). We induced the expression of the protein with 500 µM CuSO4 for 20 hrs.
Immunoprecipitation
We transfected plasmids pAc-CrPV148-3FH, pAc-CrPV108-3FH, pAc5.1-V5/HisA vector (Invitrogen) into S2 cells (1×107 cells) for 24 hour, resuspended cells in IP buffer [30mM Hepes pH 7.4, 150mM KOAc, 2mM MgOAc, 5mM DTT, 0.1% NP-40, protease inhibitor tablet (Roche)] and sonicated on ice. We centrifuged (14,000×g) and added supernatant to magnetic beads (Dynal) conjugated with anti-Flag M2 antibody. We washed beads and boiled in SDS sample buffer at 95°C for 5 min. We ran eluted sample in 4–20% gradient SDS-PAGE gel, transferred to PVDF membrane and probed with Drosophila Ago1 or Ago2 antibodies. We followed the purification profile by western blot (Supplementary Fig. 5)
Suppressor-Argonaute 2 complex purification
We resuspended two grams of either induced or uninduced stable S2 cells in 1ml IP buffer and ground into powder in a mortar and pestle under liquid nitrogen. We prepared crude extracts by adding 9ml IP buffer to ground S2 cell and homogenized for 2 min on ice. We centrifuged (14,000×g) and added supernatant to anti-Flag M2 antibody conjugated magnetic beads (Invitrogen), washed with IP buffer and eluted using IP buffer with 3X Flag peptide (400 µg ml−1)(Sigma). We added eluted sample to Talon magnetic beads (Invitrogen), washed with IP buffer and eluted in IP buffer containing 250 mM Imidazole. We stained eluted material with sliver according to manufacturer’s instruction (Invitrogen). To determine the expression profile of Ago2 and CrPV-1A in stable S2 cells we followed the one step (Flag IP) purification protocol and analyzed by western blot analysis (Supplementary Fig. 5).
Protein Identification
We resolved eluted sample on a 4–20% gradient gel and stained using coomassie brilliant blue (Sigma). We excised protein bands and destanined followed by in gel trypsin digestion. We analyzed the trypsin generated peptide fragments using modular mass spectrometric tool, which includes several MALDI-MS and MALDI-MS/MS mass spectrometer as well as an HPLC-MS/MS mass spectrometer47.
Expression and purification of recombinant proteins
We cloned CrPV-1A in to pHis-Gb1-II vector48, transformed into BL21DE3 cells and purified His-tag fusion protein using Ni-NTA resin (Qiagen). We purified GST-DCV-1A as described22. Polyclonal antibody against CrPV-1A was raised in rabbits immunized with purified recombinant proteins. Expression of full-length Drosophila Argonaute 1 in E.coli was carried out as described for CrPV-1A.
RNAi in Fly
We established transgenic flies expressing CrPV-1A in developing eye as described22. We performed eye pigment determination assays on 3 days virgin females49. We established microRNA sensor lines expressing CrPV-1A as below. We recombined P[Tub-bantam sensor]26 transgenic stocks by genetic crosses with the P[engrailed-Gal4] line27 to obtain the homozygous w1118 ; II P[engrailed-Gal4] ; III P[Tub-bantam sensor] miRNA bantam sensor line. We crossed homozygous P[pUAS-CrPV-148], P[pUAS-CrPV108] and w1118 stocks to the sensor line. We dissected wing imaginal discs from third instar larvae fixed in 4% formaldehyde, and stained with mouse monoclonal anti-GFP (Roche) to visualize sensor expression using Apotome microscopy. To observe the effect of RNAi suppressors on retro-transposons expression, we made transgenic flies expressing CrPV-1A as described50. We measured the expression levels of 412, mdg1 and 297 transposon in double heterozygous UASp>VSR; da>GAL4 and control heterozygous da>GAL4 2-days old females. We manually separated fifty heads and processed for total RNA extraction and reverse transcription. We analyzed Transposon levels by real-time PCR using primers as described 28,29,50.
Holo-RISC fractionation
We transfected 20 µg dsRNA (200 bp FLuc dsRNA) in to stable S2 cells (1×107) using Effectene reagent (Qiagen). After 48 hr, cells were induced with CuSO4 for additional 20 hrs. We performed Holo-RISC purification as described 35. We resuspended cells in hypotonic buffer (10 mM Hepes pH 7.4 and 6 mM β-mercaptoethanol), lysed by passing through insulin syringe, spun down at 14,000×g for 25 min at 4°C. We centrifuged the post-nuclear, cytoplasmic extract at 200,000 ×g for 3hrs at 4°C to separate the ribosomal pellet (P200) from supernatant (S200). We probed P200 and S200 fraction for Argonaute 2 and CrPV-1A by western blot.
Northern blotting
We treated ribosomal pellet (P200) with 1mM MgCl2 and 400mM KOAc. We centrifuged extracted soluble material at 100,000×g for 1 hr at 4°C. We treated the supernatant with SDS and proteinase K and extracted RNA with phenol. We resolved small RNA on 12% Urea-gel, transferred onto Hybond-N+ nylon membrane (Amersham Biosciences) and hybridized with Fluc sense strand riboprobe.
Recombinant Sindbis virus production and Fly injection
We generated Sindbis virus expressing RNAi suppressor CrPV-1A and DCV-1A as described15. We determined growth kinetics of viruses in BHK 21 cells. We injected five-day-old female flies in the thorax with 50 nl of virus (2500–5000 PFU) using a nanoinjector (Nanoject II, Drummond Scientific). We observed GFP expression in flies by GFP microscopy and western blot analysis. We incorporated GFP images per group of flies (4–5 each) into the Volocity software (Improvision, UK) under the measurement module. We used non-injected flies as background for further calculation. To determine the pathogenicity of DCV in transgenic flies expressing CrPV-1A and DCV-1A, we injected 100 TCID50 of DCV by intra-thoracic injection as described22.
Supplementary Material
Acknowledgments
We thank Judith Frydman (Stanford University) and member of the Andino laboratory Leonid Gitlin, Dwight Barnes, Michelle Flenniken and Adam Lauring for useful discussion for preparing the manuscript. Special thank to Ronald van Rij (Nijmegen Centre for Molecular Life Sciences, the Netherlands) and Carla Saleh (Institut Pasteur, France) for their help. We thank Dr Graham Belsham (National Veterinary Institute, Denmark) for critical reading of the manuscript and useful comments. We are grateful to Phil Zamore (UMass Medical School), Mikiko Siomi and Keita Miyoshi (Keio University School of Medicine, Japan) for providing antibody directed against Drosophila Argonaute 2, Hans Heidner for providing Sindbis virus plasmid (University of Texas, San Antonio) and Michelle Moritz (UCSF) for Drosophila embryo extract preparation. This work was financially supported by Pasteur Institute, the CNRS and by grants from ANR (AKROSS) and from ARC to C.A., and by NIH grants AI40085 and AI064738 to R.A.
Footnotes
Author contributions
A.N. and R.A. designed and interpreted most of the experiments; A.N. performed most of the experiments; B.B. and C.A. designed and performed the IR[white], GFP/bantam sensor and retrotransposon experiments in flies; M.T. and M.K. performed the fly injections; A.A. made some plasmid constructs; C.D. and A.K. carried out the mass spectrometry analysis; J.G. advised on the project. A.N., R.A., A.A. and C.A. prepared the manuscript.
References
- 1.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 2.van Rij RP, Andino R. The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 2006;24:186–193. doi: 10.1016/j.tibtech.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 7.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 8.Pazhouhandeh M, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci U S A. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasschau KD, et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 11.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 12.Gomariz-Zilber E, Thomas-Orillard M. Drosophila C virus and Drosophila hosts: a good association in various environments. Journal of Evolutionary Biology. 1993;6:677–689. [Google Scholar]
- 13.Brun P, Plus N. The Genetics and Biology of Drosophila. Vol 2d. New York: Academic Press; 1980. The viruses of Drosophila; pp. 625–702. [Google Scholar]
- 14.Jousset F-X, Plus N. Etude de la transmission horizontale et de la transmission verticale de Picornavirus de Drosophila melanogaster et de Drosophila immigrans. Ann Microbiol (Inst Pasteur) 1975;126:231–249. [PubMed] [Google Scholar]
- 15.Thomas JM, Klimstra WB, Ryman KD, Heidner HW. Sindbis virus vectors designed to express a foreign protein as a cleavable component of the viral structural polyprotein. J Virol. 2003;77:5598–5606. doi: 10.1128/JVI.77.10.5598-5606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 17.Gomariz-Zilber E, Poras M, Thomas-Orillard M. Drosophila C virus: experimental study of infectious yields and underlying pathology in Drosophila melanogaster laboratory populations. J Invertebr Pathol. 1995;65:243–247. doi: 10.1006/jipa.1995.1037. [DOI] [PubMed] [Google Scholar]
- 18.Reinganum C. The isolation of cricket paralysis virus from the emperor gum moth, Antheraea eucalypti Scott, and its infectivity towards a range of insect species. Intervirology. 1975;5:97–102. doi: 10.1159/000149886. [DOI] [PubMed] [Google Scholar]
- 19.Manousis T, Moore NF. Cricket Paralysis Virus, a Potential Control Agent for the Olive Fruit Fly, Dacus oleae Gmel. Appl Environ Microbiol. 1987;53:142–148. doi: 10.1128/aem.53.1.142-148.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 22.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn H, Palmenberg AC. Mutational analysis of the encephalomyocarditis virus primary cleavage. J Virol. 1996;70:6870–6875. doi: 10.1128/jvi.70.10.6870-6875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 27.Roignant JY, et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. Rna. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham JW, Sontheimer EJ. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- 33.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 34.Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 35.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 36.Thomas-Orillard M, Legendre S. [C virus of Drosophila and dynamics of host population] C R Acad Sci III. 1996;319:615–621. [PubMed] [Google Scholar]
- 37.Plus N, Croizier G, Jousset FX, David J. Picornaviruses of laboratory and wild Drosophila melanogaster: geographical distribution and serotypic composition. Ann Microbiol (Paris) 1975;126:107–117. [PubMed] [Google Scholar]
- 38.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 39.Mudiganti U, Hernandez R, Ferreira D, Brown DT. Sindbis virus infection of two model insect cell systems--a comparative study. Virus Res. 2006;122:28–34. doi: 10.1016/j.virusres.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 41.Voinnet O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001;17:449–459. doi: 10.1016/s0168-9525(01)02367-8. [DOI] [PubMed] [Google Scholar]
- 42.Zamore PD. RNA interference: listening to the sound of silence. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 43.Merai Z, et al. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 45.Vaidyanathan R, Scott TW. Apoptosis in mosquito midgut epithelia associated with West Nile virus infection. Apoptosis. 2006;11:1643–1651. doi: 10.1007/s10495-006-8783-y. [DOI] [PubMed] [Google Scholar]
- 46.Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blethrow JD, Tang C, Deng C, Krutchinsky AN. Modular mass spectrometric tool for analysis of composition and phosphorylation of protein complexes. PLoS ONE. 2007;2:e358. doi: 10.1371/journal.pone.0000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 49.Ephrussi B, Herold JL. Studies of Eye Pigments of Drosophila. I. Methods of Extraction and Quantitative Estimation of the Pigment Components. Genetics. 1944;29:148–175. doi: 10.1093/genetics/29.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS One. 2009;4:e5866. doi: 10.1371/journal.pone.0005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.