Summary
Strategies to induce biofilm dispersal are of interest due to their potential to prevent biofilm formation and biofilm‐related infections. Nitric oxide (NO), an important messenger molecule in biological systems, was previously identified as a signal for dispersal in biofilms of the model organism Pseudomonas aeruginosa. In the present study, the use of NO as an anti‐biofilm agent more broadly was assessed. Various NO donors, at concentrations estimated to generate NO levels in the picomolar and low nanomolar range, were tested on single‐species biofilms of relevant microorganisms and on multi‐species biofilms from water distribution and treatment systems. Nitric oxide‐induced dispersal was observed in all biofilms assessed, and the average reduction of total biofilm surface was 63%. Moreover, biofilms exposed to low doses of NO were more susceptible to antimicrobial treatments than untreated biofilms. For example, the efficacy of conventional chlorine treatments at removing multi‐species biofilms from water systems was increased by 20‐fold in biofilms treated with NO compared with untreated biofilms. These data suggest that combined treatments with NO may allow for novel and improved strategies to control biofilms and have widespread applications in many environmental, industrial and clinical settings.
Introduction
In nature, bacteria predominantly live in surface‐associated matrix‐encased communities called biofilms, which can cause significant damage in many industrial and clinical settings (Costerton et al., 1995). For example, in the water industry biofilms can block filtration membranes or induce fouling and corrosion in distribution systems (Coetser and Cloete, 2005; Pang et al., 2005). In the clinical context, it is estimated that 80% of acute and chronic infections are biofilm‐related (Hall‐Stoodley et al., 2004). Pathogens such as Pseudomonas aeruginosa, Serratia marcescens or Vibrio cholerae in single‐ or multi‐species microbial communities can form biofilms on living tissues in humans, causing for example infections of the respiratory, gastrointestinal and urinary tracts, or periodontal diseases. Biofilms also readily form on biomedical devices such as prostheses and catheters (Khardori and Yassien, 1995).
Bacteria in biofilms are generally highly tolerant to biocides, antibiotics and natural host defences, often becoming up to 10 000 times more resistant compared with their free‐swimming counterparts (Buckingham‐Meyer et al., 2007). Antimicrobials, such as antibiotics or chlorine‐based treatments, have traditionally been designed to inhibit planktonic bacteria. These treatments are inappropriate for biofilm control, as their use may be toxic to the environment, require unacceptably high cost and energy inputs or lead to fatal outcomes in clinical settings. To address the need for novel and improved measures against biofilms, a clear strategy is to study the biofilm life cycle and identify key trigger points that regulate biofilm development. Thus, several such switches that mediate surface attachment mechanisms (Valle et al., 2006), cell–cell signalling and biofilm maintenance (Hentzer et al., 2002) have been the target for biofilm control strategies in recent years. In addition, the last stage of biofilm development that describes the coordinated dispersal of biofilm cells presents several advantages with respect to biofilm control. Induction of biofilm dispersal could potentially use the microorganisms' own energy to remove established biofilms, revert cells to a planktonic phenotype and restore their vulnerability to antimicrobials.
In a recent study, the biologically ubiquitous gas molecule, nitric oxide (NO), was identified as an important factor mediating biofilm dispersal in the model organism P. aeruginosa (Barraud et al., 2006). Low, non‐toxic concentrations of NO were shown to induce a transition from the sessile, resistant biofilm mode of growth to the motile planktonic phenotype in P. aeruginosa. Furthermore, the addition of various antimicrobial compounds was found to almost completely remove remaining P. aeruginosa biofilms that were exposed to NO, suggesting a general effect of NO on P. aeruginosa physiology (Barraud et al., 2006). The involvement of NO in regulating biofilm formation and dispersal in P. aeruginosa was also supported by other studies (Darling and Evans, 2003; Van Alst et al., 2007). Nitric oxide‐mediated dispersal in P. aeruginosa biofilms appears to involve cyclic di‐GMP (N. Barraud and S. Kjelleberg, unpublished), a conserved secondary messenger, which level is regulated by diguanylate cyclases and phosphodiesterases (Ryan et al., 2006). Genes encoding for diguanylate cyclases and phosphodiesterases are widely distributed among bacteria, and are often associated with redox sensors, such as PAS (Per‐Arnt‐Sim) domains capable of sensing NO (Delgado‐Nixon et al., 2000; Römling et al., 2005). This suggests that NO‐mediated dispersal is not restricted to P. aeruginosa but may occur among various bacterial species.
Several lines of evidence support the hypothesis that NO‐mediated dispersal may be conserved across species. First, a recent study showed that staphylococcal biofilms were inhibited upon exposure to nitrite (NO2‐), a process that is thought to involve NO (Schlag et al., 2007). Second, several aspects of NO biology appear to be conserved across microorganisms, such as NO production apparatuses, either via reduction of NO2‐ during denitrification (Zumft, 1993) or via oxidation of arginine by NO synthases (Adak et al., 2002; Gusarov et al., 2008), as well as NO responsive networks (Rodionov et al., 2005). Third, NO has been suggested to be an ancient and highly conserved regulator of dispersal and life histories across eukaryotic organisms (Bishop and Brandhorst, 2003). For example, dissolution and dispersal of aggregated mycelial cells of fungi, Neurospora crassa (Ninnemann and Maier, 1996) and Candida tropicalis (Wilken and Huchzermeyer, 1999), and the amoeba Dictyostelium discoideum (Tao et al., 1997) were shown to rely on NO signalling. Taken together, these observations strongly suggest that NO may mediate biofilm dispersal across a broad range of microorganisms.
The objective of the present study was to examine the effects of NO on various biofilms of clinical and industrial significance. Single‐species biofilms of Gram‐negative and Gram‐positive bacterial species and one yeast, as well as multi‐species biofilms from water distribution and treatment systems were exposed to low doses of NO, typically in the picomolar and low nanomolar range. Low concentrations of various NO donors were found to induce dispersal of biofilm cells and reduce biofilm formation across all of the species tested. Moreover, multi‐species biofilms in water systems were observed to show decreased surface coverage and heterotrophic counts in the presence of nanomolar concentrations of the NO donors sodium nitroprusside (SNP) and disodium 1‐[2‐(carboxylato)pyrrolidin‐1‐yl]diazen‐1‐ium‐1,2‐diolate (PROLI). Pretreatment with NO significantly increased the efficacy of chlorine disinfection in removing biofilms established from chlorinated water distribution and treatment systems.
Results
Nitric oxide induces dispersal of single‐species biofilms of Gram‐negative and Gram‐positive bacteria and yeasts
The effect of NO as a broad biofilm‐dispersing agent was assessed in a range of biofilm‐forming microorganisms of industrial and/or clinical significance (Table 1). Delivery of exogenous NO to biofilms was achieved by using NO‐releasing compounds, called NO donors. Nitric oxide donors establish steady‐state levels of NO, thus mimicking endogenous NO production. Although the exact amount and location of NO liberated in vivo within biofilms from NO donors have not yet been established, approximately a 1000‐fold linear relationship between NO donor concentrations and NO steady‐state levels was measured in vitro (R2 = 0.94) by using solutions of the NO donor SNP in phosphate‐buffered saline (PBS) in the range 250 µM–1 mM (Fig. 1). Similar release profiles were observed with the nitrosothiols S‐nitroso‐N‐acetylpenicillamine (SNAP) and S‐nitroso‐L‐glutathione (GSNO) (data not shown). Thus, effective concentrations of NO delivered to the cells are estimated to be 1000 times lower than the concentration of NO donor used herein.
Table 1.
Microbial strains used in this study.
| Strain | Site of biofilm formation or infection | Source or reference |
|---|---|---|
| Gram‐negative | ||
| Serratia marcescens MG1 | Respiratory and urinary tracts | Givskov et al. (1988) |
| Vibrio cholerae 92A1552 | Gastrointestinal tract and wounds | Yildiz and Schoolnik (1998) |
| Escherichia coli BW20767 | Gastrointestinal and urinary tracts | Metcalf et al. (1996) |
| Fusobacterium nucleatum | Oral cavity | UNSW culture collection |
| Gram‐positive | ||
| Bacillus licheniformis | Food processing surfaces | UNSW culture collection |
| Staphylococcus epidermidis | Catheters and medical prostheses | UNSW culture collection |
| Yeast | ||
| Candida albicans | Oral cavity, skin | UNSW culture collection |
Figure 1.
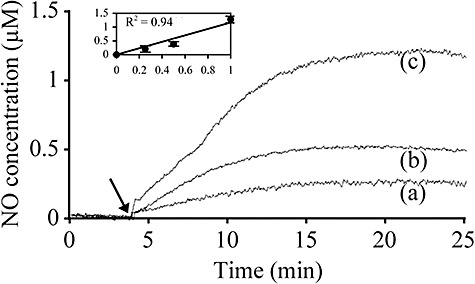
Nitric oxide release profiles from the NO donor SNP. After the NO baseline signal was stabilized for at least 30 min in the PBS solution, SNP was added (arrow) at final concentrations of (a) 250 µM, (b) 500 µM and (c) 1 mM and the amount of NO released was quantified by using the NO electrode. The inset shows the linear relationship between SNP concentration (mM, x‐axis) and NO increase (µM, y‐axis); error bars indicate standard deviation (n = 3).
Biofilms of the opportunistic pathogen S. marcescens dispersed in response to SNP at concentrations between 25 nM and 500 nM, with a 60% reduction in biofilm coverage at a concentration of 500 nM SNP (Table 2). S‐nitroso‐N‐acetylpenicillamine (100 nM) was also effective in dispersing S. marcescens biofilms. The SNP, SNAP and GSNO were effective at inducing dispersal of V. cholerae biofilms, where 500 nM SNP induced a 73% reduction in surface coverage and 1 µM SNAP and 1 µM GSNO led to a 29% and 34% reduction respectively (Table 2, P < 0.01). Escherichia coli biofilms were also dispersed by exposure to low levels of NO as demonstrated by using SNP (Table 2). In these experiments, addition of NO resulted in an increased number of bacteria in suspension as revealed by colony‐forming units (cfu) counts (data not shown). To confirm the involvement of NO in the observed effects on biofilms, the NO scavenger 2‐phenyl‐4,4,5,5‐tetramethylimidazoline‐1‐oxyl‐3‐oxide (PTIO) was used. With added PTIO, the SNP effect was reduced by 93% in S. marcescens biofilms, 72% in V. cholerae biofilms and 65% in E. coli biofilms.
Table 2.
Effects of low concentrations of NO donors on various biofilm‐forming microorganisms.
| Biofilm‐forming microorganism | Optimum NO donora concentrations | Maximum percentage removal (SE)b | |
|---|---|---|---|
| Gram‐negative | |||
| Serratia marcescens | SNP | 25–500 nM | 60.0% (±4.1%) |
| SNAP | 100 nM | 37.8% (±10.5%) | |
| Vibrio cholerae | SNP | 25–500 nM | 72.5% (±1.9%) |
| SNAP | 1 µM | 28.6% (±3.6%) | |
| GSNO | 1 µM | 33.6% (±7.4%) | |
| Escherichia coli | SNP | 500 nM | 38.1% (±8.7%) |
| Fusobacterium nucleatum | SNP | 1–10 µM | 55.6% (±5.6%) |
| Gram‐positive | |||
| Bacillus licheniformis | SNP | 100–500 nM | 93.2% (±2.0%) |
| Staphylococcus epidermidis | SNP | 10 µM | 58.6% (±2.8%) |
| Yeast | |||
| Candida albicans | SNP | 25–100 nM | 61.4% (±6.7%) |
NO donors used: SNP, sodium nitroprusside; SNAP, S‐nitroso‐N‐acetylpenicillamine; and GSNO, S‐nitroso‐L‐glutathione.
Percentage removal indicates the percentage of total biofilm surface that was removed after exposure to NO relative to control biofilms that were not exposed to NO. SE, standard error (n ≥ 3).
Biofilm culture conditions and analysis methodology for each strain are detailed in Experimental procedures.
The effect of NO exposure was also investigated on a key organism for biofilms of oral consortia, the anaerobic bacterium Fusobacterium nucleatum. The presence of SNP was found to inhibit biofilm formation by F. nucleatum (Table 2). The most effective concentrations of SNP were 1 µM and 10 µM, which achieved 48% and 55% reduction in biofilm surface coverage respectively (P < 0.01). These concentrations appear to be one order of magnitude higher than those observed with other organisms. This may reflect that, compared with other bacteria, F. nucleatum may use a distinct mechanism such as a different NO‐sensor domain to activate biofilm dispersal in response to NO signalling.
A strong effect of SNP on the stability of Gram‐positive Bacillus licheniformis biofilms was observed, with 500 nM SNP inducing greater than 90% reduction in surface coverage of the biofilms (Table 2, P < 0.001). Staphylococcus epidermidis was found to have reduced biofilm formation in the presence of low concentrations of SNP. A 59% reduction of surface coverage was achieved in the presence of 10 µM SNP.
Nitric oxide‐mediated biofilm dispersal was also tested on the fungus Candida albicans. When pre‐established C. albicans biofilms were exposed to low concentrations of SNP, it was found that extremely low concentrations of SNP, i.e. 25 nM and 100 nM, were able to induce a reduction in biofilm formation by up to 61% (Table 2, P < 0.01).
Finally, combinatorial treatments of NO and an antibiotic were assessed on V. cholerae biofilms. Tetracycline is the usual antibiotic of choice to treat V. cholerae infections, but resistance to this drug is increasing (Dromigny et al., 2002). Although tetracycline (14 µM) alone, at a concentration below the minimum inhibitory concentration (approximated at 22 µM in this study) for V. cholerae, had very limited effect on biofilms of this organism resulting in only a 21% reduction in surface coverage (Fig. 2), SNP was able to enhance tetracycline activity against biofilm cells. Addition of tetracycline after 25 nM SNP and 500 nM SNP treatment induced 67% and 65% reduction of biofilm bacteria respectively, when compared with biofilms treated with tetracycline alone. Overall, combined exposure to 500 nM SNP and tetracycline resulted in 90% reduction in biofilm coverage compared with the untreated controls (Fig. 2).
Figure 2.
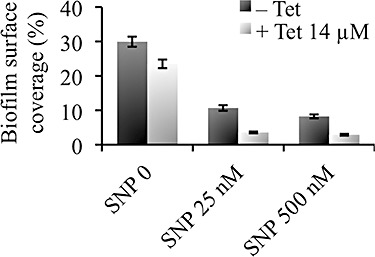
Nitric oxide effect on V. cholerae biofilm antimicrobial sensitivity. Pre‐established V. cholerae biofilms were treated for 24 h in the presence or absence of the NO donor SNP, and/or the antibiotic tetracycline (Tet) at 14 µM. Biofilm cells remaining on the slides were stained with SYTO 9 to allow analysis using fluorescence microscopy and quantification (per cent surface coverage) using digital image analysis. Data are mean values and error bars indicate standard error (n = 3).
Nitric oxide induces dispersal of multi‐species biofilms in water systems and increases the efficacy of chlorine treatments
Recycled and potable water distribution. To study the effect of NO on multi‐species biofilms formed in recycled water distribution systems, an annular reactor (AR) containing unplasticized polyvinyl chloride (uPVC) coupons was connected to a recycled water network for 3 months. After this time, biofilms were sampled and treated in the laboratory. The data demonstrate that SNP treatment was effective at removing multi‐species biofilms as revealed with both microscopy and viability analyses by performing heterotrophic cfu counts of biofilm bacteria (Fig. 3). The most efficient concentration of SNP was 500 nM, which induced dispersal of 47 ± 3% of the biofilm compared with the untreated controls, as revealed by cfu measurements of biofilm bacteria (Fig. 3B). As seen on the microscopy images, control biofilms established from recycled water distribution system harbour mature microcolonies that contain both living and dead cells. After SNP treatments, the size of biofilm aggregates (microcolonies) on the surface was considerably reduced, indicative of dispersal events (Fig. 3A). In addition, biofilms treated with SNP displayed increased number of cells (cfu) in their effluent runoff (data not shown). Furthermore, the ratios of cfu counts to biofilm surface coverage were calculated for each treatment and normalized to the control experiments. The ratios did not vary significantly: 1.0, 0.9 and 0.9 for untreated, 100 nM SNP and 500 nM SNP respectively. These data indicate that SNP treatment effectively induced the removal of viable cells from the surface.
Figure 3.
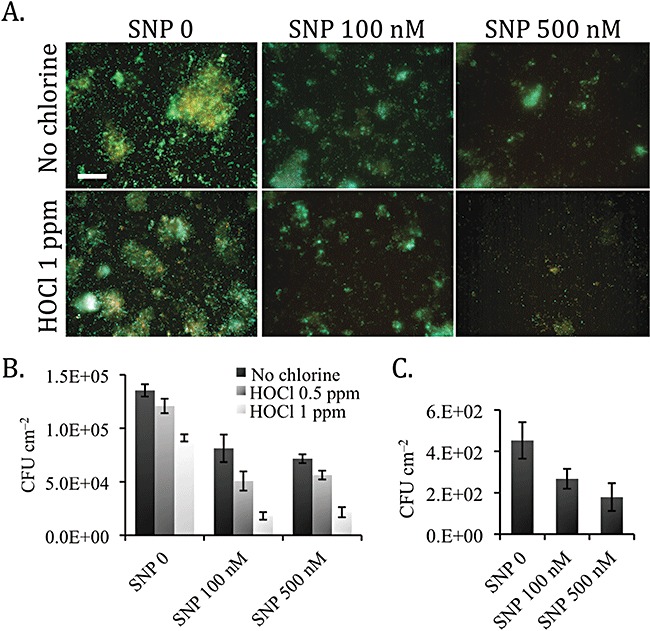
Effect of NO on multi‐species biofilms established from water distribution systems. Three‐month‐old biofilms from recycled and potable water distribution systems were exposed to 0 (control), 100 nM and 500 nM SNP for 18 h and then (recycled water biofilms) treated for 10 min with free chlorine (HOCl) at 0.5 ppm and 1 ppm and no chlorine controls. (A) The images show microscopic pictures of recycled water biofilms after 1 ppm HOCl exposure (lower panels) or no chlorine controls (upper panels) and stained with the LIVE/DEAD reagents. Live cells appear green, dead cells appear red. Bar, 50 µm. Viability analyses of the biofilms were assessed by heterotrophic colony‐forming units (cfu) measurements of (B) recycled water biofilms and (C) potable water biofilms. Data are mean values and error bars indicate standard error (n = 3).
The total number of viable bacteria in biofilms decreased after exposure to SNP and it was observed that the relative proportions of different colony morphologies on plates did not change as a result of SNP treatment. Moreover, by using denaturing gradient gel electrophoresis no significant change in the biofilm microbial community could be detected after induction of dispersal with SNP (data not shown). This suggests that exposure to SNP was not selective for specific species within the mixed community; rather, SNP treatment appears to be broadly effective across the entire microbial community. Such an effect is in agreement with the observations of NO‐mediated dispersal across a broad range of biofilm‐forming organisms (Table 2).
Biofilms exposed to 100 nM and 500 nM SNP, respectively, also exhibited increased sensitivity to chlorine (HOCl) treatments. For example, 1 ppm HOCl, which is within the recommended range (1–2 ppm) for water distribution systems, was up to 20‐fold more efficient at removing 500 nM SNP‐treated biofilms compared with control biofilms as determined by cfu counts (Fig. 3B). Overall, combined NO and chlorine treatments resulted in 85–90% removal of recycled water biofilms compared with the untreated controls. Consistent data were obtained when coupons were analysed for total biofilm surface by quantification of surface coverage (not shown). Similar results were observed in an independent replicate experiment.
Sodium nitroprusside treatment was also assessed on multi‐species biofilms established from a potable water distribution system. Control potable water biofilms generally contained 2‐log lower number of cells as seen by heterotrophic cfu measurements (typically 4.5 × 102 cfu cm−2) compared with control biofilms established in recycled water (typically 1.4 × 105 cfu cm−2). Multi‐species biofilms established in potable water distribution system were also reduced in total cfu counts upon exposure to nanomolar concentrations of SNP. The most efficient concentration of SNP was 500 nM, which induced dispersal of 60 ± 15% of the biofilm compared with the untreated controls (Fig. 3C).
Reverse osmosis filtration membrane. Biofilms established on a reverse osmosis (RO) filtration membrane connected in line with a full‐scale water filtration system for 3 months were exposed to 100 nM SNP for 1 h followed by disinfection with chlorine for 10 min. Sodium nitroprusside itself induced a 30% reduction in biofilm viability compared with untreated controls, as revealed by cfu measurements (Fig. 4A). Moreover, the efficacy of chlorine disinfection was increased by twofold when assessed on biofilms that were pretreated with SNP compared with untreated biofilms. Overall, combined exposure to SNP and chlorine induced a 94% reduction in biofilms (Fig. 4A).
Figure 4.
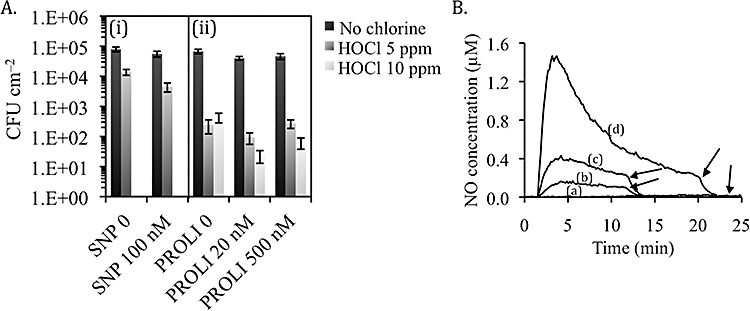
Multi‐species biofilms on a RO filtration membrane exposed to SNP or the fast NO donor PROLI in combination with chlorine. A. Reverse osmosis membrane coupons harbouring multi‐species biofilms were treated: (i) in the presence or absence of 100 nM SNP and subsequently exposed to 5 ppm HOCl for 10 min; or (ii) simultaneously in the presence or absence of 20 nM or 500 nM PROLI and/or free chlorine at 5 ppm or 10 ppm for 2 h. Biofilms were analysed by performing cfu counts. Data are mean values and error bars indicate standard error (n ≥ 3). B. Nitric oxide release profiles from PROLI in water. (a) Control, (b) 625 nM, (c) 1.25 µM and (d) 2.5 µM PROLI. Arrows indicate addition of NO scavenger PTIO (100 µM) to the solutions.
Reverse osmosis membrane biofilms were also assessed for simultaneous exposure to NO and chlorine. For these experiments, the NO donor PROLI was used. 1‐[2‐(carboxylato)pyrrolidin‐1‐yl]diazen‐1‐ium‐1,2‐diolate shows an unusual, fast release of NO in water (Fig. 4B) making it a preferred candidate for treatment of filtration membranes, systems which allow for a short exposure time of the compound only. Moreover, PROLI was previously shown to have very low potential toxicity, as tested against L929 Mouse Fibroblasts (Hetrick et al., 2008), which appears suitable for use in potable water systems. After 2 h exposure, 20 nM and 500 nM PROLI induced, respectively, 41% and 32% reduction in total biofilm surface as revealed by cfu counts (Fig. 4A). Further, treatment with PROLI dramatically increased the efficacy of chlorine at removing biofilm cells. Thus, 20 nM PROLI increased 19‐fold the efficacy of 10 ppm chlorine, and the combination of PROLI and chlorine resulted in an overall 3‐log reduction in the number of biofilm cells (Fig. 4A). PROLI 500 nM was also able to increase the efficacy of chlorine by up to sevenfold.
Discussion
The results presented in this study demonstrate that exposure to low levels of NO donors induces dispersal of diverse single‐species biofilms of Gram‐positive and Gram‐negative bacteria and yeast, as well as multi‐species biofilms from water systems. These observations suggest that NO‐mediated biofilm dispersal is widespread among biofilm‐forming microorganisms. Moreover, NO donors were also observed to increase the sensitivity of V. cholerae and multi‐species biofilms to antimicrobial treatments. This suggests that low doses of NO can have profound effects on the physiology of bacteria in biofilms and function widely as a signal mediating the transition to a planktonic‐like mode of growth, thus rendering cells more vulnerable to antimicrobials. In P. aeruginosa, transcriptomic analyses revealed that exposure to low levels of NO induces global responses in biofilm bacteria, including upregulation of genes involved in motility and energy metabolism and downregulation of adhesins and defence mechanisms (N. Barraud and S. Kjelleberg, unpublished).
Nitric oxide‐based strategies to induce biofilm dispersal involve extremely low concentrations of NO, in the picomolar to low nanomolar range that should be safe to humans and to the environment. Indeed NO gas and NO donors are currently used clinically. For example, GSNO, which is endogenously produced in mammals, was suggested for its potential use as a therapeutic treatment of respiratory diseases with doses up to 30 µmol administered with a nebulizer (Snyder et al., 2002; Que et al., 2005). Further, systemic administration of SNP solutions, of up to 650 µM SNP, was approved by the Food and Drug Administration for the treatment of hypertension in humans. At the higher concentrations required to achieve these clinical effects, NO may be toxic and can lead to pathologies such as neurologic excitotoxicity or hypotension. In contrast, our data show that NO donors are active at extremely low levels against biofilms, for example at 500 nM for SNP, a 3‐log difference compared with the concentrations used for treatment of hypertension. Hence, NO treatments should not induce any systemic toxic effect at the levels used here. In natural environments, basal NO levels appear to be below the detection limit of most measurement systems, therefore likely not to exceed 10–100 pM (Zafiriou et al., 1980), and thus are unable to induce dispersal of biofilms without exogenous addition of NO donors. From an environmental perspective, NO in aqueous solutions is quickly oxidized mostly to nitrite (NO2‐) (Ignarro et al., 1993), for which the maximum contaminant level in drinking water recommended by the Environmental Protection Agency is 1 ppm (22 µM). Again, these concentrations are several orders of magnitude above the concentrations of NO that are effective for inducing biofilm dispersal.
The findings presented in this study identify a novel and unprecedented measure to control biofilms. Nitric oxide donors were active against all biofilms tested and the data suggest that NO can be effective across multi‐species microbial communities and is not selective for any particular strain. Combinatorial treatments of low levels of NO and chlorine, which appear acceptable for use in water environments, achieved almost complete eradication of the biofilms, up to 99.97% removal. This reflects considerable progress in biofilm control practices and would allow control and removal of biofilms in water systems on a continuous basis without any disruption of industrial processes. In addition, the efficacy of NO and tetracycline, as observed in this study, again at levels that appear suitable for treatment of infections in humans, provides a great opportunity for the eradication of recalcitrant infections of V. cholerae and possibly other pathogens. This strongly suggests that NO donor compounds warrant thorough investigation as biofilm‐dispersing agents useful in the treatment of drug‐resistant biofilm infections and control of environmental biofilms.
Furthermore, the use of compounds to modulate NO activity may also be valuable for enhanced control of beneficial microbial communities in biotechnology processes; for example, for the rejuvenation of biofilms to improve productivity and stability in bioremediation or biotransformation systems (Schachter, 2003).
The development of NO‐based biofilm control biotechnology may be facilitated by information available from previous research on the use of NO for therapeutic treatments. A broad range of NO donors, which directly or indirectly release NO, or agents that increase NO bioactivity have been developed (Wang et al., 2002; Keefer, 2003). Several methods can be used to achieve controlled delivery of NO to biofilm cells in various application areas, including chemical carriers for dosing of NO donors directly in the liquid phase, or delivery at the target site, such as toothpastes for the eradication of dental biofilms. Moreover, NO donors may also be embedded in polymer coatings that can be applied onto surfaces (Reynolds et al., 2004; Frost et al., 2005).
Overall, combined treatments of low doses of NO with standard antimicrobials offer great potential for the control of biofilms in environmental, industrial and clinical settings, with clear benefits such as reduced ecological impact and reduced treatment costs. Finally, because NO appears to induce the transition from a biofilm to a free swimming phenotype, and thus reduces the antimicrobial tolerance of bacteria on surfaces, via a signalling mechanism rather than toxic effect, NO‐based biofilm control strategies would not be expected to select for resistant strains as seen with antibiotics.
Experimental procedures
Single‐species biofilms of various bacteria and the yeast C. albicans
Strains and culture conditions. The strains of bacteria and a yeast that were used in this study are listed in Table 1. Overnight cultures were grown at 30°C in Luria Bertani (LB) for B. licheniformis, E. coli, S. marcescens (formerly Serratia liquefaciens) and V. cholerae, in Tryptic Soy Broth (TSB) (BD Diagnostics) for S. epidermidis, and anaerobically in Schaedler Broth (BD Diagnostics) for F. nucleatum. The fungus C. albicans was grown at 30°C in Yeast Peptone Dextrose Broth (YPD) (Sigma).
Nitric oxide donors and amperometric NO measurements. Four NO donors were used in this study: SNP (Sigma), GSNO (MP Biomedicals), SNAP (Sigma), and disodium PROLI (Alexis Biochemicals). The NO scavenger PTIO (Sigma) was also used. Experiments were carried out where donors were freshly diluted in the respective biofilm medium, and used at final concentrations ranging from 20 nM to 10 µM as indicated. These concentrations were within the range that was previously observed to be effective at inducing biofilm dispersal in P. aeruginosa (Barraud et al., 2006). Amperometric measurements of NO release from NO donors were carried out by using a NO analyser (Apollo 4000, World Precision Instruments) with ISO‐NOP electrode calibrated using SNAP and copper sulfate according to the manufacturer's instructions.
Biofilm experiments. Biofilms of S. marcescens, V. cholerae, E. coli, B. licheniformis and S. epidermidis were cultivated for 24 h on either glass slides (V. cholerae, B. licheniformis and S. epidermidis) or uPVC slides (S. marcescens and E. coli), immersed in biofilm medium in sterile petri dishes with gentle shaking, in triplicate. The biofilm media used were 1/5 strength LB for S. marcescens, E. coli and B. licheniformis biofilms, 2M medium as previously described (Paludan‐Muller et al., 1996) for V. cholerae biofilms, and 1/5 strength TSB for S. epidermidis biofilms. The conditions used here were identified as those that were optimal for biofilm growth in the absence of any treatment for each organism. After 24 h, the supernatant was replaced with fresh medium containing the indicated concentrations of SNP, SNAP or GSNO (in addition to controls without NO donor), and 200 µM PTIO for the NO‐scavenging experiments, and the biofilms were incubated for a further 24 h. For combined NO and antibiotic assays in V. cholerae biofilms, after the initial 24 h of biofilm development, the NO donor was added in combination with 14 µM tetracycline. The tetracycline concentration used was below the minimum inhibitory concentration for V. cholerae that was previously determined to be 22 µM by monitoring the OD600 over 24 h in a microtitre plate assay with V. cholerae inoculum in 2M medium and twofold serial dilutions of tetracycline (data not shown). For biofilm analysis, the slides were rinsed and the biofilms were stained with SYTO 9 (3 µl ml−1) (Molecular Probes). Using epifluorescence microscopy (Leica model DMR), 15 selected fields of view per slide were imaged in the XY plane, at regular intervals and across the entire slide. Image analysis (ImageJ, NIH) was performed and total biofilm surface was determined as total surface coverage. The results in Table 2 are presented as the percentage of total biofilm surface reduction in cultures treated with NO relative to the total biofilm surface in control cultures that were not exposed to an NO donor. In Fig. 2, the results of combined NO and tetracycline treatments on V. cholerae biofilms are shown for each treatment condition as the percentage of biofilm surface coverage compared with the total substratum surface.
Fusobacterium nucleatum was selected as a model oral biofilm‐forming organism. Briefly, cells were grown anaerobically in Schaedler Broth to an OD600 of 0.1, at which time SNP was added. Bacteria were allowed to attach for 4 h on a sterile glass slide. Then the slides were rinsed and stained with 1% crystal violet (CV). Attached cells were enumerated microscopically by digital image capture and subsequent image analysis, and the total biofilm surface was determined as total surface coverage. Results are presented as the percentage of total biofilm surface reduction induced by NO treatments compared with the total biofilm surface in control cultures.
Candida albicans biofilms were grown for 24 h in 24‐well polystyrene plates in 1/10 strength YPD with shaking at 100 r.p.m. After 24 h, the supernatant was replaced with fresh medium containing SNP and the cells were incubated for a further 24 h. The wells were then rinsed, stained with 1% CV, and washed again thoroughly, before the CV absorbed into the biofilm was dissolved in absolute ethanol and the total biofilm surface was quantified by measurement of OD540 (Wallac‐Victor2).
Experiments were carried out in triplicate and a statistical comparison of the percentage of surface covered by biofilms was performed using analysis of variance and Tukey's multiple comparison tests.
Multi‐species biofilms established from water distribution systems
Model potable and recycled water system biofilms were grown in two continuous flow ARs (BioSurfaces Technologies) connected directly to a potable and recycled water distribution system. Sterile stainless steel and uPVC coupons were placed on the exposed face of the inner‐rotating cylinder of the ARs, which received chlorinated potable and recycled water respectively, at a rate of 30 l h−1 making the hydraulic retention time 2.2 min. Biofilms were allowed to grow on coupon surfaces for a period of 90 days. Stainless steel coupons (potable) and uPVC coupons (recycled) were transferred to sterile bioreactors in the laboratory for NO exposure. The bioreactors consisted of 1 l polypropylene beakers with bottom inlet, top outlet and magnetic stirring, covered with aluminium foil and containing modified polypropylene microscope slide racks (Kartell) that fit the coupons. Coupons were placed in three separate bioreactors where they were exposed for 18 h to a continuous flow (50 ml h−1) of 1/4 strength Ringers solution (Oxoid), pH 7.4, containing 0, 100 nM or 500 nM SNP respectively. For recycled water biofilm assays, coupons were then carefully transferred into sterile 25 ml glass vials containing 20 ml of chlorine treatments in Ringers solution that were freshly made from sodium hypochlorite (Sigma) and calibrated for free chlorine (HOCl) content by N,N‐diethyl‐p‐phenylenediamine analysis using a Pocket Colorimeter II (HACH). After 10 min of gentle shaking, the reactions were stopped by adding 100 µM sodium thiosulfate.
The coupons were processed either for microscopy analysis, in duplicate, or for viability assay by performing heterotrophic plate counts, in triplicate. For microscopy analysis, biofilms were stained using the LIVE/DEAD BacLight Kit (Molecular Probes), visualized by using fluorescent microscopy and the biofilm surface coverage was quantified by digital image analysis as described above. For viability analysis, the coupons were placed in stomacher bags containing 25 ml Ringers solution. Biofilm cells were firstly removed from the coupon surfaces by agitation of the coupon by hand, followed by sonication at 400 W for 60 s (Branson 2210) and then stomaching for 60 s (Seward Stomacher 80) to remove and homogenize the remaining biofilm. The homogenate was then serially diluted and plated on oligotrophic R2A medium agar (Oxoid), using a pour‐plate technique. The plates were incubated at 25°C for 7 days. This protocol has previously been optimized for the removal of potable water biofilms for heterotrophic cfu measurements (data not shown). Analysis of variance tests at a significance level of 95% were used to compare the impact of the various combinations of low doses of NO, and chlorine disinfectant on biofilm growth.
Multi‐species biofilms established on RO filtration membrane
A small‐scale residential RO system (Crystal Clear Purification Systems) equipped with a RO cartridge model CSM RE1518‐50 (Sae‐Han) was connected to the Ravensthorpe (Western Australia) water treatment plant for 3 months. The pressure of the inlet was approximately 60 psi (400 kPa) at a flow rate of 100 ml min−1. After this period, coupons were extracted under sterile conditions from the biofouled membranes, transferred to 3 ml Ringers solution containing NO and/or chlorine treatments in 12 well plates (Sarstedt) and incubated at 25°C with gentle shaking. For SNP experiments, coupons were exposed to 0 (controls) or 100 nM SNP for 1 h. Then coupons were rinsed in Ringers solution and treated for 10 min with 5 ppm HOCl and no chlorine controls. For PROLI experiments coupons were simultaneously exposed to the NO donor PROLI (20 nM, 500 nM and untreated controls) and HOCl (5 ppm, 10 ppm and no chlorine controls) for 2 h. After treatments, biofilms were analysed by performing heterotrophic cfu counts. Biofilm cells were removed from the membrane coupons by using a sterile swab, serially diluted, plated onto R2A agar plates and the plates were incubated at 25°C for 7 days before enumeration of cfu.
Acknowledgments
This work was funded by the Environmental Biotechnology Cooperative Research Centre (EBCRC) and the Centre for Marine Bio‐Innovation.
References
- Adak S., Aulak K.S., Stuehr D.J. Direct evidence for nitric oxide production by a nitric‐oxide synthase‐like protein from Bacillus subtilis. J Biol Chem. 2002;277:16167–16171. doi: 10.1074/jbc.M201136200. [DOI] [PubMed] [Google Scholar]
- Barraud N., Hassett D.J., Hwang S.H., Rice S.A., Kjelleberg S., Webb J.S. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C.D., Brandhorst B.P. On nitric oxide signaling, metamorphosis, and the evolution of biphasic life cycles. Evol Dev. 2003;5:542–550. doi: 10.1046/j.1525-142x.2003.03059.x. [DOI] [PubMed] [Google Scholar]
- Buckingham‐Meyer K., Goeres D.M., Hamilton M.A. Comparative evaluation of biofilm disinfectant efficacy tests. J Microbiol Methods. 2007;70:236–244. doi: 10.1016/j.mimet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Coetser S.E., Cloete T.E. Biofouling and biocorrosion in industrial water systems. Crit Rev Microbiol. 2005;31:213–232. doi: 10.1080/10408410500304074. [DOI] [PubMed] [Google Scholar]
- Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin‐Scott H.M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Darling K.E., Evans T.J. Effects of nitric oxide on Pseudomonas aeruginosa infection of epithelial cells from a human respiratory cell line derived from a patient with cystic fibrosis. Infect Immun. 2003;71:2341–2349. doi: 10.1128/IAI.71.5.2341-2349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado‐Nixon V.M., Gonzalez G., Gilles‐Gonzalez M.A. Dos, a heme‐binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- Dromigny J.A., Rakoto‐Alson O., Rajaonatahina D., Migliani R., Ranjalahy J., Mauclere P. Emergence and rapid spread of tetracycline‐resistant Vibrio cholerae strains, Madagascar. Emerg Infect Dis. 2002;8:336–338. doi: 10.3201/eid0803.010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost M.C., Reynolds M.M., Meyerhoff M.E. Polymers incorporating nitric oxide releasing/generating substances for improved biocompatibility of blood‐contacting medical devices. Biomaterials. 2005;26:1685–1693. doi: 10.1016/j.biomaterials.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Givskov M., Olsen L., Molin S. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J Bacteriol. 1988;170:5855–5862. doi: 10.1128/jb.170.12.5855-5862.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Starodubtseva M., Wang Z.Q., McQuade L., Lippard S.J., Stuehr D.J., Nudler E. Bacterial nitric‐oxide synthases operate without a dedicated redox partner. J Biol Chem. 2008;283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall‐Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hentzer M., Riedel K., Rasmussen T.B., Heydorn A., Andersen J.B., Parsek M.R. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. et al. [DOI] [PubMed] [Google Scholar]
- Hetrick E.M., Shin J.H., Stasko N.A., Johnson C.B., Wespe D.A., Holmuhamedov E., Schoenfisch M.H. Bactericidal efficacy of nitric oxide‐releasing silica nanoparticles. ACS Nano. 2008;2:235–246. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignarro L.J., Fukuto J.M., Griscavage J.M., Rogers N.E., Byrns R.E. Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from L‐arginine. Proc Natl Acad Sci USA. 1993;90:8103–8107. doi: 10.1073/pnas.90.17.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer L.K. Progress toward clinical application of the nitric oxide‐releasing diazeniumdiolates. Annu Rev Pharmacol Toxicol. 2003;43:585–607. doi: 10.1146/annurev.pharmtox.43.100901.135831. [DOI] [PubMed] [Google Scholar]
- Khardori N., Yassien M. Biofilms in device‐related infections. J Ind Microbiol. 1995;15:141–147. doi: 10.1007/BF01569817. [DOI] [PubMed] [Google Scholar]
- Metcalf W.W., Jiang W., Daniels L.L., Kim S.K., Haldimann A., Wanner B.L. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Ninnemann H., Maier J. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem Photobiol. 1996;64:393–398. doi: 10.1111/j.1751-1097.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- Paludan‐Muller C., Weichart D., McDougald D., Kjelleberg S. Analysis of starvation conditions that allow for prolonged culturability of Vibrio vulnificus at low temperature. Microbiology. 1996;142:1675–1684. doi: 10.1099/13500872-142-7-1675. [DOI] [PubMed] [Google Scholar]
- Pang C.M., Hong P., Guo H., Liu W.T. Biofilm formation characteristics of bacterial isolates retrieved from a reverse osmosis membrane. Environ Sci Technol. 2005;39:7541–7550. doi: 10.1021/es050170h. [DOI] [PubMed] [Google Scholar]
- Que L.G., Liu L., Yan Y., Whitehead G.S., Gavett S.H., Schwartz D.A., Stamler J.S. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M.M., Frost M.C., Meyerhoff M.E. Nitric oxide‐releasing hydrophobic polymers: preparation, characterization, and potential biomedical applications. Free Radic Biol Med. 2004;37:926–936. doi: 10.1016/j.freeradbiomed.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Rodionov D.A., Dubchak I.L., Arkin A.P., Alm E.J., Gelfand M.S. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:415–431. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R.P., Fouhy Y., Lucey J.F., Dow J.M. Cyclic di‐GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Gomelsky M., Galperin M.Y. C‐di‐GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Schachter B. Slimy business – the biotechnology of biofilms. Nat Biotechnol. 2003;21:361–365. doi: 10.1038/nbt0403-361. [DOI] [PubMed] [Google Scholar]
- Schlag S., Nerz C., Birkenstock T.A., Altenberend F., Gotz F. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol. 2007;189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A.H., McPherson M.E., Hunt J.F., Johnson M., Stamler J.S., Gaston B. Acute effects of aerosolized S‐nitrosoglutathione in cystic fibrosis. Am J Respir Crit Care Med. 2002;165:922–926. doi: 10.1164/ajrccm.165.7.2105032. [DOI] [PubMed] [Google Scholar]
- Tao Y.P., Misko T.P., Howlett A.C., Klein C. Nitric oxide, an endogenous regulator of Dictyostelium discoideum differentiation. Development. 1997;124:3587–3595. doi: 10.1242/dev.124.18.3587. [DOI] [PubMed] [Google Scholar]
- Valle J., Da Re S., Henry N., Fontaine T., Balestrino D., Latour‐Lambert P., Ghigo J.M. Broad‐spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc Natl Acad Sci USA. 2006;103:12558–12563. doi: 10.1073/pnas.0605399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alst N.E., Picardo K.F., Iglewski B.H., Haidaris C.G. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun. 2007;75:3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.G., Xian M., Tang X., Wu X., Wen Z., Cai T., Janczuk A.J. Nitric oxide donors: chemical activities and biological applications. Chem Rev. 2002;102:1091–1134. doi: 10.1021/cr000040l. [DOI] [PubMed] [Google Scholar]
- Wilken M., Huchzermeyer B. Suppression of mycelia formation by NO produced endogenously in Candida tropicalis. Eur J Cell Biol. 1999;78:209–213. doi: 10.1016/S0171-9335(99)80100-9. [DOI] [PubMed] [Google Scholar]
- Yildiz F.H., Schoolnik G.K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafiriou O.C., McFarland M., Bromund R.H. Nitric oxide in seawater. Science. 1980;207:637–639. doi: 10.1126/science.207.4431.637. [DOI] [PubMed] [Google Scholar]
- Zumft W.G. The biological role of nitric oxide in bacteria. Arch Microbiol. 1993;160:253–264. doi: 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]


