Summary
The production of hydrogen via microbial biotechnology is an active field of research. Given its ease of manipulation, the best‐studied bacterium Escherichia coli has become a workhorse for enhanced hydrogen production through metabolic engineering, heterologous gene expression, adaptive evolution, and protein engineering. Herein, the utility of E. coli strains to produce hydrogen, via native hydrogenases or heterologous ones, is reviewed. In addition, potential strategies for increasing hydrogen production are outlined and whole‐cell systems and cell‐free systems are compared.
Introduction
Research on sustainable biogas production is increasing due to strong public interest in protecting the global climate and a growing demand for independency from scarce fossil energy sources (Kessel, 2000; Armaroli and Balzani, 2011). Hydrogen has tremendous potential because it is the most abundant element in the universe (Dunn, 2002), is renewable, efficient and clean (Hansel and Lindblad, 1998), and is utilized for fuel cells in portable electronics, power plants and internal combustion engines (Dunn, 2002). Among the existing renewable energy sources such as ethanol and algal diesel, hydrogen continues to be very attractive because of its various means of production, non‐polluting nature and large energy content per mass (142 MJ kg−1 for H2). In addition, it is estimated that the global energy system will shift from fossil fuels to hydrogen (Dunn, 2002) as well as methane (Alves et al., 2009).
Currently, hydrogen is produced mainly by the water‐gas shift reaction (Yi and Harrison, 2005), as a by‐product of petroleum refining and chemical production (Das and Veziroglu, 2001), and by electrolysis of water (Armaroli and Balzani, 2011). However, methods such as electrolysis of water and thermocatalytic reformation of hydrogen‐rich compounds (which are non‐renewable resources) require higher energy input to generate hydrogen gas compared with biological means (Das and Veziroglu, 2001). The commercial production of hydrogen by electrolysis of water achieves an efficiency of 75%; however, this cost is currently several times higher than that produced from fossil fuels (Ewan and Allen, 2005). Also, hydrogen production dependent on fossil resources is clearly unsustainable, and all the operations associated with sequestration are energy intensive, costly, and potentially damaging to our environment (Edwards et al., 2007).
In contrast, biological methods to produce hydrogen are environmentally more attractive because they utilize microorganisms to produce hydrogen from diverse renewable resources (Edwards et al., 2007). Although there are several methods for biohydrogen production, biohydrogen may be produced through either photosynthetic or fermentative processes; in general, fermentative hydrogen production is more efficient than photosynthetic means (Yoshida et al., 2005). Thus, to date, fermentative hydrogen production has been shown to be the most promising approach because it has the advantages of (i) higher hydrogen‐production rates, (ii) independence from the availability of light, and (iii) a variety of carbon sources such as organic compounds, low‐cost wastes, and cellulosic or cellobiose substrates (Vardar‐Schara et al., 2008).
Biological synthesis and uptake of hydrogen are catalysed by hydrogenases (Hyd), which catalyse the reaction 2H+ + 2e‐ ↔ H2 (g); the hydrogenases have been classified as [NiFe], [FeFe] or [Fe] enzymes based on the type of iron sequestered by the active site of the large subunit of Hyd. [FeFe] and [NiFe] hydrogenases have similar overall structures as the active site is located within a large subunit and electrons are delivered to this centre via iron–sulfur (Fe‐S) centres located in the small subunits (Forzi and Sawers, 2007). [Fe] hydrogenases lack Fe‐S clusters and are found only in a small group of methanogenic archea (Zirngibl et al., 1992). In general, these Hyd systems are complex requiring many maturation proteins (Vardar‐Schara et al., 2008); for example, [NiFe] Hyd 3 of E. coli requires 10 maturation proteins (Forzi and Sawers, 2007), including HypABCDEF (metallochaperones for NiFe insertion) and SlyD (nickel insertion) (Drapal and Böck, 1998).
During the last 4 years, the number of research publications utilizing engineering strategies to increase hydrogen synthesis has been increasing. In this review, we summarize the research trends regarding enhanced bacterial hydrogen production by recombinant Escherichia coli strains redesigned through metabolic engineering, heterologous gene expression, adaptive evolution and protein engineering.
Utility of using Escherichia coli strains
Escherichia coli is a robust bacterium for developmental research based on genetic engineering because its whole genome sequence is available (Blattner et al., 1997) and its metabolic pathways are relatively well‐established. Also, in general, cultivation, storage and genetic manipulation (e.g. plasmid purification and transformation) for E. coli are quite technically simpler than with other strains. Specifically, the KEIO Collection, an E. coli single deletion mutant library (Baba et al., 2006), is a powerful tool since it can be used to evaluate rapidly the importance of each nonlethal gene. Furthermore, since the kanamycin resistant (KmR) gene in the knockout mutation can be eliminated by recombination between the flippase recognition target sites via the flippase recombinase protein produced from pCP20 (Cherepanov and Wackernagel, 1995), multiple deletion mutants may be rapidly constructed with little scarring by combining mutations of the Keio Collection via P1 transduction and selecting each mutation based on kanamycin resistance (Maeda et al., 2007a; 2008a). The only limitation to using P1 transduction to accumulate multiple deletions is that the mutations must not be too close. Similarly, multiple deletion strains may be made by sequential one‐step inactivations (Datsenko and Wanner, 2000) followed by kanamycin gene‐eliminating steps. In contrast, traditional methods require different selection makers (antibiotic resistant genes) to introduce multiple mutations into a single strain (Lee et al., 2005; Yoshida et al., 2006). This novel P1 transduction technique has facilitated metabolic engineering by allowing facile removal of many deleterious genes (Maeda et al., 2007a; 2008a,b). Furthermore, the ASKA Library is available, which allows each E. coli protein to be produced from a plasmid either with or without a fused GFP tag (Kitagawa et al., 2006).
Native E. coli hydrogenases
Escherichia coli has four native hydrogenases (Hyd 1, 2, 3 and 4). Hyd 1 and 2 have primarily hydrogen uptake activity (Ballantine and Boxer, 1986; King and Przybyla, 1999) [although one recent report indicates that Hyd 1 also has hydrogen synthesis activity under micro‐aerobic conditions (Kim et al., 2010)], Hyd 3 is reversible with the synthesis rate greater than the uptake rate (Maeda et al., 2007a), and Hyd 4 is probably inactive (Self et al., 2004) although gene expression has been observed during biofilm formation (Herzberg et al., 2006; Domka et al., 2007). These four E. coli hydrogenases are classified as [NiFe] hydrogenases, and they contain two cyanide molecules and a carbon monoxide molecule at the active site (Blokesch et al., 2002); Hyd 1 is encoded by hyaABCDEF (Richard et al., 1999), Hyd 2 is encoded by hybOABCDEFG (Richard et al., 1999), Hyd 3 is encoded by hycABCDEFGHI (Bagramyan and Trchounian, 2003), and Hyd 4 is encoded by hyfABCDEFGHI (Andrews et al., 1997). Bioinformatics analysis for the large subunits of E. coli hydrogenases (hyaB for Hyd 1, hybC for Hyd 2, hycE for Hyd 3, and hyfG for Hyd 4) indicates that there is comparatively high homology between Hyd 1 and 2 and between Hyd 3 and 4 (Vardar‐Schara et al., 2008). Given the hydrogen degradation and synthesis activity of these native hydrogenases, it is imperative that researchers utilizing E. coli for heterologous expression of hydrogenases work with mutants that lack native hydrogenase activity (Maeda et al., 2007a); for example, by using a hyaB hybC hycE hyfG mutant (T. Maeda and T.K. Wood, unpublished).
In E. coli, hydrogen is produced by the formate hydrogenlyase system (FHL) that consists of two enzymatic activities: a formate dehydrogenase‐H [encoded by fdhF (Axley et al., 1990)] for producing 2H+, 2e‐ and CO2 from formate and Hyd 3 for synthesizing molecular hydrogen from 2H+ and 2e‐ (Sawers et al., 1985). The FHL system may be used for the regulation of internal pH into the cells (Böck and Sawers, 1996) although this is far from clear. Regulation of FHL includes the HycA repressor (Sauter et al., 1992) and the FhlA activator (Schlensog et al., 1994). Two additional formate dehydrogenases are found in E. coli that consume formate: FDHN (formate dehydrogenase‐N) and FDHO (formate dehydrogenase‐O) (Rossmann et al., 1991). Also, FocA (Suppmann and Sawers, 1994) and FocB (Andrews et al., 1997) export formate, and nitrate reductase A (α‐subunit encoded by narG) consumes formate by converting nitrate into nitrite by using electrons produced from formate by FDHN (Bertero et al., 2003). In addition, pyruvate dehydrogenase (AceE) and pyruvate oxidase (PoxB) consume pyruvate produced from glucose metabolism via the phosphoenolpyruvate‐producing pathway (Angelides et al., 1979; Abdel‐Hamid et al., 2001) as well as pyruvate produced glucose metabolism via the succinate‐producing pathway (phosphoenolpyruvate to succinate) and lactate‐producing pathway (pyruvate to lactate) (Manish et al., 2007). Thus, these well‐established pathways motivated engineers to redirect the metabolic flux towards hydrogen production.
Metabolic engineering to enhance hydrogen production
Since hydrogen is produced through the reaction HCOO‐ + H2O ↔ H2 + HCO3‐ (Woods, 1936) by the FHL complex in well‐known pathways, metabolic engineering from formate metabolism is possible using E. coli native hydrogenases. The first improvement to enhance hydrogen production from formate in E. coli was by inactivating the HycA repressor of FHL and by overproducing the FhlA activator of FHL; strain SR13 with both changes had sevenfold higher transcription of the FHL complex (e.g. fdhF and the hyc operon) and 2.8‐fold higher hydrogen productivity than the wild‐type strain (Yoshida et al., 2005) (see Table 1 for a comparison of the strains discussed in this review).
Table 1.
Comparison of in vivo hydrogen production by engineered Escherichia coli strains.
| System | H2 production rate (reported units) | H2 production rate (converted units) | Substrate | Reference |
|---|---|---|---|---|
| Protein engineering | ||||
| Protein engineering of HycE (truncation) of E. coli | 9 µmol H2 (mg protein)−1 h−1 | 9 µmol H2 (mg protein)−1 h−1 | Formate | Maeda et al. (2008b) |
| Protein engineering of FhlA of E. coli | 7 µmol H2 (mg protein)−1 h−1 | 7 µmol H2 (mg protein)−1 h−1 | Formate | Sanchez‐Torres et al. (2009) |
| Site‐directed mutagenesis of HydA from C. reinhardtii | approximately 10 µmol H2 ml−1 | 19 µmol H2 (mg protein)−1 h−1 | Glucose | Agapakis et al. (2010) |
| Metabolic engineering through modifying multiple native genes in E. coli | ||||
| Inactivation of HycA and overexpression of FhlA | 23.6 g H2 l−1 h−1 | 254 µmol H2 (mg protein)−1 h−1 | Formate | Yoshida et al. (2005) |
| Inactivation of HyaB, HybC, HycA, FdoG and overexpression of FhlA | 113 µmol H2 (mg protein)−1 h−1 | 113 µmol H2 (mg protein)−1 h−1 | Formate | Maeda et al. (2008a) |
| Inactivation of HycA, LdhA, FrdBC and overexpression of FhlA | 13 mmol (g DCW)−1 l−1 h−1 | 26 µmol H2 (mg protein)−1 h−1 | Glucose | Yoshida et al. (2006) |
| Inactivation of HyaB, HybC, HycA, FdoG, FrdC, LdhA and AceE | 32 µmol H2 (mg protein)−1 h−1 | 32 µmol H2 (mg protein)−1 h−1 | Glucose | Maeda et al. (2007b) |
| Inactivation of Hyd 1, Hyd 2, LdhA and overexpression of truncated FhlA | 5.3 mmol H2 l−1 h−1 | 24 µmol H2 (mg protein)−1 h−1 a | Glucose | Turcot et al. (2008) |
| Inactivation of HycA, HyaAB, HybBC, LdhA and FrdAB | 31.3 mmol H2 (g DCW)−1 h−1 | 63 µmol H2 (mg protein)−1 h−1 | Glucose | Kim et al. (2009) |
| Inactivation of HyaAB, HybABC, HycA, LdhA and FrdBC | 1.9 mmol H2 (g DCW)−1 l−1 h−1 | 1.5 µmol H2 (mg protein)−1 h−1 | Glucose | Mathews et al. (2010) |
| Production of Hyd 1 | 3 ml H2 100 ml−1 | 0.8 µmol H2 (mg protein)−1 h−1 a | Glucose + formate | Kim et al. (2010) |
| Inactivation of HycA and LacI | 5.88 ml H2 OD−1 h−1 | 11 µmol H2 (mg protein)−1 h−1 | Cheese whey | Rosales‐Colunga et al. (2010) |
| Adaptive evolution | ||||
| Chemical mutagenesis and adaptive evolution | 22 µmol H2 (mg protein)−1 | 4 µmol H2 (mg protein)−1 h−1 | Glycerol | Hu and Wood (2010) |
| Heterologous gene expression | ||||
| Production of [Fe] hydrogenase from E. cloacae | 0.96 mmol h−1 | 14.5 µmol H2 (mg protein)−1 h−1 a | Glucose | Chittibabu et al. (2006) |
| Production of HoxEFUYH hydrogenase from Synechocystis sp. PCC 6803 | 22 ± 3 µmol H2 (mg protein)−1 | 4 µmol H2 (mg protein)−1 h−1 | Glucose | Maeda et al. (2007c) |
| Production of HoxEFUYH hydrogenase and the maturation proteins HypABCDEF and HoxW from Synechocystis sp. PCC 6803 | 8.4 µmol H2 l−1 | 0.004 µmol H2 (mg protein)−1 h−1 | Glucose | Wells et al. (2011) |
| Production of HydFEGA | 429.3 nmol H2 min−1 l−1 | 0.12 µmol H2 (mg protein)−1 h−1 a | Glucose | Akhtar and Jones (2008a) |
| Production of HydFEGA and inactivation of IscR | 1257.5 nmol H2 min−1 l−1 | 0.34 µmol H2 (mg protein)−1 h−1 a | Glucose | Akhtar and Jones (2008a) |
| Inactivation of IscR, production of HydFEGA hydrogenase from C. acetobutylicum, CpFdX ferredoxin from C. pasterianum and YdbK pyruvate–flavodoxin oxidoreductase from E. coli | 9.6 mmol H2 (g DCW)−1 h−1 | 19 µmol H2 (mg protein)−1 h−1 | Glucose | Akhtar and Jones (2009) |
| Production of HupSL hydrogenase from Rhodobacter sphaeroides | 19.68 µL H2 (ml culture−1) h−1 | 1.1 µmol H2 (mg protein)−1 h−1 | Glucose | Lee et al. (2010) |
| Inactivation of HycA and TatC and expression of the genes encoding ScrKYABR invertase from Bacillus subtilis | 1.38 ml H2 (mg DCW)−1 l−1 | 3.9 µmol H2 (mg protein)−1 h−1 | Sucrose | Penfold and Macaskie (2004) |
| Inactivation of IscR, production of HydFEGA hydrogenase from C. acetobutylicum, CpFdX ferredoxin from C. pasterianum and YdbK pyruvate–flavodoxin oxidoreductase from E. coli, and amyE from B. subtilis | 30 µmol H2 culture−1 | 0.65 µmol H2 (mg protein)−1 h−1 a | Starch | Akhtar and Jones (2009) |
| Single gene knockout or expression | ||||
| Inactivation of HycA | NA | 109 µmol H2 (mg protein)−1 h−1 | Formate | Yoshida et al. (2005) |
| Production of FhlA | 7 µmol H2 (mg protein)−1 h−1 | 7 µmol H2 (mg protein)−1 h−1 | Formate | Maeda et al. (2008a) |
| Inactivation of HycA | 31 ml H2 (OD)−1 l−1 | 6.3 µmol H2 (mg protein)−1 h−1 | Glucose | Penfold et al. (2003) |
| Inactivation of FocA | 14.9 µmol H2 (mg DCW)−1 | 1.8 µmol H2 (mg protein)−1 h−1 | Glucose | Fan et al. (2009) |
| Inactivation of HybC | 12.1 µmol H2 (mg DCW)−1 | 1.4 µmol H2 (mg protein)−1 h−1 | Glucose | Fan et al. (2009) |
| Inactivation of NarL | 14.4 µmol H2 (mg DCW)−1 | 1.7 µmol H2 (mg protein)−1 h−1 | Glucose | Fan et al. (2009) |
| Inactivation of Ppc | 11.2 µmol H2 (mg DCW)−1 | 1.3 µmol H2 (mg protein)−1 h−1 | Glucose | Fan et al. (2009) |
| Production of Fnr | 6.2 µmol H2 (mg DCW)−1 | 3.1 µmol H2 (mg protein)−1 h−1 | Glucose | Fan et al. (2009) |
Assuming cell turbidity is 1.
To increase hydrogen further, along with deleting hycA and overexpressing fhlA, hydrogen uptake activity was eliminated by deleting hyaB, the large subunit of Hyd 1 and hybC, the large subunit of Hyd 2 (Maeda et al., 2008a). In addition, to direct the formate metabolic flux towards hydrogen production, FDHO which converts formate into CO2 without producing hydrogen, was inactivated by deleting fdoG (Maeda et al., 2008a). The metabolically engineered strain with four mutations and one plasmid to produce activator FhlA had 141‐fold higher hydrogen productivity than the wild‐type strain and reached the theoretical yield (1 mol of hydrogen produced from 1 mol of formate) (Maeda et al., 2008a).
Since glucose is far less expensive than formate, hydrogen production from glucose was pursued based on the same principle and re‐directing the metabolic flux of formate. As expected, inactivating hydrogen uptake via Hyd 1 and 2 (hya and hyb) and the FHL repressor (hycA) enhanced hydrogen production during glucose fermentation (Penfold et al., 2003; Yoshida et al., 2006; Maeda et al., 2007b; Turcot et al., 2008; Fan et al., 2009; Kim et al., 2009; Mathews et al., 2010). Hydrogen production was improved further by expressing an FhlA variant that carries a deletion of the N‐terminal part of FhlA (Self et al., 2001; Turcot et al., 2008), whereas the original FhlA had a little effect for enhanced hydrogen production (Maeda et al., 2007b). The transcriptional activity for Hyd 3 (hyc operon) was increased twofold by the truncated FhlA, which is a formate‐independent transcriptional activator (Self et al., 2001).
Escherichia coli metabolizes glucose to phosphoenolpyruvate, phosphoenolpyruvate to pyruvate, then pyruvate to formate via pyruvate formate lyase (Bagramyan and Trchounian, 2003); succinate and lactate are co‐metabolites during glucose fermentation that are synthesized from phosphoenolpyruvate and pyruvate (Bagramyan and Trchounian, 2003) (Fig. 1). Therefore, pathways to produce succinate and lactate were silenced by deleting ppc to inactivate phosphoenolpyruvate carboxylase (Fan et al., 2009), by inactivating frdABCD encoding fumarate reductase (Yoshida et al., 2006; Maeda et al., 2007b; Kim et al., 2009; Mathews et al., 2010), by deleting ldhA encoding lactate dehydrogenase (Yoshida et al., 2006; Manish et al., 2007; Maeda et al., 2007b; Kim et al., 2009; Mathews et al., 2010), and by inactivating aceE encoding the component of pyruvate dehydrogenase (Maeda et al., 2007b). Furthermore, metabolic flux analysis revealed that the inactivation of lactate dehydrogenase and fumarate reductase directed most of the carbon from glucose to the glycolytic pathway leading to hydrogen production by the FHL system (Kim et al., 2009). Therefore, hydrogen productivity and hydrogen yield were improved by the disruption of these enzyme activities.
Figure 1.
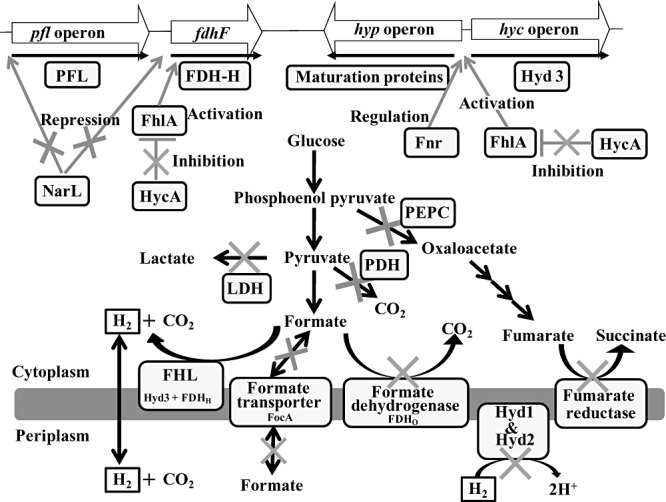
Metabolic engineering strategies in Escherichia coli. Hydrogen is produced from formate through the formate hydrogenlyase system. Abbreviations: Hyd3, hydrogenase 3; FDHH, formate dehydrogenate‐H; FDHO, formate dehydrogenase‐O; Hyd1, hydrogenase 1; Hyd2, hydrogenase 2; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase; and PFL, pyruvate formate lyase. The cross marks in the figure indicate inactivated genes.
Other deleterious genes whose inactivation leads to enhanced hydrogen production include iscR and tatABCDE (Penfold et al., 2006; Akhtar and Jones, 2008a). Fe‐S cluster assembly and incorporation into E. coli hydrogenases is an essential process, and IscR is a negative regulator for the iron sulfur cluster machinery; hence, inactivating IscR led to threefold increased hydrogen production probably by increasing the amount of active Hyd as well as by decreasing the sensitivity of Hyd to oxygen, which usually inactivates the Hyd function (Akhtar and Jones, 2008a). Hydrogen production has also been improved twofold by inactivating the twin‐arginine translocation (TAT) system, which transports folded proteins across the cytoplasmic membrane (Penfold et al., 2006). The improvement may be due to inactivating formate dehydrogenase‐N, formate dehydrogenase‐O, Hyd 1 and Hyd 2, which are translocated by TAT system. In addition, NarL, a two‐component regulator protein for the nitrate/nitrite response, and FocA encoding a formate transporter are deleterious for enhancing hydrogen production (Fan et al., 2009). In addition, producing Fnr, a DNA‐binding transcriptional global regulator, was also effective in enhancing hydrogen production (Fan et al., 2009) in agreement with the fact that the deletion of fnr gene had a negative effect for hydrogen production (Maeda et al., 2007b). So far, the best metabolically modified strains for hydrogen production that incorporate the above concepts have either seven systems inactivated (hyaB, hybC, hycA, fdoG, ldhA, frdC and aceE) (Maeda et al., 2007b), five systems inactivated (hyaAB, hybABC, hycA, ldhA and frdBC) (Mathews et al., 2010) (hycA, hya, hyb, ldhA and frdAB) (Kim et al., 2009), or three systems inactivated (hya, hyb and ldhA) and a truncated FhlA produced (Turcot et al., 2008).
Recently, recombinant E. coli BL21 expressing HyaA and HyaB or HyaABCDEF of E. coli Hyd 1 actively produced hydrogen, and the hydrogen production was enhanced twofold by the addition of formate (Kim et al., 2010), implying that hydrogen synthesis by recombinant Hyd 1 could be similar to that by the FHL complex consisting of Hyd 3 and formate dehydrogenase‐H. Also, an exhaustive search of all E. coli pathways for their impact on hydrogen production through screening the entire Keio mutant library (3985 isogenic mutants) using both glucose and formate as substrates was conducted by T. Maeda and T.K. Wood (unpublished), and several uncharacterized genes related to bacterial hydrogen production were identified. Hence, further improvements in hydrogen production are possible, and study of the four well‐known native hydrogenases continues to provide surprises.
Adapted evolution to enhance hydrogen production from renewable resources
A recent trend for bacterial hydrogen production is to utilize renewable resources; for example, glycerol accumulates as a by‐product during biodiesel manufacture (Sabourin‐Provost and Hallenbeck, 2009), and the price of crude glycerol could be reasonable and sustainable (Willke and Vorlop, 2004). Hence, converting glycerol to hydrogen is attractive. However, the specific growth rate of wild‐type E. coli on glycerol is quite low compared with that with glucose, which results in low hydrogen productivity (Murarka et al., 2008). To overcome this limitation, an efficient glycerol‐utilizing strain, HW2, was constructed. Starting with a strain harbouring the frdC mutation and by combining random chemical mutagenesis and adaptive evolution with selection based on growth on glycerol, a fivefold increase in growth on glycerol was achieved as well as a 20‐fold increase in hydrogen synthesis (Hu and Wood, 2010). Ethanol production was also increased fivefold (Hu and Wood, 2010).
Metabolic engineering has also been performed to produce hydrogen from another renewable source, cheese whey, which contains a high concentration of lactose (Rosales‐Colunga et al., 2010). In this study, hydrogen production from cheese whey was improved 20% by inactivating two proteins: HycA and LacI (LacI is a DNA‐binding transcription factor that represses the transcription of the operon involved in the transport and catabolism of lactose).
Heterologous gene expression to enhance hydrogen production in vitro
Escherichia coli remains a robust host to clone hydrogen‐related enzymes and then to utilize them in vitro. For example, an active cytoplasmic NADP‐dependent Hyd derived from Pyrococcus furiosus, an anaerobic hyperthermophile, was expressed in E. coli by cloning the four structural genes encoding the Hyd and nine genes encoding maturation proteins, HypCDABEF, HycI, SlyD and ferredoxin oxidoreductase A (FrxA) under control of the promoter of the E. coli hya operon, which has the highest transcriptional activity among the hya, hyb, hyc and hyp operons (Sun et al., 2010). Additionally, a Hyd derived from Desulfovibrio vulgaris was successfully produced in vitro by cloning its structural genes (hydAB), its maturation proteins, and the E. coli iron–sulfur cluster operon (hydEFG and ORF1‐ORF2‐iscS‐iscU‐iscA‐hscB‐hscA‐fdx‐ORF3 gene cluster) (Laffly et al., 2010).
Recombinant proteins NuoE and NuoF, which form the heterodimer of Trichomonas vaginalis hydrogenosome NADH dehydrogenase, were successfully expressed in E. coli and its reduced ferredoxin activity was characterized in vitro (Do et al., 2009). Also, a synthetic hydrogen‐producing electron transfer circuit composed of heterologously expressed [FeFe]‐Hyd, ferredoxin and pyruvate‐ferredoxin oxidoreductase was successfully expressed and investigated in vitro to insulate an electron transfer pathway to produce hydrogen coupled to the breakdown of glucose (Agapakis et al., 2010). Similarly, a synthetic ferredoxin‐dependent NAD(P)H:H2 pathway model system was also constructed in E. coli cells and hydrogen accumulation was observed with a maximum partial hydrogen pressure equivalent to a biochemically effective intracellular NADPH/NADP+ ratio of 13:1 (Veit et al., 2008). Also, an active recombinant [Fe‐Fe]‐type Hyd (HydA) of Clostridium acetobutylicum in E. coli BL21(DE3) was also achieved using three Clostridium maturation proteins (HydF, HydE and HydG); this heterologous expression resulted in hydrogen production in vitro (Akhtar and Jones, 2008b).
Heterologous gene expression to enhance hydrogen production in vivo
One of the earliest E. coli strains constructed with a recombinant Hyd was that which used a probable [FeFe] Hyd from Clostridium butyricum expressed in E. coli HK16 (this host is defective in native Hyd activity); there was roughly a threefold higher Hyd activity compared with both the wild‐type C. butyricum and to E. coli C600 (Karube et al., 1983). A [Fe]‐Hyd, HydA from Enterobacter cloacae, was also heterologously expressed as a glutathione‐HydA fusion protein in E. coli BL21 (Mishra et al., 2004; Chittibabu et al., 2006), and the recombinant E. coli strain had a yield of 3.1 mol H2 mol−1 glucose, which was higher than the E. cloacae wild‐type strain. Other improvements in hydrogen production by expressing heterologous genes in E. coli cells were found by expressing the genes necessary for sucrose transport and metabolism in E. coli strains defective in the HycA FHL repressor and TAT system to promote hydrogen production from sucrose: scrK encoding an ATP‐dependent fructokinase, scrY encoding a sucrose‐specific porin of the outer membrane, scrA encoding enzyme IIscr of the phosphotransferase system for sucrose uptake, scrB encoding an intracellular β‐d‐fructofuranoside fructohydrolase, which catalyses the hydrolysis of sucrose 6‐phosphate to β‐d‐fructose and α‐d‐glucose 6‐phosphate, and scrR encoding the negative repressor of the scr regulon (Penfold and Macaskie, 2004).
The first cloning of a cyanobacterial [Ni‐Fe]‐Hyd, HoxEFUYH derived from Synechocystis sp. PCC 6803, yielded surprising results (Maeda et al., 2007c). The hydrogen yield from the resultant strain was enhanced 41‐fold, and the results of a DNA microarray analysis demonstrated that the expression of the four hydrogenases of E. coli were not affected by the expression of the cyanobacterial HoxEFUYH. However, the mechanism for the large increase in hydrogen productivity was due to the unexpected finding that the heterologous Hyd inhibited hydrogen uptake activity through Hyd 1 and 2 rather than producing hydrogen (Maeda et al., 2007c). Recently, hydrogen production by this cyanobacterial [Ni‐Fe]‐Hyd was achieved at low rates in E. coli (Wells et al., 2011) by producing not just the five structural proteins HoxEFUYH but by also producing seven ORFs encoding the maturation proteins HypABCDEF and HoxW that are independent of the E. coli FHL pathway for hydrogen production. This resulted in hydrogen production at a rate of 4 nmol H2 (mg protein)−1 h−1.
Other improvements in bacterial hydrogen production through heterologous gene expression in E. coli were by overexpressing a [Fe‐Fe]‐Hyd, HydA derived from Ethanoligenes harbinense (Zhao et al., 2010), and a HupSL Hyd consisting of small and large subunits of Hyd isolated from Rhodobacter sphaeroides (Lee et al., 2010). Interestingly, hydrogen production by recombinant HupSL in E. coli was greater than in the original strain, Rhodobacter sphaeroides (Lee et al., 2010) in that hydrogen‐producing activity of the Hyd was negligible (Koku et al., 2002).
Hydrogen production via the [Fe‐Fe]‐type Hyd (HydA) of Clostridium acetobutylicum was achieved in vivo in E. coli BL21(DE3) by expressing E. coli YdbK, a probable pyruvate–flavodoxin oxidoreductase, and Clostridium pasteurianum[4Fe‐4S]‐ferredoxin together with Clostridium acetobutylicum HydF, HydE, HydG and HydA (Akhtar and Jones, 2009). In a background that includes the iscR mutation, the hydrogen yield from glucose increased twofold, and hydrogen production from starch was also achieved by heterologously expressing amyE encoding an alpha‐type amylase derived from Bacillus subtilis (Akhtar and Jones, 2009).
Protein engineering to enhance hydrogen production
Unlike metabolic engineering and heterologous‐gene cloning strategies, protein engineering studies to improve Hyd enzyme function have not been developed extensively. The primary reason may be that there have been few high‐throughput methods to readily measure Hyd activity (either directly or indirectly) since for protein engineering, a beneficial screening method is critical to find an improved Hyd variant among many candidate variants fabricated through error‐prone PCR (epPCR), DNA shuffling, or saturation mutagenesis. There are several relatively easy methods to assay Hyd activity; for example, a facile method to detect formate concentrations consumed by the E. coli FHL by using potassium permanganate (Maeda and Wood, 2008) but this method is not a direct measure of Hyd activity. Also, there is a method to directly assay hydrogen gas by a chemical reaction (Bagramyan and Martirosov, 1989), but this method is not easily adapted to a high‐throughput screen. Furthermore, many groups measure hydrogen uptake by methyl viologen (Krasna, 1979) and reason that Hyd enzymes are reversible so improvements in uptake should lead to improved hydrogen formation; however, this approach is not optimal since you usually get what you screen for and little else.
The best high throughput screens to date are (i) a chemochromic membrane, which detects hydrogen produced from single colonies via a blue‐colour change, which is triggered by binding of hydrogen to a thin‐film WO3 sensor attached to the membrane (Seibert et al., 1999); and (ii) a high‐throughput screening assay apparatus, which detects hydrogen directly based on the chemical reaction between a water‐soluble tetrazolium colour indicator and hydrogen (Schrader et al., 2008). These tools should prove quite useful for protein engineering.
The first study to evolve a hydrogenase, [Fe‐Fe] HydA from Clostridia sp., utilized DNA shuffling of the hydrogenases of C. acetobutylicum and C. saccharobutylicum (Nagy et al., 2007); however, no screening method was used and little improvement was obtained. The first protein engineering using a high‐throughput screening method (via chemochromic membranes) evolved the large subunit of E. coli Hyd 3, HycE, by epPCR, DNA shuffling, and saturation mutagenesis; the best epPCR variant, epHycE95, had amino‐acid changes S2T, Y50F, I171T, A291V, T366S, V433L, M444I and L523Q, which led to 17‐fold better hydrogen productivity compared with wild‐type HycE (Maeda et al., 2008b). DNA shuffling using the best three variants from epPCR created a HycE variant with a truncation at Y464, and the truncated HycE had 23‐fold higher hydrogen production compared with wild‐type HycE. Saturation mutagenesis at T366, which was identified via a common residue change in two epPCR variants, led to 30‐fold higher activity compared with the wild‐type HycE (Maeda et al., 2008b).
Hydrogen production has also been enhanced by protein engineering of FhlA, the FHL transcriptional activator of transcription of the fdhF (formate dehydrogenase‐H), hyc (Hyd 3), hyp and hydN‐hypF (maturation proteins) operons. Through random mutagenesis using epPCR over the whole gene as well as over the N‐terminal region of FhlA, a variant was identified (Q11H, L14V, Y177F, K245R, M288K and I342) with ninefold increased hydrogen production (Sanchez‐Torres et al., 2009). As expected, the mechanism by which the engineered FhlA variant improved hydrogen production was via increased transcription of all of the genes activated by FhlA (Sanchez‐Torres et al., 2009).
Protein engineering of the [Fe‐Fe]‐type Hyd HydA1 from Chlamydomonas reinhardtii was performed using an in vitro screen (Stapleton and Swartz, 2010) based on hydrogen uptake (using methyl viologen). The HydA1 variants were constructed through a SIMPLEX (SIngle‐Molecule PCR‐Linked EXpression) method (Rungpragayphan et al., 2003), which enables variants to be synthesized in vitro directly from the epPCR products in a cell‐free protein synthesis system. One variant with fourfold higher activity than the original enzyme was obtained (Stapleton and Swartz, 2010).
Lastly, site‐directed mutagenesis of HydA from C. reinhardtii was performed to arrange the ferredoxin‐binding surface of HydA (Agapakis et al., 2010). Based on a previous structural model for the interaction between Hyd and its cognate [2Fe‐2S]‐ferredoxin, four amino acids at the 2nd, 5th, 119th and 126th codons were selected and since ferredoxin is rich in negatively charged residues, these amino acids were replaced with lysines to confer a positive charge at the Hyd binding surface. As a result, it was demonstrated that the mutations in HydA, E5K and D126K, were important for enhanced hydrogen production (Agapakis et al., 2010).
Cell‐free systems versus whole‐cell systems for hydrogen production
To date, almost all bacterial hydrogen production is based on whole‐cell systems; however, in vitro hydrogen production by cell‐free systems has gained attention. In general, hydrogenases are sensitive to oxygen (Vincent et al., 2005) and this includes the E. coli hydrogenases (Glick et al., 1980). Exceptions are the hydrogenases from Ralstonia eutropha (Van der Linden et al., 2004) and Aquifex aeolicus (Pandelia et al., 2010), which are somewhat oxygen‐resistant; hence, hydrogenase‐related experiments are conducted under anaerobic conditions. Therefore, whole‐cell systems are more robust since it is easier to recover hydrogenase activity with intact cells than via cell‐free systems. The next problem to surmount in in vitro systems is the maturation of most hydrogenases (e.g. nickel insertion, iron–sulfur formation or C‐terminus cleavage of hydrogenases). Therefore, many genes have to be expressed in an active state in cell‐free systems. For example, the heterologous expression of a Pyrococcus furiosus Hyd required 13 genes (4 structural genes and 9 maturation genes) (Sun et al., 2010). Furthermore, the stability of complex hydrogenases and their maturation enzymes in cell‐free systems needs to be addressed as well as the scale at which these systems may be operated (gram rather than nanogram quantities required). Lastly, purification of a large number of complex enzymes and cofactors like NADP+ may not be cost effective.
In contrast to whole‐cell systems, cell‐free hydrogen production systems have the advantages that synthetic pathways to produce more hydrogen may be more readily conceived and their efficiencies are much greater. The first group to show such high efficiencies exist for cell‐free systems utilized 11 enzymes from the pentose phosphate cycle along with a hydrogenase from Pyrococcus furiosus to generate 11.6 mol of hydrogen per mol glucose or 97% of the maximum stoichiometric yield (Woodward et al., 2000). Next, Zhang et al. (2007) extended this system by using 13 enzymes in vitro to convert starch into hydrogen using basically the same system with two additional enzymes to process glucan.
Perspectives
Microbial biotechnology is a promising approach to enhance bacterial hydrogen production (De Genève and Fernández, 2008; Ramos et al., 2008) as it should lead to biotechnology processes that utilize substrates efficiently, have high capacities, have fast sugar transport, tolerate inhibitors and end products, and have high metabolic fluxes and ultimately produce a single fermentation product such as hydrogen (Alper and Stephanopoulos, 2009). To date, there are several engineered E. coli strains that have overcome two major technical problems: (i) slow hydrogen production (Yoshida et al., 2005; Maeda et al., 2008a; Kim et al., 2009); and (ii) low hydrogen yield (Chittibabu et al., 2006; Yoshida et al., 2006; Maeda et al., 2008a), and these strains hold promise for further improvements in hydrogen production since there remain uncharacterized deleterious proteins as well as beneficial ones for hydrogen production in E. coli. To date, utilizing native E. coli hydrogenases has resulted in higher rates of hydrogen production (Table 1).
It has been estimated (Vardar‐Schara et al., 2008) that the annual cost to maintain 1 kW of electricity generated from hydrogen may be ∼ $172 000 when formate is used [assuming $12 per kg formate as 1 mol H2 per mol formate) and may be ∼ $4600 when glucose is used (assuming $0.22 per kg glucose and 1.8 mol H2 per mol glucose (Mathews et al., 2010)]. However, the current annual cost for 1 kW of electricity in the residential sector is ∼ $1000, which is generated mainly from fossil fuels (69%) (Booth and Spangler, 2011). Hence, a combination of increasing the hydrogen yield and reducing the cost of the substrate is required to obtain a 4.6‐fold decrease in cost for hydrogen. Calculated another way, for using hydrogen as fuel for transportation, the US Department of Energy targeted a hydrogen cost goal by 2015 of $2.00–3.00 per gallon of gasoline equivalent (gge) (DOE, 2005). Considering that the energy content of 1 kg of H2 is equivalent to 1 gallon of gasoline (DOE, 2005), the estimated feedstock cost to produce hydrogen is ∼ $11 gge−1[based on 1.8 mol H2 per mol glucose (Mathews et al., 2010)]. Hence, again, about a fourfold reduction in cost is required. One way to reduce costs is to utilize low cost renewable resources for hydrogen production since feedstock costs account for 60% of overall costs for hydrogen production (Zhang, 2009). Since the maximum hydrogen yield in native E. coli is 2 mol of hydrogen from 1 mol of glucose (Hallenbeck and Ghosh, 2009), additional metabolic engineering is required to utilize by‐products such as acetic acid to increase the hydrogen yield. In addition, protein engineering strategies (Maeda et al., 2008b) can continue to improve enzymes for in vivo/in vitro biological hydrogen production and to stabilize hydrogenases (e.g. increase oxygen tolerance).
Given the current limitations of cell‐free systems (enzyme and cofactor costs, enzyme instability, oxygen inactivation, requirements for maturation proteins, etc.), it appears whole‐cell systems are more likely to be used in the near future to generate hydrogen. The proposal (Zhang, 2009) to utilize a minimal microbe (Glass et al., 2006) to produce a synthetic hydrogen‐producing pathway or to create a hydrogen‐producing strain by total synthesis of the whole genomic sequence as has been done previously (Gibson et al., 2008) may provide breakthroughs for bacterial hydrogen production using whole‐cell systems in that it would allow one to design a synthetic system that uses all of the electron equivalents for hydrogen production in order to increase efficiency (i.e., the yield) of whole‐cell systems. Additional engineering is also required to create cells to produce hydrogen from renewable resources such as kitchen garbage (containing mainly sugar substrates) and waste wood (targeting cellulosic substrate) since this is where these strains are mostly to be utilized first. Therefore, significant improvements in reaction rates as well as yields in engineered E. coli strains are expected.
Acknowledgments
This research was supported by the National Science Foundation (CBET‐0753702) and the Japan Society for the Promotion of Science (22780070 and 21–7007).
References
- Abdel‐Hamid A.M., Attwood M.M., Guest J.R. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology. 2001;147:1483–1498. doi: 10.1099/00221287-147-6-1483. [DOI] [PubMed] [Google Scholar]
- Agapakis C.M., Ducat D.C., Boyle P.M., Wintermute E.H., Way J.C., Silver P.A. Insulation of a synthetic hydrogen metabolism circuit in bacteria. J Biol Eng. 2010;4:3. doi: 10.1186/1754-1611-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M.K., Jones P.R. Deletion of iscR stimulates recombinant clostridial Fe‐Fe hydrogenase activity and H2‐accumulation in Escherichia coli BL21(DE3) Appl Microbiol Biotechnol. 2008a;78:853–862. doi: 10.1007/s00253-008-1377-6. [DOI] [PubMed] [Google Scholar]
- Akhtar M.K., Jones P.R. Engineering of a synthetic hydF‐hydE‐hydG‐hydA operon for biohydrogen production. Anal Biochem. 2008b;373:170–172. doi: 10.1016/j.ab.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Akhtar M.K., Jones P.R. Construction of a synthetic YdbK‐dependent pyruvate:H2 pathway in Escherichia coli BL21(DE3) Metab Eng. 2009;11:139–147. doi: 10.1016/j.ymben.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Alper H., Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol. 2009;7:715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- Alves M.M., Pereira M.A., Sousa D.Z., Cavaleiro A.J., Picavet M., Smidt H., Stams A.J.M. Waste lipids to energy: how to optimize methane production from long‐chain fatty acids (LCFA) Microb Biotechnol. 2009;2:538–550. doi: 10.1111/j.1751-7915.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.C., Berks B.C., McClay J., Ambler A., Quail M.A., Golby P., Guest J.R. A 12‐cistron Escherichia coli operon (hyf) encoding a putative proton‐translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- Angelides K.J., Akiyama S.K., Hammes G.G. Subunit stoichiometry and molecular weight of the pyruvate dehydrogenase multienzyme complex from Escherichia coli. Proc Natl Acad Sci USA. 1979;76:3279–3283. doi: 10.1073/pnas.76.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armaroli N., Balzani V. The hydrogen issue. ChemSusChem. 2011;4:21–36. doi: 10.1002/cssc.201000182. [DOI] [PubMed] [Google Scholar]
- Axley M.J., Grahame D.A., Stadtman T.C. Escherichia coli formate‐hydrogen lyase. Purification and properties of the selenium‐dependent formate dehydrogenase component. J Biol Chem. 1990;265:18213–18218. [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagramyan K.A., Martirosov S.M. Formation of an ion transport supercomplex in Escherichia coli. An experimental model of direct transduction of energy. FEBS Lett. 1989;246:149–152. doi: 10.1016/0014-5793(89)80272-8. [DOI] [PubMed] [Google Scholar]
- Bagramyan K., Trchounian A. Structural and functional features of formate hydrogen lyase, an enzyme of mixed‐acid fermentation from Escherichia coli. Biochem Mosc. 2003;68:1159–1170. doi: 10.1023/b:biry.0000009129.18714.a4. [DOI] [PubMed] [Google Scholar]
- Ballantine S.P., Boxer D.H. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur J Biochem. 1986;156:277–284. doi: 10.1111/j.1432-1033.1986.tb09578.x. [DOI] [PubMed] [Google Scholar]
- Bertero M.G., Rothery R.A., Palak M., Hou C., Lim D., Blasco F. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. et al. [DOI] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M. The complete genome sequence of Escherichia coli K‐12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. et al. [DOI] [PubMed] [Google Scholar]
- Blokesch M., Paschos A., Theodoratou E., Bauer A., Hube M., Huth S., Böck A. Metal insertion into NiFe‐hydrogenases. Biochem Soc Trans. 2002;30:674–680. doi: 10.1042/bst0300674. [DOI] [PubMed] [Google Scholar]
- Böck A., Sawers G. Cellular and molecular biology. In: Neidhardt F.C., Curtiss J.R., Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., editors. ASM Press; 1996. pp. 262–282. , and . In Escherichia coli and Salmonellaet al (eds). Washington, DC, USA: , pp. [Google Scholar]
- Booth W., Spangler M.R. USEI. US Department of Energy; 2011. Electric Power Annual 2009; pp. 54–66. [Google Scholar]
- Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp‐catalyzed excision of the antibiotic‐resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Chittibabu G., Nath K., Das D. Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL‐21. Process Biochem. 2006;41:682–688. [Google Scholar]
- Das D., Veziroglu T.N. Hydrogen production by biological processes: a survey of literature. Int J Hydrogen Energy. 2001;26:13–28. [Google Scholar]
- Datsenko K.A., Wanner B.L. One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Genève J., Fernández M. New avenues for microbial biotechnology: the beginning of a golden era. Microb Biotechnol. 2008;1:104–106. doi: 10.1111/j.1751-7915.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do P.M., Angerhofer A., Hrdy I., Bardonova L., Ingram L.O., Shanmugam K.T. Engineering Escherichia coli for fermentative dihydrogen production: potential role of NADH‐ferredoxin oxidoreductase from the hydrogenosome of anaerobic protozoa. Appl Biochem Biotechnol. 2009;153:21–33. doi: 10.1007/s12010-008-8508-5. [DOI] [PubMed] [Google Scholar]
- DOE. 2005. DOE announces new hydrogen cost goal. [WWW document]. URL http://www.hydrogen.energy.gov/news_cost_goal.html.
- Domka J., Lee J., Bansal T., Wood T.K. Temporal gene‐expression in Escherichia coli K‐12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Drapal N., Böck A. Interaction of the Hydrogenase Accessory Protein HypC with HycE, the Large Subunit of Escherichia coli Hydrogenase 3 during Enzyme Maturation. Biochemistry. 1998;37:2941–2948. doi: 10.1021/bi9720078. [DOI] [PubMed] [Google Scholar]
- Dunn S. Hydrogen futures: toward a sustanable energy system. Int J Hydrogen Ener. 2002;27:235–264. [Google Scholar]
- Edwards P.P., Kuznetsov V.L., David W.I. Hydrogen energy. Philos Transact A Math Phys Eng Sci. 2007;365:1043–1056. doi: 10.1098/rsta.2006.1965. [DOI] [PubMed] [Google Scholar]
- Ewan B.C.R., Allen R.W.K. A figure of merit assessment of the routes to hydrogen. Int J Hydrogen Energy. 2005;30:809–819. [Google Scholar]
- Fan Z., Yuan L., Chatterjee R. Increased hydrogen production by genetic engineering of Escherichia coli. PLoS ONE. 2009;4:e4432. doi: 10.1371/journal.pone.0004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzi L., Sawers R.G. Maturation of [NiFe]‐hydrogenases in Escherichia coli. Biometals. 2007;20:565–578. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Benders G.A., Andrews‐Pfannkoch C., Denisova E.A., Baden‐Tillson H., Zaveri J. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. et al. [DOI] [PubMed] [Google Scholar]
- Glass J.I., Assad‐Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M. Essential genes of a minimal bacterium. Proc Natl Acad Sci USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.R., Wang P.Y., Schneider H., Martin W.G. Identification and partial characterization of an Escherichia coli mutant with altered hydrogenase activity. Can J Biochem. 1980;58:361–367. doi: 10.1139/o80-047. [DOI] [PubMed] [Google Scholar]
- Hallenbeck P.C., Ghosh D. Advances in fermentative biohydrogen production: the way forward? Trends Biotechnol. 2009;27:287–297. doi: 10.1016/j.tibtech.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Hansel A., Lindblad P. Toward optimization of cyanobacteria as biotechnologically relevant producers of molecular hydrogen, a clean and renewable energy source. Appl Microbiol Biotechnol. 1998;50:153–160. [Google Scholar]
- Herzberg M., Kaye I.K., Peti W., Wood T.K. YdgG (TqsA) controls biofilm formation in Escherichia coli K‐12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Wood T.K. An evolved Escherichia coli strain for producing hydrogen and ethanol from glycerol. Biochem Biophys Res Commun. 2010;391:1033–1038. doi: 10.1016/j.bbrc.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Karube I., Urano N., Yamada T., Hirochika H., Sakaguchi K. Cloning and expression of the hydrogenase gene from Clostridium butyricum in Escherichia coli. FEBS Lett. 1983;158:119–122. doi: 10.1016/0014-5793(83)80689-9. [DOI] [PubMed] [Google Scholar]
- Kessel D.G. Global warming – facts, assessment, countermeasures. J Petrol Sci Eng. 2000;26:157–168. [Google Scholar]
- Kim S., Seol E., Oh Y.K., Wang G.Y., Park S. Hydrogen production and metabolic flux analysis of metabolically engineered Escherichia coli strains. Int J Hydrogen Energy. 2009;34:7417–7427. [Google Scholar]
- Kim J.Y.H., Jo B.H., Cha H.J. Production of biohydrogen by recombinant expression of [NiFe]‐ hydrogenase 1 in Escherichia coli. Microb Cell Fact. 2010;9:54. doi: 10.1186/1475-2859-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King P.W., Przybyla A.E. Response of hya expression to external pH in Escherichia coli. J Bacteriol. 1999;181:5250–5256. doi: 10.1128/jb.181.17.5250-5256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Ara T., Arifuzzaman M., Ioka‐Nakamichi T., Inamoto E., Toyonaga H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K‐12 ORF Archive): unique resources for biological research. DNA Res. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Koku H., Eroğlu İ., Gündüz U., Yücel M., Türker L. Aspects of the metabolism of hydrogen production by Rhodobacter sphaeroides. Int J Hydrogen Energy. 2002;27:1315–1329. [Google Scholar]
- Krasna A.I. Hydrogenase: properties and applications. Enzyme Microb Technol. 1979;1:165–172. [Google Scholar]
- Laffly E., Garzoni F., Fontecilla‐Camps J.C., Cauazza C. Maturation and processing of the recombinant [FeFe] hydrogenase from Desulfovibrio vulgaris Hildenborough (DvH) in Escherichia coli. Int J Hydrogen Energy. 2010;35:10761–10769. [Google Scholar]
- Lee S.J., Lee D.Y., Kim T.Y., Kim B.H., Lee J., Lee S.Y. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl Environ Microbiol. 2005;71:7880–7887. doi: 10.1128/AEM.71.12.7880-7887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Lee H.J., Park J.M., Lee J.H., Park J.S., Shin H.S. Bacterial hydrogen production in recombinant Escherichia coli harboring a HupSL hydrogenase isolated from Rhodobacter sphaeroides under anaerobic dark culture. Int J Hydrogen Energy. 2010;35:1112–1116. et al. [Google Scholar]
- Maeda T., Wood T.K. Formate detection by potassium permanganate for enhanced hydrogen production in Escherichia coli. Int J Hydrogen Energy. 2008;33:2409–2412. [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Appl Microbiol Biotechnol. 2007a;76:1035–1042. doi: 10.1007/s00253-007-1086-6. [DOI] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007b;77:879–890. doi: 10.1007/s00253-007-1217-0. [DOI] [PubMed] [Google Scholar]
- Maeda T., Vardar G., Self W.T., Wood T.K. Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC Biotechnol. 2007c;7:25. doi: 10.1186/1472-6750-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2008a;1:30–39. doi: 10.1111/j.1751-7915.2007.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Protein engineering of hydrogenase 3 to enhance hydrogen production. Appl Microbiol Biotechnol. 2008b;79:77–86. doi: 10.1007/s00253-008-1416-3. [DOI] [PubMed] [Google Scholar]
- Manish S., Venkatesh K.V., Banerjee R. Metabolic flux analysis of biological hydrogen production by Escherichia coli. Int J Hydrogen Energy. 2007;32:3820–3830. [Google Scholar]
- Mathews J., Li Q.Z., Wang G.Y. Characterization of hydrogen production by engineered Escherichia coli strains using rich defined media. Biotechnol Bioprocess Eng. 2010;15:686–695. [Google Scholar]
- Mishra J., Khurana S., Kumar N., Ghosh A.K., Das D. Molecular cloning, characterization, and overexpression of a novel [Fe]‐hydrogenase isolated from a high rate of hydrogen producing Enterobacter cloacae IIT‐BT 08. Biochem Biophys Res Commun. 2004;324:679–685. doi: 10.1016/j.bbrc.2004.09.108. [DOI] [PubMed] [Google Scholar]
- Murarka A., Dharmadi Y., Yazdani S.S., Gonzalez R. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microbiol. 2008;74:1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L.E., Meuser J.E., Plummer S., Seibert M., Ghirardi M.L., King P.W. Application of gene‐shuffling for the rapid generation of novel [FeFe]‐hydrogenase libraries. Biotechnol Lett. 2007;29:421–430. doi: 10.1007/s10529-006-9254-9. et al. [DOI] [PubMed] [Google Scholar]
- Pandelia M.E., Infossi P., Giudici‐Orticoni M.T., Lubitz W. The oxygen‐tolerant hydrogenase I from Aquifex aeolicus weakly interacts with carbon monoxide: an electrochemical and time‐resolved FTIR study. Biochemistry. 2010;49:8873–8881. doi: 10.1021/bi1006546. [DOI] [PubMed] [Google Scholar]
- Penfold D.W., Macaskie L.E. Production of H2 from sucrose by Escherichia coli strains carrying the pUR400 plasmid, which encodes invertase activity. Biotechnol Lett. 2004;26:1879–1883. doi: 10.1007/s10529-004-6035-1. [DOI] [PubMed] [Google Scholar]
- Penfold D.W., Forster C.F., Macaskie L.E. Increased hydrogen production by Escherichia coli strain HD701 in comparison with the wild‐type parent strain MC4100. Enzyme Microb Technol. 2003;33:185–189. [Google Scholar]
- Penfold D.W., Sargent F., Macaskie L.E. Inactivation of the Escherichia coli K‐12 twin‐arginine translocation system promotes increased hydrogen production. FEMS Microbiol Lett. 2006;262:135–137. doi: 10.1111/j.1574-6968.2006.00333.x. [DOI] [PubMed] [Google Scholar]
- Ramos J.L., Fernández M., Solano J., Pizarro‐Tobías P., Daniels C. Environmental microbiology meets microbial biotechnology. Microb Biotechnol. 2008;1:443–445. doi: 10.1111/j.1751-7915.2008.00068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D.J., Sawers G., Sargent F., McWalter L., Boxer D.H. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology. 1999;145:2903–2912. doi: 10.1099/00221287-145-10-2903. [DOI] [PubMed] [Google Scholar]
- Rosales‐Colunga L.M., Razo‐Flores E., Ordoñez L.G., Alatriste‐Mondragón F., De León‐Rodríguez A. Hydrogen production by Escherichia coliΔhycAΔlacI using cheese whey as substrate. Int J Hydrogen Energy. 2010;35:491–499. [Google Scholar]
- Rossmann R., Sawers G., Böck A. Mechanism of regulation of the formate‐hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol. 1991;5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- Rungpragayphan S., Nakano H., Yamane T. PCR‐linked in vitro expression: a novel system for high‐throughput construction and screening of protein libraries. FEBS Lett. 2003;540:147–150. doi: 10.1016/s0014-5793(03)00251-5. [DOI] [PubMed] [Google Scholar]
- Sabourin‐Provost G., Hallenbeck P.C. High yield conversion of a crude glycerol fraction from biodiesel production to hydrogen by photofermentation. Bioresour Technol. 2009;100:3513–3517. doi: 10.1016/j.biortech.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Torres V., Maeda T., Wood T.K. Protein engineering of the transcriptional activator FhlA to enhance hydrogen production in Escherichia coli. Appl Environ Microbiol. 2009;75:5639–5646. doi: 10.1128/AEM.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., Bohm R., Bock A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Sawers R.G., Ballantine S.P., Boxer D.H. Differential expression of hydrogenase isoenzymes in Escherichia coli K‐12: evidence for a third isoenzyme. J Bacteriol. 1985;164:1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlensog V., Lutz S., Böck A. Purification and DNA‐binding properties of FHLA, the transcriptional activator of the formate hydrogenlyase system from Escherichia coli. J Biol Chem. 1994;269:19590–19596. [PubMed] [Google Scholar]
- Schrader P.S., Burrows E.H., Ely R.L. High‐throughput screening assay for biological hydrogen production. Anal Chem. 2008;80:4014–4019. doi: 10.1021/ac702633q. [DOI] [PubMed] [Google Scholar]
- Seibert M., Flynn T., Benson D., Tracy E., Ghirardi M. Development of selection and screening procedures for rapid identification of H2‐producing algal mutants with increased O2 tolerance. In: Zaborsky O.R., Benemann J.R., Matsunaga T., Miyake J., San Pietro A., editors. Plenum Press; 1999. pp. 227–234. [Google Scholar]
- Self W.T., Hasona A., Shanmugam K.T. N‐terminal truncations in the FhlA protein result in formate‐ and MoeA‐independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology. 2001;147:3093–3104. doi: 10.1099/00221287-147-11-3093. [DOI] [PubMed] [Google Scholar]
- Self W.T., Hasona A., Shanmugam K.T. Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J Bacteriol. 2004;186:580–587. doi: 10.1128/JB.186.2.580-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J.A., Swartz J.R. A cell‐free microtiter plate screen for improved [FeFe] hydrogenases. PLoS ONE. 2010;5:e10554. doi: 10.1371/journal.pone.0010554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Hopkins R.C., Jenney F.E., McTernan P.M., Adams M.W. Heterologous expression and maturation of an NADP‐dependent [NiFe]‐hydrogenase: a key enzyme in biofuel production. PLoS ONE. 2010;5:e10526. doi: 10.1371/journal.pone.0010526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppmann B., Sawers G. Isolation and characterization of hypophosphite‐resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- Turcot J., Bisaillon A., Hallenbeck P.C. Hydrogen production by continuous cultures of Escherchia coli under different nutrient regimes. Int J Hydrogen Energy. 2008;33:1465–1470. [Google Scholar]
- Van der Linden E., Burgdorf T., Bernhard M., Bleijlevens B., Friedrich B., Albracht S.P.J. The soluble [NiFe]‐hydrogenase from Ralstonia eutropha contains four cyanides in its active site, one of which is responsible for the insensitivity towards oxygen. J Biol Inorg Chem. 2004;9:616–626. doi: 10.1007/s00775-004-0555-y. [DOI] [PubMed] [Google Scholar]
- Vardar‐Schara G., Maeda T., Wood T.K. Metabolically‐engineered bacteria for producing hydrogen via fermentation. Microb Biotechnol. 2008;1:107–125. doi: 10.1111/j.1751-7915.2007.00009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit A., Akhtar M.K., Mizutani T., Jones P.R. Constructing and testing the thermodynamic limits of synthetic NAD(P)H:H2 pathways. Microb Biotechnol. 2008;1:382–394. doi: 10.1111/j.1751-7915.2008.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent K.A., Parkin A., Lenz O., Albracht S.P., Fontecilla‐Camps J.C., Cammack R. Electrochemical definitions of O2 sensitivity and oxidative inactivation in hydrogenases. J Am Chem Soc. 2005;127:18179–18189. doi: 10.1021/ja055160v. et al. [DOI] [PubMed] [Google Scholar]
- Wells M.A., Mercer J., Mott R.A., Pereira‐Medrano A.G., Burja A.M., Radianingtyas H., Wright P.C. Engineering a non‐native hydrogen production pathway into Escherichia coli via a cyanobacterial [NiFe] hydrogenase. Metab Eng. 2011;13:445–453. doi: 10.1016/j.ymben.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Willke T., Vorlop K.D. Industrial bioconversion of renewable resources as an alternative to conventional chemistry. Appl Microbiol Biotechnol. 2004;66:131–142. doi: 10.1007/s00253-004-1733-0. [DOI] [PubMed] [Google Scholar]
- Woods D.D. Hydrogenlyases: the synthesis of formic acid by bacteria. Biochem J. 1936;30:515–527. doi: 10.1042/bj0300515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward J., Orr M., Cordray K., Greenbaum E. Biotechnology: enzymatic production of biohydrogen. Nature. 2000;405:1014–1015. doi: 10.1038/35016633. [DOI] [PubMed] [Google Scholar]
- Yi K.B., Harrison D.P. Low‐pressure sorption‐enhanced hydrogen production. Ind Eng Chem Res. 2005;44:1665–1669. [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. Enhanced hydrogen production from formic acid by formate hydrogen lyase‐overexpressing Escherichia coli strains. Appl Environ Microbiol. 2005;71:6762–6768. doi: 10.1128/AEM.71.11.6762-6768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. Enhanced hydrogen production from glucose using ldh‐ and frd‐inactivated Escherichia coli strains. Appl Microbiol Biotechnol. 2006;73:67–72. doi: 10.1007/s00253-006-0456-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y.H.P. A sweet out‐of‐the‐box solution to the hydrogen economy: is the sugar‐powered car science fiction? Energy Environ Sci. 2009;2:272–282. [Google Scholar]
- Zhang Y.‐H.P., Evans B.R., Mielenz J.R., Hopkins R.C., Adams M.W.W. High‐yield hydrogen production from starch and water by a synthetic enzymatic pathway. PLoS ONE. 2007;2:e456. doi: 10.1371/journal.pone.0000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Xing D.F., Zhang L., Ren N.Q. Characterization and overexpression of a [FeFe]‐hydrogenase gene of a novel hydrogen‐producing bacterium Ethanoligenens harbinense. Int J Hydrogen Energy. 2010;35:9598–9602. [Google Scholar]
- Zirngibl C., Van Dongen W., Schwörer B., Von Bünau R., Richter M., Klein A., Thauer R.K. H2‐forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron–sulfur clusters in methanogenic archaea. Eur J Biochem. 1992;208:511–520. doi: 10.1111/j.1432-1033.1992.tb17215.x. [DOI] [PubMed] [Google Scholar]


