Summary
Vaccines are the most effective tools to prevent infectious diseases and to minimize their impact on humans or animals. Despite the successful development of vaccines that are able to elicit potent and protective immune responses, the majority of vaccines have been so far developed empirically and mechanistic events leading to protective immune responses are often poorly understood. This hampers the development of new prophylactic as well as therapeutic vaccines for infectious diseases and cancer. Biological correlates of immune‐mediated protection are currently based on standard readout such as antibody titres and ELISPOT assays. The development of successful vaccines for difficult settings, such as infectious agents leading to chronic infection (HIV, HCV. . .) or cancer, calls for novel ‘readout systems’ or ‘correlates’ of immune‐mediated protection that would reliably predict immune responses to novel vaccines in vivo. Systems biology offers a new approach to vaccine design that is based upon understanding the molecular network mobilized by vaccination. Systems vaccinology approaches investigate more global correlates of successful vaccination, beyond the specific immune response to the antigens administered, providing new methods for measuring early vaccine efficacy and ultimately generating hypotheses for understanding the mechanisms that underlie successful immunogenicity. Using functional genomics, specific molecular signatures of individual vaccine can be identified and used as predictors of vaccination efficiency. The immune response to vaccination involves the coordinated induction of master transcription factors that leads to the development of a broad, polyfunctional and persistent immune response integrating all effector cells of the immune systems.
Immune response and vaccination principles
Vaccines are currently most effective treatments in preventing a number of infectious diseases and minimizing their impact on human or animal populations. A vaccine is a biological preparation that improves immunity to a particular disease upon administration to an animal/human. A vaccine typically contains one or several antigens that resemble a disease‐causing microorganism, and is often made from weakened or killed forms of the microbe or its derived antigenic proteins or its toxins + adjuvant. The antigens stimulate the body's immune system to recognize the agent as foreign, induces specific immune responses (immunogenicity). Vaccines can be prophylactic (e.g. to prevent or reduce the effects of a future infection by any natural pathogen), or therapeutic (e.g. vaccines against cancer). An ideal vaccine must have particular biological and physical characteristics, and vaccine development must gain from new technology advances (Levine and Sztein, 2004).
Immune responses induced by vaccine (Fig. 1)
Figure 1.
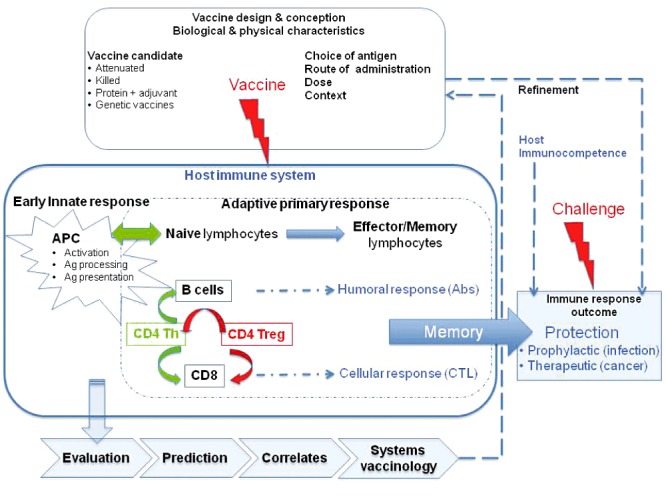
Systems vaccinology: from vaccine conception and design to protection of the organism. The goal of a good vaccine is to obtain a sustained immune memory in T‐ and/or B‐cell compartments, allowing the host to respond more rapidly and with more efficiency to gain the race against an infectious challenge (prophylactic vaccines) or a tumour (therapeutic vaccines). Note that the design of the vaccine and the immunocompetence of the host are determinant for triggering an effective protection. After designing a vaccine, systems vaccinology is helpful to evaluate the host complex immune response at various levels of biological organization (from molecule to organism), considering actors from the innate or adaptive immune system. This approach establishes predictions, correlates or models that help to validate and refine the vaccine.
Following vaccination, both the innate and the adaptive immune systems synergize to elicit an immune response. Indeed, after vaccine inoculation, antigen‐presenting cells – notably dendritic cells – take up antigens and traffic to the draining lymph nodes where they present processed antigens to naïve CD4+ and CD8+ T lymphocytes (CTL). Naïve T cells are stimulated to proliferate and differentiate into effector and memory T cells. Activated, effector and memory CD4+ T cells provide help to B cells to mount antibody responses, and to naïve CD8+ T cells to enhance their antigen‐driven clonal expansion and differentiation into cytotoxic CTL. Most antigens and vaccines trigger both B‐ and T‐cell responses, such that there is no rationale in opposing antibody production (humoral immunity) and T‐cell responses (cellular immunity). Nevertheless, the nature of the vaccine exerts a direct influence on the type of immune effectors that are predominantly elicited to mediate protective efficacy. The quality of the vaccine‐induced immune response depends on several factors, e.g. nature of antigen, route of administration, quality of antigen presentation, vaccine preparation adjuvants, timing between challenges. Moreover, the state of immunocompetence of subjects receiving the vaccine preparation can modify response efficacy due to their altered lymphocyte repertoires or antigen‐presenting cell potential (Goriely and Goldman, 2007; Chen et al., 2009).
Recently, the importance of the innate immune response has been recognized in determining the orientation of the quality and quantity of the adaptive immune response. Indeed, the discovery of pathogen recognition receptors such as Toll‐like receptor, expressed at the surface of most immune system cells and triggered in a specific manner upon ligation of various classes of pathogen‐associated molecular patterns, places this initial recognition event as a key to the overall immune response. This context accounts for the antigen (carried in a vaccine preparation) to be taken up by the innate immune system cells, namely dendritic cells, which will in turn present this antigen to T lymphocytes and initiate the adaptive immune response. Clearly, this double‐step recognition system adds to the complexity of the activation and regulation of the immune system, and places the innate response and associated inflammatory processes as key actors of the immune response when considering vaccine development (Medzhitov, 2010; Schenten and Medzhitov, 2011). Therefore, in addition to the choice of antigen, the context of vaccine inoculation has to be carefully evaluated.
Immune responses required for protection
The immune mechanisms that induce immune protection are variable depending on the nature of pathogen and on the available TCR and BCR lymphocyte repertoires of the host. To be effective a vaccine should be capable of eliciting a number of features:
Favourable activation of antigen‐presenting cells to initiate antigen processing and presentation to T cells;
Broad activation of T and B cells to give a high yield of memory cells;
Generation of memory B cells, production of antibodies, in response to antigen‐specific B‐cell activation that neutralize infectious agents by binding specifically to their surface. In addition, antibodies can target the invading pathogen for destruction by either complement or antibody‐dependent cellular cytotoxicity;
Generation of memory CTL to limit the spread of infectious agents by recognizing and killing infected cells or secreting specific antiviral cytokines;
Generation of memory CD4+ T cells that do not prevent but participate to the reduction, control and clearance of pathogens by producing cytokines that support activation and differentiation of B cells and CTL;
Generation of memory T cells to several epitopes, to overcome MHC variations across the population and limit immune response escape of the pathogen;
Limit the recruitment/expansion of regulatory T lymphocytes (Treg), concomitant to the induction of vaccine‐specific immune responses; and
Long‐term immune memory persistence, often related to persistent antigen presentation and chronic infection.
Before the 1990s, most vaccination programs have been developed and evaluated based on the efficacy of vaccine to induce high titres of antibodies and vaccine programs have not focused on the quality of the T‐cell response to provide protection against disease. Noteworthy, most successful vaccines currently in use were developed with little understanding of cellular immune responses nor memory responses.
Evaluation of vaccine efficacy
To generate vaccine‐mediated protection is a complex challenge when the rules and identity of vaccine‐induced immune correlates of protection are poorly characterized. Vaccine protective efficacy is primarily conferred by the induction of antigen‐specific antibodies. However, the peak of vaccine‐induced antibody titres (i.e. antibody response quantity) does not solely explain antibody‐mediated protection. The quality of such antibody responses (e.g. their avidity, epitope mapping, glycosylation, diversity. . .), as well as that of the associated T‐cell response, have been identified as determining factors of high‐affinity antibody responses and efficient immune memory. Different types of memory T cells (central‐memory and effector‐memory) have been identified based on their functional and migratory properties and therefore contribute differently to the long‐lasting cellular immunity (Zielinski et al., 2011).
New methods have emerged to assess a growing number of vaccine‐associated immune parameters, raising questions relative to the optimal markers to study and their correlation with vaccine‐induced protection. The primary vaccine efficacy surrogate markers have traditionally been the antibody titre to vaccine antigens or the measurement of antibody function such as antiviral neutralizing activity. More recently, the measurement of T‐cell function in conjunction, with or without antibody measurements, has been used to assess vaccine efficacy (e.g. measurement of epitope immunoreactivity at the individual cell level using the ELISPOT, simultaneous measurement of intracellular cytokine production and cell phenotype using flow cytometry, binding of tetramers to cell surface receptors). New biomarkers evaluate T‐cell functions (e.g. memory, helper, effector), as well as T‐cell interactions with other cells of the immune system such as dendritic or antigen presenting cells. Notably, as exemplified by the development of multicolour flow cytometry (see below), these methods assess vaccine efficacy at the individual immune cell level rather than measuring the total immune response.
Current ways to monitor vaccine immunogenicity provide very limited information and substantial efforts are required to propose comprehensive immunological assessments. Indeed, induction of antigen‐specific memory immune responses does not imply that these antibodies, memory cells or cytokines represent surrogates – or even correlates – of vaccine efficacy. Moreover, such potential surrogate markers for vaccine efficacy are vaccine‐dependent, therefore not necessarily applicable to all vaccines. These considerations highlight the requirement to develop new methods for measuring early vaccine efficiency. Thus, an integrated evaluation of vaccine efficiency must consider various multiscale and multiparametric variables:
potential efficiency of the host (age‐, disease‐ or treatment‐related immunodeficiency);
antigenicity of the vaccine preparation;
cellular and humoral immunogenicity; and
effective immune protection of the organism.
Investigations should be done at the various scales of the organism response (protection) to the lower levels (i.e. cell populations, cell, gene). The goal is to establish correlates between protection and cellular and molecular responses: mucosal response, local antibody production, timely B‐ and T‐cell responses, appropriate effector or regulatory biological pathways. Systems immunology provides new tools to assess such responses, to investigate the dynamics and repertoire modifications following an immunization, to better understand the mechanisms of cell activation, and derive models of efficient vaccine‐induced immune responses.
Rationale of vaccine evaluation
A central goal of vaccine research is to identify when an (early) vaccine‐induced immune response is predictive of later efficient protection from infection or disease. Correlates of protection after vaccination are often relative to various factors such as pathogen prevalence levels, host factor deficiency or HLA polymorphism. An immune correlate can be used for guiding vaccine development and refinement, for predicting vaccine efficacy in different settings, and for guiding vaccination policies and regulatory decisions.
However, traditional immunomonitoring methods used to depict vaccine immunogenicity are not suitable to predict vaccine efficacy. For example, assays for antibodies are often based on binding of antigen, when binding antibodies do not necessarily have functions. It is also obvious that cell‐mediated immune functions are critical in protecting against intracellular infections, and in almost all diseases, CD4+ cells are necessary to help B‐cell development. The best case of cellular immunity is the Bacillus Calmette‐Guérin (BCG) vaccine against tuberculosis. All attempts to develop better protection against tuberculosis are based on improving cellular responses to BCG vaccine, but at the moment, no true correlate is known. Production of IFNγ by CD4+ T cells is necessary to prevent disease after exposure but is not an adequate correlate of BCG vaccine‐induced protection (Black et al., 2002; Mittrücker et al., 2007).
The possibility of finding some way to define the key, early correlates of vaccination would therefore be an invaluable tool but represents a considerable challenge. In particular, the ability to read out such information early after vaccination would greatly modify the development and evaluation of new vaccines. To this end, the expanding knowledge on the molecular mechanisms of immune responses, the availability of high throughput genomic and proteomic technologies and the development of integrative Systems Biology offer new approaches for modelling vaccine‐induced immune responses and open the possibility to establish predictive signatures of effective responses.
In this line, a wealth of complementary immunomonitoring technologies has emerged in order to follow a number immunological parameters related to measure vaccine‐induced immune responses. These include the evaluation of (i) innate immune responses, e.g. dendritic cell activation (Pulendran, 2004), inflammatory response, complement activation, (ii) adaptive antibody responses, e.g. antibody and B‐cell immune responses (Bondada and Robertson, 2003), neutralizing antibodies (Ochsenbauer and Kappes, 2009), non‐neutralizing antibodies, antibody‐dependent cell cytotoxicity, (iii) adaptive T‐cell immune responses, e.g. CTL activity, T‐cell specificity using tetramer/peptides, cytokine production (Seder et al., 2008), and (iv) lymphocyte repertoire diversity, e.g. Immunoscope/CDR3 spectratyping, TCR/Ig rearrangement quantification, TCR/Ig deep sequencing (Boudinot et al., 2008; Boyd et al., 2009). Importantly, the rapid progress in flow cytometry implies that an increasing number of parameters can be looked at simultaneously, which is highly relevant for a more comprehensive assessment of lymphocyte characteristics, in particular their multifunctional profile. The challenge is to globally analyse these multiscale multiparametric high‐throughput data in order to extract characteristic signatures of immune response and vaccine efficiency, and thus predictors of protection against further infectious challenges.
Current state‐of‐the‐art of systems vaccinology
In face of the infinite diversity of pathogenic microbes, existing or yet to come, the immune system has evolved many different weapons and tactics, some global and others specific. Seen from an operational perspective, the immune system is a dynamic and responsive tissue, made of a very large set of diverse, circulating, though interconnected cells with loop/circuit types of interactions.
A better understanding of immune responses can benefit from global approaches aimed not only at studying the individual components involved, but also at studying and modelling (i) the complex interactions between these components and (ii) most importantly their spatial and temporal aspects. Systems biology is developing the tools to tackle this type of complexity, based on analysing large data sets with non‐supervised methods and/or through modelling, to extract/generate statistically significant results, irrespective of preconceived hypotheses (Benoist et al., 2006; Germain et al., 2011). The systems biology framework should provide novel analyses of immune responses, identifying response‐specific signatures and assessing their predictive value. The principle of this approach is to integrate high‐throughput data, e.g. any omics, and produce a model of the immune response triggered.
Historically, progress in vaccine development has come in waves produced by technological revolutions. Current developments translate vaccinology as a combinatorial science, which studies the diversity of pathogens and the complexity of the immune system, throughout screening or immunoinformatic tools. The future advances for vaccine development will be based on taking a systems biology approach to the immune system, leading to the creation of a virtual or in silico immune system capable of complex simulations (Flower and Timmis, 2007; Coward et al., 2010; Germain et al., 2011).
Immunoinformatics modelling and vaccine development
A major goal of immunoinformatics is to develop tools for computational vaccinology and accelerate development of new vaccines. Current approaches applied to vaccinology aim at predicting vaccine immunogenicity allowing its advancement into clinic without the uncertainties of the current vaccine development processes.
Reverse vaccinology
Reverse vaccinology involves the in silico screening of a pathogen entire genome to identify genes encoding proteins with the attributes of good vaccine targets. This reverse approach takes advantage of the increasing availability of whole pathogen genome sequences, either single pathogenic isolate or pan‐genomes (the genomic information from several isolates) of a pathogenic species. Indeed, the genome sequence provides an exhaustive catalogue of virtually all protein antigens that the pathogen can express at any time. Reverse vaccinology thus begins with bioinformatics analysis to identify antigens in silico that are then tested experimentally. This sequence is a reversal of the standard workflow in which analysis requiring culturing the organism comes initially and bioinformatics analysis subsequently. This approach, used originally against meningococcus, allows fast identification of candidate antigen as target for vaccination and provides new solutions for those vaccines that have been difficult or impossible to develop (Rappuoli, 2001; Sette and Rappuoli, 2010). Several curated databases are now developing that provide comprehensive information about experimentally validated antigens, e.g. Protegen (Yang et al., 2011), IEDB (Zhang et al., 2008), AntigenDB (Ansari et al., 2010).
Immunomics
Immunomics or computational vaccinology makes use of modelling of antigen processing and presentation in order to support the T‐cell epitopes mapping. Web‐accessible computational methods have been developed for each of the different antigen processing steps including proteasome cleavage, transport by the transporter associated with antigen processing, binding of peptides to MHC molecules and presentation on the cell surface. (Brusic et al., 2004; De Groot, 2006; Flower, 2007; Flower et al., 2010). For example, PEPVAC (promiscuous epitope‐based VACine) is a tool optimized for the formulation of multi‐epitope vaccines with broad population coverage, using HLA binding profile matrices coupled to filtering for immunoproteasome cleavage using probabilistic modelling (Reche and Reinherz, 2005). OptiTope offers a step‐by‐step interface to assist immunologists in designing epitope‐based vaccines. It relies on an original algorithm that maximizes the overall immunogenicity of an epitope set (Toussaint and Kohlbacher, 2009). Using such prediction tools, novel T‐cell epitopes have been discovered across various targets, including pathogen antigens, cancer antigens, autoantigens and allergens.
A new complementary immunoproteomics technologies called immunosignaturing has developed based on the measuring of serum antibody reactivity spectrum against random sequence peptide arrays, and applied to defining immunosignatures of serum antibody responses against infection or vaccine (Legutki et al., 2010). Although such global technologies pose analysis and interpretation challenges, computer‐aided statistical modelling schemes are developing (Brown et al., 2011). Immunomics is now leading to vaccine informatics combining immunoinformatics algorithms and resources to predict T‐ and B‐cell immune epitopes to in silico protein immunogenicity prediction, systematic transcriptomics and proteomics gene expression analyses, data and literature mining, and Vaccine Ontology formalism in order to offer computer‐based strategies for automated vaccine development (He et al., 2010).
Vaccinomics
A new era of genomic vaccinology and computational prediction methods comes out, enabling systematic screening of multiple complete genomes of pathogens, together with analysis of the variability of pathogens and/or HLA complex. Vaccinomics, a branch of omics, encompasses the fields of immunogenetics and immunogenomics applied to understanding the mechanisms of heterogeneity in immune responses to vaccines (Poland et al., 2008; Poland and Oberg, 2010). It investigates heterogeneity in host genetic markers that results in variations in vaccine‐induced immune responses, with the aim of predicting and minimizing vaccine failures or adverse events. An important study in the field of HIV vaccine development has been recently released concerning a genome‐wide association analysis in a multi‐ethnic cohort of HIV‐1 controllers and progressors. Three hundreds and thirteen genome‐wide significant single‐nucleotide polymorphisms were identified, all located in the MHC locus. Careful sequence analysis and statistical validation identify some specific amino acids of the HLA‐B peptide‐binding groove as accounting for the major genetic impact of host control of HIV‐1 infection (The International HIV Controllers Study, 2010).
Systems vaccinology
While genomics has been successfully used to identify new vaccine antigens, the systems biology framework is also a promising tool for evaluating vaccine‐induced immune responses, identifying response‐specific signatures and assessing their predictive value.
A first proof of concept was brought by Pulendran and colleagues who applied a systems biology approach to study the immune response induced by the yellow fever vaccine, one of the most successful vaccines ever developed (Querec et al., 2009). Their strategy involved immunology, genomics and bioinformatics in order to gain a global picture of the nearly 30 000 genes, proteins and cells participating in immune responses to vaccination. Using this approach, the investigators identified gene expression signatures in the blood a few days after vaccination that could predict, with up to 90% accuracy, the strength of the immune response to the yellow fever vaccine. Sékaly and colleagues made similar observations using functional genomics coupled to polychromatic flow cytometry, showing a strong and coordinated initial response that determines the ensuing efficient polyfunctional and lasting adaptive response (Gaucher et al., 2008). Both studies underline a strong correlation between the early innate immunity‐related events and protective vaccine response. The consistency of these predictive signatures across several trials, for both CD8+ T cell and antibody responses to the yellow fever vaccine, raises the possibility that these rules or their components might have broad applicability for different types of immunogens designed to protect against diverse pathogens.
In this line, the group of Pulendran has recently published a second systems biology‐based study looking at the innate and adaptive immune responses in healthy adults receiving either trivalent inactivated or live attenuated influenza vaccines. Their findings identify early innate response‐related molecular signatures that predict with 90% accuracy later antibody titres. Interestingly, their study unravels a so far unknown role of the calcium/calmodulin‐dependent protein kinase type IV in controlling antigen‐specific antibody production, bringing new insights into vaccine response mechanisms (Nakaya et al., 2011).
Even if some of the predictor genes are common to the Yellow fever and Influenza vaccine studies, there remains to determine whether such signatures could be predictive across various vaccine setups when others would be vaccine‐specific. However, the comparison of two different Influenza vaccines with similar clinical effectiveness but a different capacity to induce antigen‐specific antibody responses led to the identification of early predictive signatures that correlate with antibody titre. Therefore, systems biology approaches permit the observation of a global picture of vaccine‐induced immune responses at an early time point after vaccination. These gene expression signatures of early innate immune activation predict the ensuing adaptive immune responses. Moreover, the key genes in the predictive signatures are not all related to inflammatory immune responses. Thus, in addition to providing a potential tool for the forward assessment of vaccine efficacy, the findings from this systems approach provide a starting point for the development of new hypotheses aimed at elucidating the parameters that control memory T cell and antibody production. Similar studies are now extending to other available successful vaccines aiming at building a reference immunome database of vaccine‐induced responses as compared with baseline measurements before vaccination, as advocated by R. Germain (Germain, 2010). This concerns, for example, HIV (Fonseca et al., 2011) or adjuvant research (Pulendran, 2004; Lindqvist et al., 2011). Another area in which systems biology can bring additional insight is in reinvestigating failed vaccine candidates, such as the Merck MRKAd5 HIV vaccine (Sekaly, 2008), or looking for rules to hypo‐ or non‐responsiveness to vaccines of some recipients, e.g. Pneumococcal and Herpes zoster vaccine response in the elderly (Clinical Trials NCT01307449 & NCT01331161), Hepatitis B virus vaccine non‐responders (US Human & Health Services Grant 1U19AI089987‐01).
Rational development of novel genetic vaccines
The complex combinatorial nature of molecular mechanisms that regulate immune system function has, in the past, limited our ability to fully predict immune responses. By bringing together high‐throughput experimental methods and information technology, our ability to decipher complex interactions that occur in the immune system has significantly improved. The current developments in computational vaccinology, including systemic models of vaccine responses, aim to establish such immune correlates and thus to accelerate the development of effective vaccines. Considerable efforts and specific research program are currently developed with this specific aim. In this line a number of research initiatives have been supported, such as VIOLIN (Vaccine Investigation and Online Information Network, http://www.violinet.org) integrates in a dedicated database of curated vaccine experimental data, a vaccine target prediction algorithm and a vaccine ontology. In the same line, DyNAVacS is a web‐based integrative tool to assist researchers designing and optimizing their DNA vaccine design (Harish et al., 2006).
In the past years, we have contributed to new genetic vaccine design within the scope of CompuVac, a European FP6 integrated project devoted to (i) rational development of a novel platform of genetic vaccines and (ii) standardization of vaccine evaluation, assembled a platform of viral vectors and virus‐like particles and developed standardized protocols and database to comparatively evaluate vector platform efficacy. Vaccine Vector candidate were systematically evaluated for their T‐cell and B‐cell immune response induction in parallel to measure early transcriptome changes in spleen dendritic cells 6 h after vaccine inoculation. Following a typical modelling scheme using unsupervised and supervised algorithms, we have systematically extracted statistically coherent molecular signatures from our transcriptome data across all vaccine vectors analysed. We then constructed a signature‐based random forest T‐cell response prediction model starting from a training set. This prediction model was then validated on an independent test set showing that spleen dendritic cell transcriptome changes only 6 h after vaccine injection are predictive of antigen‐specific T‐cell expansion at day 12 (N. Dérian, B. Bellier, W. Chaara, H.P. Pham, A. Six and D. Klatzmann, in preparation).
Mathematical and computer modelling
Vaccine design and evaluation should also gain from mathematical/in silico models for host/pathogen interactions, and from immune response modelling (see for reviews Cohn and Mata, 2007; Flower and Timmis, 2007). For example, one can consider ImmSim, a cellular automata‐based simulator of immune responses used to compare the behaviour of 64 virtual viruses with various speeds of growth, infectivity level and lethal load. Protection against infection conferred by different vaccine strategies could be tested and showed how different viruses are more susceptible to either antibody or T‐cell‐mediated responses (Kohler et al., 2000). Recently, C‐ImmSim has been developed to extend the original agent‐based model of Seiden and Celada with a simulator engine that represents pathogens, in addition to lymphocyte receptors, and makes use of bioinformatics methods for T‐ and B‐cell epitope prediction. The authors produce various simulations of classical immunization experiments, investigation of the role of MHC haplotype heterozygosity during influenza infection and of the emergence of high‐affinity clonal expansion during chronic infection, thus offering a means to in silico better understanding of immune responses (Rapin et al., 2010). On another line, models of influenza viral epitope spread over years, their spatial dissemination and antisera responses will certainly guide the design of more adapted vaccines (Park et al., 2009). More generally, mathematical and computer modellers have developed strategies to represent immune components with the languages of statecharts (Vainas et al., 2011) or to provide visual simulations of their behaviour with multi‐agent technologies (Chavali et al., 2008). The proof of concept that complex mathematical modelling can be automatically generated from graphical communication medium designed by biologists (McEwan et al., 2011) opens new potential for the development of efficient and predictive models in vaccinology.
Conclusion
Despite their great success, mechanisms describing how effective vaccines stimulate protective immune responses are poorly known. A major challenge in vaccinology is to prospectively determine vaccine efficacy. High‐throughput technologies, such as gene expression profiling, multiplex analysis of cytokines and multiparametric flow cytometry, imaging combined with computational modelling offer new perspectives. Recent advances in systems biology have now broad implications for vaccinology. Elucidation of clusters of signatures that correlate with vaccine immunogenicity should facilitate not only the rapid screening of vaccines but also the formulation of new hypotheses on how vaccines mediate long‐term protective immune responses. The realization of these challenges could finally lead to the development of new tools, as vaccine chip including limited number of gene probe sets that can identify predictive signatures for all the correlates of immunogenicity and protection. The key to success relies on good interactions between multidisciplinary experts of immunology, vaccinology, computer science, bioinformatics, biostatistics. . . The recent development and financial support of systems biology institutes and transnational programmes contribute to make it possible (Benoist et al., 2006; Hood et al., 2008; Schubert, 2011). Systems vaccinology (Fig. 2), thus offers great promise for future translation of basic immunology research advances into successful vaccines. Complementary to the ‘classical’ approach to vaccine development, it should speed up vaccine development and vaccine candidate selection in addition to bringing new hypotheses to the underlying mechanisms to efficient vaccine‐induced immune responses.
Figure 2.

A systems biology‐based vaccine design scheme. Top‐down and bottom‐up modelling strategies are complementary to generate new hypotheses that can be tested in in silico simulations, as well as classical bench or field studies. In the end, identified vaccine response efficacy signatures lead to the validation of optimized vaccine candidates.
Acknowledgments
This work was supported by Université Pierre et Marie Curie, Centre National de la Recherche Scientifique, Institut pour la Recherche Médicale, and by grants from the European Union [grant numbers LSHB‐CT‐04‐005246 (EC‐FP6‐COMPUVAC), LSBH‐CT‐06‐018933 (EC‐FP6‐Clinigene)].
References
- Ansari H.R., Flower D.R., Raghava G.P. AntigenDB: an immunoinformatics database of pathogen antigens. Nucleic Acids Res. 2010;38:D847–D853. doi: 10.1093/nar/gkp830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Germain R.N., Mathis D. A plaidoyer for ‘systems immunology’. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- Black G.F., Weir R.E., Floyd S., Bliss L., Warndorff D.K., Crampin A.C. BCG‐induced increase in interferon‐gamma response to mycobacterial antigens and efficacy of BCG vaccination in Malawi and the UK: two randomised controlled studies. Lancet. 2002;359:1393–1401. doi: 10.1016/S0140-6736(02)08353-8. et al. [DOI] [PubMed] [Google Scholar]
- Bondada S., Robertson D.A. Assays for B lymphocyte function. In: Coligan J.E., Bierer B.E., Margulies D.H., Shevach E.M., Strober W., editors. John Wiley & Sons; 2003. pp. 333–334. [Google Scholar]
- Boudinot P., Marriotti‐Ferrandiz M.E., Pasquier L.D., Benmansour A., Cazenave P.A., Six A. New perspectives for large‐scale repertoire analysis of immune receptors. Mol Immunol. 2008;45:2437–2445. doi: 10.1016/j.molimm.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Boyd S.D., Marshall E.L., Merker J.D., Maniar J.M., Zhang L.N., Sahaf B. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Stafford P., Johnston S., Dinu V. Statistical Methods for Analyzing Immunosignatures. BMC Bioinformatics. 2011;12:349. doi: 10.1186/1471-2105-12-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusic V., Bajic V.B., Petrovsky N. Computational methods for prediction of T‐cell epitopes – a framework for modelling, testing, and applications. Methods. 2004;34:436–443. doi: 10.1016/j.ymeth.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Chavali A.K., Gianchandani E.P., Tung K.S., Lawrence M.B., Peirce S.M., Papin J.A. Characterizing emergent properties of immunological systems with multi‐cellular rule‐based computational modeling. Trends Immunol. 2008;29:589–599. doi: 10.1016/j.it.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Chen W.H., Kozlovsky B.F., Effros R.B., Grubeck‐Loebenstein B., Edelman R., Sztein M.B. Vaccination in the elderly: an immunological perspective. Trends Immunol. 2009;30:351–359. doi: 10.1016/j.it.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., Mata J. Quantitative modeling of immune responses. Immunol Rev. 2007;216:5–8. doi: 10.1111/j.1600-065X.2006.00492.x. [DOI] [PubMed] [Google Scholar]
- Coward J., Germain R.N., Atlan‐Bonnet G. Perspectives for computer modeling in the study of T cell activation. Cold Spring Harb Perspect Biol. 2010;2:a005538. doi: 10.1101/cshperspect.a005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot A.S. Immunomics: discovering new targets for vaccines and therapeutics. Drug Discov Today. 2006;11:203–209. doi: 10.1016/S1359-6446(05)03720-7. [DOI] [PubMed] [Google Scholar]
- Flower D., Macdonald I., Ramakrishnan K., Davies M., Doytchinova I. Computer aided selection of candidate vaccine antigens. Immunome Res. 2010;6:S1. doi: 10.1186/1745-7580-6-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower D.R. Immunoinformatics and the in silico prediction of immunogenicity. An introduction. Methods Mol Biol. 2007;409:1–15. doi: 10.1007/978-1-60327-118-9_1. [DOI] [PubMed] [Google Scholar]
- Flower D.R., Timmis J. Springer; 2007. , and In Silico Immunology. New York, USA: [Google Scholar]
- Fonseca S.G., Procopio F.A., Goulet J.P., Yassine‐Diab B., Ancuta P., Sékaly R.P. Unique features of memory T cells in HIV elite controllers: a systems biology perspective. Curr Opin HIV AIDS. 2011;6:188–196. doi: 10.1097/COH.0b013e32834589a1. [DOI] [PubMed] [Google Scholar]
- Gaucher D., Therrien R., Kettaf N., Angermann B.R., Boucher G., Filali‐Mouhim A. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R.N. Ron Germain: towards a grand unified theory – interview by Amy Maxmen. J Exp Med. 2010;207:266–267. doi: 10.1084/jem.2072pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R.N., Meier‐Schellersheim M., Nita‐Lazar A., Fraser I.D.C. Systems biology in immunology: a computational modeling perspective. Annu Rev Immunol. 2011;29:527–585. doi: 10.1146/annurev-immunol-030409-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely S., Goldman M. From tolerance to autoimmunity: is there a risk in early life vaccination? J Comp Pathol. 2007;137:S57–S61. doi: 10.1016/j.jcpa.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Harish N., Gupta R., Agarwal P., Scaria V., Pillai B. DyNAVacS: an integrative tool for optimized DNA vaccine design. Nucleic Acids Res. 2006;34:W264–W266. doi: 10.1093/nar/gkl242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Rappuoli R., De Groot A.S., Chen R.T. Emerging vaccine informatics. J Biomed Biotechnol. 2010;2010:218590. doi: 10.1155/2010/218590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L., Rowen L., Galas D.J., Aitchison J.D. Systems biology at the Institute for Systems Biology. Brief Funct Genomic Proteomic. 2008;7:239–248. doi: 10.1093/bfgp/eln027. [DOI] [PubMed] [Google Scholar]
- Kohler B., Puzone R., Seiden P.E., Celada F. A systematic approach to vaccine complexity using an automaton model of the cellular and humoral immune system. I. Viral characteristics and polarized responses. Vaccine. 2000;19:862–876. doi: 10.1016/s0264-410x(00)00225-5. [DOI] [PubMed] [Google Scholar]
- Legutki J.B., Magee D.M., Stafford P., Johnston S.A. A general method for characterization of humoral immunity induced by a vaccine or infection. Vaccine. 2010;28:4529–4537. doi: 10.1016/j.vaccine.2010.04.061. [DOI] [PubMed] [Google Scholar]
- Levine M.M., Sztein M.B. Vaccine development strategies for improving immunization: the role of modern immunology. Nat Immunol. 2004;5:460–464. doi: 10.1038/ni0504-460. [DOI] [PubMed] [Google Scholar]
- Lindqvist M., Nookaew I., Brinkenberg I., Samuelson E., Thörn K., Nielsen J., Harandi A.M. Unraveling molecular signatures of immunostimulatory adjuvants in the female genital tract through systems biology. PLoS ONE. 2011;6:e20448. doi: 10.1371/journal.pone.0020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan C.H., Bersini H., Klatzmann D., Thomas‐Vaslin V., Six A. In: Stepney S., Welch P.H., Andrews P.S., Ritson C.G., editors. Luniver press; 2011. , and ) A computational technique to scale mathematical models towards complex heterogeneous systems. CoSMoS 2011 Proceedings on the 2011 workshop on Complex Systems Modelling and Simulation, and (eds). . ISBN 978‐1‐905986‐32‐3. [Google Scholar]
- Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Mittrücker H.W., Steinhoff U., Köhler A., Krause M., Lazar D., Mex P. Poor correlation between BCG vaccination‐induced T cell responses and protection against tuberculosis. Proc Natl Acad Sci USA. 2007;104:12434–12439. doi: 10.1073/pnas.0703510104. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya H.I., Wrammert J., Lee E.K., Racioppi L., Marie‐Kunze S., Haining W.N. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer C., Kappes J.C. New virologic reagents for neutralizing antibody assays. Curr Opin HIV & AIDS. 2009;4:418–425. doi: 10.1097/COH.0b013e32832f011e. [DOI] [PubMed] [Google Scholar]
- Park A.W., Daly J.M., Lewis N.S., Smith D.J., Wood J.L., Grenfell B.T. Quantifying the impact of immune escape on transmission dynamics of influenza. Science. 2009;326:726–728. doi: 10.1126/science.1175980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A., Oberg A.L. Vaccinomics and bioinformatics: accelerants for the next golden age of vaccinology. Vaccine. 2010;28:3509–3510. doi: 10.1016/j.vaccine.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland G.A., Ovsyannikova I.G., Jacobson R.M. Personalized vaccines: the emerging field of vaccinomics. Expert Opin Biol Ther. 2008;8:1659–1667. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B. Modulating vaccine responses with dendritic cells and Toll‐like receptors. Immunol Rev. 2004;199:227–250. doi: 10.1111/j.0105-2896.2004.00144.x. [DOI] [PubMed] [Google Scholar]
- Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin N., Lund O., Bernaschi M., Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology, a genome‐based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- Reche P.A., Reinherz E.L. PEPVAC: a web server for multi‐epitope vaccine development based on the prediction of supertypic MHC ligands. Nucleic Acids Res. 2005;33:W138–W142. doi: 10.1093/nar/gki357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D., Medzhitov R. The control of adaptive immune responses by the innate immune system. Adv Immunol. 2011;109:87–124. doi: 10.1016/B978-0-12-387664-5.00003-0. [DOI] [PubMed] [Google Scholar]
- Schubert C. Systems immunology: complexity captured. Nature. 2011;473:113–114. doi: 10.1038/nj7345-113a. [DOI] [PubMed] [Google Scholar]
- Seder R.A., Darrah P.A., Roederer M. T‐cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- Sekaly R.P. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med. 2008;205:7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HIV Controllers Study. The Major Genetic Determinants of HIV‐1 Control Affect HLA Class I Peptide Presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint N.C., Kohlbacher O. OptiTope − a web server for the selection of an optimal set of peptides for epitope‐based vaccines. Nucleic Acids Res. 2009;37:W617–W622. doi: 10.1093/nar/gkp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainas O., Harel D., Cohen I.R., Efron S. Reactive animation: from piecemeal experimentation to reactive biological systems. Autoimmunity. 2011;44:271–281. doi: 10.3109/08916934.2010.523260. [DOI] [PubMed] [Google Scholar]
- Yang B., Sayers S., Xiang Z., He Y. Protegen: a web‐based protective antigen database and analysis system. Nucleic Acids Res. 2011;39:D1073–D1078. doi: 10.1093/nar/gkq944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang P., Kim Y., Haste‐Andersen P., Beaver J., Bourne P.E. Immune epitope database analysis resource (IEDB‐AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski C.E., Corti D., Mele F., Pinto D., Lanzavecchia A., Sallusto F. Dissecting the human immunologic memory for pathogens. Immunol Rev. 2011;240:40–51. doi: 10.1111/j.1600-065X.2010.01000.x. [DOI] [PubMed] [Google Scholar]


