Summary
In silico analysis of the Bifidobacterium breve UCC2003 genome predicted two distinct loci, which encode three different restriction/modification systems, each comprising a modification methylase and a restriction endonuclease. Based on sequence homology and observed protection against restriction we conclude that the first restriction endonuclease, designated BbrI, is an isoschizomer of BbeI, the second, BbrII, is a neoschizomer of SalI, while the third, BbrIII, is an isoschizomer of PstI. Expression of each of the B. breve UCC2003 methylase‐encoding genes in B. breve JCM 7017 established that BbrII and BbrIII are active and restrict incoming DNA. By exploiting knowledge on restriction/modification in B. breve UCC2003 we successfully increased the transformation efficiency to a level that allows the reliable generation of mutants by homologous recombination using a non‐replicative plasmid.
Introduction
The commensal gut microbiota has long been appreciated for its influence on gut health (reviewed by O'Hara and Shanahan, 2006; Turroni et al., 2008). Bifidobacteria constitute a specific group of mostly commensal bacteria, which inhabit the gastrointestinal tract (GIT) of mammals, including the human GIT, where they are estimated to represent 3–6% of the adult faecal flora (Ventura et al., 2004; Saxelin et al., 2005; Zoetendal and Vaughan, 2006). The presence of bifidobacteria in the human GIT has been associated with many beneficial health effects, such as the prevention of diarrhoea, amelioration of lactose intolerance and immunomodulation (reviewed by Leahy et al., 2005). Indeed, the health benefits of probiotic bacteria such as bifidobacteria have been shown to extend beyond the GIT (Lenoir‐Wijnkoop et al., 2007). These many positive attributes have led to the widespread incorporation of bifidobacteria as live components of commercial health‐promoting probiotic foods. Despite these commercial and scientific interests, fundamental knowledge is still scarce regarding the exact molecular mechanisms by which bifidobacteria contribute to host health and well‐being. Such scientific knowledge is essential to scientifically explain the purported health benefits, and consequently support the inclusion of such bacteria as probiotics in functional foods.
The genome sequences on Bifidobacterium longum subsp. longum NCC2705 (Schell et al., 2002), B. longum subsp. longum DJ010A (Lee et al., 2008), B. adolescentis ATCC15703 (Suzuki et al., 2006), B. adolescentis L2‐32 (Fulton et al., 2007), B. dentium ATCC27678 (Sudarsanam et al., 2008) and B. animalis subsp. lactis HN019 (Collett et al., 2008) have recently become available and have contributed very significantly to advancing our knowledge on bifidobacterial genetics and metabolism. However, the availability of a genome sequence is merely a first step towards a better understanding of a specific probiotic property, and unravelling the molecular mechanisms by which bifidobacteria bring about positive host responses demands the availability of suitable molecular tools. To date, relatively few molecular tools for bifidobacteria have been developed, which explains why the genetics of these microbes is rather poorly understood, certainly when compared with other bacteria of industrial importance.
Available genetic tools for bifidobacteria include bifidobacterial plasmids, which were first reported by Sgorbati and colleagues (1982). In recent years significant effort has focused on identifying and sequencing plasmids from bifidobacteria, and exploiting some of these native bifidobacterial replicons for the creation of Escherichia coli–Bifidobacterium shuttle vectors (Lee and O'Sullivan 2006; Alvarez‐Martín et al., 2007; Cronin et al., 2007; Sangrador‐Vegas and colleagues, 2007). A limitation of many of these shuttle vectors is the low transformation efficiency of many of the bifidobacteria tested, coupled in some cases with segregational instability (Lee and O'Sullivan, 2006).
The observed differences in transformation efficiency among different strains of bifidobacteria may be attributed, at least in part, to restriction/modification (R–M) systems, which are ubiquitous among prokaryotes and generally comprise of a restriction endonuclease (REase) and cognate methyltransferase (MTase) (Murray, 2002; Tock and Dryden, 2005). R–M systems are believed to serve primarily as defensive instruments that protect prokaryotic cells against invading DNA such as promiscuous plasmids or infecting bacteriophage. R–M systems are classified into four groups (designated type I, II, III and IV) on the basis of their subunit composition, co‐factor requirement, recognition sequence structure and the cleavage site relative to the recognition sequence (Roberts et al., 2003). Type I R–M systems consist of three different subunits, HsdM, HsdR and HsdS, that are responsible for modification, restriction and sequence recognition respectively. Type I REases require ATP, MG2+ and AdoMet for activity. In general they interact with two asymmetrical bi‐partite recognition sites, translocate the DNA in an ATP hydrolysis‐dependent manner and cut the DNA distal to the recognition sites, approximately half‐way between two sites (Murray, 2002). Typically, in a type II R–M system the REase recognizes and cleaves within a short (4–8 bp) palindromic DNA sequence. Protection of ‘self’ DNA from restriction occurs by methylation using an MTase, which modifies specific adenosyl or cytosyl residues within the sequence recognized by the corresponding REase (Kobayashi, 2001; Pingoud et al., 2005). Type III R–M systems consist of two subunits, Mod, responsible for DNA recognition and modification, and Res, responsible for DNA cleavage. Active nucleases require ATP and MG2+ for activity and are stimulated by AdoMet. The holoenzyme, composed of two Res and two Mod subunits, interacts with two unmodified asymmetric target sites positioned in inverse orientations with respect to each other and cuts the DNA close to one recognition site (Janscak et al., 2001). Type IV R–M systems are specified by either one of two structural genes encoding proteins with specificities for methylated, hydroxymethylated or glucosyl‐hydroxymethylated bases in the target DNA molecule (Roberts et al., 2003).
REase activity in Bifidobacterium was first described by Khosaka and colleagues (1982) and to date a total of 23 bifidobacterial proven or putative R–M systems have been identified, as listed on the REBASE website (http://rebase.neb.com/rebase). BbeI, the first bifidobacterial REase to be described, was isolated from Bifidobacterium breve YIT4006, recognizing and cleaving the sequence 5′‐GGCGC↓C‐3′. However, two copies of the BbeI recognition sequence are required for full endonuclease activity (Khosaka et al., 1982). Subsequently Khosaka and colleagues (1983) reported on the identification of the REases BinSI and BinSII from B. longum subsp. infantis S76e. BinSI is an isoschizomer of EcoRII (recognizing and cleaving the sequence 5′↓CCWGG‐3′), while BinSII exhibits the same restriction specificity as BbeI (5′‐GGCGC↓C‐3′). BinI was isolated from B. longum subsp. infantis 659, and recognizes the asymmetric pentanucleotide sequence 5′‐GGATCNNNN↓N‐3′ (Khosaka and Kiwaki, 1984). Skrypina and colleagues (1988) showed that four out of 12 bifidobacterial strains exhibited REase activity, of which two, BadI from B. adolescentis LVA1 and BbfI from B. bifidum LVA3, are isoschizomers of XhoI (5′‐C↓TCGAG‐3′), while the REases Bbf7411I from B. bifidum 7411 and Bla7920I from B. lactentis 7920 are neoschizomers of BspMII (5′‐T↓CCGGA‐3′). Hartke and colleagues (1996) identified two REases from B. longum subsp. longum BL2: BloI is an isoschizomer of XhoII (5′‐R↓GATCY‐3′), while BloII is an isoschizomer of PstI (5′‐CTGCA↓G‐3′).
In the current study we report on the identification and preliminary characterization of three R–M systems encoded on the genome of B. breve UCC2003. Circumventing these R–M systems allowed the development of a reliable method for the creation of gene disruptions in B. breve UCC2003.
Results
Sequence, genetic organization and amino acid analysis of the BbrI, BbrII and BbrIII R–M systems from B. breve UCC2003
Two loci, predicted to encode three different R–M systems, were identified from the annotation of the genome sequence of B. breve UCC2003 (S. Leahy. M. O'Connell Motherway, J. Moreno Munoz, G.F. Fitzgerald, D. Higgins and D. van Sinderen, unpubl. results) and designated BbrI, BbrII and BbrIII (Fig. 1A). The G+C content for each system is 58% which is in agreement with the approximately 60% G+C content for bifidobacteria (Ventura et al., 2007). The first gene of the BbrI R–M system, bbrIM, codes for a protein (M.BbrI; 43.2 kDa) with 60% and 53% identity to cytosine‐specific MTases from Clavibacter michiganesis and Photorhabdus luminescens respectively; M.BbrI also contains the six highly conserved motifs characteristic of known 5′‐methylcytosine MTases (Kumar et al., 1984) (Fig. 1B). The cytosine‐specific MTases from C. michiganesis and P. luminescens are known to methylate of the sequence 5′‐GGC(m5)GCC‐3′, which is also the recognition sequence of the BbeI REase identified by Khosaka and colleagues (1982) from B. breve YIT4006. The protein product of the second ORF, bbr0215, exhibits 94% identity to a hypothetical protein encoded by B. longum subsp. longum NCC2705 (Schell et al., 2002). The third gene of the BbrI gene cluster, bbrIR, is separated from bbr0215 by remnants of an insertion sequence element. The bbrIR gene encodes a protein (30 kDa) exhibiting low homology (33%) to various type II R–M system restriction subunits and for this reason it is predicted to represent the restriction component of the BbrI R–M system, probably an isoschizomer of BbeI.
Figure 1.

A. Schematic representation of R–M systems encoded by B. breve UCC2003. Each arrow indicates an ORF. Predicted protein function is indicated by M (modification) or R (restriction) in the gene name. The percentage amino acid (aa) identity is indicated. B. Alignment of the six highly conserved motifs of MTases M.BbrI and M.BbrII from B. breve UCC2003 with annotated cytosine MTases from B. dentium ATCC27678, B. adolescentis ATCC15703 and B. longum NCC2705 and M.AluI and M.EcoRII. Highly conserved aa are marked with an asterisk above the sequence and indicated in bold, while aa are shaded in grey if at least four of the depicted protein sequences contain an identical residue at a particular position. C. Sequence alignment of M.BbrIII and M.PstI. Amino acids are shaded if both of the depicted proteins contain identical residues at corresponding positions. The boxed sequences CMIs, CMI, CMII and CMIII represent the four highly conserved regions of N6‐adenine MTases.
The R–M systems BbrII and BbrIII are located adjacent to each other on the genome of UCC2003 (Fig. 1A). The first gene of the BbrII R–M system, bbrIIM, codes for a 349‐amino‐acid protein (38.8 kDa), exhibiting 47% identity to the HgiDII cytosine‐specific MTase (Düsterhöft and Kröger, 1991). As was the case for M.BbrI, the six highly conserved motifs of cytosine‐specific MTases are present in M.BbrII (Fig. 1B). The encoded product of the bbrIIR gene is a 695‐amino‐acid protein (79.4 kDa) exhibiting 40% identity to R.HgiDII, which recognizes the sequence 5′‐G↓TCGAC‐3′. This is the same recognition sequence as that of SalI; however, M.SalI is a N6‐adenosine MTase, while M.BbrII and M.HgiDII are predicted to be cytosine‐specific MTases. R.BbrII therefore is assumed to represent a neoschizomer of SalI.
The third identified R–M system on the genome of B. breve UCC2003, BbrIII, is predicted to encode an isoschizomer of PstI and BloII, the latter representing a REase identified from B. longum subsp. longum BL2 (Hartke et al., 1996). The first gene, bbrIIIM, encodes a 315‐amino‐acid protein (36.3 kDa), which shares 32% identity with M.PstI, an N6‐adenosine MTase (Walder, Walder et al., 1984). The four conserved motifs characteristic of N6‐adenosine‐methyltransferase, CMIs, CMI, CMII and CMIII (Timinskas et al., 1995), can be identified in M.BbrIII (Fig. 1C). The second gene bbrIIIR encodes a 355‐amino‐acid protein (36.6 kDa), exhibiting 38% identity to the REase Pst1 (5′‐CTGCA↓G‐3′).
Assessment of R–M activity in B. breve UCC2003
To establish if the identified R–M systems are functional in B. breve UCC2003 and whether they affect transformation efficiency of this strain, the transformation frequency of two E. coli–bifidobacterial shuttle vectors, pPKCM7 and pAM5 (Table 1), was determined when these plasmids had been isolated either from B. breve UCC2003 (DNA protected from R–M) or from E. coli JM101 (DNA sensitive to R–M). 200 ng quantities of each of these plasmid DNAs isolated from these two different hosts was used to transform B. breve UCC2003 by electroporation. Transformants were selected on RCA supplemented with chloramphenicol (Cm) in case of plasmid pPKCM7, or tetracycline (Tet) in case of plasmid pAM5, and enumerated following anaerobic incubation at 37°C for 48 h (Fig. 2). For each plasmid there was a 500‐fold higher transformation efficiency of the plasmid DNA isolated from B. breve UCC2003 as compared with the DNA isolated from E. coli, thus indicating that one or more of the identified R–M systems encoded by B. breve UCC2003 is functional and contributes to the efficiency at which plasmids can be introduced in this strain.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| EC101 | Cloning host, repA+ kmr | Law et al. (1995) |
| JM109 | F' traD36 proAB lacIqZ M15 recA1 relA1 endA1 thi hsdR17 | |
| JM101 | supE, thi (lacproAB) (F' traD36 proAB lacIqZ M15 | Yanisch‐Perron et al. (1985) |
| BM101 | E. coli JM101with bbrIIM and bbrIIIM integrated in the chromosome and transcribed by an IPTG‐inducible lac promoter | This study |
| B. breve strains | ||
| NCFB 2257 | Isolate from infant intestine | NCFB |
| NCFB 2258 | Isolate from infant intestine | NCFB |
| NCFB 8815 | Isolate from nursling stool | NCFB |
| NCFB 11815 | Isolate from infant intestine | NCFB |
| Yakult | Isolate from infant intestine | Oishi et al. (2008) |
| LMG 13208 | Isolate from infant intestine | UCC |
| JCM 7017 | Isolate from infant intestine | JCM |
| JCM 7019 | Isolate from infant intestine | JCM |
| UCC2004 | Isolate from nursling stool | UCC |
| UCC2003 | Isolate from nursling stool | Mazéet al. (2007) |
| UCC2003‐galA‐744 | pORI19‐tet‐G744 insertion mutant of UCC2003 | This study |
| UCC2003‐gaIA‐476 | pORI19‐tet‐G476 insertion mutant of UCC2003 | This study |
| UCC2003‐apuB‐939 | pORI19‐tet‐apuB insertion mutant of UCC2003 | This study |
| Plasmids | ||
| pNZ8048 | Cmr, nisin‐inducible translational fusion vector | de Ruyter et al. (1996) |
| pNZ‐M.BbrI | pNZ8048 derivative containing bbrIM (bbr0216) | This study |
| pNZ‐M.BbrII | pNZ8048 derivative containing bbrIIM (bbr1121) | This study |
| pNZ‐M.BbrIII | pNZ8048 derivative containing bbrIIIM (bbr1119) | This study |
| pNZ‐M.BbrII + M.BbrIII | pNZ8048 derivative containing bbrIIM and bbrIIIM | This study |
| pAM5 | pBC1‐puC19‐Tcr | Alvarez‐Martín et al. (2007) |
| pPKCM7 | pblueCm harbouring rep pCIBA089 | Cronin et al. (2007) |
| pREP4 | Low‐copy‐number LacI expressing pQE60 companion plasmid | Qiagen |
| pQE60 | AmpR overexpression vector | Qiagen |
| pQE60 M.BbrII + M.BbrIII | pQE60 derivative containing bbrIIM and bbrIIIM transcriptionally fused to IPTG‐inducible promoter | This study |
| pKVB2 | Tcr, Kmr containing internally deleted E. coli glgB gene 11.7 kb | Kiel et al. (1987) |
| pKVB2‐M.BbrII‐M.BbrIII | pKVB2 derivative containing bbrIIM and bbrIIIM transcriptionally fused to IPTG‐inducible promoter | This study |
| pORI19 | Emr, repA‐, ori+, cloning vector | Law et al. (1995) |
| pORI19‐tet‐G744 | Internal 744 bp fragment of galA and tetW cloned in pORI19 | This study |
| pORI19‐tet‐G476 | Internal 476 bp fragment of galA and tetW cloned in pORI19 | This study |
| pORI19‐tet‐apuB | Internal 939 bp fragment of apuB and tetW cloned in pORI19 | This study |
JCM, Japan Collection of Microorganisms; NCFB, National Collection of Food Bacteria, Reading, UK; UCC, University College Cork, Cork, Ireland.
Figure 2.

Transformation efficiency of pPKCM7 or pAM5 DNA isolated from E. coli (grey bars) or B. breve UCC2003 (black bars).
BbrI, BbrII and BbrIII represent three R–M systems
In order to verify the prediction that M.BbrI, M.BbrII and M.BrrIII represent distinct MTases that protect, based on their similarities to characterized R–M systems, DNA sequences cut by BbeI, SalI and PstI, respectively, genomic DNA of B. breve UCC2003 was restricted with these enzymes and analysed by agarose gel electrophoresis. The results obtained showed that B. breve UCC2003 genomic DNA is protected from restriction with BbeI and PstI, but not SalI (Fig. 3A).
Figure 3.
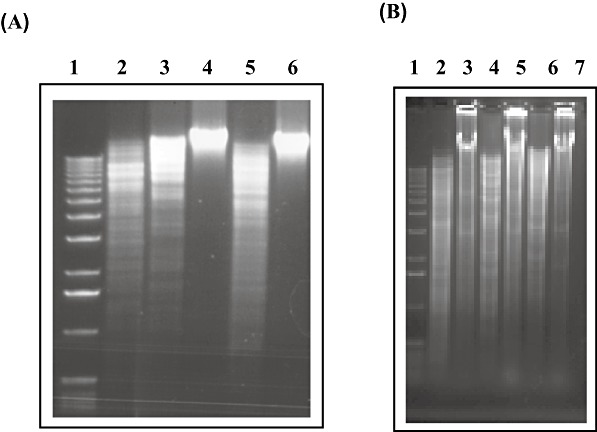
A. Restriction analysis of total DNA from B. breve UCC2003. Lane 1, molecular weight marker X (Roche). Lanes 2–6: total B. breve UCC2003 DNA restricted with lane 2, BamHI; lane 3, HindIII; lane 4, BbeI; lane 5, SalI; lane 6, PstI. B. Restriction analysis of total DNA from B. breve JCM7017 harbouring pNZ8048, pNZ‐M.BbrI, pNZ‐M.BbrII or pNZ‐M.BbrIII. Lane 1, molecular weight marker X (Roche). Lane 2, JCM7017 harbouring pNZ8048 restricted with BbeI; lane 3, JCM7017 harbouring pNZ‐M.BbrI restricted with BbeI; lane 4, JCM7017 harbouring pNZ8048 restricted with SalI; lane 5, JCM7017 harbouring pNZ‐M.BbrII restricted with SalI; lane 6, JCM7017 harbouring pNZ8048 restricted with PstI; lane 7, JCM7017 harbouring pNZ‐M.BbrIII restricted with PstI. The restrictions were analysed on a 1% agarose gel followed by staining with ETBR.
To establish the precise MTase activity of each of the predicted MTase‐encoding genes in B. breve UCC2003, bbrIM, bbrIIM, bbrIIIM and their corresponding upstream regions (presumed to contain their native promoters) were amplified by PCR and cloned in pNZ8048 to generate pNZ‐M.BbrI, pNZ‐M.BbrII and pNZ‐M.BbrIII respectively (see Experimental procedures and Table S1). These plasmids, as well as the control plasmid pNZ8048, were introduced into B. breve JCM 7017, whose genomic DNA is susceptible to BbeI, PstI and SalI restriction (data not shown). Restriction analysis revealed that genomic DNA of B. breve JCM 7017 expressing M.BbrI, M.BbrII or M.BbrIII were protected from restriction with BbeI, SalI or PstI, respectively, while genomic DNA of B. breve JCM7017 harbouring pNZ8048 was restricted by all three enzymes (Fig. 3B). Collectively these results demonstrate that B. breve UCC2003 encodes three MTases that methylate within the sequences, 5′‐GGCGCC‐3′ (for M.BbrI), 5′‐GTCGAC‐3′ (for M.BbrII) and 5′‐CTGCAG‐3′ (for M.BbrIII).
To establish if the methylase activities associated with the BbrI and BbrIII R–M systems were present in other B. breve strains, genomic DNA from nine additional B. breve strains was restricted with BbeI or PstI (Table S2). Only for three strains, B. breve UCC2004, NCFB 2258 and NCFB 8815, the DNA was protected from restriction with BbeI. In addition, DNA from B. breve NCFB 8815 was also protected from restriction with PstI. Genomic DNA from the remaining six strains was restricted by these two enzymes. This would indicate that different strains of B. breve exhibit quite a variety of different R–M activities.
To determine the individual effect of each R–M system on the transformation frequency of B. breve UCC2003, we first introduced plasmid pAM5, which harbours one PstI, two SalI and three BbeI sites, into B. breve JCM7017 strains harbouring either pNZ8048, pNZ‐M.BbrI, pNZ‐M.BbrII or pNZ‐M.BbrIII. The methylation of the pAM5 DNA at the appropriate sequence in each of the methylase expressing strains was confirmed by restriction analysis (results not shown) prior to introducing 200 ng of each plasmid preparation into B. breve UCC2003 by electroporation. The number of transformants was determined after 48h of anaerobic incubation at 37°C on RCA with tetracycline selection (Fig. 4). pAM5 DNA isolated from JCM7017 expressing M.BbrIII allowed an almost 1000‐fold higher transformation frequency as compared with pAM5 isolated from E. coli or JCM 7017 harbouring pNZ8048. A 10‐ and 5‐fold higher transformation efficiency was observed for pAM5 isolated from JCM7017 expressing M.BbrII and M.BbrI respectively. The transformation frequency obtained with pAM5 DNA isolated from JCM 7017 expressing M.BbrIII was comparable to the transformation frequency obtained with pAM5 plasmid DNA isolated from B. breve UCC2003. However, in the latter case DNA preparations contain just the pAM5 plasmid, while in the former case the DNA preparation would have contained a mixture of pAM5 and pNZ‐M.BbrIII. These results demonstrate that the BbrIII restriction endonuclease (isoschizomer of PstI) is highly active in B. breve UCC2003 and that the activity of this restriction endonuclease appears to represent the main limitation to the genetic accessibility of B. breve UCC2003, at least for plasmid pAM5.
Figure 4.
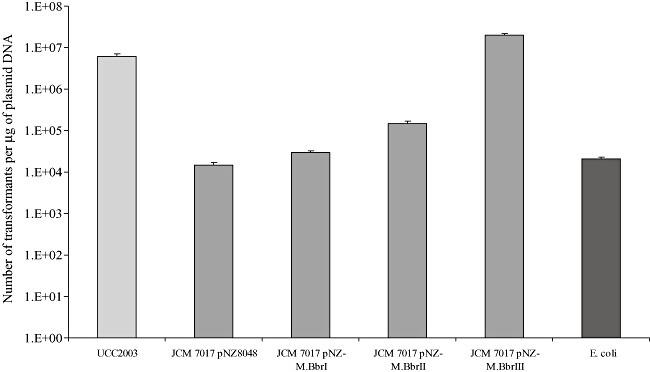
Transformation efficiency of B. breve UCC2003 using pAM5 plasmid DNA isolated from UCC2003, B. breve JCM7017 harbouring pNZ8048, pNZ‐M.BbrI, pNZ‐M.BbrII, pNZ‐M.BbrIII or E. coli.
Expression of M.BbrII and M.BbrIII in E. coli and methylation of plasmid DNA
From the data presented above it was clear that all three REases BbrI, BbrII and BbrIII are active in B. breve UCC2003. In order to enhance transformation efficiencies of B. breve UCC2003 by prior methylation of plasmid DNA, two E. coli strains expressing both M.BbrII and M.BbrIII were constructed. In the first, E. coli pNZ‐M.BbrII‐M.BbrIII, two of the bifidobacterial methylases were expressed on plasmid pNZ8048 (see Experimental procedures and Table S1). As expected, chromosomal (and plasmid) DNA from E. coli strain EC101 harbouring pNZ‐M.BbrII‐M.BbrIII is protected from restriction with PstI. The second E. coli strain, BM1, harbours bbrIIM and bbrIIIM under the control of an IPTG‐inducible promoter integrated into the glgB gene on the E. coli JM101 chromosome (see Experimental procedures). Upon induction with 10 mM IPTG total DNA from E. coli BM1 is protected from restriction with PstI (Fig. S1A). However, complete protection from SalI restriction was not observed (results not shown) and this may be due to the lower level expression of bbrIIM from the E. coli chromosome as compared with expression from plasmid pNZ‐M.BbrII‐M.BbrIII. In addition, SalI can restrict hemi‐methylated DNA, therefore the observed restriction by SalI may be a reflection of incomplete methylation.
To evaluate the effect of methylation of plasmid DNA on transformation efficiency, pAM5 was introduced into E. coli pNZ‐M.BbrII‐M.BbrIII and E. coli BM1 by electroporation. Expression of M.BbrII and M.BbrIII in BM1 harbouring pAM5 was effected by the addition of 10 mM IPTG prior to the isolation of plasmid DNA. Plasmid preparations of E. coli harbouring pNZ‐M.BbrII‐M.BbrIII or E. coli BM1 were then used for B. breve UCC2003 transformation. pAM5 DNA isolated from E. coli harbouring pNZ‐M.BbrII‐M.BbrIII gave a 1000‐fold higher transformation frequency as compared with pAM5 from E. coli pNZ8048 while plasmid DNA isolated from E. coli BM1 gave a 50‐fold higher transformation frequency. pAM5 DNA isolated from EC101 pNZ‐M.BbrII‐M.BbrIII gave transformation efficiencies comparable to those obtained with plasmid DNA isolated from B. breve (Fig. S1B).
Disruption of the galA and apuB genes of B. breve UCC2003
In order to establish if methylation of a non‐replicating plasmid by the B. breve UCC2003 MTases would increase transformation efficiency to a sufficiently high level that would allow site‐specific homologous recombination, two genes, galA and apuB, were selected as mutational targets. The galA and apuB genes encode an endogalactanase and an amylopullulanase, respectively, which are involved in extracellular polysaccharide metabolism by B. breve UCC2003 (Hinz et al., 2005; Ryan et al., 2006; O'Connell Motherway et al., 2008). To establish if gene disruption could be achieved using homologous recombination, DNA fragments of 476 and 744 bp, representing internal fragments of the galA gene, and a 939 bp internal fragment of the apuB gene were cloned in pORI19 and provided with a tetracycline resistance marker, generating plasmids pORI19‐tet‐G744, pORI19‐tet‐G476 and pORI19‐tet‐apuB respectively (see Experimental procedures). These plasmids, being derivatives of pORI19, cannot replicate in B. breve UCC2003 as they lack a functional replication protein (Law et al., 1995). These pORI19 derivatives were introduced into E. coli EC101 harbouring pNZ‐M.BbrII‐M.BbrIII to facilitate methylation, and preparations of the resulting methylated pORI19‐derived plasmids were then introduced into B. breve UCC2003 by electroporation. Tetracycline‐resistant transformants were isolated at a frequency of 50 per µg of transformed DNA when greater than 700 bp of homologous DNA was used. The number of potential integrants was slightly reduced when the smaller region of homologous DNA was used. All transformants obtained were expected to carry galA or apuB gene disruptions, while no such transformants were obtained when unmethylated pORI19 constructs were introduced into B. breve UCC2003. The suspected chromosomal integration of the pORI constructs was verified by colony PCR on a selection of Tetr transformants using a forward primer upstream of the region of integration and a reverse primer based on pORI19 (galAp1 and pORI19A, or apuBp1 and pORI19B) (results not shown). Southern hybridizations confirmed the assumed integration of the individual pORI‐derived plasmids by homologous recombination. For the presumed galA disruptions of B. breve UCC2003, Southern hybridizations were performed using SphI‐digested genomic DNA and employing a 2.6 kb PCR fragment encompassing galA as a probe. SphI was selected for the genomic digests since there are no corresponding restriction sites within the galA sequence. The galA fragment hybridized to a 6.1 kb fragment of UCC2003 genomic DNA, while in the UCC2003 derivatives with a presumed pORI‐tet‐G476 or pORI‐tet‐G744 integration this band was absent, and expected hybridization signals of 10.5 kb and 557 bp, or 10.8 kb and 848 bp, respectively, were observed (Fig. 5). For two of each of the UCC2003 mutant strains examined the galA probe also hybridized to a 5.3 kb or 5.5 kb Sph1 fragment for the pORI19‐tet‐G476 and pORI19‐tet‐G744 integrants respectively [Fig. 5B(i), lanes 4 and 5; Fig. 5B(ii), lanes 5 and 6]. These hybridization signals indicate that duplication of pORI19‐tet‐galA plasmids had occurred after integration of the plasmid into the bacterial chromosome in these mutant strains. For the suspected apuB integrants of strain UCC2003, Southern hybridizations were performed using BamHI‐digested genomic DNA and a 1 kb probe encompassing an internal fragment of apuB. The apuB fragment hybridized to a 3.6 kb fragment of UCC2003 genomic DNA. For the apuB mutant strains the anticipated hybridization signals of 2.1 and 7.2 kb were obtained (Fig. S2).
Figure 5.

A. Schematic representation of the relevant regions of the B. breve UCC2003 and UCC2003‐galA‐476 and UCC2003‐galA‐744 (in brackets) chromosomes. Chromosomal DNA is represented by a thin line, the galA gene is represented by a black arrow, the internal galA fragment is indicated by a solid grey line and pORI19 is indicated by a boxed line. SphI sites relevant to the Southern hybridization analysis are indicated. B. Southern hybridization analysis of SphI chromosomal DNAs of (i) B. breve UCC2003 (lane 2) and four representative B. breve UCC2003‐galA‐476 mutants (lanes 3–6); (ii) B. breve UCC2003 (lane 2) and four representative B. breve UCC2003‐galA‐744 mutants (lanes 3–6). The hybridization signals for molecular weight marker X (Roche) are in lane 1 and the molecular weight of the relevant hybridization signals are indicated to the left of the panel. A PCR product of 2.6 kb encompassing galA was used as a probe for the hybridization.
Collectively these results demonstrate that methylation of plasmid DNA by the B. breve UCC2003 MTases M.BbrII and M.BbrIII in E. coli circumvents the BbrII and BbrIII REase activities in B. breve UCC2003 and allows a sufficiently high transformation efficiency so as to allow reliable homologous recombination in B. breve UCC2003. In addition, these data illustrate that chromosomal integration in B. breve UCC2003 can be achieved with less than 500 bp of homologous DNA.
Phenotypic analysis of the B. breve UCC2003 plasmid integrants
In order to verify the expected phenotypic consequences of the created gene disruptions in galA and apuB, strains B. breve UCC2003, and individual representatives of B. breve UCC2003 mutants generated by insertion of pORI19‐tet‐G744 or pORI19‐tet‐G476, designated here as UCC2003‐galA‐476 and UCC2003‐galA‐744, respectively, were analysed for their ability to grow on galactan as the sole carbohydrate source (Fig. 6A). Similarly, B. breve UCC2003 and a derivative with an integrated pORI19‐tet‐apuB (designated UCC2003‐apuB‐939) were analysed for the ability to grow on starch, amylopectin, glycogen or pullulan as the sole carbohydrate source (Fig. 6B). In contrast to the wild‐type B. breve UCC2003, the B. breve UCC2003‐galA‐476 or UCC2003‐galA‐744 mutant strains failed to grow on potato galactan, while comparable growth of the parent and galA mutant strains was observed when glucose was the sole carbohydrate source. In a similar manner it was shown that B. breve UCC2003‐apuB‐939 failed to grow on starch, amylopectin, glycogen or pullulan, which contrasted with observed good growth on these substrates by the parent strain. Comparable growth for parent and mutant strains was observed when glucose was used as the sole carbohydrate source. These results confirm that the chromosomal plasmid integrations in UCC2003 cause a demonstrable phenotype and clearly illustrate the importance of the extracellular enzymes specified by galA and apuB in the metabolism of specific high‐molecular‐weight polysaccharides by B. breve UCC2003.
Figure 6.

A. Growth profile analysis of B. breve UCC2003 (white), B. breve UCC2003‐galA‐476 (grey) and B. breve UCC2003‐galA‐744 (black) in modified Rogosa broth supplemented with potato galactan or glucose. B. Growth profile analysis of B. breve UCC2003 (white) and B. breve UCC2003‐apuB (black) in modified Rogosa broth supplemented with starch, amylopectin, glycogen, pullulan or glucose as indicated.
Discussion
Bifidobacterial strains demonstrate substantial variability in the efficiency of transformation by plasmids from E. coli, while many strains exhibit complete resistance to transformation (Lee and O'Sullivan, 2006). Progress in the evaluation of probiotic factors in bifidobacteria has been slow due to the lack of efficient and versatile systems for genetic manipulation (Ventura et al., 2004). While quite a number of E. coli–bifidobacterial shuttle vectors have been developed, it has been noted that widespread application of these plasmids among bifidobacterial species is limited (Lee and O'Sullivan, 2006).
As shown here, R–M systems are one of the major obstacles hindering progress in the genetic accessibility and analysis of B. breve UCC2003, and are likely to do this in other (bifido)bacteria as well. Convincing evidence to support this notion can be obtained from the available bifidobacterial genome sequences. Genes specifying R–M systems can be identified in all sequenced bifidobacterial genomes. The genomes of B. longum subsp. longum NCC2705 (Schell et al., 2002) and B. longum subsp. longum DJ010A (Lee et al., 2008) both harbour a single type I R–M system, two type II R–M systems and one type IV R–M system. The type II REases specified by blo_1473 and bld_0356 are predicted to be isoschizomers of EcoRII, which restricts within the sequence ↓CCWGG, while the REases specified by blo_564 and bln_1359 are predicted to be isoschizomers of Sau3A1, which recognizes the sequence ↓GATC. The recognition sequence of the type I and type IV R/M systems in the sequenced B. longum genomes are unknown. The genome of B. adolescentis ATCC15703 (Suzuki et al., 2006) specifies two MTase subunits and six REase subunits. The restriction subunits specified by bad_1283 and bad_1232 are predicted to be isoschizomers of KpnII and Sau3AI, respectively, while the remaining four are as yet unknown. The sequenced genomes of B. dentium ATCC27678 (Sudarsanam et al., 2008) and B. animalis HN019 (Collett et al., 2008) both harbour a single type II R–M system, where the REase is predicted to be an isoschizomer of AvaII, which recognizes the sequence G↓GWCC (Sutcliffe and Church, 1978). Based on the results obtained for B. breve UCC2003, it is tempting to speculate that exploiting the MTases encoded by the aforementioned sequenced bifidobacterial strains would allow the transformation efficiencies of these strains to be improved. For bifidobacterial strains that are particularly recalcitrant to transformation or where the complete genome sequence is not known it may be possible to methylate plasmid DNA isolated from E. coli by incubating the DNA with crude cell extracts of the Bifidobacteria in the presence of S‐adenosylmethionine thereby possibly improving the transformation efficiency.
An alternative method that would circumvent bifidobacterial R–M systems would be to introduce plasmid DNA by conjugation. To date conjugation has not been conclusively demonstrated for the genus Bifidobacterium. Until recently the only evidence supporting the possibility of conjugation in bifidobacteria was the identification of genes encoding proteins potentially involved in the conjugation process on various bifidobacterial plasmids. Putative relaxase‐encoding genes have been identified on plasmids pJK36 and pJK50 from B. longum subsp. longum (Park et al., 1999; 2000), while homologues of septal DNA translocator (Tra) proteins have been identified on the B. breve plasmid pCIBb1 (O'Riordan and Fitzgerald, 1999) and the B. pseudocatenulatum plasmid p4M (Gibbs et al., 2006). Recently, Shkoporov and colleagues (2008) sequenced three plasmids of bifidobacterial origin: pB44 from B. longum, pB90 from B. bifidum and pB21a from B. breve. Both pB44 and pB90 harbour genes encoding potential mobilization functions while pB21A encodes a putative Tra protein. These proteins were exploited in efforts to achieve conjugation in bifidobacteria, and although antibiotic‐resistant, PCR‐positive and thus putative transconjugants were obtained, plasmid transfer has as yet not been demonstrated.
The difficulties associated with obtaining sufficiently high transformation efficiencies so as to allow insertional mutagenesis in B. breve UCC2003 through homologous recombination led us to believe that R–M systems were the barrier that needed to be overcome in order to achieve this. In the present study we describe three different R–M systems specified by the genome of UCC2003: BbrI, an isoschizomer of BbeI; BbrII, a neoschizomer of SalI; and BbrIII, an isoschizomer of PstI. Restriction analysis of chromosomal DNA from UCC2003 showed that the DNA is protected from restriction with BbeI and PstI, but not SalI. The observed restriction of DNA by SalI can be explained by M.SalI being a N6‐adenosine‐methylase, while M.BbrII is predicted to be cytosine‐specific MTase, which may therefore not confer (full) protection against SalI restriction. However, the finding that M.BbrII does provide full protection against SalI restriction when it is expressed from a multicopy plasmid in B. breve JCM 7017 would indicate that M.BbrII in such circumstances is more abundant, thereby eliciting complete methylation and concomitant protection of the DNA. The three R–M systems identified in B. breve UCC2003 do not appear to be highly conserved among B. breve strains, just one strain examined in this study, B. breve NCIMB 8815, was shown to exhibit protection of BbrI and BbrIII recognition sites indicating that this species and indeed the genus Bifidobacterium is likely to harbour a very diverse range of R–M activities.
The contribution of each R–M system in impeding plasmid transformation of B. breve UCC2003 was determined and established that all three systems impact on transformation efficiency, with BbrIII, at least under the circumstances used here, providing the biggest hurdle to incoming DNA. To facilitate methylation of plasmid DNA by M.BbrII and M.BbrIII, thereby enhancing the transformation frequency of B. breve UCC2003, two E. coli strains were constructed, where bbrIIM and bbrIIIM were expressed in different ways, either from their own promoter on plasmid pNZ8048 or from an IPTG‐inducible promoter on the E. coli chromosome. The observed higher transformation efficiency for pAM5 DNA isolated from E. coli pNZ‐M.BbrII‐M.BbrIII may be attributed to the high copy number of pNZ8048 plasmids in E. coli and resulting higher expression levels of the MTases as compared with expression from single copy on the E. coli chromosome in E. coli BM1.
Having established that the use of M.BbrII‐ and M.BbrIII‐methylated plasmid DNA results in a significantly increased transformation efficiency of B. breve UCC2003, we conclusively showed that gene disruptions in B. breve UCC2003 can be created using a non‐replicating and M.BbrII‐ and M.BbrIII‐methylated plasmid. We have previously produced a gene disruption in the apuB gene of B. breve UCC2003 by adaptation of the lactococcal two plasmid homologous recombination system (O'Connell Motherway et al., 2008). However, in our hands this system was very tedious, time‐consuming and not reliable (O'Connell Motherway et al., 2008; our unpublished results). Therefore, insertional mutagenesis of the apuB gene was deemed an appropriate control to evaluate the validity and reliability of the plasmid methylation strategy. By M.BbrII‐M.BbrIII‐mediated methylation of plasmid DNA in E. coli prior to transformation into B. breve UCC2003, gene disruptions not only in apuB, but also in galA were successfully and reliably created, as verified by genetic and phenotypic analyses.
This, to the best of our knowledge, therefore represents the first reliable system for creating insertional mutation in a member of the genus Bifidobacterium. The ability to achieve chromosomal integration of a non‐replicative plasmid with less than 500 bp of homologous DNA also opens the opportunity for the creation of a bank of B. breve UCC2003‐derived mutants carrying random chromosomal integrations, which in turn will provide a range of possibilities to further advance fundamental knowledge on the physiology, biochemistry and genetics of this strain. Such information will obviously be relevant to other bifidobacteria and will be crucial to understand the health‐promoting properties that have been attributed to various members of this genus.
Experimental procedures
The description of the experimental procedures resides in Appendix S1 in Supporting information.
Acknowledgments
This research was financially supported by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. A. Restriction analysis of E. coli JM101 and two representative JM101 bbrIIM and bbrIIIM methylase integration strains. Lane 1, molecular weight marker X (Roche). Lanes 2-4, Pst1 digest of total DNA isolated from JM101 following induction with 0, 1 or 10 mM IPTG. Lanes 5-7 and lane 8-10, PstI digests of total DNA isolated from two representative JM101 bbrIIM and bbrIIIM methylase integration strains after induction with 0, 1 or 10 mM IPTG. B. Transformation efficiency of B. breve UCC2003 with pAM5 plasmid DNA isolated from B. breve UCC2003, E. coli pNZM. BbrII-M.BbrIII, E. coli BM1 or E. coli pNZ8048.
Fig. S2. A. Schematic representation of the relevant regions of the B. breve UCC2003 and UCC2003-apuB-939 chromosome. Chromosomal DNA is represented by a thin line, the apuB gene is represented by a black arrow, the internal apuB fragment is indicated by a solid grey line and pORI19 is indicated by a boxed line. BamHI sites relevant to the Southern hybridization analysis are indicated.
B. Southern hybridization analysis of BamHI-digested chromosomal DNAs of B. breve UCC2003 (lane 1) and four representative B. breve UCC2003-apuB-939 mutants (lanes 2-5). The molecular weight of the relevant hybridization signals are indicated to the left of the panel. The internal 1 kb PCR amplicon of apuB was used as a probe for the hybridization.
Table S1. Oligonucleotide primers used in this study.
Table S2. Restriction analysis of genomic DNA from B. breve strains with BbeI and PstI.
Appendix S1. Experimental procedures.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alvarez‐Martín P., O'Connell‐Motherway M., Van Sinderen D., Mayo B. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol. 2007;76:1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
- Collett M.A., Depree K.M., Rand C.J., Mason C., Stanton J.‐A.L. Bifidobacterium animalis subsp. lactis HN019, whole genome shotgun sequence. NCBI Database. 2008 [Google Scholar]
- Cronin M., Knobel M., O'Connell‐Motherway M., Fitzgerald G.F., Van Sinderen D. Molecular dissection of a bifidobacterial replicon. Appl Environ Microbiol. 2007;73:7858–7866. doi: 10.1128/AEM.01630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düsterhöft A., Kröger M. Cloning, sequence and characterization of m5C‐methyltransferase‐encoding gene, hgiDIIM (GTCGAC), from Herpetosiphon giganteus strain Hpa2. Gene. 1991;106:87–92. doi: 10.1016/0378-1119(91)90569-w. [DOI] [PubMed] [Google Scholar]
- Fulton L., Clifton S., Fulton B., Xu J., Minx P., Pepin K.H. 2007. et al.Bifidobacterium adolescentis (L2‐32) genome sequence. NCBI Database.
- Gibbs M.J., Smeianov V.V., Steele J.L., Upcroft P., Efimov B.A. Two families of rep‐like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol Biol Evol. 2006;6:1097–1100. doi: 10.1093/molbev/msj122. [DOI] [PubMed] [Google Scholar]
- Hartke A., Benachour A., Bouibonnes P., Auffray Y. Characterisation of a complex restriction/modification system detected in a Bifidobacterium longum strain. Appl Microbiol Biotechnol. 1996;45:132–136. [Google Scholar]
- Hinz S.W., Pastink M.I., Van Den Broek L.A., Vincken J.P., Voragen A.G. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl Environ Microbiol. 2005;71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P., Sandmeier U., Szczelkun M.D., Bickle T.A. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J Mol Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- Khosaka T., Kiwaki M. BinI: a new site‐specific endonuclease from Bifidobacterium infantis. Gene. 1984;31:251–255. doi: 10.1016/0378-1119(84)90217-8. [DOI] [PubMed] [Google Scholar]
- Khosaka T., Sakurai T., Takahashi H., Saito H. A new site‐specific endonuclease BbeI from Bifidobacterium breve. Gene. 1982;17:117–122. doi: 10.1016/0378-1119(82)90063-4. [DOI] [PubMed] [Google Scholar]
- Khosaka T., Kiwaki M., Rak B. Two site‐specific endonucleases BinSI and BinSII from Bifidobacterium infantis. FEBS Lett. 1983;163:170–174. doi: 10.1016/0014-5793(83)80812-6. [DOI] [PubMed] [Google Scholar]
- Kiel J.A., Vossen J.P., Venema G. A general method for the construction of Escherichia coli mutants by homologous recombination and plasmid segregation. Mol Gen Genet. 1987;207:294–301. doi: 10.1007/BF00331592. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. Behaviour of restriction‐modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R.J., Wilson G.G. The DNA (cytosine‐5) methyltransferases. Nucleic Acids Res. 1984;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J., Buist G., Haandrikman A., Kok J., Venema G., Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy S.C., Higgins D.G., Fitzgerald G.F., Van Sinderen D. Getting better with bifidobacteria. J Appl Microbiol. 2005;98:1303–1315. doi: 10.1111/j.1365-2672.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- Lee J.H., O'Sullivan D.J. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl Environ Microbiol. 2006;72:527–535. doi: 10.1128/AEM.72.1.527-535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Karamychev V.N., Kozyavkin S.A., Mills D., Pavlov A.R., Pavlova N.V. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir‐Wijnkoop I., Sanders M.E., Cabana M.D., Caglar E., Corthier G., Rayes N. Probiotic and prebiotic influence beyond the intestinal tract. Nutr Rev. 2007;65:469–489. doi: 10.1111/j.1753-4887.2007.tb00272.x. et al. [DOI] [PubMed] [Google Scholar]
- Mazé A., O'Connell‐Motherway M., Fitzgerald G.F., Deutscher J., Van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N.E. Immigration control of DNA in bacteria: self versus non‐self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- O'Connell Motherway M., Fitzgerald G.F., Neirynck S., Ryan S., Steidler L., Van Sinderen D. Characterisation of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Sato T., Yokoi W., Yoshida Y., Ito M., Sawada H. Effect of probiotics, Bifidobacterium breve and Lactobacillus casei, on bisphenol A exposure in rats. Biosci Biotechnol Biochem. 2008;72:1409–1415. doi: 10.1271/bbb.70672. [DOI] [PubMed] [Google Scholar]
- O'Riordan K., Fitzgerald G.F. Molecular characterization of a 5.75 kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of the mode of replication. FEMS Microbiol Lett. 1999;174:285–294. doi: 10.1111/j.1574-6968.1999.tb13581.x. [DOI] [PubMed] [Google Scholar]
- Park M.S., Shin D.W., Lee K.H., Ji G.E. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology. 1999;145:585–592. doi: 10.1099/13500872-145-3-585. [DOI] [PubMed] [Google Scholar]
- Park M.S., Shin D.W., Lee K.H., Ji G.E. Characterisation of plasmid pKJ36 from Bifidobacterium longum and construction of an E. coli–Bifidobacterium shuttle vector. J Microbiol Biotechnol. 2000;10:310–320. [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V., Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ruyter P.G., Kuipers O.P., De Vos W.M. Controlled gene expression systems for Lactococcus lactis with the food‐grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.M., Fitzgerald G.F., Van Sinderen D. Screening for and identification of starch‐, amylopectin‐, and pullulan‐degrading activities in bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrador‐Vegas A., Stanton C., Van Sinderen D., Fitzgerald G.F., Ross R.P. Characterization of plasmid pAS479 from Bifidobacterium pseudolongum subsp. globosum and its use for expression vector construction. Plasmid. 2007;58:140–147. doi: 10.1016/j.plasmid.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Saxelin M., Tynkkynen S., Sandholm T.M., De Vos W.M. Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol. 2005;16:204–211. doi: 10.1016/j.copbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Schell M.A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgorbati B., Scardovi V., Leblanc D.J. Plasmids in the genus Bifidobacterium. J Gen Microbiol. 1982;128:2121–2131. doi: 10.1099/00221287-128-9-2121. [DOI] [PubMed] [Google Scholar]
- Shkoporov A.N., Efimov B.A., Khokhlova E.V., Steele J.L., Kafarskaia L.I., Smeianov V.V. Characterization of plasmids from human infant Bifidobacterium strains: sequence analysis and construction of E. coli–Bifidobacterium shuttle vectors. Plasmid. 2008;60:136–148. doi: 10.1016/j.plasmid.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Skrypina N.A., Kramarov V.M., Liannaia A.M., Smolianinov V.V. Restriction endonucleases from Bifidobacteria. Mol Gen Mikrobiol Virusol. 1988;5:15–16. [PubMed] [Google Scholar]
- Sudarsanam P., Ley R., Guruge J., Turnbaugh P.J., Mahowald M., Liep D., Gordon J. 2008. , and ) Draft genome sequence of Bifidobacterium dentium (ATCC 27678). NCBI Database.
- Sutcliffe J.G., Church G.M. The cleavage site of the restriction endonuclease Ava II. Nucleic Acids Res. 1978;5:2313–2319. doi: 10.1093/nar/5.7.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Tsuda Y., Kanou N., Inoue T., Kumazaki K., Nagano S. 2006. et al.Bifidobacterium adolescentis ATCC 15703 complete genome sequence. NCBI Database.
- Timinskas A., Butkus V., Janulaitis A. Sequence motifs characteristic for DNA (cytosine‐N4) and DNA (adenine0N6) methyltransferases. Classification of all DNA methyltransferases. Gene. 1995;157:3–11. doi: 10.1016/0378-1119(94)00783-o. [DOI] [PubMed] [Google Scholar]
- Tock M.R., Dryden D.T.F. The biology of restriction and anti‐restriction. Curr Opin Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Turroni F., Ribbera A., Foroni E., Van Sinderen D., Ventura M. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek. 2008;94:35–50. doi: 10.1007/s10482-008-9232-4. [DOI] [PubMed] [Google Scholar]
- Ventura M., Van Sinderen D., Fitzgerald G.F., Zink R. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek. 2004;86:205–223. doi: 10.1023/B:ANTO.0000047930.11029.ec. [DOI] [PubMed] [Google Scholar]
- Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G.F., Chater K.F., Van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R.Y., Walder J.A., Donelson J.E. The organization and complete nucleotide sequence of the PstI Restriction‐Modification System. J Biol Chem. 1984;259:8015–8026. [PubMed] [Google Scholar]
- Yanisch‐Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoetendal E.G., Vaughan E. E., De Vos W.M. A microbial world within us. Mol Microbiol. 2006;59:1639–1650. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A. Restriction analysis of E. coli JM101 and two representative JM101 bbrIIM and bbrIIIM methylase integration strains. Lane 1, molecular weight marker X (Roche). Lanes 2-4, Pst1 digest of total DNA isolated from JM101 following induction with 0, 1 or 10 mM IPTG. Lanes 5-7 and lane 8-10, PstI digests of total DNA isolated from two representative JM101 bbrIIM and bbrIIIM methylase integration strains after induction with 0, 1 or 10 mM IPTG. B. Transformation efficiency of B. breve UCC2003 with pAM5 plasmid DNA isolated from B. breve UCC2003, E. coli pNZM. BbrII-M.BbrIII, E. coli BM1 or E. coli pNZ8048.
Fig. S2. A. Schematic representation of the relevant regions of the B. breve UCC2003 and UCC2003-apuB-939 chromosome. Chromosomal DNA is represented by a thin line, the apuB gene is represented by a black arrow, the internal apuB fragment is indicated by a solid grey line and pORI19 is indicated by a boxed line. BamHI sites relevant to the Southern hybridization analysis are indicated.
B. Southern hybridization analysis of BamHI-digested chromosomal DNAs of B. breve UCC2003 (lane 1) and four representative B. breve UCC2003-apuB-939 mutants (lanes 2-5). The molecular weight of the relevant hybridization signals are indicated to the left of the panel. The internal 1 kb PCR amplicon of apuB was used as a probe for the hybridization.
Table S1. Oligonucleotide primers used in this study.
Table S2. Restriction analysis of genomic DNA from B. breve strains with BbeI and PstI.
Appendix S1. Experimental procedures.


