Summary
The oleochemical industry is currently still dominated by conventional chemistry, with biotechnology only starting to play a more prominent role, primarily with respect to the biosurfactants or lipases, e.g. as detergents, or for biofuel production. A major bottleneck for all further biotechnological applications is the problem of the initial mobilization of cheap and vastly available lipid and oil substrates, which are then to be transformed into high‐value biotechnological, nutritional or pharmacological products. Under the EU‐sponsored LipoYeasts project we are developing the oleaginous yeast Yarrowia lipolytica into a versatile and high‐throughput microbial factory that, by use of specific enzymatic pathways from hydrocarbonoclastic bacteria, efficiently mobilizes lipids by directing its versatile lipid metabolism towards the production of industrially valuable lipid‐derived compounds like wax esters (WE), isoprenoid‐derived compounds (carotenoids, polyenic carotenoid ester), polyhydroxyalkanoates (PHAs) and free hydroxylated fatty acids (HFAs). Different lipid stocks (petroleum, alkane, vegetable oil, fatty acid) and combinations thereof are being assessed as substrates in combination with different mutant and recombinant strains of Y. lipolytica, in order to modulate the composition and yields of the produced added‐value products.
Lipids and biotechnology
Lipids constitute a large and diverse group of compounds, which all are united by their hydrophobic nature and consequently their solubility in non‐polar organic solvents. Lipids and related lipophilic compounds are found, e.g. in animal and plant fats and oils, as waxes, phospholipids, eicosonoids, terpenes, and lipid‐soluble steroids and vitamins. The main sources for the industrial use of lipids are animal fats and plant oils, with the latter ones being forecasted for 2008–2009 to be produced at a scale of 418 million tonnes worldwide (http://www.abareconomics.com/interactive/08ac_Sept/htm/oilseeds.htm).
The chemical industry has developed a number of processes for the chemical conversion of lipids into various valuable industrial end products. An overview on the plethora of chemical conversion processes that is being worked upon can be obtained from the 2008 Euro Fed Lipid Congress abstracts book (Oils, Fats, and Lipids in the third millennium, 7–10 September, 2008, Book of abstracts). For example, lipids can serve as raw materials for the production of functional polymers such as polyurethanes, polyamides and polyesters by subjecting unsaturated vegetable oils (e.g. soy or canola) to ozonolysis followed by a hydrogenation step (Narine, 2008). Or, lipid‐derived fatty acid monomers are being modified by adding specific functional groups prior to their incorporation into specific polymers, such as polyesters and polyamides (Behr and Rothstock, 2008). Hydrocarbons for use as hydrocarbon‐based biofuels can be synthesized from lipids by thermal and catalytic decarboxylation (Stadlbauer et al., 2008). Rare functionalized unsaturated fatty acids can be produced from calendula and tung oils (Biermann et al., 2007), while modified fatty acids with reduced trans‐fat content can be obtained from vegetable oils via metal‐based catalysis (Golden et al., 2008). These and other innovative strategies for the production of added‐value lipids typically still depend on costly and energy‐consuming chemical reactions.
Biotechnology on the other hand provides a wide range of industrial applications that cannot easily be achieved by conventional chemistry. Although the production of biofuels from oils via lipase catalysis currently represent a major focus in the application of biotechnology to lipid transformation (Liu et al., 2008; Watanabe and Shimada, 2008), new types of bioconversions are emerging, which complement pure enzymatic catalysis now by whole cell‐based biocatalytic (WCB) conversion strategies. The former group of enzymatic bioconversions include, apart from lipase catalysed conversions, e.g. the biosynthesis of very‐long‐chain PUFAs employing desaturases and elongases (Hoppe et al., 2008) or the rational application of acyl‐CoA acyltransferases to modify the relative composition of sunflower oils, aiming at lower trans‐fats content during hydrogenation process while enhancing the proportion of stearic fat (Martinez‐Force et al., 2008). WCB on the other hand are employed for the production of conjugated PUFAs using Bifidobacterium species (Gorissen et al., 2008), for the production of biosurfactants from vegetable oils, often refinery wastes, using different bacteria and yeast species (Bernarski et al., 2004; Raza et al., 2007; Sarubbo et al., 2007; reviewed in Van Bogaert et al., 2007), but also for the targeted production of fine chemicals, such as vanillin from rice bran oil by using Aspergillus niger and Pycnoporus cinnabarinus (Zheng et al., 2007).
Yeast strains seem to be particularly suited for the conversion of lipids. Some prominent yeast species, the so‐called oleaginous yeasts that in fact were originally isolated from lipid substrates, have evolved sophisticated metabolic pathways that allow them to thrive on a variety of lipid substrates (reviewed in Thevenieau et al., 2009;2010). Yet, only few examples have been reported this far on the use of these yeasts in lipid bioconversion: Yarrowia lipolytica as well as some species of the genus Candida were employed for the lipid‐derived production of biosurfactants and of gamma‐decalactone, a high‐value aroma substance (Endrizzi and Belin, 1995; Wachéet al., 2001). The notable under‐utilization of oleaginous yeast for lipid bioconversion may correspond to the prominent research focus on conventional yeasts such as Saccharomyces cerevisiae, which are thus much better studied in general terms. Yet, since recently the sequence of the oleaginous yeast Y. lipolytica has been published (Dujon et al., 2004), the foundations were laid to make this yeast a prime model organism for studying lipid metabolism in oleaginous yeasts. In this review we describe targeted efforts in utilizing Y. lipolytica for the bioconversion of lipids into diverse high‐value products, by combining the yeast's lipid‐metabolizing potential with biosynthetic capacities derived from oil‐ and lipid‐degrading bacteria.
Y. lipolytica as a versatile fermentation platform
Yarrowia lipolytica is considered a ‘non‐conventional’ yeast primarily due to its ability to grow on a variety of substrates that are rich in lipids such as certain dairy and meat products (reviewed in Wolf, 2003; Beopoulos et al., 2010). Variant strains have also been isolated from soil, sewage and oil‐polluted environments (reviewed in Beopoulos et al., 2010), where in the latter case its ability to grow on a variety of aliphatic hydrocarbons has been exploited for the bioremediation of oily and greasy waste waters (Oswal et al., 2002; Lanciotti et al., 2005). Yarrowia is considered a GRAS (generally recognized as safe) organism that has already been used for some biotechnological applications like, e.g. citric acid production and heterologous protein expression; however, none of these production processes has exploited lipids as substrates (Madzak et al., 2004; Forster et al., 2007). On the other hand, this yeast has been used to produce neutral lipids consisting of triacylglycerols (TAGs) and sterol esters from oleic acid as substrate (Mlickova et al., 2004), which accumulate as intracellular lipid bodies making up for about 80% of the cell dry weight (reviewed in Fickers et al., 2005; Beopoulos et al., 2009). Yarrowia's ability to accumulate lipids is now being explored, e.g. to produce in a targeted way either oils rich in saturated fatty acids (Papanikolaou et al., 2003), or mono‐ and poly‐unsaturated oils, by corresponding genetic engineering of the yeast (Beopoulos et al., 2008).
Yarrowia's versatile lipid metabolism allows it, in response to the prevailing metabolic conditions, to either degrade, store or modify exogenous lipid substrates via distinct compartmentalized metabolic routes (reviewed in Fickers et al., 2005). Yarrowia lipolytica shows remarkable adaptations to lipid substrates in its ability to actually communicate with lipids by producing extracellular protrusions that serve to gather lipid droplets at the cell's surface, as well as to store significant amounts of lipids intracellularly (Mlickova et al., 2004). Recent research efforts directed at elucidating its lipid metabolism (reviewed in Fickers et al., 2005) together with available genomic data (Dujon et al., 2004) make it now possible to intercept its branched lipid metabolism for biotechnological purposes. Figure 1 demonstrates the scope of Y. lipolytica's storage capacity when grown on lipids (triolein) or lipid precursors (oleic acid) as compared with a rich non‐lipid carbon source (glucose), and for different genetic backgrounds, i.e. the wild type (WT), a mutant deficient in lipid production (JMY 1877), and an ‘obese’ mutant (JMY 1367), where metabolic routes competing with lipid production are blocked.
Figure 1.
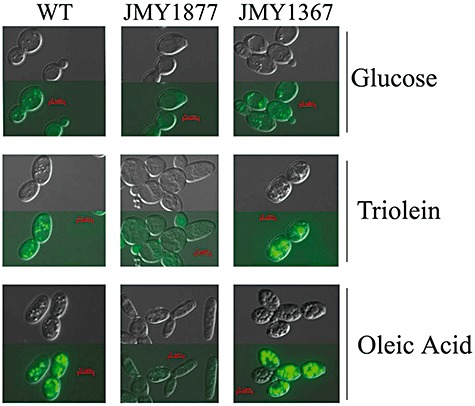
Scanning electron and fluorescence microscope visualization of lipid accumulation and lipid body formation depending on genotype of Y. lipolytica cells grown on glucose (YPD), triglyceride (YNB‐Triolein) and fatty acid (YNB‐Oleic acid). Strains used are the wild type (WT), a quadruple lipid body‐deficient mutant deleted for all acyl transferases (Δdga1, Δlro1, Δare1and Δare2) (JMY1877) (A. Beopoulos, R. Haddouche, P. Kabran, T. Chardot and J.‐M. Nicaud, in preparation) and an «obese» lipid‐overproducing mutant strain deleted for GUT2 and the POX genes (JMY1367) (Δpox1–6 andΔgut2) (Beopoulos et al., 2008). Upper panel (no staining) and lower panel (lipid are stained with lipidToxTMgreen neutral strain).
Yarrowia lipolytica may gradually develop into a new yeast model organism for industrial use with gradually enhanced understanding of its genetics and physiology. A wide range of tools have by now been developed that allow genetic manipulation of Yarrowiato be done with the same ease as for S. cerevisiae(Fickers et al., 2003; Madzak et al., 2004). A large set of additional mutations to more specifically alter this yeast's lipid metabolism are currently being engineered by Dr Nicaud and colleagues at INRA (R. Haddouche, in preparation). These strains then serve as tool box for the rational redesign of Yarrowia's lipid flux with modified β‐oxidation, TAG biosynthesis and ω‐oxidation thus allowing for the production of lipid precursors or for their conversion into other added‐value derivatives. Additional engineering efforts can further optimize and tailor Yarrowia's lipid metabolism to create new branch points for interception, and to increase metabolic flux along these engineered routes.
Hydrocarbonoclastic bacteria as sources for novel enzymes
The term ‘Hydrocarbonoclastic bacteria’ refers to group of marine bacteria that are capable of utilizing aliphatic hydrocarbons as their sole carbon and energy source. These bacteria are present in low abundance in marine environments but bloom after an oil spill (Golyshin et al., 2003). One species, Alcanivorax borkumensis, appears to dominate microbial communities associated with oil‐polluted waters and is assumed to be mostly responsible for the removal of marine oil contaminations (Golyshin et al., 2003). This bacterium is particularly efficient at degrading a wide range of aliphatic hydrocarbons, which initiated research aimed at finding enzymes that could be used for bioremediation, biocatalysis and biosynthesis. Earlier completed genome sequence of A. borkumensis (Schneiker et al., 2006) and its first genome and proteome analyses revealed multiple alkane degradation pathways as well as metabolic routes for the accumulation of carbon storage compounds from scavenged alkanes (Sabirova et al., 2006). More recently, genome sequences of other hydrocarbonoclastic marine bacteria [Marinobacter aquaeolei, Idiomarina loihiensis, Oleospira antarctica (Marinobacter and Oleospira are currently being sequenced)] have become available, which provides additional sources for the identification of novel enzyme functions for the biocatalytic conversion of lipophilic compounds into valuable products, such as putative hydrolases and oxygenases.
Marine hydrocarbonoclastic bacteria have not only evolved efficient mechanisms for the degradation of aliphatic hydrocarbons, but also for the formation of storage compounds enabling them to survive periods of poor nutrient conditions (reviewed in Steinbüchel et al., 1991). Alcanivorax has been shown to accumulate significant amounts of wax esters (WEs) and TAGs (Kalscheuer et al., 2007), while Marinobacter hydrocarbonoclasticus is known to make WEs from recalcitrant isoprenoid alcohols that are released from accumulated (bacterio)chlorophylls in marine sediments (Rontani et al., 1999). Other yet undiscovered hydrocarbonoclastic bacteria may have evolved other scavenging mechanisms that allow them to utilize other types of hydrocarbons and make new types of storage compounds, and hence it is anticipated that targeted screening and metagenomics efforts focussed on this group of marine bacteria will uncover new metabolic functions that can be exploited for the conversion of hydrocarbons and lipids into high‐value products.
High‐value target compounds
Research efforts under the project described here all aim at producing high‐value fine chemicals, carotenoids and biopolymers from bulk materials like cheap animal fats or vegetable oils. Some key target compounds are described in the following.
Wax esters are esters of long‐chain aliphatic alcohols and fatty acids (Fig. 2) and are used as high‐performance lubricants for engines, transmission and hydraulic systems (Lea, 2002). They are also used in cosmetics, foods and pharmaceuticals. Currently, these compounds are produced primarily from mineral oils, although the production of biodegradable lubricants from renewable sources increasingly attracts interest (Goodstein, 2004). However, as the most accessible natural sources of WEs, whale sperm oil and jojoba oil, are too expensive for wide range use, researchers have explored microbial sources for the synthesis of bio‐WEs and thus some Gram‐negative bacteria such as, e.g. Acinetobacter, Alcanivorax and Marinobacter species have been reported to synthesize different types of WEs as carbon storage compounds (Kalscheuer, 2010).
Figure 2.
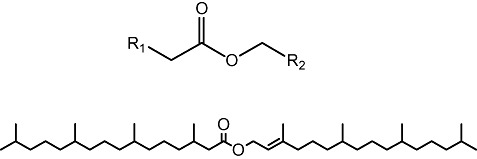
Fatty acid‐fatty alcohol (top) and isoprenoid WE (bottom).
Polyhydroxyalkanoates (PHAs) are biodegradable biopolymers synthesized by bacteria and used as bioplastics, e.g. for a variety of packaging and medical applications. Chemically being composed of 3‐hydroxy fatty acid monomers, these polymers can be classified as short‐chain‐length (SCL), medium‐chain‐length (MCL) or long‐chain length (LCL) PHAs. SCL‐PHAs contain monomers with a chain length of three to five carbons, while MCL‐PHAs contain 3‐hydroxy acid monomers with 6–16 carbon chains, and long‐chain‐length PHAs contain monomers longer than with 16 carbon atoms (Fig. 3). Currently, the best‐characterized PHA is the SCL‐PHA polyhydroxybutyrate PHB (four‐carbon‐chain length) which can be produced in an industrial scale. However, PHB is a rather brittle and inflexible biopolymer which compromised in its range of applications. To increase the utility of these biopolymers it is necessary to produce PHAs with different monomer compositions, e.g. longer chain length for improved mechanical properties.
Figure 3.
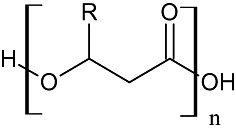
Short‐ and medium‐chain‐length PHAs. SCL‐PHA's: polyhydroxybutyrate (C4), polyhydroxyvalerate (C5). MCL‐PHA's: poly(3‐hydroxyhexanoate) (C6), poly(3‐hydroxyoctanoate), poly(3‐hydroxydecanoate) (C10), poly(3‐hydroxydodecanoate) (C12), poly(3‐hydroxytetradecanoate) (C14), and poly(3‐hydroxyhexadecanoate) (C16).
Yeasts with their more active metabolism yielding PHA precursors appears to be a better choice, as compared with bacterial hosts, for large‐scale PHA production, because of their ability to grow on a wider range of low‐cost feedstocks and because their sizes allow easier recovery during downstream processing (Terentiev et al., 2004). Recently attempts have therefore been aimed to produce PHAs in S. cerevisiae, Pichia pastoris and Arxula adeninivorans by overexpressing either Pseudomonas aeruginosaPhaC synthase or the entire phbABC cluster of Ralstonia eutropha (Poirier et al., 2001; 2002; Terentiev et al., 2004). However, PHA production rates so far obtained in yeasts were low compared with those obtained with bacteria. Yarrowia lipolytica specialized in efficient mobilization, storage, and conversion of lipids into PHA, is expected to be more appropriate yeast as heterologous host for PHA production.
Hydroxylated fatty acids (HFAs) have been used as chiral building blocks for the synthesis of a number of fine chemicals (Seebach et al., 1986; Lee et al., 2002) (Fig. 4). Moreover, some of HFAs have high potential for antimicrobial agents or biomedical applications (Hou, 2008). Enantiomerically pure HFAs can be derived from PHAs by chemical hydrolysis, which is a not very cost‐effective production route. Alternatively, HFAs can be produced from PHAs by enzymatic depolymerization using PHA depolymerases that in PHA‐producing bacteria catalyse the mobilization of storage compounds for growth (Ren et al., 2005). The most desirable process for HFAs production, however, would be conversion of 3‐hydroxy acyl‐CoA intermediates from microbial β‐oxidation of fatty acids into R‐3‐hydroxy acyl acids. By employing genetic engineering of Yarrowia, we will target 3‐hydroxy intermediates of β‐oxidation into these valuable compounds.
Figure 4.
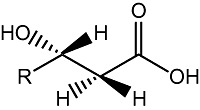
R‐3‐HFA.
Carotenoids represent a large group of structurally diverse pigments synthesized by microorganisms and plants (Fig. 5). Carotenoids are used commercially as food colorants, feed supplements, nutraceuticals and for cosmetic and pharmaceutical purposes. Of the more than 600 different carotenoids identified in nature, only a small number can be synthesized in useful quantities by chemical synthesis, extraction from their natural sources or microbial fermentation (reviewed in Lee and Schmidt‐Dannert, 2002).
Figure 5.
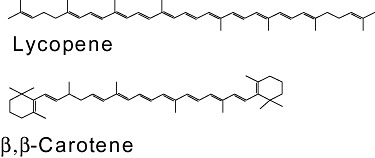
Examples of carotenoid structures.
Over the last decade genes encoding the enzymes of the carotenoid biosynthetic pathways leading to carotenoids with C30, C40 or C50 carbon backbones have been cloned from microorganisms and plants. Biosynthesis of these pigments in non‐carotenogenic hosts such as Escherichia colior Yarrowia requires first the extension of its isoprenoid pathway with a gene encoding geranylgeranyl diphosphate synthase (CrtE), which catalyses head‐to‐tail condensation of an additional C5 isoprenoid unit to the general isoprenoid precursor farnesyl diphosphate (C15) to yield geranylgeranyl diphosphate (C20, GGDP). An additional recombinant enzyme, phytoene synthase (CrtB), is then required for the head‐to‐head condensation of two GGDP molecules to the colourless C40 carotenoid phytoene. To obtain coloured carotenoids in E. coli, a phytoene desaturase (CrtI) needs to be coexpressed to synthesize the orange‐red acyclic carotenoid lycopene. Extension of this core pathway with known enzymes that catalyse cyclization, glucosylation and diverse oxygenation reactions allows the production of an array of acyclic and cyclic carotenoids and xanthophylls in E. coli.Production of C30 carotenoids occurs from FPP via a head‐to‐head condensation reaction catalysed by CrtM and desaturation of the resulting dehydrosqualene by CrN to diaponeurosporene or diapolycopene. Expression of these enzymes in the host E. colifacilitated their characterization and allowed the recombinant biosynthesis of diverse cyclic and acyclic carotenoids in E. coli(Schmidt‐Dannert et al., 2000). Dr Schmidt‐Dannert's group at the University of Minnesota has extensively explored combinatorial, evolutionary and genomic strategies for the production of diverse carotenoid structures and carotenoid cleavage compounds in engineered microbial hosts (Schmidt‐Dannert, 2001; Mijts et al., 2004; Marasco et al., 2006). Many of these new carotenoid structures have valuable pigment and antioxidant properties. However, despite successes in creating these diverse recombinant carotenoid pathways, there is a pressing need to find ways of producing carotenoid structures (most cannot be chemically synthesized) in sufficient quantities for commercial applications.
Polyenic polymers are new types of specialized, conducting polymers (Fig. 6) – also referred to as electronic plastics – have been developed for applications in an emerging class of ‘plastic’ opto‐electronic devices such as lasers, ultra fast image processors, thin‐film transistors, highly sensitive plastic photodiodes and photodiode arrays and all‐polymer integrated circuits. These new polymers, like other commonly used plastics, are chemically synthesized from petroleum‐derived building blocks. Biological routes to functional materials independent petroleum‐based economy could instead be described as a biorefinery process in which biomass components are converted into high‐value compounds.
Figure 6.
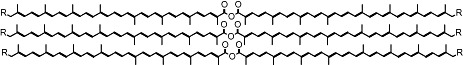
Proposed structure of stacked polyenic carotenoid ester chains.
Nature produces an array of stable conducting organic molecules designed and optimized to carry out electron transport and light‐harvesting functions, the latter via photoinduced electron transfer. The largest class of molecules with delocalized electron systems – carotenoids – is synthesized from polycondensated isoprenoid building blocks. The conjugated polyisoprenoids resemble the first chemically synthesized electronic polymer polyacetylene, but contain additional methyl‐groups along the chain and are in an all‐trans conformation, which exert a stabilizing effect on the molecule and increases electrical conductivity. Dr Schmidt‐Dannert's group has developed a unique biosynthetic platform for the production of end‐group functionalized polyenic building blocks in metabolically engineered E. colicells (Mijts et al., 2005).
Microbial genome mining for novel biosynthetic enzymes together with combinatorial biosynthesis and in vitro evolution of biosynthetic enzymes was used to engineer E. colistrains that produce fully conjugated isoprenoid chains containing carboxylic acid, aldehyde or alcohol functions at both terminal ends. The produced conjugated and oxygenated isoprenoid chains (C30 and C40 carotenoids) are extremely stable (in fact, the compounds appear self‐assemble and their unusual stability may be due to chain stacking) and represent ideal building blocks for the synthesis of conducting polymers. Their light‐absorbing properties make them ideally suited for photoinduced electron transfer, a much‐desired property of electronic plastics.
LipoYeasts project
The project's concept is illustrated on Fig. 7. The LipoYeasts project combines biosynthetic potential of hydrocarbonoclastic and other hydrocarbon and lipid‐degrading bacteria of various lipid‐contaminated environments with the lipid‐mobilizing potential of the oleaginous yeast Y. lipolytica to produce valuable lipid‐derived by‐products, such as poly‐ and wax esters, HFAs, carotenoids and carotenoid esters. For this purpose, the lipid metabolism of Yarrowiais to be adjusted in such a way, that it creates a high pool of lipid‐derived compounds, precursors of high‐value products. The main metabolic routes to be optimized are β‐ and ω‐oxidations and lipid accumulation. More specifically, we will concentrate on the construction of strains with reduced capacity in the storage of fatty acid into lipid bodies and with reduced capacity to degrade fatty acids by either β‐ or ω‐oxidation, as shown in Fig. 7.
Figure 7.
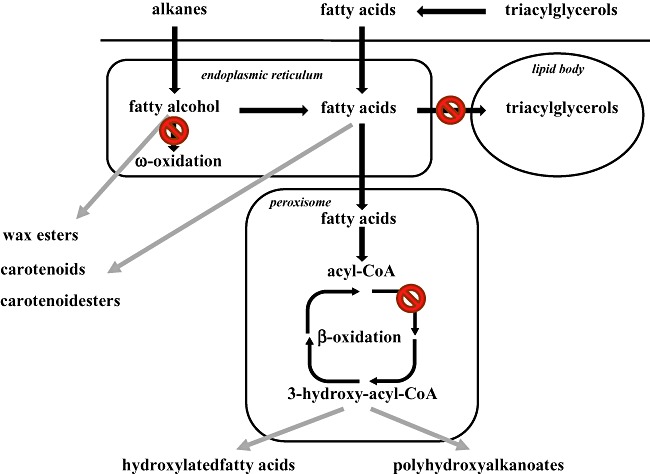
Metabolic engineering of Yarrowia lipolytica for production of added‐value products. Lipids such as triacylglycerols (TAGs), fatty acids and n‐alkanes are used as carbon sources. They get hydrolysed (TAGs) extracellularly and get transported into the cell, where fatty acids get either stored as TAGs in lipids bodies or get further oxidized in ω‐ or β ‐oxidation cycles. Storage of fatty acids as TAGs will be blocked. β‐Oxidation cycle provides precursors for hydroxylated fatty acids (HFAs) and polyhydroxyalkanoates (PHAs). Modifications of Pox1–6 genes catalysing the first step of β‐oxidation will be explored for modulating monomer composition of the produced HFAs and PHAs. Finally, the ω‐oxidation will be also blocked to avoid the production of undesired dicarboxylic fatty acids. The resulted pool of fatty acids and fatty acid alcohols will be used for the production of WEs, carotenoids or carotenoid esters by expressing the corresponding bacterial biosynthetic genes.
To develop Y. lipolytica into a fermentation platform for the conversion of lipid feed stocks into commercially interesting products, we propose the following complementary focus areas (WPs) of research and development: (i) engineering of metabolic precursor pools in Y. lipolytica for the production of value added products from lipids, (ii) conversion of metabolic precursor pools in Yarrowiato value‐added products by overexpressing heterologous biosynthetic enzymes, (iii) discovery and characterization of novel aliphatic enzyme activities by metagenomic screening of marine hydrocarbonoclastic microbial communities, (iv) pilot process development and identification of commercial applications, and (v) project coordination and IPR management.
Already available for this project are expression vectors and several deletion strains of Yarrowia as well as a collection of genes encoding enzymes for the production of WEs, 3‐HFAs, PHAs and carotenoids. We can therefore immediately begin with WP1 to improve lipid precursor pools in Yarrowia and implementation of the proposed heterologous biosynthetic capabilities into Yarrowia (WP2). Parallel to these efforts we will add new enzyme functions identified in metagenomic libraries to our toolbox of lipid‐modifying enzymes (WP3) for expression in Yarrowia. Once the first recombinant yeast producer strains have been generated, pilot processes for mid‐scale production will be developed and applications for produced compounds identified.
Conclusion
Current imbalance between chemical technologies and biotechnologies to make use of bulk fats and oils is addressed by this collaborative project. It aims to create new possibilities to convert lipids into valuable products by using the oleaginous yeast Y. lipolytica as a lipid‐mobilizing platform. This research will also hopefully bridge the gap between enzyme discovery studies based on the enormous amount of genomic information generated via genome sequencing projects on one hand and availability of easily genetically modifiable and well‐studied lipid‐mobilizing yeast on the other hand. By combining all these valuable and rather separated experiences together, we plan to make the practical use of this unique combination of available information and strains, by constructing yeast cell factories for the production of different high‐value industrial compounds. Thus we hope to expand the knowledge: (i) in yeast metabolism by studying fluxes of fatty acids intermediates, (ii) of recently discovered but not fully characterized enzymes derived from hydrocarbonoclastic bacteria and to understand their function by their heterologous expression in Yarrowia, (iii) in enzyme mining and in establishment and improvement of screening protocols applied to metagenomes, (iv) in new enzymes and their unique properties, and (v) in novel applicational aspects of the produced compounds. The consortium collectively has all tools and expertise at their disposal to meet this goal via close interaction between the partners. The progress of the work will be posted on the consortium webpage (http://www.lipoyeasts.ugent.be/) inviting a wide range of feedback from the scientific community.
Acknowledgments
The authors of the article would like to acknowledge European Commission for its financial support in the frame of the project LipoYeasts (contract number 213068).
References
- Behr A., Rothstock S. 2008. p. 15.
- Beopoulos A., Mrozova Z., Thevenieau F., Le Dall M.T., Hapala I., Papanikolaou S. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A., Cescut J., Haddouche R., Uribelarrea J.L., Molina‐Jouve C., Nicaud J.M. Yarrowia lipolytica as a model for bio‐oil production. Prog Lipid Res. 2009;48:375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Beopoulos A., Desfougeres T., Sabirova J., Zinjarde S., Neuveglise C., Nicaud J.M. The hydrocarbon‐degrading oleaginous yeast Yarrowia lipolytica. In: Timmis K.N., editor. Springer‐Verlag; 2010. pp. 2111–2121. [Google Scholar]
- Bernarski W., Adamczak M., Tomasik J., Plaszczyk M. Application of oil refinery waste in the biosynthesis of glycolipids by yeast. Bioresour Technol. 2004;95:15–18. doi: 10.1016/j.biortech.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Biermann U., Butte W., Eren T., Haase D., Metzger J.O. Regio‐ and stereoselective Diels‐Alder additions of maleic anhydride to conjugated triene fatty acid methyl esters. Eur J Org Chem. 2007;23:3859–3862. [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. et al. [DOI] [PubMed] [Google Scholar]
- Endrizzi A., Belin J.M. Bioconversion of methyl‐ricinoleate to 4‐hydroxy‐decanoic acid and to gamma‐decalactone by yeasts of the genus Candida. J Basic Microbiol. 1995;35:285–292. doi: 10.1002/jobm.3620350503. [DOI] [PubMed] [Google Scholar]
- Fickers P., Dall M.T., Gaillardin C., Thonart P., Nicaud J.M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Methods. 2003;55:727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Fickers P., Benetti P.H., Wache Y., Marty A., Mauersberger S., Smit M.S., Nicaud J.M. Hydrophobic substrate utilization by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005;5:527–543. doi: 10.1016/j.femsyr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Forster A., Aurich A., Mauersberger S., Barth G. Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl Microbiol Biotechnol. 2007;75:1409–1417. doi: 10.1007/s00253-007-0958-0. [DOI] [PubMed] [Google Scholar]
- Golden T., Leahy J.J., Curtin T. 2008. p. 59.
- Golyshin P.N., Martins Dos Santos V.A., Keiser O., Ferrer M., Sabirova Y.S., Lunsdorf H. Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon‐degrading bacterium that plays a global role in oil removal from marine systems. J Biotechnol. 2003;106:215–220. doi: 10.1016/j.jbiotec.2003.07.013. et al. [DOI] [PubMed] [Google Scholar]
- Goodstein D. WWNorton; 2004. [Google Scholar]
- Gorissen L., Raes K., Leroy F., De Vuyst L., De Smet S. 2008. p. 78. , and ) Production of conjugated linoleic acid and conjugated linolenic acid by Bifidobacterium species. 6th Euro Fed Lipid Congress, Book of abstracts, p.
- Hoppe K., Hoffman M., Wagner M., Abbadi A., Fulda M., Feussner I. 2008. p. 75.
- Hou C.T. New bioactive fatty acids. Asia Pac J Clin Nutr. 2008;1:192–195. [PubMed] [Google Scholar]
- Kalscheuer R. Genetics of wax ester and triacylglycerol biosynthesis in bacteria. In: Timmis K.N., editor. Springer‐Verlag; 2010. pp. 527–535. [Google Scholar]
- Kalscheuer R., Stöveken T., Malkus U., Reichelt R., Golyshin P.N., Sabirova J.S. Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol. 2007;189:918–928. doi: 10.1128/JB.01292-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R., Gianotti A., Baldi D., Angrisani R., Suzzi G., Mastrocola D., Guerzoni M.E. Use of Yarrowia lipolytica strains for the treatment of olive mill wastewater. Bioresour Technol. 2005;96:317–322. doi: 10.1016/j.biortech.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Lea C.W. European development of lubricants derived from renewable resources. Industrial Lubrication and Tribology. 2002;54:268–274. [Google Scholar]
- Lee P.C., Schmidt‐Dannert C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol. 2002;60:1–11. doi: 10.1007/s00253-002-1101-x. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Park S.H., Lee Y., Lee S.H. Production of chiral and other valuable compounds from microbial polyesters. In: Doi Y., Steinbüchel A., editors. Wiley‐VCH; 2002. pp. 375–387. [Google Scholar]
- Liu D., Du W., Liu H., Sun Y. 2008. p. 44.
- Madzak C., Gaillardin C., Beckerich J.M. Heterologous protein expression and secretion in the non‐conventional yeast Yarrowia lipolytica: a review. J Biotechnol. 2004;109:63–81. doi: 10.1016/j.jbiotec.2003.10.027. [DOI] [PubMed] [Google Scholar]
- Marasco E.K., Vay K., Schmidt‐Dannert C. Identification of carotenoid dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J Biol Chem. 2006;281:31583–31593. doi: 10.1074/jbc.M606299200. [DOI] [PubMed] [Google Scholar]
- Martinez‐Force E., Salas J.J., Venegas‐Caleron M., Troncoso‐Ponce M.A., Fernandez‐Garcia J.D., Gonzales‐Mellado D. 2008. p. 31. et al) Designing sunflower oils for healthy foods. 6th Euro Fed Lipid Congress, Book of abstracts, p.
- Mijts B.N., Lee P.C., Schmidt‐Dannert C. Carotenoid pathways. Methods Enzymol. 2004;388:315–329. doi: 10.1016/S0076-6879(04)88025-X. [DOI] [PubMed] [Google Scholar]
- Mijts B.N., Lee P.C., Schmidt‐Dannert C. Identification of a carotenoid oxygenase synthesizing acyclic xanthophyls: combinatorial biosynthesis and directed evolution. Chem Biol. 2005;12:453–460. doi: 10.1016/j.chembiol.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Mlickova K., Roux E., Athenstaedt K., d’Andrea S., Chardot T., Nicaud J.M. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2004;70:3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narine S.S. 2008. p. 17.
- Oswal N., Sarma P.M., Zinjarde S.S., Pant A. Palm oil mill effluent treatment by a tropical marine yeast. Bioresour Technol. 2002;85:35–37. doi: 10.1016/s0960-8524(02)00063-9. [DOI] [PubMed] [Google Scholar]
- Papanikolaou S., Muniglia L., Chevalot I., Aggelis G., Marc I. Accumulation of a cocoa‐butter‐like lipid by Yarrowia lipolytica cultivated on agro‐industrial residues. Curr Microbiol. 2003;46:124–130. doi: 10.1007/s00284-002-3833-3. [DOI] [PubMed] [Google Scholar]
- Poirier Y., Erard N., Petetot J.M. Synthesis of polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae by using intermediates of fatty acid β‐oxidation. Appl Environ Microbiol. 2001;67:5254–5260. doi: 10.1128/AEM.67.11.5254-5260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Erard N., MacDonald‐Comber Petetot J. Synthesis of polyhydroxyalkanoate in the peroxisome of Pichia pastoris. FEMS Microbiol Lett. 2002;207:97–102. doi: 10.1111/j.1574-6968.2002.tb11035.x. [DOI] [PubMed] [Google Scholar]
- Raza Z.A., Rehman A., Khan M.S., Khalid Z.M. Improved production of biosurfactant by a Pseudomonas aeruginosa mutant using vegetable oil refinery wastes. Biodegradation. 2007;18:115–121. doi: 10.1007/s10532-006-9047-9. [DOI] [PubMed] [Google Scholar]
- Ren Q., Grubelnik A., Hoerler M., Ruth K., Hartmann R., Felber H., Zinn M. Bacterial poly(hydroxyalkanoates) as a source of chiral hydroxyalkanoic acids. Biomacromolecules. 2005;6:2290–2298. doi: 10.1021/bm050187s. [DOI] [PubMed] [Google Scholar]
- Rontani J.F., Bonin P.C., Volkman J.K. Production of wax esters during aerobic growth of marine bacteria on isoprenoid compounds. Appl Environ Microbiol. 1999;65:221–230. doi: 10.1128/aem.65.1.221-230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova J.S., Ferrer M., Regenhardt D., Timmis K.N., Golyshin P.N. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J Bacteriol. 2006;188:3763–3773. doi: 10.1128/JB.00072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarubbo L.A., Farias C.B., Campos‐Takaki G.M. Co‐utilization of canola oil and glucose on the production of a surfactant by Candida lipolytica. Curr Microbiol. 2007;54:68–73. doi: 10.1007/s00284-006-0412-z. [DOI] [PubMed] [Google Scholar]
- Schmidt‐Dannert C. Directed evolution of single proteins, metabolic pathways and viruses. Biochemistry. 2001;40:13125–13136. doi: 10.1021/bi011310c. [DOI] [PubMed] [Google Scholar]
- Schmidt‐Dannert C., Umeno D., Arnold F.H. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18:750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- Schneiker S., Martins dos Santos V., Bartels D., Bekel T., Brecht M., Buhrmester J. Insights into marine oil‐degradation: the genome sequence of Alcanivorax borkumensis, a cosmopolitan and efficient hydrocarbon‐degrading bacterium. Nat Biotechnol. 2006;24:997–1004. doi: 10.1038/nbt1232. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebach D., Chow H.F., Jackson R.F.W., Sutter M.A., Thaisrivongs S., Zimmermann J. (+)‐11,11′‐di‐O‐methylelaiophylidene. Preparation from elaiophylin and total synthesis from (R)‐3‐hydroxybutyrate and (S)‐malate. Liebigs Annalen der Chemie. 1986;7:1281–1308. [Google Scholar]
- Stadlbauer E.A., Weber B., Bojanowski S., Sajjad H., Stengl S. 2008. p. 14.
- Steinbüchel A. Polyhydroxyalkanoic acids. In: Byrom D., editor. MacMillan Publishers; 1991. p. 123. [Google Scholar]
- Terentiev Y., Breuer U., Babel W., Kunze G. Non‐conventional yeasts as producers of polyhydroxyalkanoates – genetic engineering of Arxula adeninivorans. Appl Microbiol Biotechnol. 2004;64:376–381. doi: 10.1007/s00253-003-1498-x. [DOI] [PubMed] [Google Scholar]
- Thevenieau F., Nicaud J.M., Gaillardin C. Applications of the non‐conventional yeast Yarrowia lipolytica. In: Kunze G., Satyanarayana T., editors. Springer; 2009. pp. 590–613. [Google Scholar]
- Thevenieau F., Beopoulos A., Desfougeres T., Sabirova J., Albertin K., Zinjarde S., Nicaud J.M. Uptake and assimilation of hydrophobic substrates by the oleaginous yeast Yarrowia lipolytica. In: Timmis K.N., editor. Springer‐Verlag; 2010. pp. 1513–1527. [Google Scholar]
- Van Bogaert I.N., Saerens K., De Muynck C., Develter D., Soetaert W., Vandamme E.J. Microbial production and application of sophorolipids. Appl Microbiol Biotechnol. 2007;76:23–34. doi: 10.1007/s00253-007-0988-7. [DOI] [PubMed] [Google Scholar]
- Waché Y., Aguedo M., Choquet A., Gatfield I.L., Nicaud J.M., Belin J.M. Role of β‐oxidation enzymes in γ‐decalactone production by the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2001;67:5700–5704. doi: 10.1128/AEM.67.12.5700-5704.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Shimada Y. 2008. p. 46. , and ) Solvent‐free production of BDF using Candida Antarctica lipase stepwise alcoholysis. 6th Euro Fed Lipid Congress, Book of abstracts, p.
- Wolf K., Breunig K., Barth G., editors. Springer‐Verlag; 2003. [Google Scholar]
- Zheng L., Zheng P., Sun Z., Bai Y., Wang J., Guo X. Production of vanillin from waste residue of rice bran oil by Aspergillus nigerand Pycnoporus cinnabarinus. Bioresour Technol. 2007;98:1115–1119. doi: 10.1016/j.biortech.2006.03.028. [DOI] [PubMed] [Google Scholar]


