Summary
The potential for using marine microbes for biodiscovery is severely limited by the lack of laboratory cultures. It is a long‐standing observation that standard microbiological techniques only isolate a very small proportion of the wide diversity of microbes that are known in natural environments from DNA sequences. A number of explanations are reviewed. The process of establishing laboratory cultures may destroy any cell‐to‐cell communication that occurs between organisms in the natural environment and that are vital for growth. Bacteria probably grow as consortia in the sea and reliance on other bacteria for essential nutrients and substrates is not possible with standard microbiological approaches. Such interactions should be considered when designing programmes for the isolation of marine microbes. The benefits of novel technologies for manipulating cells are reviewed, including single cell encapsulation in gel micro‐droplets. Although novel technologies offer benefits for bringing previously uncultured microbes into laboratory culture, many useful bacteria can still be isolated using variations of plating techniques. Results are summarized for a study to culture bacteria from a long‐term observatory station in the English Channel. Bacterial biodiversity in this assemblage has recently been characterized using high‐throughput sequencing techniques. Although Alphaproteobacteria dominated the natural bacterial assemblage throughout the year, Gammaproteobacteria were the most frequent group isolated by plating techniques. The use of different gelling agents and the addition of ammonium to seawater‐based agar did lead to the isolation of a higher proportion of Alphaproteobacteria. Variation in medium composition was also able to increase the recovery of other groups of particular interest for biodiscovery, such as Actinobacteria.
The need for pure cultures of environmentally relevant marine bacteria and archaea
Marine microbes offer great opportunities for biodiscovery (Bull et al., 2000; Glöckner and Joint, 2010), yet that potential has yet to be realized. Despite a huge microbial diversity, there is a lack of laboratory cultures of the microbes that are most abundant in the environment that severely limits development of biodiscovery research. It has been known for more than 30 years that many more bacteria are present in the surface ocean than can be cultured by the traditional microbiological approach of plating a sample onto selective media (Hobbie et al., 1977). For microbial ecologists, the priority has been to develop methods to characterize the large number of bacteria that were revealed by epifluorescence microscopy of, for example, a DAPI‐stained sample. The greatest progress occurred following the introduction of molecular biology techniques into marine microbial ecology, and these methods have fundamentally changed our understanding of microbes in the natural environment. As a consequence of developments leading to polymerase chain reaction (PCR) and novel sequencing capabilities, large and rapidly expanding databases exist containing 16S sequences from all marine provinces. We now have more precise knowledge of the groups of bacteria present in seawater and, despite the numerous biases of molecular methods, we also have a better estimation of relative abundance. Indeed, one species of marine bacteria (Candidatus‘Pelagibacter ubique’, or SAR11) is considered to be the most abundant organism on the planet. It has been suggested that it accounts for 25% of the bacterial 16S rRNA genes recovered and it has been found in almost every seawater sample surveyed. Morris and colleagues (2002) suggested that, on average, 35% of the cells in the ocean surface layer are SAR11, and in some samples they may reach densities of 450 000 cells ml−1.
SAR11 illustrates the problem that has faced microbial ecologists for decades – that the most abundant microbes in the seas are not those that can be readily cultured by standard microbiological procedures. Indeed, the organisms that are abundant in seawater are often not related to any microbes that are currently in culture and there are no representative cultures of many of the widespread and abundant Bacteria and Archaea in the sea. Microbial diversity is now recognized to be huge. Achtman and Wagner (2008) pointed out that there are only 7031 microbial species whose taxonomy has been validated and that have been validly described. Classical taxonomic approaches have failed to give a full and accurate picture of the microbial diversity in the ocean. Many new candidate divisions are based solely on 16S rRNA gene sequences, with an increasing gap between the phyla that are known only from molecular sequences and those that have cultured representatives (Curtis et al., 2002). Up until 1987, all bacterial divisions that were then recognized had cultured representatives. Twenty years later, more than 100 bacterial divisions have been proposed but only 30 possess a cultivated representative (Achtman and Wagner, 2008).
There is little doubt that the establishment of pure cultures of representatives of all bacterial divisions is one of the major challenges of modern microbiology. It is also an essential prerequisite for the development of marine biodiscovery. While it is possible to access the genetic information in uncultured organisms through genomics (Glöckner and Joint, 2010; Heidelberg et al., 2010), it is clear that the potential of any organism can best be achieved by having that particular organism available for experimentation in the laboratory.
Even in this age of high‐throughput DNA sequencing, cultures are still essential. They provide almost the only way to discover the physiology of microbes – to establish which organic substrates are used, to determine what secondary metabolites might be released, or biotransformations might be possible. Indeed, a culture is required for a full taxonomic characterization – and to give a name to an organism. So cultures remain an essential requirement, not only for biodiscovery, but also for marine microbial ecologists if we are to understand the role of microbes in the Earth System.
Barriers to isolation of cultures
The low culturability of marine microbes is well known and has been particularly severe in the marine environment. For a long time, this was referred to as the ‘great plate count anomaly’ (Staley and Konopka, 1985) because of the several orders of magnitude difference between the number of colonies that developed on laboratory medium and total number of bacteria that could be counted by epifluorescence microscopy of DAPI‐stained samples. Only a small fraction of naturally occurring microbial assemblages were cultured on conventional selective media (Skinner et al., 1952; Amann et al., 1995) and standard plating technique recovered a very small proportion, 0.001–1% of the total assemblage (Kogure et al., 1979; Staley and Konopka, 1985; Amann et al., 1995).
It is nevertheless relevant to ask if those microbes that have been readily cultivated could be considered to be a representative subset of the whole microbial biodiversity. If so, they could be construed to reflect the functional and phylogenetic diversity present in every natural habitat and there would be less concern that more cultures are not available. However, this does not appear to be the case. What is clear (and this is the experience of many laboratories) is that those bacteria that are readily cultured are not those that are abundant in 16S sequence databases for marine waters. A later section of this article will summarize attempts to bring bacteria into culture from the English Channel. In general, those cultivated tended to be representatives of the Gammaproteobacteria, whereas Alphaproteobacteria are most abundant in the natural environment, as revealed by 16S clone libraries and by the latest methodologies involving 16S tag pyrosequencing (Gilbert et al., 2009). It is clear that in order to better understand the microbes that are present in natural assemblages, as well as to provide access to novel strains that might have biotechnological potential, it is essential that more bacteria from a wider phylogenetic range are brought into culture.
Why has it been so difficult to culture the most abundant bacteria? There are many possible reasons. (i) Laboratory culture may destroy the interactions that occur between organisms in the natural environment: the fastest growing species may overwhelm those that divide only very slowly, thus leading to an imbalance of cell‐to‐cell communications: or inhibitory compounds may be produced that result in the inactivation of the cells by other microbes in the immediate vicinity. (ii) Marine bacteria may be unable to grow on the substrate or combination of substrates provided: because so few marine bacteria are in culture, we have little knowledge of organic substrates or concentrations that may be used in the sea. (iii) Virus infection may prevent growth in culture; this may be either infection with phage or the change to the lytic cycle of temperate phages when nutrients are supplied to starved bacterial cells. (iv) The (relatively) high concentrations of substrate required for detectable growth in the laboratory may of themselves be toxic, particularly for marine bacteria that have evolved under oligotrophic conditions. (v) Common laboratory practices tend to neglect the first round of cultures in liquid media that apparently display no visible growth, due to poor cell density detection methods.
Approaches to solving the challenge of cultivation
It is worth considering the fundamental principles that microbiologists use in trying to establish cultures. The aim is to separate a single viable cell from other bacteria that are present in an assemblage and to maintain that physical separation while the cell divides to form a culture. This separation is traditionally done on agar plates or by dilution to extinction in liquid cultures. A major drawback of this approach is that rapidly growing bacteria can outgrow slow‐growing bacteria on an agar plate or in liquid culture and that it is difficult to retrieve slow‐growing organisms. But it must also be recognized that the conditions that bacteria experience in these cultures are a long way removed from those under which pelagic bacteria are growing in the ocean. In the sea, except in some coastal environments, organic substrate concentrations are extremely low and there is competition with other bacteria in the assemblage.
Even if we know substrate requirements of an isolate of a particular species, this does not mean that all members of that species will utilize the same substrates. Indeed differences in substrate utilization can be a useful way to distinguish different clades or isolates. For example, Ivars‐Martínez and colleagues (2008) found that ability to utilize organic substrates was a useful way to distinguish different isolates of the ubiquitous bacterium Alteromonas macleodii. Using multi‐locus sequence analysis and a detailed statistical analysis of nine loci for each of 23 isolates, they showed that it was possible to identify different clades within the single species. In general, strains clustered with the depth in the water column from which the isolate originated. This study suggests that niche‐specific factors may be important in determining the physiology of the bacterial cell.
Individual bacterial species have a restricted complement of enzymes (particularly if the genome is small) and an essential substrate for one bacterial species may be a metabolic by‐product of another species. The traditional microbiological approach of selective culture does not allow for such interactions. When developing culturing strategies, there has generally been a lack of attention to the complex networks of interactions that occur in natural microbial assemblages. These may originate from mutualistic interactions, such as those derived from syntrophic relationships, or from cell‐to‐cell communications. It is now known that even the simplest unicellular organisms can interact with other organisms through cell‐to‐cell signalling, or quorum sensing (Joint et al., 2007; Williams et al., 2007). It may be that such cell‐to‐cell signalling may be required to trigger growth of other co‐occurring bacteria. None of these processes can operate in standard microbiological isolation procedures that aim to develop a culture from a single cell.
One reason why isolation of the most abundant bacteria in the sea has been so difficult is that we have an insufficient knowledge of the microbes themselves and, importantly, of the organic substrates that they use in the sea. There is circularity here. We need cultures to be able to investigate the basic metabolism and physiology that allows these microbes to grow, but we lack this fundamental information because we do not possess cultures of relevant bacteria and archaea. With better understanding of natural assemblages, we may be able to mimic in the laboratory, the conditions that apply in the ocean.
Alain and Querellou (2009) have recently reviewed the problem of culturability and suggest that lack of patience may be another important factor in the failure to grow many bacteria and archaea. They point out that many experiments have shown that lengthy periods of incubation are required and can significantly improve cultivation success, particularly for microbes that originate from oligotrophic habitats where a non‐growing or dormancy state may be the norm. Transition from a ‘non‐growing’ to a ‘growing’ state in a synthetic laboratory medium is obviously a critical event but is poorly understood. Adaptation to laboratory growth conditions may well be a very slow process. Alain and Querellou (2009) also point out that the lag phase of bacterial growth may be variable and that this variability might depend on the history of the cells – whether they are healthy, stressed or damaged cells. The difference between conditions in the sea and in vitro may require de novo synthesis of enzymes to enable growth in the synthetic medium. Patience has often been rewarded by very long‐term incubations. For example, in demonstrating methane formation from long‐chain alkanes under anaerobic conditions, Zengler and colleagues (1999) only observed gas formation in the presence of hexadecane after a 4‐month incubation of the enrichment culture. Extremely slow growth or very low cell densities are often observed following isolation of marine bacteria.
Patience was clearly required to obtain the first laboratory cultures of SAR11. Connon and Giovannoni (2002) developed the concept of applying the extinction to dilution approach by using very low‐nutrient media in microtitre dishes. This approach simplified handling and enabled a high‐throughput screening to be developed based on fluorescence microscopy (Connon and Giovannoni, 2002). The result was a significant improvement on traditional dilution to extinction methods because there was rapid and sensitive detection of growth, which led to improved culture efficiency. In particular, the method was suitable for the isolation of slow‐growing bacteria and led Rappé and colleagues (2002) to isolate the first representatives of the SAR11 clade. Although this approach was developed a number of years ago, it has not solved the basic problem that the majority of bacteria in the sea have not been isolated. As discussed above, it is likely that bacteria may not grow well in laboratory culture because growth of a single bacterial clade in pure cultures destroys the complex relationships that occur in the natural environment. Two important interactions are cell‐to‐cell communication, which can moderate the activity of a mixed population of bacteria, and the maintenance of consortia.
Quorum sensing
In the last decade, it has become clear that bacteria can communicate with each other – that is, communicate in the sense that gene expression of all members of a population can be simultaneously regulated. The process of communication has been called quorum sensing (QS) and involves the production of signalling molecules that diffuse between cells, hence affecting the whole population. QS is population density dependent and sufficient bacteria have to be present for the process to be effective. QS has been shown to be involved in an increasing number of microbial processes in diverse Gram‐negative and Gram‐positive bacteria found in soils and both the marine and freshwater environments. Activities under QS control include secondary metabolite production (Griffin et al., 2004), motility (Daniels et al., 2004), symbiosis (Wisniewski‐Dye and Downie, 2003), nodulation (Sanchez‐Contreras et al., 2007), conjugal plasmid transfer, biofilm maturation (Parsek and Greenberg, 2005) and virulence and have been described in numerous bacterial genera (Williams et al., 2007). An analysis of all bacterial genomes that have been sequenced to date indicates that between 5% and 25% of the genome is involved in QS. A number of chemically distinct families of QS signal molecules have been identified. The most intensively investigated are the N‐acylhomoserine lactone (AHL) family in Gram‐negative bacteria and the peptide autoinducers of Gram‐positive bacteria. QS contributes to environmental adaptation in a number of different ways. For example, in the genus Pseudomonas, AHLs are involved in the elaboration of virulence determinants in pathogenic species, and AHLs are thought to be important in biofilm maturation (Diggle et al., 2002).
Quorum sensing depends on the synthesis of small molecules that diffuse in and out of bacterial cells and which increase in concentration as the bacterial population density increases. At a critical threshold concentration, a target sensor kinase or response regulator is activated, resulting in gene expression. Such concerted action benefits the population by, for example, maintaining biofilm mode of growth, or providing collective defence against competitors. Joint and colleagues (2002) have also shown that QS crosses the prokaryotic–eukaryotic boundary. Bacterial assemblages are capable of interacting, through the production and detection of cell‐to‐cell signal molecules so that the individual bacteria behave in an analogous way to multicellular organisms, exhibiting very complex interactions (Joint et al., 2007). Although great progress in the study of QS has been made with laboratory cultures, we do not really understand interactions between different, individual bacterial species in the natural environment. It remains an untested hypothesis that QS could play a role in the growth of individual bacterial clades and may influence the ability to grow in laboratory culture.
Practically, however, it is not just a question of adding signal molecules and hoping that bacteria will miraculously grow. Many factors are involved. There is rapid turnover of signal molecules. For example, Joint and colleagues (2002) found that the signalling capacity of AHLs was rapidly lost in seawater because the homoserine ring is unstable at seawater pH. More significantly, Tait and colleagues (2009) have recently shown that some bacterial species will selectively destroy signal molecules produced by other species. So it may not be easy to find the right conditions for QS to influence the isolation of novel clades. Perhaps co‐culture with a well‐established AHL‐producing bacterium might indicate which novel bacteria might be brought into culture as a result of optimizing the QS response. Clearly much research is required but, given the importance of QS to a wide range of bacteria, more consideration should be given to exploiting the considerable knowledge on QS and applying that knowledge to the isolation of environmentally relevant bacteria.
Consortia
Another mechanism for interaction that may be crucial to successful isolation of marine bacteria involves metabolic consortia. In a microbial consortium, the metabolism of one species results in the production of a compound, or series of compounds, that can be metabolized by other species. One of the first demonstrations of microbial consortia was by Slater and Bull (1982) who used continuous culture to isolate a bacterial assemblage from soil that was capable of breaking down the herbicide 2‐chloroproprionamide (2CPA). The interesting observation was that different experiments always resulted in the isolation of the same six species of bacteria. The herbicide was broken down as a result of co‐metabolism by four of the bacteria in the consortium. The other two species appeared to play no role in the catabolism of the herbicide and they existed on the by‐products of metabolism of the core set of four species. A similar investigation was performed more recently by Bell and colleagues (2005) who measured the overall respiration of mixed cultures composed of an increasing number of individual bacterial isolates. The more complex the bacterial assemblage was in terms of species richness, the higher was the respiration of the bacterial community.
These experiments demonstrate that bacteria are capable of concerted action – apparently driven by the different metabolic capabilities of different species. Clearly, these types of processes must operate in the natural environment and it must be common for one bacterium to depend on other bacteria for essential metabolites. The traditional microbiological approach of aiming to isolate a single bacterial species by definition destroys consortia. By specifically targeting the isolation of consortia, rather than single species, it may be possible to bring many more environmentally relevant bacteria into laboratory culture.
One appropriate approach that has been used to isolate consortia has involved incubation in diffusion chambers. This separates the bacterial assemblage of interest from a source of nutrient, using a semi‐permeable membrane (Pörtner and Märkl, 1998; Kaeberlein et al., 2002). The growing culture is maintained within a chamber, nutrients diffuse through to the growing culture and, just as important, inhibitory substances that are the end‐products of metabolism also diffuse away. The result can be high densities of bacterial cells in the diffusion culture. Different types of membrane‐based systems have been developed to grow microbial communities, either directly in the field in natural habitats or in laboratory systems that simulate a microbe's natural environment (e.g. Kaeberlein et al., 2002; Plugge and Stams, 2002; Ferrari et al., 2005; 2008; Bollmann et al., 2007). These cultivation systems have had some success in isolating previously uncultured bacteria from soils, the sea and activated sludge. It has also been suggested that growth of environmental isolates in diffusion chambers might aid subsequent isolation onto classical solid media. Bollmann and colleagues (2007) found that there was adaptation within the culture that allowed the cells to grow on agar plates; alternatively, there was selection for cells that could more readily grow in high cell densities. Clearly the approach of diffusion culture could also be beneficial for the development of metabolic consortia, and has the advantage that different combinations of substrates could be added to the media at different times in the growth phase of the consortium.
Given this experience and the developing knowledge on QS and metabolic consortia, perhaps we should not be surprised that traditional microbiological methods have not been effective in isolating environmentally relevant bacteria and archaea. Most traditional culture methods rely on techniques that in the best case disrupt cell‐to‐cell communication for a short time. All cells dependent on signal exchanges with other cells of the same or different species will be unable to divide. New approaches are required to solve the fundamental problem of separating individual bacteria from the other cells in the assemblage, of manipulating and maintaining them under benign conditions for relatively long periods while cell growth occurs.
Microencapsulation – a novel approach to culture isolation
Medical biochemists and immunologists are also interested in the separation of cell lines (cancer cells, T cells, stem cells) and they have developed a wide range of techniques that should be of value to microbial ecologists. Zengler and colleagues (2002; 2005) were among the first to utilize one of these techniques. It involved micro‐droplet encapsulation in an agarose matrix to isolate and analyse cell types independent of their characteristics and activities. The application of this method in microbial ecology resulted in a very promising and novel approach to the isolation of marine bacteria. In essence, Zengler and colleagues prepared gel micro‐droplets by dispersing agarose, mixed with water from the natural environment that contained bacteria, into a non‐aqueous phase (oil) to form an emulsion. By rapidly cooling the emulsion, the molten agarose solidified and, due to the consistent stirring, formed micro‐droplets, a proportion of which contained single bacterial cells. The advantage of the approach is that the micro‐droplets are physically distinct and, because they are much larger than bacterial cells, they can be manipulated, e.g. centrifuged, pipetted, incubated and sorted by high‐speed cell sorters. Zengler and colleagues were able to adjust the ratio of number of bacteria in their initial sample to the number of agarose drops formed so that 5–10% of the micro‐droplets contained a single cell.
Encapsulation into agarose fulfils two functions. It physically separates an individual bacterial cell from other cells. It also provides the matrix for development of a microcolony because the agarose is porous and nutrients can diffuse into the growing colony and waste metabolites can diffuse out. Moreover, the physical constraints within the gel matrix do not prevent bacteria from replication and growth. The first use of this technique in the area of environmental microbiology resulted in the isolation of a number of bacteria belonging to 16S rRNA gene clades which contain no previously cultivated representatives (Zengler et al., 2002). Moreover, these isolates were not detected within the 16S rRNA gene clone library produced from the same water sample (167 clones of the clone library were screened); so encapsulation was conducive to the growth of fastidious bacteria. The great potential and sensitivity of the method is that it allows the detection and isolation not only of the most abundant, but also very scarce bacteria. This is entirely due to the physical separation and containment within the gel micro‐droplets, which permits the simultaneous and relatively non‐competitive growth of both slow‐ and fast‐growing microorganisms in media, thereby preventing overgrowth by the fast‐growing microorganisms – the ‘microbial weeds’ (Eilers et al., 2000).
A further distinct advantage of encapsulation is that it allows the incubation of a diversity of bacteria within the same medium (i.e. a consortium), while still being maintained in a clonal state (i.e. a culture) within the gel micro‐droplet. Metabolites excreted by some bacteria may stimulate, or indeed be essential for the growth of others. Or other molecules may be involved in cell‐to‐cell communication and trigger the growth of dormant bacteria (e.g. the resuscitation promotion factor on actinomycetes: Kell and Young, 2000). It is increasingly common to find examples of co‐culture leading to the growth of novel bacteria.
The reasons why this method, despite its obvious advantages, has not been routinely used in microbiological laboratories are not clear. One is the limited adoption of high‐throughput methods in laboratories used to traditional techniques. A more important limitation is the second stage in the isolation process when positive clonal cultures are separated from micro‐droplets and distributed into microtitre plates for secondary culture. During this step, cultures are static and interspecies cell‐to‐cell communication is abolished. In order to improve the recovery of previously uncultivated microbial species, it seems necessary to investigate innovative methods to circumvent both circularities mentioned: (i) lack of knowledge impairs the ability to culture but cultures are required to increase our knowledge and (ii) isolation is required to get isolates (trivial) but is often detrimental to cultivability. The coupling of community cultures with secondary clonal cultures in microbioreactors has been suggested as a possible approach to this problem (Alain and Querellou, 2009).
Expected contributions to biodiscovery by novel marine bacteria
Pharmaceutical companies have invested heavily in combinatorial chemistry as the route to the development of new drugs. However, this approach has not delivered the anticipated new drugs, with the result that many large pharmaceutical companies now brought many fewer compounds to market, or are in the pipeline, than anticipated. There are signs of a return to research into natural products, which has been so successful in the past (Bull and Stach, 2007). Natural products are the most important anticancer and anti‐infective agents, and many have reached the market without chemical modification. It is likely that Nature, and natural selection, which has operated for 3.7 billion years in the Earth, has optimized bioactive compounds through that long time period of evolution. There are few generalizations that support the case for biodiscovery research using marine microbes. It appears that approaches that rely on the production of very large numbers of compounds (i.e. combinatorial chemistry) are unlikely to deliver new drugs and other products (the approach has certainly not lived up to promise so far); the chemical diversity found in natural products is much greater than that achieved by synthetic chemistry; novel organisms may well provide compounds that can act as molecular scaffolds and which may have versatile binding properties. But the major challenges to natural product drug research are that it is labour intensive to isolate and fully characterize active compounds from extracts and that it requires a great deal of effort to produce adequate quantities of active compounds. But most of all, there may have been too much emphasis on easily cultured organisms and novel microbes are required. Before the emergence of metagenomics and functional screening of clone libraries from environmental samples, the traditional approach for biodiscovery involved the screening of culture collections. The ability to culture strains, and to characterize their metabolism and products clearly remains an important advantage. The methods are tried and tested and most of the products in use today in microbial biotechnology were obtained from isolated strains.
Isolation of bacteria from the English Channel – a case study
Although we have emphasized in this article that new approaches are required to isolate environmentally relevant bacteria, it is still possible to recover novel bacteria using modifications of the traditional microbiologist's approach. In this section, we will present the results of a study that attempted to obtain a large number of new cultures of marine bacteria that might have potential for biodiscovery, but which relied to a large extent on colony formation on plates.
The study site was an observatory off the southern coast of the UK – the Western Channel Observatory (WCO) in the English Channel ( http://www.westernchannelobservatory.org.uk/). This region has been the subject of intensive research for more than 100 years and there is a large database of environmental and biological data for the region (Southward et al., 2005). It therefore provides an ideal environment from which to attempt to isolate novel bacteria. Although data on bacteria are only available for the last decade, bacteria are abundant and assemblages are diverse. Moreover, the bacterial data can be readily placed within the environmental context, which is defined by physical measurements such as temperature and salinity, chemical measurements such as nutrient concentrations and biological measurements on phytoplankton and zooplankton. A very detailed analysis of the bacterial assemblage has recently been completed which describes seasonal changes in bacterial diversity over a 12‐month period (Gilbert et al., 2009). The study used the latest molecular approaches of 16S tag pyrosequencing developed by Sogin and colleagues (2006) to describe how bacterial populations vary throughout the year.
Gilbert and colleagues (2009) determined microbial diversity on 12 occasions using this high‐throughput sequence analysis. More than 180 000 16S tag sequences were generated, which revealed more than 7000 genera. Interestingly, one in every 25 reads could be attributed to a new genus, confirming that there is a huge uncharacterized level of microbial diversity in the sea. The results also showed that novel microbes were present and abundant in environments that can be readily sampled within a few hours from shore by research vessel. Biodiscovery does not have to mean expeditions to exotic and far away locations. Although Gilbert and colleagues (2009) did a large amount of sequencing, it was clear that even state‐of‐the‐art sequencing technology is not able to adequately describe the total diversity present at any one time point. The total data set contained 17 673 unique sequences but there was large seasonal variation. Only 93 (0.5%) of the 16S sequences were found at all time points, but these were the dominant bacteria and accounted for 50% of the total reads sequenced. As would be expected, the most abundant phylum was Proteobacteria, which comprised the majority of all sequenced reads. As in other marine provinces, the ubiquitous SAR11 clade (belonging to the Alphaproteobacteria) was again dominant and SAR11 16S sequences were found in ∼12% of the total sequenced reads. But there was large seasonality in the less abundant bacteria. About 78% of all operational taxonomic units (OTUs) were only found at one time point and 67% were only found once. This indicates that there is a large assemblage of rare bacteria which are very transient. This extensive data set on bacterial biodiversity in the coastal waters of the English Channel demonstrates that there are very many novel bacteria that have yet to be described – and offer potential for biodiscovery research.
Of the dominant bacterial groups described by Gilbert and colleagues (2009), there was considerable seasonal variability but, averaged over the whole year, Alphaproteobacteria accounted for ∼40% of the total 16S sequences, Betaproteobacteria were ∼5% and Gammaproteobacteria were ∼20%; another major group was the Bacteroidetes (∼20%). Actinobacteria are a group that have been a particular focus for biodiscovery studies because of the large number of useful products that have already been derived from this group (Bull et al., 2000). Most Actinobacteria have been isolated from soils and sediments, but in the study by Gilbert and colleagues (2009), Actinobacteria accounted for ∼5% of the 16S sequences derived from the surface water column.
The experiments to isolate novel bacteria began 5 years before the 16S tag sequence analysis of the bacterial assemblage in the English Channel, so we did not have detailed knowledge on bacterial diversity at the beginning of the study. At that time, PCR‐based and DGGE analysis suggested that Alphaproteobacteria were dominant, so this group was a particular target. Actinobacteria were also a target because of their important history in biodiscovery (Bull and Stach, 2007). The isolation approach taken was to compare low‐nutrient media with a range of commercially available microbiological media. The aim was to isolate a large number of bacteria in pure culture, to characterize them using 16S sequencing and to screen these isolates for a number of features of interest. To date, a number of unusual enzyme activities have been discovered that have commercial application (and will be protected by patents).
The media used all contained seawater which was collected from two offshore stations in the English Channel – station L4 and E1 (Southward et al., 2005) – and then filtered through 0.2‐µm‐pore‐size Nuclepore filters or Whatman GFF glass fibre filters and stored at room temperature in the dark until used for media preparation. It has been suggested that some marine bacteria do not grow well on microbiological media that are solidified with agar. Therefore different gelling agents were used in the isolation media. Unamended filtered seawater (i.e. that relied on organic substrates naturally present in seawater) was used to form solid media with the following gelling agents: 1.5% agar, 1% agar, 1% agarose and 1.0% Noble agar (which formed a sloppy agar). A number of organic substrates were also tested: casein, colanic acid, succinoglycan, laminarin and xanthan (all added at a concentration of 10 mg ml−1). Ammonium chloride and sodium nitrite were added to filtered seawater to test the effect of inorganic nitrogen sources on growth. A few commercial media were also used: R2A (a ‘low‐nutrient’ medium originally developed by Reasoner et al., 1979, to detect coliforms in potable water), and selective media for the isolation of Actinomycetes (Difco) and Vibrio species (Difco) and ‘Marine Agar’ (Difco) which is based on the medium of Zobell (1941). Finally a few plates were supplemented with extracts from marine bacteria grown in the laboratory, with the aim of testing if there were complex nutritional requirements that were likely to be met only by organic matter originating from whole cells.
Table 1 summarizes the results obtained. It should be emphasized that this was not an experiment designed to test different isolation approaches and media. The numbers of samples in each treatment reflect the needs of a biodiscovery programme, rather than an investigation of the efficacy of microbiological media. A fragment of the 16S rRNA gene of each of the isolates was sequenced and some degree of identity could be ascribed to the isolates. In some cases, 16S sequences were > 97% similar to cultured representative, and might have been identical or at least very similar to established cultures. But in the majority of cases, 16S sequences showed greater differences to cultured representatives and are probably novel isolates. On the basis of 16S sequence, each isolate has been allocated to one of the major bacterial groups (Table 1). It is clear that some media were more successful than others, particularly in isolating Alphaproteobacteria and Actinobacteria, two particular groups of interest for biodiscovery programmes. As would be expected, Gammaproteobacteria was the most common group to be isolated. Overall, of 651 isolates obtained in this study, 60% were Gammaproteobacteria, 14% were Alphaproteobacteria, 10% were Actinobacteria and 9% were Bacteroidetes. However, some of the media were more successful in isolating bacteria from groups other than Gammaproteobacteria.
Table 1.
Bacterial groups isolated by plating seawater from the English Channel on selective media.
| SWa | R2Ab | 1% agarc | Agarosed | Noble agare | Actinomycetef | Vibriog | Marine agarh | +NH4+i | +NO2‐j | Caseink | Bacterial extractl | Specific substratem | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alphaproteobacteria (93 isolates) | 5 | 5 | 15 | 24 | 6 | 5 | 2 | 24 | 3 | 4 | |||
| Betaproteobacteria (1 isolate) | 1 | ||||||||||||
| Gammaproteobacteria (391 isolates) | 41 | 18 | 19 | 55 | 14 | 96 | 8 | 20 | 24 | 20 | 12 | 13 | 51 |
| Actinobacteria (70 isolates) | 2 | 1 | 8 | 16 | 11 | 29 | 1 | 1 | |||||
| Bacteroidetes (57 isolates) | 6 | 6 | 11 | 16 | 3 | 5 | 6 | 3 | |||||
| Firmicutes (23 isolates) | 4 | 1 | 2 | 12 | 1 | 3 | |||||||
| Environmental sequences (16 isolates) | 1 | 1 | 1 | 3 | 1 | 6 | 3 |
Filtered seawater + 2.5% agar.
R2A media.
Filtered seawater + 1% agar (sloppy agar).
Filtered seawater + agarose.
Filtered seawater + Noble agar.
Actinomycete selection agar (Difco).
Vibrio selection agar (Difco).
Marine agar (Difco).
Filtered seawater + 1.5% agar + ammonium chloride (10 mmol l−1).
Filtered seawater + 1.5% agar + sodium nitrite (10 mmol l−1).
Filtered seawater + 1.5% agar + casein (10 mg ml−1).
Filtered seawater + 1.5% agar + extract from an uncharacterized bacterial culture.
Specific substrates used were colonic acid, succinoglycan, laminarin and xanthan (10 mg ml−1).
Figure 1 shows the proportion of five major groups isolated on six media. The bacterial groups are Alphaproteobacteria, Gammaproteobacteria, Actinobacteria, Bacteroidetes and Firmicutes. Sloppy (1%) agar and agarose were better than 1.5% agar for isolating both Alphaproteobacteria and Actinobacteria (Fig. 1). In the case of 1% agar, 28% of the 54 isolates were Alphaproteobacteria and 15% were Actinobacteria. Similar ratios (25% and 16%) were achieved using agarose as gelling agent. One other medium was even more successful in isolating Alphaproteobacteria; seawater solidified with 1.5% agar and supplemented with ammonium chloride (+NH4+ medium) resulted in 41% of the isolates being Alphaproteobacteria.
Figure 1.
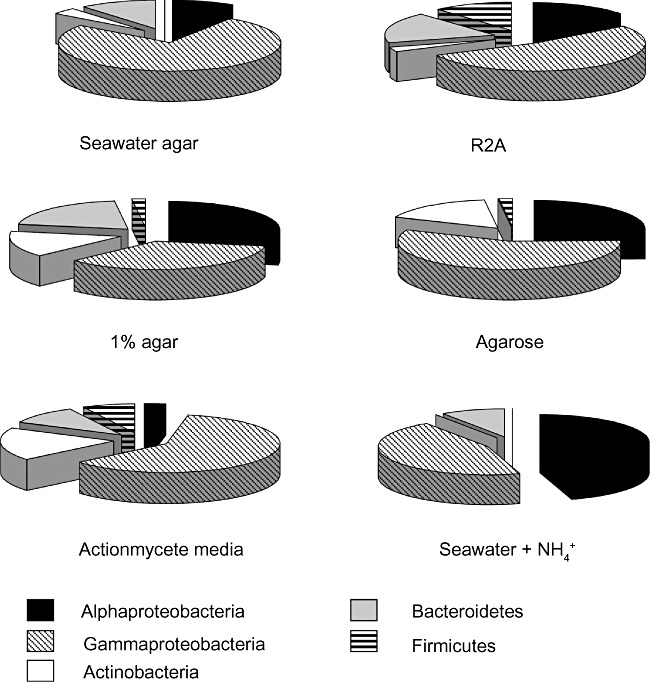
Proportion of major bacterial groups isolated on different media. Seawater agar – filtered seawater + 1.5% agar; R2A media; 1% agar – filtered seawater + 1% agar; Agarose – filtered seawater + agarose; Actinomycete media – actinomycetes selection agar (Difco); Seawater + NH4+– filtered seawater + ammonium chloride.
The commercial selective media showed the usual range of success of this method (Table 1). Of 161 cultures isolated on Actinomycete medium, 29 (18%) were Actinobacteria. The most abundant group was again the Gammaproteobacteria (60%) and this medium was also successful in selecting for members of the Bacteroidetes (10%) and Firmicutes (7%) groups. Gammaproteobacteria were 73% of the bacteria isolated on the seawater + 1.5% agar and on the commercial Marine agar medium. Therefore, neither of these media would appear to be suitable for isolating those bacteria that are most abundant in the environment – the Alphaproteobacteria.
In terms of the groups that were targeted in this study, the most successful medium for isolating Alphaproteobacteria was the +NH4+ medium, 41% of isolates being Alphaproteobacteria, followed by 1% agar (28%) and agarose (25%). For Actinobacteria, three media (the commercial Actinomycete medium, 1% agar and agarose) resulted in 15–18% of the isolates being Actinobacteria.
Conclusions – challenges to microbial cultivation
It is clear that, although the vast majority of bacteria and archaea in the ocean have yet to be brought into culture, they could provide a great deal of novel organisms for biodiscovery programmes. Novel technologies, such as encapsulation into gel micro‐droplets and development of consortia, offer considerable advantages over standard microbiological approaches and should result in the isolation and culturing of many previously uncultured microbes. It is also clear from the study reported here that the standard microbiological approach of isolating single cells on microbiological media still has much to offer. In the case of bacteria from the English Channel, the addition of ammonium chloride resulted in many more Alphaproteobacteria being isolated on filtered seawater solidified with 1.5% agar. Perhaps a large fraction of the Alphaproteobacteria is not able to utilize complex organic nitrogen compounds and require a source of ammonium ions. So, isolation on solid microbiological media is still worth exploring in any biodiscovery project.
However, some important groups will not be brought into culture using this approach. Not a single culture of the SAR11 clade was established using any of the media used in this study, even though SAR11 is by far the most dominant clade in the English Channel accounting for ∼12% of 16S sequences (Gilbert et al., 2009). Since Rappé and colleagues (2002) first established cultures, SAR11 has proved to be a very difficult bacterium to grow in the laboratory. Recently, Tripp and colleagues (2008) suggested that SAR11 requires a source of exogenous reduced sulfur compounds and does not have a complete complement of metabolic functions. They found that growth of laboratory cultures could be enhanced with methionine and dimethylsulfoniopropionate (DMSP), but the resulting cell densities still remained very low (c. 107 cells ml−1, compared with 1.1 × 106 ml−1 for culture in unmodified seawater). However, many basic questions remain about this clade (Joint, 2008) – how can this bacterium be so successful that it dominates many diverse marine provinces – yet is so difficult to grow in laboratory culture?
The SAR11 clone also illustrates a more fundamental problem and that is how to uniquely characterize a bacterial species or OTU. Lateral gene transfer is very common in natural environments. Gilbert and colleagues (2008) have recently described a fosmid clone from the English Channel that contains a 16S rRNA gene with high sequence similarity to that of the SAR11 clade. Yet more than half of the fosmid clone showed no sequence similarity to the published genome sequence of SAR11 (Candidatus‘P. ubique’ HTCC1062). So this SAR11 clone in the English Channel had acquired genes from other bacteria and presumably had a more diverse phenotype than might be expected based on 16S rRNA taxonomy. Lateral gene transfer might mean that SAR11 genomes could be very diverse in different marine provinces. If this is true of SAR11, it is probably also true of most other marine bacteria and archaea. The detailed analysis of 23 isolates of A. macleodii by Ivars‐Martínez and colleagues (2008) demonstrates considerable variation in one bacterial species and it was to identify several different clades. Another recent example of the difficulties of characterizing a ‘species’ is the suggestion by Cho and Giovannoni (2004) that there was a unique group of oligotrophic marine Gammaproteobacteria (OMG). This group of Gammaproteobacteria was introduced because isolates only grew on low‐nutrient (i.e. oligotrophic) media and they belonged to independent phylogenetic clades, based on their 16S rRNA gene sequences. However, Mühling and colleagues (2008) subsequently discovered a number of environmental 16S clones from coastal nutrient‐rich environments that grouped within these phylogenetic OMG clades, indicating that this phylogenetic group is not confined exclusively to the oligotrophic environment. Unfortunately, we do not have representative ‘OMG’ isolates available from these coastal environments and, thus, cannot compare their physiology to that of those from the oligotrophic region of the oceans.
It is important to emphasize that a 16S rRNA gene sequence alone is probably not sufficient to uniquely identify any microbe in the sea – and certainly will not be an indicator of phenotype. In terms of a biodiscovery programme, bacteria with identical 16S sequences may have very different bioactivities. Although 16S sequences have been incredibly important for the development of microbial ecology, it is much more important to design biodiscovery research in terms of a function of interest, rather than on the basis of 16S sequence or operational taxonomy.
Another important problem relates to the culture of the very large number of marine bacteria that are associated with particles – on some cases, this is the major part of the assemblage (Clarke and Joint, 1986). Simulating oganic particles or patches of concentrated substrates (dissolved and particulate organic matter: DOM, POM) and the surrounding ‘phycospheres’ (Bell and Mitchell, 1972; Azam and Ammerman, 1984; Bowen et al., 1993) and ‘detritospheres’ (Biddanda and Pomeroy, 1988) may be important for the successful culture of many attached bacteria. If autoclaved seawater is used in isolation procedures, we are likely to destroy, and certainly alter, a large fraction of the compounds (e.g. DOM, POM) that make the fluid viscous and which may serve as either source for nutrients or surface area for growth or both. Filtration will be less detrimental to organic molecules than autoclaving, but it will remove particles (e.g. POM, but also inorganic particles) that may be required by some bacteria as surfaces for growth. One approach could be to ‘clone’ these particles, i.e. isolate individual intact particles including their inhabitants. In order to allow the organisms to multiply, we also need to obtain particles of the same type, but free of microorganisms. It is not obvious how bacteria that occupy the surface might be removed, or at least killed, to provide a substratum for new ‘tenants’ without altering the surface structure and/or properties of POM and other particles. Prolonged desiccation and possibly even gamma radiation of these particles may be a first step to test. However, interspecific interactions of the ‘resident’ bacteria on a particle also influence the colonization of ‘newcomers’ (Grossart et al., 2003). If these interactions are truly syntrophic, it may only be possible to isolate some of these bacteria as mixed cultures.
Encapsulation in porous agarose micro‐drops already goes a long way towards providing surfaces, while still allowing that nutrients and DOM are reaching the cells. However, this surface may not provide an appropriate structure or material for the growth of some microbes, in particular, if nutrients or substrates are being leached out of the natural particulate surface. The effect of various solidifying agents in the isolation of bacteria has been well documented – and this study provides further evidence for this. For example, Tamaki and colleagues (2005) demonstrated that a very different set of phylogenetic groups of microbes were isolated from a freshwater sediment when gellan gum was used as gelling agent as compared with agar. Notably, Betaproteobacteria and Planctomycetes were clearly favoured by gellan gum (Tamaki et al., 2005). Although there are too few studies to draw clear conclusions, it still appears that the application of different gelling agents is a worthwhile undertaking to target a wide genetic range of microorganisms.
It is clear that the development of biodiscovery research will require innovative approaches to be developed to significantly improve culture methods. These should be based on the growing knowledge of cell‐to‐cell communication, on data produced by molecular and metagenomic approaches and on high‐throughput procedures developed by researchers into eukaryotic cells.
Acknowledgments
This research was partially supported through grants to I.J. from the EU – project Microbial Marine Communities Diversity: from Culture to Function (MIRACLE) (EVK3‐CT‐2002‐00087) – and from the Natural Environment Research Council (UK) – Grant No. NE/B505770/1. We also acknowledge contributions to the isolation of marine bacteria from Dr Karen Tait and Dr Darren Clark (Plymouth Marine Laboratory).
Acknowledgements
IJ and JQ also acknowledge funding from the European Commission to attend the Joint EC‐NSF_CIESM Workshop on Marine genomics: at the interface of Marine Microbial Ecology and Biotechnological Applications, held in October 2008 (http://ec.europa.eu/research/biotechnology/ec‐us/docs/monaco.pdf).
References
- Achtman M., Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Alain K., Querellou J. Cultivating the uncultured: limits, advances and future challenges. Extremophiles. 2009;13:583–594. doi: 10.1007/s00792-009-0261-3. [DOI] [PubMed] [Google Scholar]
- Amann R.I., Ludwig W., Schleifer K.‐H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam F., Ammerman J.W. Cycling of organic matter by bacterioplankton in the pelagic marine ecosystems: microenvironmental considerations. In: Fasham M.J.R., editor. Plenum Publishing Corp.; 1984. pp. 345–360. [Google Scholar]
- Bell W., Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull. 1972;143:265–277. [Google Scholar]
- Bell T., Newman J.A., Silverman B.W., Turner S.L., Lilley A.K. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- Biddanda B.A., Pomeroy L.R. Microbial aggregation and degradation of phytoplankton‐derived detritus in seawater. I. Microbial succession. Mar Ecol Prog Ser. 1988;42:79–89. [Google Scholar]
- Bollmann A., Lewis K., Epstein S.S. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 2007;73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen J.D., Stolzenbach K.D., Chisholm S.W. Simulating bacterial clustering around phytoplankton cells in a turbulent ocean. Limnol Oceanogr. 1993;38:36–51. [Google Scholar]
- Bull A.T., Stach J.E.M. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Bull A.T., Ward A.C., Goodfellow M. Search and discovery strategies for biodiscovery: the paradigm shift. Microbiol Mol Biol Rev. 2000;64:573–606. doi: 10.1128/mmbr.64.3.573-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.C., Giovannoni S.J. Cultivation and growth characteristics of a diverse group of oligotrophic marine gammaproteobacteria. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K.R., Joint I.R. Methodology for estimating numbers of free‐living and attached bacteria in estuarine water. Appl Environ Microbiol. 1986;51:1110–1120. doi: 10.1128/aem.51.5.1110-1120.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connon S.A., Giovannoni S.J. High‐throughput methods for culturing microorganisms in very low nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis T.P., Sloan W.T., Scannell J.W. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R., Vanderleyden J., Michiels J. Quorum sensing and swarming in bacteria. FEMS Microbiol Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Diggle S.P., Winzer K., Lazdunski A., Williams P., Cámara M. Advancing the quorum in Pseudomonas aeriginosa and the regulation of N‐acylhomoserine lactone production and virulence gene expression. J Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H., Pernthaler J., Glöckner F.O., Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari B., Binnerup S.J., Gillings M. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol. 2005;71:8714–8720. doi: 10.1128/AEM.71.12.8714-8720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari B.C., Winsley T., Gillings M., Binnerup S. Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc. 2008;3:1261–1269. doi: 10.1038/nprot.2008.102. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Mühling M., Joint I. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique’ genome. ISME J. 2008;2:790–793. doi: 10.1038/ismej.2008.49. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Field D., Swift P., Newbold L., Oliver A., Smyth T. Seasonal succession of microbial communities in the Western English Channel using 16S rRNA‐tag pyrosequencing of the V6 region. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. et al. [DOI] [PubMed] [Google Scholar]
- Glöckner F.O., Joint I. Marine microbial genomics in Europe: current status and perspectives. Microbiol Biotechnol. 2010;3:523–530. doi: 10.1111/j.1751-7915.2010.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin A.S., West S.A., Bucking A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- Grossart H.‐P., Kiørboe T., Tang K., Ploug H. Bacterial colonization of particles: growth and interactions. Appl Environ Microbiol. 2003;69:3500–3509. doi: 10.1128/AEM.69.6.3500-3509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg K.B., Gilbert J.A., Joint I. Marine genomics: at the interface of marine microbial ecology and biodiscovery. Microbiol Biotechnol. 2010;3:531–543. doi: 10.1111/j.1751-7915.2010.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J.E., Daley R.J., Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;5:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivars‐Martínez E., D'Auria G., Rodríguez‐Valera F., Sánchez‐Porro C., Ventosa A., Joint I., Mühling M. Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol Ecol. 2008;17:4161–4175. doi: 10.1111/j.1365-294x.2008.03883.x. [DOI] [PubMed] [Google Scholar]
- Joint I. Unravelling the enigma of SAR11. ISME J. 2008;2:455–456. doi: 10.1038/ismej.2008.30. [DOI] [PubMed] [Google Scholar]
- Joint I., Tait K., Callow M.E., Callow J.A., Milton D., Williams P., Cámara M. Cell‐to‐cell communication across the procaryote–eucaryote boundary. Science. 2002;298:1207. doi: 10.1126/science.1077075. [DOI] [PubMed] [Google Scholar]
- Joint I., Downie J.A., Williams P. Bacterial conversations: talking, listening and eavesdropping. An introduction. Philos Trans R Soc Lond B Biol Sci. 2007;362:1115–1117. doi: 10.1098/rstb.2007.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T., Lewis K., Epstein S.S. Isolating, uncultivable' microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Kell D.B., Young M. Bacterial dormancy and culturability: the role of autocrine growth factors – commentary. Curr Opin Microbiol. 2000;3:238–243. doi: 10.1016/s1369-5274(00)00082-5. [DOI] [PubMed] [Google Scholar]
- Kogure K., Simidu U., Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Morris R.M., Rappé M.S., Connon S.A., Vergin K.L., Siebold W.A., Carlson C.A., Giovannoni S.J. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Mühling M., Woolven‐Allen J.A., Murrell J.C., Joint I. Improved group‐specific PCR primers for DGGE analysis of the genetic diversity of complex microbial communities. ISME J. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- Parsek M.R., Greenberg E.P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Plugge C.M., Stams A.J.M. Enrichment of thermophilic syntrophic anaerobic glutamate‐degrading consortia using a dialysis membrane reactor. Microb Ecol. 2002;43:379–387. doi: 10.1007/s00248-001-0047-3. [DOI] [PubMed] [Google Scholar]
- Pörtner R., Märkl H. Dialysis cultures. Appl Microbiol Biotechnol. 1998;50:403–414. doi: 10.1007/s002530051312. [DOI] [PubMed] [Google Scholar]
- Rappé M.S., Conan S.A., Vergin K.L., Giovannoni S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–631. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Reasoner D.J., Blannon J.C., Geldreich E.E. Rapid seven‐hour fecal coliform test. Appl Environ Microbiol. 1979;38:229–236. doi: 10.1128/aem.38.2.229-236.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Contreras M., Bauer W.D., Gao M., Robinson J.B., Downie J.A. Quorum‐sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos Trans R Soc Lond B Biol Sci. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner F.A., Jones P.C.T., Mollison J.E. A comparison of a direct‐ and a plate‐counting technique for the quantitative estimation of soil micro‐organisms. J Gen Microbiol. 1952;6:261–271. doi: 10.1099/00221287-6-3-4-261. [DOI] [PubMed] [Google Scholar]
- Slater J.H., Bull A.T. Environmental microbiology: biodegradation. Philos Trans R Soc Lond B Biol Sci. 1982;297:575–597. [Google Scholar]
- Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southward A.J., Langmead O., Hardman‐Mountford N.J., Aiken J., Boalch G.T., Dando P.R. Long‐term oceanographic and ecological research in the western English Channel. Adv Mar Biol. 2005;47:1–105. doi: 10.1016/S0065-2881(04)47001-1. et al. [DOI] [PubMed] [Google Scholar]
- Staley J.T., Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Tait K., Williams P., Cámara M., Williamson H., Gan Chan K., Joint I. Quorum sensing signal molecule synthesis and turnover in polymicrobial marine biofilms modulates communication with algal zoospores. Environ Microbiol. 2009;11:1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x. [DOI] [PubMed] [Google Scholar]
- Tamaki H., Sekiguchi Y., Hanada S., Nakamura K., Nomura N., Matsumura M., Kamagata Y. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation‐based techniques. Appl Environ Microbiol. 2005;71:2162–2169. doi: 10.1128/AEM.71.4.2162-2169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp H.J., Kitner J.B., Schwalbach M.S., Dacey J.W.H., Wilhelm L.J., Giovannoni S.J. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- Williams P., Winzer K., Chan W.C., Cámara M. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski‐Dye F., Downie J.A. Quorum sensing in Rhizobium. Antonie Van Leeuwenhoek. 2003;81:397–407. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- Zengler K., Richnow H.H., Rossello‐Mora R., Michaelis W., Widdel F. Methane formation from long‐chain alkanes by anaerobic microorganisms. Nature. 1999;401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
- Zengler K., Toledo G., Rappé M., Elkins J., Mathur E.J., Short J.M., Keller M. Cultivating the uncultured. Proc Natl Acad Sci USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K., Walcher M., Clark G., Haller I., Toledo G., Holland T. High‐throughput cultivation of microorganisms using microcapsules. Methods Enzymol. 2005;397:124–130. doi: 10.1016/S0076-6879(05)97007-9. et al. [DOI] [PubMed] [Google Scholar]
- Zobell C. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res. 1941;4:42–75. [Google Scholar]


