Summary
Marine Crenarchaeota, ubiquitous and abundant organisms in the oceans worldwide, remain metabolically uncharacterized, largely due to their low cultivability. Identification of candidate genes for bicarbonate fixation pathway in the Cenarchaeum symbiosum A was an initial step in understanding the physiology and ecology of marine Crenarchaeota. Recent cultivation and genome sequencing of obligate chemoautotrophic Nitrosopumilus maritimus SCM1 were a major breakthrough towards understanding of their functioning and provide a valuable model for experimental validation of genomic data. Here we present the identification of multiple key components of 3‐hydroxipropionate/4‐hydroxybutyrate cycle, the fifth pathway in carbon fixation, found in data sets of environmental sequences representing uncultivated superficial and bathypelagic Crenarchaeota from Sargasso sea (GOS data set) and KM3 (Mediterranean Sea) and ALOHA (Atlantic ocean) stations. These organisms are likely to use acetyl‐CoA/propionyl‐CoA carboxylase(s) as CO2‐fixing enzyme(s) to form succinyl‐CoA, from which one molecule of acetyl‐CoA is regenerated via 4‐hydroxybutyrate cleavage and another acetyl‐CoA to be the pathway product. The genetic distinctiveness and matching sympatric abundance imply that marine crenarchaeal genotypes from the three different geographic sites share similar ecophysiological properties, and therefore may represent fundamental units of marine ecosystem functioning. To couple results of sequence comparison with the dark ocean primary production, dissolved inorganic carbon fixation rates were measured at KM3 Station (3000 m depth, Eastern Mediterranean Sea), i.e. at the same site and depth used for metagenomic library construction.
Introduction
The bathypelagic and abyssal zones of the world oceans are important and less understood microbial ecosystems on Earth. Most marine microbiological studies so far have been devoted to the photic zone (0–200 m), which is generally considered as the ecosystem where the most primary production occurs. Participation of microbial communities of deeper water bodies in carbon cycling was referred until recently exclusively to degradation of complex organic molecules, and thus the heterotrophy was coined as the predominant lifestyle for deep‐sea prokaryotes.
Although archaea were previously characterized as extremophiles, mesophilic Crenarchaeota are now recognized to be an ubiquitous component of marine plankton (DeLong, 2006). Moreover, this group of archaea is among the most widely distributed and abundant groups of microorganisms on the planet, with the marine ‘Group 1’ clade alone comprising over 20% of picoplankton in the world ocean (Herndl et al., 2005; Wuchter et al., 2006). Considering the vast dimensions of the oceans, occupying more than two‐thirds of the Planet's surface and reaching an average depth of 3800 m, marine Crenarchaeota obviously play a fundamental role in global organic and inorganic matter biochemical cycling. These organisms are estimated to number at least 1028 cells in total, if not more (Whitman et al., 1998; Nicol and Schleper, 2006). But, because of our inability to cultivate them, our understanding on their physiology and biogeochemical function remained mostly speculative.
Being elusive to isolation and growth in pure cultures, deep‐sea archaea are thus poorly understood in terms of their metabolic features and only two complete genomes from mesophilic Marine Group 1.1a Crenarchaeota are available to date, that of Nitrosopumilus maritimus SCM1 (Könneke et al., 2005), and that of the uncultivated sponge symbiont Cenarchaeum symbiosum A (Hallam et al., 2006a). The genomic analyses suggested that deep‐sea ammonium‐oxidizing archaea may also be ecologically important for carbon cycling, as they appear to be autotrophic (Herndl et al., 2005; Hallam et al., 2006a,b; Ingalls et al., 2006; Yakimov et al., 2007; 2009). Indeed, both available genomes possess genes for the newly described 3‐hydroxypropionate/4‐hydroxybutyrate CO2‐fixation pathway (Hallam et al., 2006a,b; Berg et al., 2007). This autotrophic cycle was originally discovered in the phototrophic bacterium Chloroflexus aurantiacus and involves the carboxylation of acetyl‐CoA to malonyl‐CoA by the biotin‐dependent acetyl‐CoA carboxylase. Because Archaea contain only trace amounts of fatty acids, if any, in their lipids, acetyl‐CoA carboxylase cannot serve as the key enzyme of fatty acid synthesis but is rather involved into the CO2‐fixation pathway. The Global Ocean Sampling (GOS) database contains a high proportion of acetyl‐CoA carboxylase large subunit genes (accA), comparable with that of RuBisCO large subunit, suggesting that this archaeal‐specific pathway is widely distributed in oceans (Berg et al., 2007; Rusch et al., 2007; Thauer, 2007). Compared with recently developed molecular tools to monitor the occurrence and diversity the archaeal amoA gene of the ocean (Francis et al., 2005; Agoguéet al., 2008; Mosier and Francis, 2008; Schleper, 2008; De Corte et al., 2009), environmental monitoring of genetic determinants of marine crenarchaeal autotrophy is still to be developed. As far as acetyl‐CoA carboxylase gene was proposed to be an indicative signature for CO2 fixation capacity in Crenarchaeota (Berg et al., 2007; 2010), the first attempt to couple the deep‐sea ‘dark’ ocean primary production with distribution of archaeal accA‐like was performed in the bathypelagic zone of Tyrrhenian Sea (Central Mediterranean) (Yakimov et al., 2009). The found number of accA‐like gene copies covered 75% of CARD‐fluorescent in situ hybridization (FISH) counted crenarchaeal cells, thus suggesting that absolute dominance of the deep‐sea planktonic Crenarchaeota possess the genetic capacities to autotrophy.
Here we report on the comparative analyses of N. maritimus SCM1 and C. symbiosum A genomic DNA sequences with environmental genome sequences from Global Ocean Survey scaffolds and other publicly available metagenomic data sets of bathypelagic microbial communities from the depth of 3000 m (Ionian Sea, Mediterranean) (Martín‐Cuadrado et al., 2007; Martin‐Cuadrado et al., 2008) and 4000 m (North Pacific Subtropical Gyre) (Konstantinidis et al., 2009), to determine the presence and distribution of highly conserved functional genes with the potential to mediate carbon assimilation in marine planktonic Crenarchaeota.
Sequence analysis was supported with the dark ocean primary production data. Dissolved inorganic carbon (DIC) fixation rates were measured at the same site (and depth) used for metagenomic library construction, namely at KM3 Station (3000 m in depth, Eastern Mediterranean) (Martín‐Cuadrado et al., 2007; Martin‐Cuadrado et al., 2008).
Results and discussion
KM3 Station description
The Ionian Sea is identified as the region from the Sicily Strait to the Cretan passage. In this transition basin, different water masses (e.g. Modified Atlantic Water, MAW; Levantine Intermediate Water, LIW; Eastern Mediterranean Deep Water, EMDW) undergo transformations along their pathways between the Eastern and Western Mediterranean (Malanotte‐Rizzoli et al., 1997). Water temperature at the depth of 3010 m was 13.92°C and salinity 38.76 PSU. Nitrate and phosphate concentrations exhibited the common depth‐increasing trend reaching at the depth of 3010 m the concentration of 3.75 ± 0.54 and 0.13 ± 0.02 µmol l−1 respectively. Concentration of nitrite remained rather constant with depth and did not excess 0.04 ± 0.01 µmol l−1. Such a high N:P molar ratio (29.15) observed at the bottom layer is typical for younger EMDW of Aegean origin (Kress et al., 2003). Oxygen concentration profile was also almost constant (130 ± 4.7 µmol l−1) with depth and no pronounced oxygen minimum layer was found.
Bathypelagic archaeal community composition at KM3 Station
At the depth of 3010 m total prokaryote number was estimated as 9.7 ± 0.15 × 104 DAPI‐stained cells ml−1. The mean contribution of Archaea to total prokaryotic abundance (i.e. FISH‐stained cells) was 45 ± 7%. The archaeal diversity was monitored by PCR amplification of 16S rRNA gene sequences by using general archaeal primers A20F and A958R (Hallam et al., 2003). A total of 96 clones were sequenced (700 bp in average) and assigned to four different patterns (97% cut‐off). As it typically reported for deep‐sea microbial communities, the bathypelagic zone of KM3 Station was significantly enriched by Crenarchaeota belonging to Marine Group 1 (91% of all clones sequenced) while the contribution of members of recently identified pSl‐12 clade or group 1A (DeLong et al., 2006; Mincer et al., 2007) was about 15 times lower and did not exceed 6%. The remaining 3% of clones belonged to the Euryarchaeota of the Marine Group II (Fig. 1). A similar trend in archaeal community composition was previously described for deep‐sea compartment of Pacific and Atlantic Oceans (Mincer et al., 2007; De Corte et al., 2009). Remarkably, the majority of the clones were matching with the 16S rRNA gene sequences of KM3 metagenomic library constructed from this site 5 years before our sampling (Martín‐Cuadrado et al., 2007; Martin‐Cuadrado et al., 2008). This finding suggests that these water masses are inhabited by a very stable and well‐established archaeal community.
Figure 1.
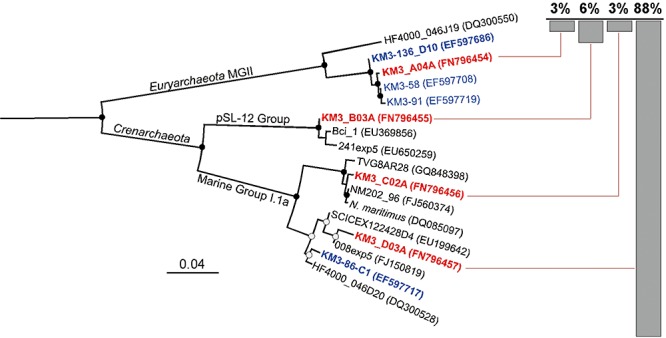
Phylogenetic tree of archaeal 16S rRNA genes amplified from the pool of environmental DNA recovered from 3010 m at KM3 Station. The tree was constructed by Neighbour‐Joining method and Jukes–Cantor distance matrix using MacVector 11.0.2 and a total of 550 non‐ambiguously aligned positions. Non‐parametric bootstrapping was performed upon 1000 replicates. Bootstrap values > 70 and ≥ 80 are shown as empty and filled circles respectively. The tree was rooted with Alteromonas genoviensis strain I96 16S ribosomal RNA gene (FJ040187). KM3 water column sequences obtained in this work and previously (Martín‐Cuadrado et al., 2007) are shown in red and blue respectively. Additionally, the 16S rDNA sequences found in KM3 fosmid library are shown in bold.
Estimation of the dark ocean primary production rates and quantitative PCR analysis of crenarchaeal accA‐like gene distribution
Several independent studies have estimated the global dark ocean DIC fixation rate of 4–8 × 1011 kg C year−1 (Herndl et al., 2005). This newly fixed carbon represents a substrate for a largely unknown deep‐sea food‐web including microbial and metazoan members. Direct link between the deep‐sea CO2 fixation and the identity of the pivotal organisms or assemblages involved in this process has not been established so far. Although the compound‐specific stable isotope analysis on crenarchaeal membrane lipids indicated that these lipids might come from chemoautotrophic, inorganic carbon‐fixing organisms (Pearson et al., 2001; Wuchter et al., 2003; Ingalls et al., 2006).
Recently, the ability of archaea to take up inorganic carbon was used as a proxy to estimate archaeal cell production and to compare this parameter with total prokaryotic production measured via leucine incorporation. The estimated archaeal production in the mesopelagic and bathypelagic North Atlantic depends on latitudinal trends and contributes between 13% and 84% to the total prokaryotic production. Thus, it was shown that planktonic archaea are actively growing in the dark ocean and may play a significant role in the oceanic carbon cycle (Herndl et al., 2005).
Using 14C‐bicarbonate addition we estimated the rates of prokaryotic primary production at KM3 Station of the water column below photic zone (from 200 m to the bottom). To convert bicarbonate incorporation into microbial biomass production, we assumed that inorganic carbon is the sole carbon source of autotrophic prokaryotes and that radioactive and natural bicarbonates are taken up by these organisms at the same rate. According to Fig. 2, prokaryotic dark primary production declined two times from the 200 m layer to the depth of 1500 m, namely from 0.41 ± 0.03 mg C m−3 day−1 to 0.20 ± 0.02 mg C m−3 day−1 and then slightly increased to 0.25–0.30 mg C m−3 day−1 near the sea bottom. Integrating these values to all aphotic water column below 200 m down to the sea bed we estimated the mean dark ocean production at KM3 Station as 810 mg C m−2 day−1. This value is almost three times higher than the rates of photosynthetic biomass production of 270 mg C m−2 day−1 calculated for this area of Eastern Mediterranean Sea in a different study (Vidussi et al., 2001).
Figure 2.
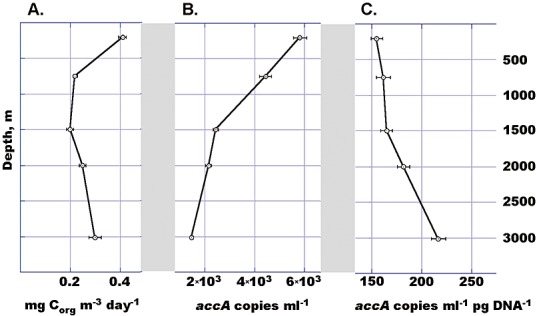
Prokaryotic dark ocean primary production rates (A); quantitative PCR‐determined gene copy numbers of crenarchaeal accA at different depths of KM3 water column given as absolute (B) and normalized against the total DNA (C). All error bars indicate standard deviation from a mean of three replicate measurements.
Six mechanisms are known today by which autotrophic organisms fix carbon (Huber et al., 2008; Berg et al., 2010). Due to sensitivity of the crucial enzymes to oxygen, only two pathways, namely 3‐hydroxypropionate/4‐hydroxybutyrate (3‐HP/4‐HB) and Calvin–Benson–Bassham cycles, likely might occur in oxygenated deep‐sea water (Thauer, 2007; Berg et al., 2010). Specially developed and previously tested primer pairs (Spiridonova et al., 2004; Yakimov et al., 2007) were used to amplify the fragments of bacterial genes encoding the large subunit of red‐ and green‐like form I RubisCO (cbbL), the key enzyme of Calvin–Benson–Bassham cycle. No amplification products were observed below the depth of 200 m, thus suggesting the absence of Calvin cycle‐operating prokaryotes in the deep‐sea water column of KM3 Station. To explore the potential link between dark autotrophic fixation of CO2 and occurrence of crenarchaeal genes encoding crucial enzymes in 3‐HP/4‐HB cycle, the abundance of accA‐like genes was monitored by quantitative PCR analysis previously described elsewhere (Yakimov et al., 2009). Although the crenarchael accA gene copy numbers collected in bathypelagic depths amounted to 30–35% of that detected in shallow water, the subsequent normalization of this gene copies to the amount of total recovered DNA produced a depth‐increasing profile of accA distribution (Fig. 2C). The pool of accA‐like gene copies found at the depth of 3010 m in KM3 Station (14 790 ± 655 copies ml−1) was comparable to the number of FISH‐counted crenarchaeal cells (25 950 ± 360 cells ml−1). Thus, taking into account that accA gene presents in single copy in two sequenced genomes of marine Crenarchaeota, the number of accA‐harbouring cells covered 54–60% of all crenarchaeal cells detected by FISH. This provides further evidence that bathypelagic Crenarchaeota are important contributors to the dark ocean primary production and could play a pivotal role in yet underestimated scale of carbon turnover in the deep‐sea ecosystems.
Metagenomic insight into carbon assimilation and central metabolic pathways in deep‐sea Crenarchaeota
It is generally acknowledged that mesophilic Crenarchaeota are chemoautotrophs using aerobic oxidation of ammonia as a mode of energy production required to sustain the fixation of inorganic carbon via 3‐hydroxypropionate cycle, with acetyl‐CoA/propionyl‐CoA carboxylase (Acc) being the key carboxylating enzyme (Könneke et al., 2005; Hallam et al., 2006b; Berg et al., 2007; Agoguéet al., 2008). The modified variant of this cycle was proposed for Metallosphaera, Sulfolobus, Archaeoglobus and Cenarchaeum species. In this system, one acetyl‐CoA and two bicarbonate molecules were reductively converted via 3‐hydroxypropionate to succinyl‐CoA with participation of methylmalonyl‐CoA mutase. This intermediate was reduced to 4‐hydroxybutyrate and converted into two acetyl‐CoA‐molecules via 4‐hydroxybutyryl‐CoA dehydratase (see Fig. S1 for details). Whether mesophilic deep‐sea Crenarchaeota use yet unknown pathway different from 3‐hydroxypropionate/4‐hydroxybutyrate CO2 fixation route remains to be shown (Berg et al., 2007). Whatever the case, the genomes of two autotrophic representatives (Nitrosopumilus and Cenarchaeum spp.) contain the genes for characteristic enzymes of this cycle, while lacking the key genes for other autotrophic pathways (Hallam et al., 2006b; Berg et al., 2007).
Acetyl(propionyl)‐CoA carboxylase
Using available public databases of deep‐sea (meta)genomes, we attempted to search for crenarchael‐specific genes potentially encoding the key enzymes of 3‐hydroxypropionate/4‐hydroxybutyrate (3‐HP/4‐HB) cycle, namely acetyl(propionyl)‐CoA carboxylase, methylmalonyl‐CoA mutase and 4‐hydroxybutyryl‐CoA dehydratase. As shown above, these enzymes were postulated to be crucial for the fixation of inorganic carbon via 3‐HP/4‐HB cycle (Berg et al., 2007). blastx analysis was conducted with N. maritimus and C. symbiosum genomic fragments containing these genes queried against > 10 kb contigs identified in the environmental database. So far, only two large‐scale metagenomic analyses of deep‐sea microbial communities were carried out at the North‐Pacific Subtropical Gyre ALOHA (4000 m) and at the Eastern Mediterranean KM3 (3000 m) stations (Martín‐Cuadrado et al., 2007; Konstantinidis et al., 2009). The crenarchaeal‐specific scaffolds available from the GOS database greater than 6.0 kb were also included into the analysis. In N. maritimus and C. symbiosum genomes, genes encoding acetyl(propyonyl)‐CoA subunits were arranged in a single operon accACB. Four ALOHA contigs (HF4000_ANIW97P9, HF4000_ANIW141M18, HF4000_APKG6D3 and HOTS_Contig54507) and two GOS scaffolds (JCVI_SCAF_1096627389063 and ACCY01042531) contained an intact acc operon with subunit organization identical to that of sequenced N. maritimus and C. symbiosum genomes. Comparison of these contigs revealed a high degree of conservation between the regions more distal to acc operon, including a large nuo operon immediately downstream predicted to encode the complete set of NADH‐ubiquinone oxidoreductase subunits (Fig. 3A). These operons are separated by the insertion of the gene ahpC coding for putative alkyl hydroperoxide reductase, enzyme responsible for direct reduction of organic hyperoxides in reduced dithiol form.
Figure 3.
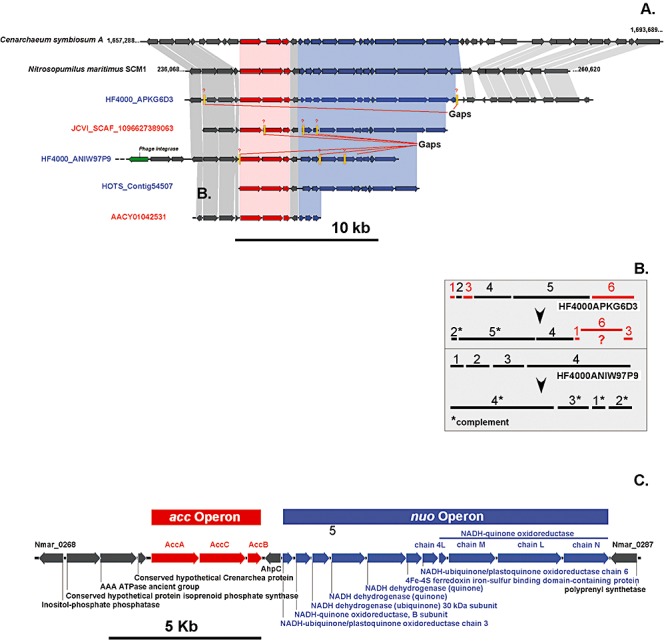
Comparison of genomic fragments containing acetyl(propionyl)‐CoA carboxylase (accA) operon in N. maritimus and C. symbiosum with five related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (4000 m WGS ALOHA database, blue). A. Genes shared in common between genomic fragments are connected by: red (acc operon), blue (nuo operon) and grey (rest of the genes) shaded boxes. B. Re‐assembling of individual reads in HF4000_ANIW97P9 and HF4000_APKG6D3 is depicted in grey insert. C. Genomic fragments shared significant conservation of gene content and order in all analysed sequences.
Upstream of the acc operon, at least four open reading frames encoding for AAA‐type ATPase, inositol phosphate phosphatase and two crenarchaeal‐specific conserved hypothetical proteins were found to be present in all analysed contigs. Thus, the genomic fragment of almost 19 kb containing acc operon was identified sharing significant conservation of gene content and order in open reading frames (Fig. 3C). Moreover, based on paired‐end analysis of individual reads assembled in HF4000_ANIW97P9 and HF4000_APKG6D3 contigs, some errors have been identified in their original presentation (Fig. 3B). Comparison of deduced amino acid sequences encoded in this genome fragment revealed that all environmental contigs were more closely related to N. maritimus than to C. symbiosum. Unfortunately, all available Mediterranean KM3 fosmid sequences were selected because they contained rRNA operons (Martin‐Cuadrado et al., 2008). Therefore acc gene sequences could not be retrieved from KM3 fosmid library. This is very likely, bearing in mind that rRNA and accA operons in N. maritimus and C. symbiosum genomes are fairly distant and separated by 656.6 and 463.0 kb respectively.
Methylmalonyl‐CoA mutase
To identify homologues of the methylmalonyl‐CoA mutase subunits predicted in N. maritimus and C. symbiosum genomes, targeted searches were conducted against ALOHA, KM3 and GOS database and at least one contig from each of them was found (Fig. 4A). Comparison of genomic context around the mmm gene cluster in C. symbiosum A and GOS scaffold AACY020561430 revealed little conservation of genes adjacent to mmm locus, whereas the mmm‐containing genomic fragment greater than 19 kb in N. maritimus was identified to share a significant homology with fosmids recovered from bathypelagic zones of both Mediterranean Sea and Atlantic ocean (Fig. 4B). However, only 195 aa C‐terminal fragment of putative methylmalonyl‐CoA mutase large subunit is present at 5′‐terminus of ALOHA fosmid HF4000_APKG7F19. Besides methylmalonyl‐CoA mutase subunits, the highly syntenic block of 15 predicted open reading frames was found to contain genes encoding putative methylmalonyl‐CoA epimerase, phosphoenolpyruvate synthase and dihydroxyacetone 3‐phosphate isomerase. First enzyme converts (2R)‐methylmalonyl‐CoA to (2S)‐methylmalonyl‐CoA (substrate for methylmalonyl‐CoA mutase), while two remaining enzymes are involved in glyceraldehyde 3‐phosphate formation. As proposed elsewhere, this triosephosphate synthesis might be coupled with 3‐HP/4‐HB pathway following the equation (Berg et al., 2007; Fig. S1):
Figure 4.
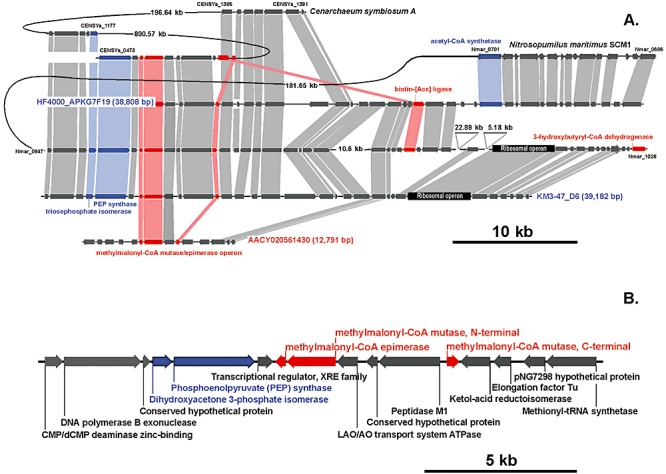
Comparison of the genomic fragments containing methylmalonyl‐CoA mutase/epimerase operon in N. maritimus and C. symbiosum with three related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (ALOHA and KM3 database, blue). A. Genes shared by genomic fragments are connected by shaded boxes: red (3‐HP/4‐HB cycle), blue (triosephosphate formation pathway) and grey (rest of the genes). B. The core genomic fragment of 19 kb shared significant conservation in gene content and order in all analysed sequences.
 |
In addition to core components of 3‐HP/4‐HB cycle, a biotin‐[Acc] ligase required for the assembly and activation of the carboxylase complex was identified in ALOHA and Mediterranean fosmids HF4000_APKG7F19 and KM3‐34‐D9 respectively. Both genes revealed high synteny with biotin ligase birA of N. maritimus SCM1 and were found to be less similar to birA of C. symbiosum A.
4‐Hydroxybutyryl‐CoA dehydratase
Genomes of N. maritimus SCM1 and C. symbiosum A contain genes for 4‐hydroxybutyryl‐CoA dehydratase, a [4Fe–4S] cluster and FAD‐containing enzyme, which catalyses the elimination of water from 4‐hydroxybutyryl‐CoA by a ketyl radical mechanism yielding crotonyl‐CoA (Martins et al., 2004). Although originally this enzyme was described in a few strict anaerobes that use it in fermentation, recently 4‐hydroxybutyryl‐CoA dehydratase has been postulated to be a pivotal enzyme in CO2‐fixing 3H‐P/4H‐B cycle in autotrophic thermophilic and, possibly, also in mesophilic Crenarchaeota (Berg et al., 2007; 2010; Huber et al., 2008;).
The gene for 4‐hydroxybutyryl‐CoA dehydratase was found in GOS metagenome and ALOHA fosmids, whereas no matches were observed in KM3 metagenome. Since the distance between this gene and rRNA operon in N. maritimus SCM1 is about 716.5 kb, it was not expected to find it among SSU rRNA‐containing fosmids from KM3. Two almost identical (95–100% nucleotide sequence identity) ALOHA fosmids HF4000_APKG7F11 and HF4000_APKG3H9 contained a stretch of open reading frames highly syntenic at the amino acid level to a 40‐kb‐long genomic fragment downstream of dehydratase gene identified in N. maritimus SCM1 (Fig. 5A). Based on this conservation, paired‐end analysis of individual reads assembled in HF4000_APKG7F11 contig revealed some assembly errors (Fig. 5B). Further analysis of the genomic context around the gene encoding 4‐hydroxybutyryl‐CoA dehydratase in C. symbiosum A and Sargasso Sea scaffold AACY020562346 revealed little conservation of genes adjacent to this locus (Fig. 5A). As a consequence, all analysed sequences possess only a 7.4‐kb‐long common genomic fragment containing nine predicted open reading frames (Fig. 5C). Three genes within this fragment code for conserved hypothetical and are therefore of potential interest in the context of 3‐HP/4‐HB pathway.
Figure 5.
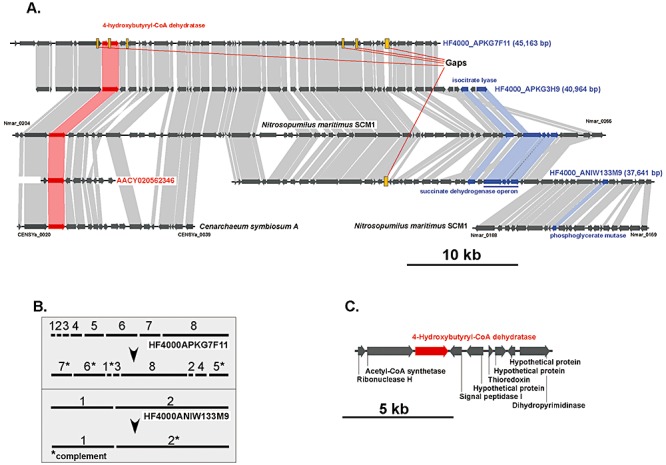
Comparison of genomic fragments harbouring 4‐hydroxybutyryl‐CoA dehydratase in N. maritimus and C. symbiosum with four related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (4000 m WGS ALOHA database, blue). A. Genes shared in common between genomic fragments are connected by shaded boxes: red (3‐HP/4‐HB cycle), blue (triosephosphate formation and glyoxylate pathway) and grey (rest of the genes). B. Re‐assemblage of individual reads in HF4000_APKG7F11 and HF4000_ANIW133M9 is depicted in grey insert. C. The genomic fragments share significant conservation of gene content and order in all analysed sequences.
Conserved homologues in 10 kb genomic fragment downstream of 4‐HB dehydratase were found in third ALOHA contig HF4000_ANIW133M9. Besides striking homology between these contigs and N. maritimus, 3′‐terminus of this genomic fragment contained genes encoding nearly identical succinate dehydrogenase subunits and isocitrate lyase (Fig. 5A). The presence of these genes along with those for succinate dehydrogenase and fumarate hydratase suggests the eventual capacity for biomass generation and for utilization of acetyl‐CoA via glyoxylate cycle (Fig. S1).
Recently, crenarchaeal 4‐hydroxybutyryl‐CoA dehydratase was shown to be robust towards oxygen, making the 3‐HP/4‐HB cycle fully oxygen‐tolerant. On the other hand, this enzyme and succinate dehydrogenase are ferredoxin‐dependent. The usage of this low‐potential electron donor required anoxia or at least very low oxygen tension in cytosol (Berg et al., 2007; 2010; Huber et al., 2008). We have checked the genome of N. maritimus for presence of O2‐scavenging enzymes such as superoxide dismutases (Sod) and thioredoxins and have found at least seven genes encoding for two putative iron/manganese SodA‐like proteins and five thiol‐specific antioxidants respectively. The presence of such a set of the genes involved in antioxidant defence suggests the capacity of N. maritimus cells to maintain their cytosolic compartments in a relatively reduced state needed for the activity of oxygen‐sensitive enzymes and low‐potential electron donors. All types of genes involved in antioxidant defence in N. maritimus are found in marine environmental metagenomes from the GOS database, whereas type 2 of SodA‐like enzyme and thioredoxin type 5 are missing in both C. symbiosum genome and deep‐sea ALOHA fosmid library (Fig. S2). None of the KM3 fosmids were found to bear the genes related to sodA and thiol‐specific antioxidants.
Taken together, these observations suggest that marine Crenarchaeota may have a genetic potential to encode a modified version of 3‐HP/4‐HB CO2 fixation pathway. Produced acetyl‐CoA further is incorporated into biomass via triosephosphate formation (Berg et al., 2007) and/or glyoxylate shunt. Biochemical and physiological studies remain necessary to fully validate this hypothesis and to establish specific pathways.
Conclusions
Analysis of (meta)genome data sets from bathypelagic environments revealed that uncultivated Crenarchaeota possess multiple component of 3‐hydroxipropionate/4‐hydroxybutyrate cycle, the fifth pathway of carbon fixation. These organisms likely use acetyl‐CoA/propionyl‐CoA carboxylase(s) as CO2‐fixing enzyme(s) to form succinyl‐CoA, from which one molecule of acetyl‐CoA is regenerated via 4‐hydroxybutyrate cleavage and another acetyl‐CoA is the carbon fixation product. Genomic context analysis of genes for 3‐hydroxipropionate/4‐hydroxybutyrate cycle (acetyl‐CoA carboxylase, methylmalonyl‐CoA mutase and 4‐hydroxybutyryl‐CoA dehydratase) share striking homology between Atlantic Mediterranean and Sargasso Sea lineages. The identification for the first time of the genes encoding for putative isocitrate lyase, the crucial enzyme in glyoxylate cycle, suggests that marine Crenarchaeota have the potential to employ glyoxylate metabolism in cell biomass formation and acetyl‐CoA regeneration.
Experimental procedures
Study site and sampling
The sampling was carried out in the Eastern Mediterranean Sea in September 2008 using the RV URANIA (National Council of Research, Italy). Twenty‐four litres of superficial, meso‐ and bathypelagic seawater were collected by using 12 l Niskin bottles mounted on a General Oceanics conductivity‐temperature‐depth rosette sampler down to a maximum depth of 3010 m (sea bottom at 3243 m depth) at the KM3 Station (Ionian Sea, Eastern Mediterranean) (36°29′98″N, 15°39′97″E). Dissolved oxygen concentrations were determined with a SBE oxygen sensor mounted on the conductivity‐temperature‐depth and nutrient concentrations (i.e. phosphate, nitrate and nitrite) with a nutrient auto‐analyser (Bran & Luebbe Autoanalyzer II, Norderstedt, Germany).
Direct cell count and FISH
To determine the abundance of bacteria and archaea in collected samples, three oligonucleotide probes were used: EUB338I (Amann et al., 1990), ARCH915 (Amann et al., 1992) and Cren512 (Juergens et al., 2000). One hundred and twenty millilitres of seawater collected at the depth of 3010 m was filtered onto 0.20‐µm‐pore‐size polycarbonate filters supported by 0.45‐µm‐pore‐size cellulose nitrate filters and then was immediately fixed with formaldehyde (final concentration 4% v/v) for 4 h, washed twice with 1× PBS (phosphate‐buffered saline), fixed in 1 ml ethanol: 1× PBS (1:1), air dried and stored at 4°C until further treatment. FISH analysis with CY3‐labelled oligonucleotide probes (final concentration, 50 ng ml−1) and DAPI (4′,6‐diamidino‐2‐phenylindole) staining were conducted according to Glockner and colleagues (1999). Filter sections on slides were embedded in Citiflour AF1 antifadent (Plano, Wetzar, Germany), covered with a coverslip and inspected using the Axiophot epifluorescence microscope (Carl Zeiss, Jena, Germany) equipped with filters 02 (DAPI), 10 (FLUOS, DTAF) and 20 (Cy3), a mercury‐arc lamp and an AxioCam digital camera. For each hybridization approach and sample, at least 750 DAPI‐stained cells in 30 randomly chosen counting squares were manually counted. The number of FISH‐stained cells was calculated by counting probe‐specific positive signals related to DAPI counts. Counting signals were always corrected by subtracting signals obtained with the negative control probe NON338 (Wallner et al., 1993).
DNA extraction
For molecular analysis, 20 l of seawater samples (3000 m deep) were filtered through Sterivex filter cartridge (Millipore, Millford, MA, USA). Sterivex filters were filled with 100 µl of TE buffer (pH 8.0) containing lysozyme (5 mg ml−1) and 300 µl of lysis buffer QRL1 (Qiagen, Milan, Italy) and stored at −20°C until processing in laboratory. The cartridges of the Sterivex filters were cracked open and the filters and the lysis buffer transferred into 50 ml sterile centrifuge tubes. Genomic DNA was isolated from the filters using RNA/DNA mini kit (Qiagen, Milano, Italy). The extraction was carried out according to the manufacturer's instructions; the quality of DNA samples was examined by agarose gel electrophoresis and concentrations were determined using the NanoDrop ND‐1000 Spectrophotometer (Wilmington, DE, USA).
16S rRNA gene amplification, cloning, sequencing and phylogenetic analysis
Total DNA collected at 3010 m depth were used for the amplification of 16S rDNA. The Archaea‐specific primers set was used to amplify 16S rDNA genes. The primers used were A20F (5′‐TTCCGGTTGATCCYGCCRG) and A958R (5′‐YCCGGCGTTGAMTCCAATT) (Hallam et al., 2003). The reaction was carried out in a MasterCycler 5331 Gradient (Eppendorf) as follows: initial denaturation at 94°C for 5 min, 35 cycles of 1 min at 94°C, 1 min at 50°C and 2 min at 72°C; final extension step of 10 min at 72°C. PCR products and genomic DNA were visualized on a 1% agarose gel, specific bands were cut out and purified using QIAquick gel extraction kit (Qiagen). The purified PCR products were ligated into pGEM‐T Easy plasmid vector (Promega) and transformed by electroporation into the Escherichia coli DH10B cells (Invitrogen). Transformants were selected on LB containing 100 µg ml−1 ampicillin under blue‐white selection. The clone libraries were screened by direct colony PCR amplification. The amplification conditions were as above. Before sequencing, QIAquick 96 PCR Purification Kit (Qiagen) was used for the purification of PCR products. Sequencing reactions were performed using the ABI Prism Big Dye 3.1 Terminator Cycle sequencing kit and an ABI Prism 3100 Avant (Applied Biosystems), DyeEx 96 Kit (Qiagen) was used for removal of unincorporated dye terminators from the products. Sequence analysis of 96 clones (550‐bp‐long fragments in average) was performed as previously described (Yakimov et al., 2007; 2009). Briefly, all sequences were edited with MacVector Software version 11.0.2 (Rajagopal, 2000) (Accelrys, San Diego, CA) and compared with DNA sequences in the public domain through the Basic Local Alignment Search Tool (blast) (Altschul et al., 1997). Phylogenetic trees were constructed using the Neighbour‐Joining method and Jukes–Cantor distance matrix; 1000 bootstrap re‐samplings were performed to estimate the reproducibility of the tree. Random order input of sequences, single jumbling and the global rearrangement option were used. The produced tree was sufficiently robust, thus secondary structure of 16S rDNA sequences were not taken into consideration.
Dark primary production
Dark CO2 fixation was estimated by the incorporation of [14C]bicarbonate (10 µCi ml−1, Amersham) in 40 ml of samples. Each experiment was performed in triplicate on board of RV Urania. Samples and one formaldehyde‐fixed blank control were incubated in the dark at in situ temperature (13°C) for 7 days. After incubation, the samples were fixed with 2% formaldehyde and then filtered through 0.1 µm polycarbonate filters (Millipore). Filters were washed three times with 10 ml of ultra‐filtered (0.1 µm) seawater, placed in scintillation vials and exposed to concentrated HCl vapour for 12 h to eliminate any residue of radioactive bicarbonate and then were stored in −20°C until counted. The scintillation vials were filled with 5 ml of Ultima Gold (PerkinElmer) scintillation liquid and the filters were counted in a Wallac 1414 analyser. The values obtained in disintegrations per minute were normalized against the values of the abiotic control. The resulting mean disintegrations per minute of the samples were converted into organic carbon produced over time and corrected for the natural DIC concentration (2 mM).
Quantitative PCR analyses of environmental DNA templates and estimation of accA gene copies
The crenarchaeal accA‐like gene abundance was evaluated by TaqMan(R) gene expression assay. The primers used were AccAF573 (5′‐GTTYGTYACDGGDCCYGAYG‐3′) and AccAR279 (5′‐TGATRTRRTCCATRCAHTCRTA‐3′), while the TaqMan probe was AccTaq183 (5′‐TTTCRWTBGAYGAWYTDGGTGGAGCWA‐3′) (Yakimov et al., 2009). 5′‐6‐FAM‐ and 3′‐TAMRA‐labelled TaqMan probe was obtained from PE Applied Biosystems. The reactions were performed in an ABI 7500 Fast Real‐Time PCR System thermocycler. The DNA templates originating from all compartments sampled were tested in triplicates along with a ‘No Template Control’. The mixtures for quantitative PCRs and the reaction conditions were as described by Yakimov and colleagues (2009). Quantitative PCR amplification was analysed with automatic setting of the baseline and threshold values, using the relative standard curve method. Standard curves for the accA gene quantification were generated by serial dilution ranging from 107 until 10 copies of plasmid containing accA gene (453‐bp‐long fragment). The plasmid was originated from the pGEM T‐Easy Vector II harbouring the crenarchaeal accA‐like gene (AM901411), extracted using PureLink™ HiPure Plasmid (Invitrogen, UK) and quantified using a Nanodrop ND‐1000 Spectrophotometer (Wilmington, DE, USA).
Comparative analysis of marine metagenomic libraries
Sequences of three N. maritimus SCM1 genes, crucial in 3‐HP/4‐HB autotrophic cycle, were queried against the NCBI non‐redundant (nr) protein database using both blastp and blastx search with a cut‐off value of < 1e‐50. Only top blast high‐scoring pairs derived from marine metagenomic libraries and C. symbiosum A genome sequences were considered for further analyses and tabulated according to the NCBI taxonomic identifier for each query. For comparative analysis, reciprocal blastn and tblastx searches between the different fosmids, scaffolds and genomic fragments were carried out, leading to the identification of regions of similarity, insertions and rearrangements. To allow the interactive visualization of genomic fragment comparisons, the MacVector (version 11.0.2) was used.
Nucleotide accession numbers
16S rRNA gene sequence data have been submitted in the DDBJ/EMBL/GenBank databases under Accession No. FN796454–FN796457.
Acknowledgments
We thank the captain and the crew of RV Urania for their expert handling of our equipment during the cruise and for highly productive technical assistance. This study was supported by the European Science Foundation Programme EuroCORES/EuroDEEP under MIDDLE project (06‐EuroDEEP‐FP‐004).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. The 3-hydroxypropionate/4-hydroxybutyrate cycle of autotrophic CO2 fixation in Crenarchaeota, as proposed by Berg and colleagues (2007; 2010). Two proposed pathways of CoA regeneration via glyoxylate cycle and glyceraldehyde 3-phosphate synthesis are also shown. The enzymes, indispensable for the depicted pathways, are evidenced in red. Enzymes: 1, acetyl (propionyl)-CoA carboxylase; 2, malonyl-CoA reductase (NADPH); 3, malonate semialdehyde reductase (NADPH); 4, 3-hydroxypropionate-CoA ligase (AMP-forming); 5, 3-hydroxypropionyl-CoA dehydratase; 6, acryloyl-CoA reductase (NADPH); 7, methylmalonyl-CoA epimerase; 8, methylmalonyl-CoA mutase; 9, succinyl-CoA reductase (NADPH or reduced methyl viologen); 10, succinic semialdehyde reductase (NADPH); 11, 4-hydroxybutyrate-CoA ligase (AMP-forming); 12, 4-hydroxybutyryl-CoA dehydratase; 13, crotonyl-CoA hydratase; 14 (S)-3-hydroxybutyryl-CoA dehydrogenase (NAD+); 15, acetoacetyl-CoA b-ketothiolase; 16, malate synthase; 17, malate dehydrogenase (NAD+); 18, citrate synthase; 19, aconitase; 20, isocitrate lyase; 21, pyruvate synthase; 22, pyruvate, water dikinase [phosphoenolpyruvate (PEP) synthase] (AMP-forming); 23, enolase; 24, phosphoglycerate mutase; 25, 3-phosphoglycerate kinase; 26, glyceraldehyde 3-phosphate dehydrogenase (NADH).
Fig. S2. Phylogenetic tree of representative superoxide dismutase (SodA) and thioredoxin proteins. The Neighbour-Joining tree with Poisson correction of distances is rooted with redoxin domain-containing protein of Burkholderia ambifaria MC40-6 (YP_001811805). Phylogenetic trees constructed by other algorithms (maximum parsimony, distance matrix and maximum likelihood) resulted in identical topologies for principal clade grouping. The scale bar represents 0.5 substitutions per site. Numbers at nodes indicate the percentage bootstrap values for the clade of this group calculated for 1000 replications. Only values above 70% are shown. The tree represents full peptide sequences from N. maritimus and C. symbiosum (shown as Nmar_ and CENSYa_ gene numbers), from ALOHA_4000 m database (red) and two first high-ranking sequences from GOS database (blue).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agogué H., Brink M., Dinasquet J., Herndl G.J. Major gradients in putatively nitrifying and non‐nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped blast and psi‐blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R., Stahl D.A. Combination of 16S rRNA‐targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R.I., Zarda B., Stahl D.A., Schleifer K.H. Identification of individual prokaryotic cells by using enzyme‐labeled, rRNA‐targeted oligonucleotide probes. Appl Environ Microbiol. 1992;58:3007–3011. doi: 10.1128/aem.58.9.3007-3011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I.A., Kockelkorn D., Buckel W., Fuchs G. A 3‐hydroxypropionate/4‐hydrdoxybutyrate autotrophic carbon dixide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- Berg I.A., Ramos‐Vera W.H., Petri A., Huber H., Fuchs G. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology. 2010;156:256–269. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- De Corte D., Yokokawa T., Varela M.M., Agogué H., Herndl G.J. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J. 2009;3:147–158. doi: 10.1038/ismej.2008.94. [DOI] [PubMed] [Google Scholar]
- DeLong E.F. Archaeal mysteries of the deep revealed. Proc Natl Acad Sci USA. 2006;103:6417–6418. doi: 10.1073/pnas.0602079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E.F., Preston C.M., Mincer T., Rich V., Hallam S.J. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. et al. [DOI] [PubMed] [Google Scholar]
- Francis C.A., Roberts K.J., Beman J.M., Santoro A.E., Oakley B.B. Ubiquity and diversity of ammonia‐oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner F.O., Fuchs B.M., Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S.J., Girguis P.R., Preston C.M., Richardson P.M., DeLong E.F. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane‐oxidizing archaea. Appl Environ Microbiol. 2003;69:5483–5491. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S.J., Konstantinidis K.T., Putnam N., Schleper C., Watanabe Y., Sugahara J. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006a;103:18296–18301. doi: 10.1073/pnas.0608549103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S.J., Mincer T.J., Schleper C., Preston C.M., Roberts K., Richardson P.M. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006b;4:e95. doi: 10.1371/journal.pbio.0040095. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndl G.J., Reinthaler T., Teira E., Van Aken H., Veth C., Pernthaler A. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H., Gallenberg M., Jahn U., Eylert E., Berg I.A., Kockelkorn D. A dicarboxylate/4‐hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci USA. 2008;105:7851–7856. doi: 10.1073/pnas.0801043105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalls A.E., Shah S.R., Hansman R.L., Aluwihare L.I., Santos G.M., Druffel E.R.M., Pearson A. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens G., Glockner F., Amann R., Saano A., Montonen L., Likolammi M. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol Ecol. 2000;34:45–56. doi: 10.1111/j.1574-6941.2000.tb00753.x. et al. [DOI] [PubMed] [Google Scholar]
- Könneke M., Bernhard A.E., De La Torre J.R., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia‐oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Konstantinidis K.T., Braff J., Karl D.M., DeLong E.F. Comparative metagenomic analysis of a microbial community residing at a depth of 4000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress N., Manca B.B., Klein B., Deponte D. Continuing influence of the changed thermohaline circulation in the eastern Mediterranean on the distribution of dissolved oxygen and nutrients: physical and chemical characterization of the water masses. J Geophys Res. 2003;108:8109. [Google Scholar]
- Malanotte‐Rizzoli P., Manca B., Ribera M., Theocharis A., Bergamasco A., Bregant D. ) A synthesis of the Ionian Sea hydrography, circulation and water mass pathways during POEM‐Phase I. Prog Oceanogr. 1997;39:153–204. et al. [Google Scholar]
- Martín‐Cuadrado A.B., López‐García P., Alba J.C., Moreira D., Monticelli L., Strittmatter A. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE. 2007;2:e914. doi: 10.1371/journal.pone.0000914. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Cuadrado A.B., Rodriguez‐Valera F., Moreira D., Alba J.C., Ivars‐Martínez E., Henn M.R. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. et al. [DOI] [PubMed] [Google Scholar]
- Martins B.M., Dobbek H., Cinkaya I., Buckel W., Messerschmidt A. Crystal structure of 4‐hydroxybutyryl‐CoA dehydratase: radical catalysis involving a [4Fe–4S] cluster and flavin. Proc Natl Acad Sci USA. 2004;101:15645–15649. doi: 10.1073/pnas.0403952101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer T.J., Church M.J., Taylor L.T., Preston C., Karl D.M. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. et al. [DOI] [PubMed] [Google Scholar]
- Mosier A.C., Francis C.A. Relative abundance and diversity of ammonia‐oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Nicol G.W., Schleper C. Ammonia‐oxidizing Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 2006;14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Pearson A., McNichol A.P., Benitez‐Nelson B.C., Hayes J.M., Eglington T.I. Origins of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound‐specific 14C analysis. Geochim Cosmochim Acta. 2001;65:3123–3137. [Google Scholar]
- Rajagopal I. SOFTWARE: genomics made easy. Science. 2000;290:474. [Google Scholar]
- Rusch D.B., Halpern A.L., Sutton G., Heidelberg K.B., Williamson S., Yooseph S. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C. Metabolism of the deep. Nature. 2008;456:712–714. doi: 10.1038/456712a. [DOI] [PubMed] [Google Scholar]
- Spiridonova E.M., Berg I.A., Kolganova T.V., Ivanovskii R.N., Kuznetsov B.B., Turova T.P. An oligonucleotide primer system for amplification of the ribulose‐1,5‐bisphosphate carboxylase/oxygenase genes of bacteria of various taxonomic groups. Mikrobiologiia. 2004;73:377–387. [PubMed] [Google Scholar]
- Thauer R.K. A fifth pathway of carbon fixation. Science. 2007;318:1732–1733. doi: 10.1126/science.1152209. [DOI] [PubMed] [Google Scholar]
- Vidussi F., Claustre H., Manca B., Luchetta A., Marty J.‐C. Phytoplankton pigment distribution in relation to upper thermocline circulation in the eastern Mediterranean Sea during winter. J Geophys Res. 2001;106:19939–19956. [Google Scholar]
- Wallner G., Amann R., Beisker W. Optimizing fluorescent in situ hybridization with rRNA‐targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- Whitman W.B., Coleman D.C., Wiebe W.J. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C., Schouten S., Boschker H.T.S., Damstè J.S.S. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- Wuchter C., Abbas B., Coolen M.J., Herfort L., Van Bleijswijk J., Timmers P. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov M.M., La Cono V., Denaro R., D'Auria G., Decembrini F., Timmis K.N. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J. 2007;1:743–755. doi: 10.1038/ismej.2007.83. et al. [DOI] [PubMed] [Google Scholar]
- Yakimov M.M., La Cono V., Denaro R. A first insight into the occurrence and expression of functional amoA and accA genes of autotrophic and ammonia‐oxidizing bathypelagic Crenarchaeota of Tyrrhenian Sea. Deep Sea Research II. 2009;56:748–754. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The 3-hydroxypropionate/4-hydroxybutyrate cycle of autotrophic CO2 fixation in Crenarchaeota, as proposed by Berg and colleagues (2007; 2010). Two proposed pathways of CoA regeneration via glyoxylate cycle and glyceraldehyde 3-phosphate synthesis are also shown. The enzymes, indispensable for the depicted pathways, are evidenced in red. Enzymes: 1, acetyl (propionyl)-CoA carboxylase; 2, malonyl-CoA reductase (NADPH); 3, malonate semialdehyde reductase (NADPH); 4, 3-hydroxypropionate-CoA ligase (AMP-forming); 5, 3-hydroxypropionyl-CoA dehydratase; 6, acryloyl-CoA reductase (NADPH); 7, methylmalonyl-CoA epimerase; 8, methylmalonyl-CoA mutase; 9, succinyl-CoA reductase (NADPH or reduced methyl viologen); 10, succinic semialdehyde reductase (NADPH); 11, 4-hydroxybutyrate-CoA ligase (AMP-forming); 12, 4-hydroxybutyryl-CoA dehydratase; 13, crotonyl-CoA hydratase; 14 (S)-3-hydroxybutyryl-CoA dehydrogenase (NAD+); 15, acetoacetyl-CoA b-ketothiolase; 16, malate synthase; 17, malate dehydrogenase (NAD+); 18, citrate synthase; 19, aconitase; 20, isocitrate lyase; 21, pyruvate synthase; 22, pyruvate, water dikinase [phosphoenolpyruvate (PEP) synthase] (AMP-forming); 23, enolase; 24, phosphoglycerate mutase; 25, 3-phosphoglycerate kinase; 26, glyceraldehyde 3-phosphate dehydrogenase (NADH).
Fig. S2. Phylogenetic tree of representative superoxide dismutase (SodA) and thioredoxin proteins. The Neighbour-Joining tree with Poisson correction of distances is rooted with redoxin domain-containing protein of Burkholderia ambifaria MC40-6 (YP_001811805). Phylogenetic trees constructed by other algorithms (maximum parsimony, distance matrix and maximum likelihood) resulted in identical topologies for principal clade grouping. The scale bar represents 0.5 substitutions per site. Numbers at nodes indicate the percentage bootstrap values for the clade of this group calculated for 1000 replications. Only values above 70% are shown. The tree represents full peptide sequences from N. maritimus and C. symbiosum (shown as Nmar_ and CENSYa_ gene numbers), from ALOHA_4000 m database (red) and two first high-ranking sequences from GOS database (blue).


