Summary
Clostridium cellulovorans, an anaerobic and mesophilic bacterium, degrades native substrates in soft biomass such as corn fibre and rice straw efficiently by producing an extracellular enzyme complex called the cellulosome. Recently, we have reported the whole‐genome sequence of C. cellulovorans comprising 4220 predicted genes in 5.10 Mbp [Y. Tamaru et al., (2010) J. Bacteriol., 192: 901–902]. As a result, the genome size of C. cellulovorans was about 1 Mbp larger than that of other cellulosome‐producing clostridia, mesophilic C. cellulolyticum and thermophilic C. thermocellum. A total of 57 cellulosomal genes were found in the C. cellulovorans genome, and they coded for not only carbohydrate‐degrading enzymes but also a lipase, peptidases and proteinase inhibitors. Interestingly, two novel genes encoding scaffolding proteins were found in the genome. According to KEGG metabolic pathways and their comparison with 11 Clostridial genomes, gene expansion in the C. cellulovorans genome indicated mainly non‐cellulosomal genes encoding hemicellulases and pectin‐degrading enzymes. Thus, by examining genome sequences from multiple Clostridium species, comparative genomics offers new insight into genome evolution and the way natural selection moulds functional DNA sequence evolution. Our analysis, coupled with the genome sequence data, provides a roadmap for constructing enhanced cellulosome‐producing Clostridium strains for industrial applications such as biofuel production.
Introduction
Consistent with the advantages of cellulosic feedstocks in terms of purchase price, potential fuel yield and environmental attributes, all scenarios known to us that foresee energy production from biomass on a scale sufficient to have large impacts on energy sustainability and security rely primarily on cellulosic biomass (Lynd et al., 2008). Although the desirable features of cellulosic biomass as a bioenergy feedstock are well known, biofuel production by fermentation is based today on plant feedstocks, from which sugars are more easily obtained, such as agricultural crop residues, grasses, wood and municipal solid waste. Process improvements associated with conversion of cellulosic biomass to sugars include the following: increasing cellulose hydrolysis yield (from 80% to 90%), halving cellulase loading (from 25 mg enzyme per gram cellulose to 12.5 mg enzyme per gram cellulose), eliminating pretreatment and incorporating consolidated bioprocessing such that enzyme production, hydrolysis and fermentation occur in a single process step (Lynd et al., 2005). Thus, the target for the enzymes of various microorganisms is the plant cell wall, which comprises many different polysaccharides, proteins and aromatic substances arranged as fibres with cross‐linkers (Doi, 2008). The plant cell wall composition and structure varies between plant species, between tissues of a single species, and even among individual cells. In fact, the complex structure of the plant cell wall consists of cellulose fibres linked with hemicellulose, pectin and lignin. The biotechnological potential of polysaccharolytic enzymes has resulted in the isolation and characterization of a large number of anaerobic, Gram‐positive, spore‐forming bacteria, the majority of which have been allocated to the genus Clostridium. Among clostridia, the cellulosomes produced by Clostridium species are particularly designed for efficient degradation of plant cell wall polysaccharides. The component parts of the multi‐component complex are integrated by virtue of a unique family of integrating modules, the cohesins and the dockerins, whose distribution and specificity dictate the overall cellulosome architecture (Bayer et al., 2008). The cellulosomes are characterized by the presence of two general components: (i) the non‐enzymatic scaffolding protein(s) with enzyme‐binding sites called cohesins, and (ii) a variety of cellulosomal enzymes with dockerins, which interact with the cohesins in the scaffolding protein. In the simplest system, there is a single scaffolding protein (scaffoldin) with a number of cohesins and a cellulose binding domain. The enzymatic subunits are bound to the scaffolding through the interaction of the cohesins and dockerins to form the cellulosome (Doi, 2008).
The cellulosome system in Clostridium cellulovorans has been studied extensively for the last 20 years and has resulted in providing basic information about mesophilic cellulosomes. This organism was isolated from a wood‐chip pile and is an anaerobic spore‐forming bacterium whose optimal growth temperature is 37°C (Sleat et al., 1984). It has the ability to utilize cellulose, xylan, pectin, cellobiose, glucose, fructose, galactose and mannose as carbon sources for growth. Its fermentation products include H2, CO2, acetate, butyrate, formate, lactate and ethanol. When grown in the presence of cellulose, electron micrographs have shown that large protuberances are present on its cell surface (Blair and Anderson, 1998), while little or no protuberances are evident when cells are grown in the presence of glucose or cellobiose (Blair and Anderson, 1999). The protuberances contain a large number of cellulosomes whose molecular mass is about 1000 kDa (Shoseyov and Doi, 1990). The C. cellulovorans cellulosomal enzymes that have been identified to date include a large gene cluster that encodes the proteins for CbpA‐ExgS‐EngH‐EngK‐HbpA‐EngL‐ManA‐EngM‐EngN (Foong et al., 1991; Liu and Doi, 1998; Tamaru and Doi, 2000; Tamaru et al., 2000) and genes for endoglucanases EngB (Foong and Doi, 1992) and EngE (Tamaru and Doi, 1999), mannanase ManA (Tamaru and Doi, 2000), pectate lyase A (Tamaru and Doi, 2001), and xylanases XynA (Kosugi et al., 2002) and XynB (Han et al., 2004a) that are dispersed throughout the genome. Thus, the cellulosomal enzymes from C. cellulovorans can degrade plant cell wall polysaccharides such as cellulose, xylan, mannan and pectin. Regulation of the expression of the cellulosomal genes is evident at the transcriptional level. Coordinate expression of cellulase and hemicellulase genes was observed in the presence of cellulose as the carbon source, as well as catabolite repression when cells were grown in glucose or cellobiose (Han et al., 2003). It was also shown that the presence of xylan or pectin as the carbon source enhanced the expression of cellulosomal xylanase and pectate lyase, as well as several non‐cellulosomal enzymes (Han et al., 2004b). In fact, pectin‐grown cells produced enzymes that were most effective in converting plant cells into protoplasts (Tamaru et al., 2002). In addition, mixed carbon substrates induced a wider variety of enzymes than a single carbon source, such as cellobiose, pectin and xylan. Therefore, it is evident that the expression of cellulosomal genes can be modified during growth on different carbon substrates such that optimal levels of certain enzymes will be attained.
So far, 20 genome sequencing projects of Clostridium species have been done or are ongoing by the Department of Energy Joint Genome Institute (JGI) since 2002. By the JGI, whole‐genome sequences of cellulosome‐producing Clostridium species, i.e. thermophilic C. thermocellum ATCC27405 and mesophilic C. cellulolyticum H10, have been sequenced in 2007 and 2009 respectively. In this study, we attempted the whole‐genome sequencing of C. cellulovorans by using the next‐generation DNA sequencers in order to compare not only cellulosomal genes but also non‐cellulosomal ones among cellulosome‐producing clostridia. In addition, since the essential carbohydrate‐related genes associated with metabolic pathways are annotated in clostridia, we analysed the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways in the C. cellulovorans genome and its comparison with 11 other clostridia whose genomes have been completely sequenced. Our findings reveal that the C. cellulovorans genome contained a minimum number of cellulosomal genes among the three cellulosome‐producing clostridia. Furthermore, since the C. cellulovorans genome included a large number of genes encoding non‐cellulosomal enzymes, the genome expansion of C. cellulovorans included genes more related to degradation of polysaccharides such as hemicelluloses and pectin than to cellulose.
Results
Features of the C. cellulovorans genome
The C. cellulovorans 743B (ATCC 35296) genome consists of 5 102 706 bp in 20 scaffolds (Tamaru et al., 2010). A total of 4220 polypeptide‐encoding open reading frames (ORFs) was identified using CRITICA, while 4297 ORFs were identified using Glimmer 2. The number of identical ORFs between CRITICA and Glimmer 2 was 2773. Sixty‐three tRNAs and 33 anti‐codons were also identified using tRNAScan‐SE (Lowe and Eddy, 1999). In comparison of the genome sizes, the C. cellulovorans genome (5.10 Mbp) was over 1 Mbp larger than the other genomes of cellulosomal clostridia and the number of predicted genes (4220 by CRITICA) was largest among them (Table 1). In addition, although the genome size of C. cellulolyticum (4.07 Mbp) was a little larger than that of C. acetobutylicum (3.94 Mbp), the number of genes (3390 by Glimmer) in C. cellulolyticum was smaller than that (3672 by Glimmer) in C. acetobutylicum. On the other hand, the G+C content in C. cellulovorans was 31.1% and similar to that (30.9%) in C. acetobutylicum, while the G+C contents in C. cellulolyticum and C. thermocellum were 37.7% and 39.0% respectively.
Table 1.
General features of cellulosomal clostridial genomes compared with that of C. cellulovorans.
| Organism | GenBank Accession No. | Genome size (Mb) | No. of genes | No. of cellulosomal genes | % GC |
|---|---|---|---|---|---|
| C. cellulovorans 743B | DF093537‐DF093556 | 5.10 | 4220 | 57 | 31.1 |
| C. acetobutylicum ATCC 824 | AE001437 | 3.94 | 3672 | 12 | 30.9 |
| C. cellulolyticum H10 | CP001348 | 4.07 | 3390 | 65 | 37.4 |
| C. thermocellum ATCC 27405 | CP000568 | 3.84 | 3191 | 84 | 39.0 |
A protein blast search against the database of Clusters of Orthologous Groups of proteins indicated that 4171 genes were encoded by 4220 predicted coding sequences using CRITICA, while 4098 genes were observed from 4297 predicted coding sequences using Glimmer 2. On the other hand, a protein blast search against the NCBI database indicated that 4184 genes were encoded by 4220 predicted coding sequences using CRITICA, while 4071 genes were observed from 4297 predicted coding sequences using Glimmer 2. Furthermore, a search of KEGG metabolic pathways revealed that we assigned 741 distinct EC numbers to 1179 (28% in 4220 genes) proteins by CRITICA mapped to KEGG pathways (Table 1), while 729 distinct EC numbers were assigned to 1095 (25% in 4297 genes) proteins by Glimmer 2 mapped to KEGG pathways. On the other hand, cellulosome‐producing clostridia such as C. cellulolyticum and C. thermocellum have already been analysed in the KEGG database. In the case of C. cellulolyticum, they assigned 619 distinct EC numbers to 846 (25% in 3390 genes) proteins, while they assigned 706 distinct EC numbers to 1073 (34% in 3191 genes) proteins in C. thermocellum (Table 1). These results indicated that the ratio of the proteins related to metabolic pathways in thermophilic C. thermocellum was larger than those in mesophilic clostridia such as C. cellulolyticum and C. cellulovorans, although the number of encoded genes in the C. thermocellum genome was the smallest among the three clostridia.
Cellulosomal genes among clostridial genomes were identified and classified as cohesin‐containing scaffolding proteins and dockerin‐containing proteins. So far, the scaffolding proteins for constructing cellulosomes were found in C. acetobutylicum (Sabathe et al., 2002), C. cellulolyticum (Pagès et al., 1999), C. cellulovorans (Shoseyov et al., 1992), C. josui (Kakiuchi et al., 1998) and C. thermocellum (Gerngross et al., 1993). In the case of the C. cellulovorans genome, a total of 57 cellulosomal genes were found, which consisted of 53 dockerin‐containing proteins and four cohesin‐containing scaffolding proteins (Table 2). Two scaffolding proteins, CbpB and CbpC, consisting of a carbohydrate‐binding module (CBM) of family 3, a surface–layer homology domain and a cohesin domain, were newly found and tandemly localized in the C. cellulovorans genome (Fig. 1), while there were no such scaffolding proteins in other cellulosomal clostridia.
Table 2.
Cellulosomal genes in the C. cellulovorans genome.
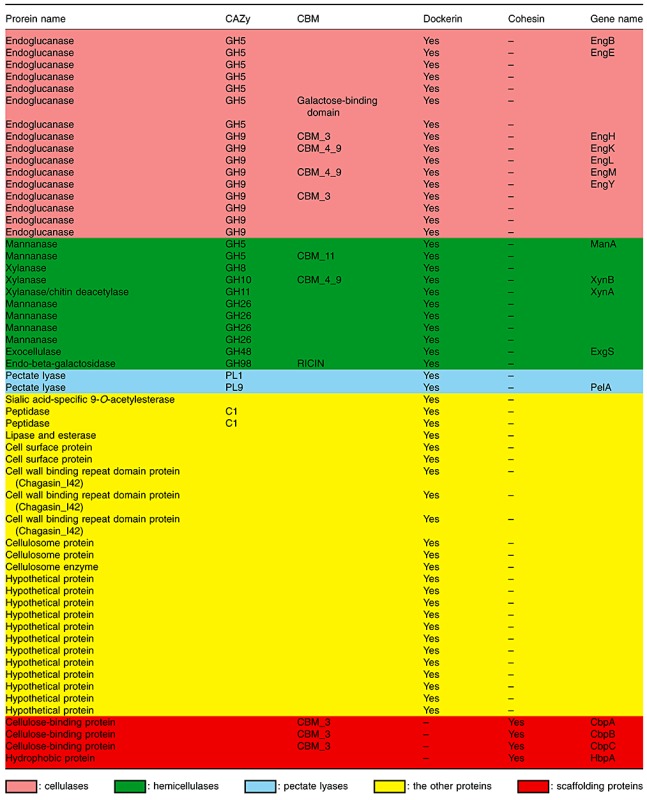
Figure 1.
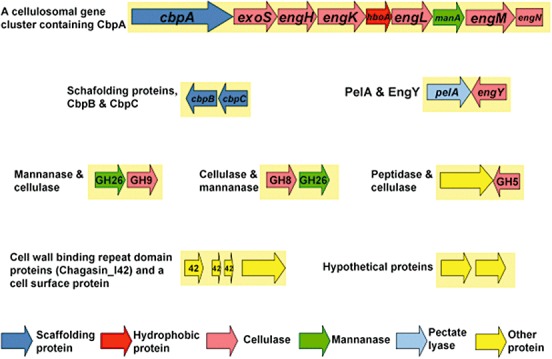
Cellulosome‐related gene clusters in the C. cellulovorans genome.
Carbohydrate‐active enzymes in C. cellulovorans and other cellulosome‐producing clostridia
Carbohydrate‐active enzymes (CAZymes) are categorized into different classes and families in the CAZy database. CAZymes that cleave, build and rearrange oligo‐ and polysaccharides play a central role in the biology of cellulosome‐producing clostridia such as C. cellulovorans and are key to optimizing biomass degradation by these species. Furthermore, the profile of CAZyme genes found in the cellulosome‐producing clostridia suggested a specific biological role. Table 3 shows the total number of carbohydrate‐active enzyme genes encoding glycosyl hydrolases (GHs), glycosyl transferases (GTs), polysaccharolytic lyases (PLs) and carbohydrate esterases (CEs) in the C. cellulovorans, C. cellulolyticum and C. thermocellum genomes respectively. Compared with the three genomes, the PL genes are not involved in the KEGG pathways among these three cellulose‐producing clostridia. Moreover, since the number of the genes encoding KEGG pathways (17%) in the C. cellulovorans genome was obviously larger than those in C. cellulolyticum and C. termocellum, C. cellulovorans differs in its content of proteins related to sugar utilization and metabolic pathways. In particular, consistent with its natural role, C. cellulovorans has several protein families related to degrading plant tissue, such as pectate lyases, exopolygalacturonate lyases, a pectin methylesterase and pectin esterases.
Table 3.
Comparison of the total number of the carbohydrate‐active enzyme genes encoding in the cellulosome‐producing clostridia.
| Organism | GHs | GTs | PLs | CEs | Pathway genes |
|---|---|---|---|---|---|
| C. cellulovorans 743B | 80 (53%) | 25 (17%) | 12 (8%) | 7 (5%) | 27 (17%) |
| C. cellulolyticum H10 | 85 (64%) | 19 (15%) | 4 (3%) | 14 (11%) | 9 (7%) |
| C. thermocellum ATCC 27405 | 63 (50%) | 34 (27%) | 4 (3%) | 13 (10%) | 11 (10%) |
GHs, glycosyl hydrolases; GTs, glycosyl transferases; PLs, polysaccharolytic lyases; CEs, carbohydrate esterases.
Cellulosomal and non‐cellulosomal enzymes in C. cellulovorans and other cellulosome‐producing clostridia
Given the relative importance of the polysaccharolytic enzyme family to the biotechnology community, we performed a detailed analysis of the CAZome of the C. cellulovorans cellulosome and non‐cellulosomal enzymes and compared them with the corresponding gene subsets from cellulosome‐producing clostridia for which genome sequences are available. We have extended this analysis to all the polysaccharide‐degrading enzyme genes (GHs and PLs) in the C. cellulovorans genome and found that in total, 80 of the 92 (87%) GH genes and 12 of the 92 (13%) PL genes were classified as cellulosomal or non‐cellulosomal enzyme genes. In the C. cellulolyticum genome, 85 of the 89 (96%) GH genes and 4 of the 89 (5%) PL genes were either cellulosomal or non‐cellulosomal genes, while 63 of the 67 (94%) GH genes and 4 of the 67 (6%) PL genes were cellulosomal or non‐cellulosomal genes in the C. thermocellum genome. Interestingly, both C. cellulolyticum and C. thermocellum had PLs only in cellulosomal genes, while two cellulosomal and 10 non‐cellulosomal PL genes were found in the C. cellulovorans genome.
Among 53 cellulosomal genes encoding dockerin‐containing proteins in the C. cellulovorans genome, a total of 29 genes coded for cellulolytic, hemicellulolytic and pectin‐degrading enzymes (Table 2). Compared with the genome‐sequenced species within cellulosomal clostridia, the proteome of C. cellulovorans focusing on dockerin‐containing proteins showed representation of many proteins with known functions. In detail, there are 16 cellulase genes belonging to families GH5, GH9 and GH48, six mannanase genes belonging to families GH5 and GH26, three xylanase genes belonging to families GH8, GH10 and GH11, an endo‐beta‐galactosidase gene belonging to family GH98, and two pectate lyase genes belonging to families PL1 and PL9. On the other hand, in addition to the genes encoding GHs and pectate lyases, genes encoding a sialic acid‐specific 9‐O‐acetylesterase, a lipase, peptidases, protease inhibitors (Chagasin I42) and cell surface proteins were found; there were seven genes encoding unknown and hypothetical proteins (Table 2).
Discussion
A more complete picture of life on, and even in, earth has recently become possible by extracting and sequencing DNA from environmental samples, a process called environmental genomics or metagenomics (Chivian et al., 2008). Moreover, functional divergence, as manifested in the accumulation of non‐synonymous substitutions in the genes encoding carbohydrate‐active enzymes, differs among lineages in a manner seemingly related to the genome size (Hehemann et al., 2010). On the other hand, the high cost of converting biomass to sugars is the primary factor impeding establishment of a cellulosic biofuels industry (Himmel et al., 2007; Lynd et al., 2008). In particular, metabolic engineering of microorganisms responsive to the needs of cellulosic ethanol production has received considerable attention and effort over the last two decades with utilization of xylose and other non‐glucose sugars also as a major focus (Shaw et al., 2008).
So far, it has been reported that 16S rDNA analysis of phylogenetic diversity among the polysaccharolytic clostridia revealed that C. cellulovorans belonged to cluster I while the other group includes cellulosome‐producing Clostridium species, i.e. C. cellulolyticum and C. thermocellum (Rainey and Stackebrandt, 1993). On the other hand, since the scaffolding protein CipA in C. acetobutylicum is a pseudogene, this bacterium does not produce cellulosomes. The genome sequence in C. cellulovorans was not similar to that in cellulosomal clostridia but was similar to other Clostridium species. Although it has been reported that cellulosomal gene clusters were non‐randomly distributed among mesophilic clostridia, there were several gene clusters containing two or three cellulosomal subunits in the C. cellulovorans genome (Fig. 1). Moreover, cellulosomal gene clusters were conserved only in mesophilic clostridia (Bayer et al., 2008; Doi, 2008). Furthermore, these cellulosomal genes were randomly distributed in the C. cellulovorans genome except for the cellulosomal genes related to a large cellulosomal cluster, whereas two large cellulosomal gene clusters were found in the C. cellulolyticum genome (Fig. 2). Even though the organization of genes encoding cellulosome subunits differs among mesophilic cellulolytic clostridia, there is nonetheless a clear similarity, particularly when looking at the cluster of genes following the main scaffoldin gene. Such a cluster is not found in C. thermocellum. This would suggest that the cellulosomes of the mesophilic clostridia, including the ‘ghost’ cellulosome of C. acetobutylicum, may have arisen from a common ancestral gene cluster. Such a cluster may have been transferred horizontally to C. cellulovorans, since its 16S RNA sequence puts it on a branch different from that harbouring C. cellulolyticum, whereas C. thermocellum, whose 16S RNA is more closely related to that of C. cellulolyticum, has its polysaccharidase genes organized quite differently.
Figure 2.
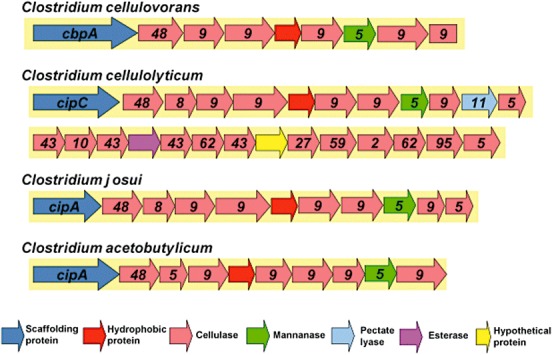
Cellulosome‐related gene clusters in the genome of mesophilic clostridia.
As compared with the two other clostridia, a smaller fraction of the polysaccharidases encoded by the C. cellulovorans genome is likely to be part of the cellulosome. Two proteins encoding CbpB and CbpC consisting of a putative cell surface‐bound polypeptide with a cohesin and a CBM were newly discovered. Moreover, no such scaffolding proteins exist in other cellulolytic clostridia including C. cellulolyticum and C. thermocellum, although other cell‐bound proteins with single cohesin domains are found in C. thermocellum (OlpA and OlpC), albeit without a CBM (Salamitou et al., 1994; Pinheiro et al., 2009). More recently, it has been reported that the RsgI‐like factor (anti‐sI) can interact with both the target polysaccharide substrate (e.g. cellulose) via its C‐terminal domain (CBM) and the cytoplasmic sI‐like factor via its N‐terminal subdomain (Kahel‐Raifer et al., 2010). In fact, we found several genes encoding RsgI‐like domains in the C. cellulovorans genome (data not shown).
Analysis of KEGG metabolic pathways revealed some interesting aspects among cellulosome‐producing clostridia. There were many metabolic enzyme genes in mesophilic C. cellulolyticum and C. cellulovorans related to fructose, mannose and galactose metabolism, while there were smaller number of those genes in thermophilic C. thermocellum. Moreover, although there was a xylose isomerase gene in C. cellulolyticum and C. cellulovorans, this gene was not present in the C. thermocellum genome. Therefore, it seems that the strategies for degradation of polysaccharides and utilization of pentoses and hexoses differ among cellulosome‐producing clostridia. On the other hand, according to the ratios between cellulosomal GHs + PLs and non‐cellulosomal GHs + PLs, C. cellulovorans possessed a larger number of non‐cellulosomal genes than celluosomal ones. Interestingly, the ratio of C. cellulotyticum was about a half of these genes, while the ratio of C. thermocellum was opposite to that of C. cellulovorans and had a larger number of cellulosomal genes than non‐cellulosomal ones (Table 4). In addition, both C. cellulolyticum and C. thermocellum in their cellulosomal genes had three and four CE genes, respectively, although C. cellulovorans had no cellulosomal CEs (Table 2). Moreover, while C. cellulolyticum contained two pectate lyases and two rhamnogalacturonan lyases, C. thermocellum had three pectate lyases and one rhamnogalacturonan lyase. More interestingly, although 12 PLs were found in the C. cellulovorans genome, 10 PLs were non‐cellulosomal genes and there was no rhamnogalacturonan lyase gene. On the other hand, there were no non‐cellulosomal PLs in C. cellulolyticum and C. thermocellum (Table 5). Thus, these findings suggest that the strategies of plant cell wall degradation are completely different between mesophilic and thermophilic clostridia. Namely, although the C. cellulovorans cellulosome is simplest compared with the C. cellulolyticum and C. thermocellum cellulosomes and enough to degrade plant cell walls (Tamaru et al., 2002), there are many kinds of non‐cellulosomal genes in the genome. Although there are larger numbers of cellulosomal genes in C. cellulolyticum and C. thermocellum than those in C. cellulovorans, both C. cellulolyticum and C. thermocellum cellulosomes consisted of more than half of hemicellulase and pectin‐degrading enzyme genes. Therefore, based on the complete degradation of plant cell walls, these findings indicated that C. cellulovorans would expect to take good aim at the synergistic effects between the cellulosome and many non‐cellulosomal enzymes, while both C. cellulolyticum and C. thermocellum would instead equip their cellulosomes with many kinds of cellulases and non‐cellulases.
Table 4.
Cellulosomal and non‐cellulosomal genes encoding GHs and PLs in the cellulosome‐producing clostridia.
| Organism | Cellulosomal GHs + PLs | Non‐cellulosomal GHs + PLs | Cellulosomal GHs + PLs/non‐cellulosomal GHs + PLs |
|---|---|---|---|
| C. cellulovorans 743B | 29 | 63 | 0.46 |
| C. cellulolyticum H10 | 47 | 42 | 1.1 |
| C. thermocellum ATCC 27405 | 53 | 14 | 3.8 |
GHs, glycosyl hydrolases; PLs, polysaccharolytic lyases.
Table 5.
Polysaccharolytic enzyme genes encoding GHs and PLs in the cellulosome‐producing clostridia.
| Organism | Total GHs + PLs | Cellulosomal GHs and PLs | Non‐cellulosomal GHs and PLs | ||
|---|---|---|---|---|---|
| GHs | PLs | GHs | PLs | ||
| C. cellulovorans 743B | 92 (100%) | 27 (29%) | 2 (2%) | 53 (58%) | 10 (11%) |
| C. cellulolyticum H10 | 89 (100%) | 43 (48%) | 4 (5%) | 42 (47%) | 0 (0%) |
| C. thermocellum ATCC 27405 | 67 (100%) | 49 (73%) | 4 (6%) | 14 (21%) | 0 (0%) |
GHs, glycosyl hydrolases; PLs, polysaccharolytic lyases.
Many of the genes encoding CAZymes are in the C. cellulovorans genome. In a previous study, nine known genes whose products are involved in cellulose and hemicellulose degradation were shown to be colocated in a cellulosomal gene cluster. In addition, although there are two large cellulosomal gene clusters in the C. cellulolyticum genome, there is only one large gene cluster in the C. cellulovorans genome (Fig. 2). On the other hand, CAZymes that cleave, build and rearrange oligo‐ and polysaccharides play a central role in the biology of polysaccharolytic clostridia such as C. cellulovorans and are key to optimizing biomass degradation by these species. Given the relative importance of this protein family to the biotechnology community, we performed a detailed examination of the CAZome of C. cellulovorans and compared it with the corresponding gene subsets from cellulosome‐producing clostridia, i.e. C. cellulolyticum and C. thermocellum for which genome sequences are available in the NCBI database (Table 1). Although one might expect that C. cellulovorans– an efficient plant polysaccharide degrader and important model of the degradation system – would contain expansions of genes whose products are involved in digesting cell wall compounds, it has surprisingly many genes encoding GHs and PLs. With a total of 92 GHs + PLs encoding genes, there was a middle number of 80 GHs among the three cellulosome‐producing clostridia (Table 3). On the other hand, most of the genes encoding GHs and PLs were GHs in both C. cellulolyticum and C. thermocellum. In particular, there was an endo‐beta‐galactosidase gene belonging to GH98 only in the C. cellulovorans cellulosome. More interestingly, there was a chitinase gene belonging to GH18 in the C. cellulolyticum and C. thermocellum cellulosomes, while a non‐cellulosomal chitinase gene was found in C. cellulovorans. In addition, there are five chitinase genes in the C. cellulolyticum genome and the C. thermocellum genome possessed three chitinase genes. It may be worth comparing the various carbohydrases with one another, particularly since they belong to a limited number of GHs and PLs families. Furthermore, it seems that this may be relevant to the origin and history of their diversity against the plant cell wall.
Although many mechanistic questions surrounding reconstruction of the cellulosome remain to be addressed, this study reveals a turning point of strategy for complete degradation of plant cell wall polysaccharides. Since we have obtained much information about the C. cellulovorans genome, many kinds of genes encoding polysaccharolytic enzymes are now available. Therefore, we hope to elucidate the synergy effects of plant cell wall degradation between the cellulosome and non‐cellulosome enzymes with different plant biomass, and to develop the total system for consolidated bioprocessing for the next‐generation biorefinery.
Experimental procedures
Genome sequencing
We sequenced a total length of 101 749 598 bp and analysed 381 514 reads by Genome Sequencer FLX 454./Roche sequencing (Margulies et al., 2005) (GS‐FLX version) to highly over‐sample the genome (20× coverage) and generated 123 892 paired‐end sequence tags, to enable the assembly of all tags using the GS De Novo Assembler version 1.1.03.24 (Roche Diagnostics), Genome Analyzer II and Sequencing kit 36‐Cycle Run (Illumina). Finally, we assembled 30 scaffolds (sets of ordered and oriented 601 contigs; total length of 5 123 527 bp) to generate approximately 5.1 Mbp of nearly contiguous C. botulinum E3 strain Alaska E43 (Accession No. NC_010723) complete genome sequence. We analysed a number of predicted genes encoded by the C. cellulovorans genome using CRITICA (version 1.05b) (Badger and Olsen, 1999) and Glimmer 2 (version 2.10) (Delcher et al., 1999) and to find regions in proteins with known functions. We annotated and classified according to Gene Otology (Ashburner et al., 2000). In silico Molecular Cloning Genomic Edition Ver. 3.0.26 software (In silico Biology, Japan) was used for individual genomic analysis.
Clostridial CAZyome comparisons
The search for carbohydrate‐active modules (GHs, GTs, PLs and CEs) and their associated CBMs in the C. cellulovorans genome was performed exactly as for the daily updates of the Carbohydrate‐Active enZYme (CAZy) database (http://www.cazy.org/) (Coutinho and Henrissat, 1999). The resulting fragments were assembled and formatted as a sequence library for blast searches. Accordingly, each protein from C. cellulovorans (and other clostridial proteomes) was searched via blast against the library of approximately 160 carbohydrate‐related enzymes using a database size parameter identical to that of the NCBI non‐redundant database. Manual analysis involved examination of the alignment of the model with the various members of each family (whether of catalytic or non‐catalytic modules), with a search of the conserved signatures and motifs characteristic of each family (Martinez et al., 2008). The presence of the catalytic machinery was verified for borderline cases whenever known in the family. The analysis of the sequence‐based families of GHs and PLs shows that those families rarely coincide with a single substrate (or product) specificity (Stam et al., 2005).
KEGG pathway analysis
In order to identify whether the carbohydrate‐active enzyme genes encoding GHs and PLs are related to metabolic pathways, the predicted proteins from the C. cellulovorans genome were searched for a database of KEGG metabolic pathway (http://www.genome.jp/kegg/pathway.html) (Kanehisa et al., 2004) and the obtained data were compared with 11 clostridial pathways, such as C. acetobutylicum ATCC 824, C. beijerinckii NCIMB 8052, C. botulinum A3 Loch Maree, C. botulinum B Eklund 17B, C. botulinum E3 str. Alaska E43, C. cellulolyticum H10, C. novyi NT, C. kluyveri DSM 555, C. tetani E88 and C. thermocellum ATCC 27405. All proteins that gave an expectation value lower than 1 × E−1 were automatically kept and manually analysed.
Accession number
The C. cellulovorans genome sequence data have been deposited in GenBank under accession numbers DF093537–DF093556.
Acknowledgments
We thank Sumitomo Corporation, Tokyo, Japan, for financial support of this work. Bioinformatics work was performed under the auspices of the Dragon Genomics Center, Takara Bio Co. Ltd, Japan.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J.H., Olsen G.J. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:514–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- Bayer E.A., Lamed R., White B., Flint H.J. From cellulosomes to cellulosomics. Chem Rec. 2008;8:364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- Blair B.G., Anderson K.L. Comparison of staining techniques for scanning electron microscopic detection of ultrastructural protuberances on cellulolytic bacteria. Biotech Histochem. 1998;73:107–113. doi: 10.3109/10520299809140514. [DOI] [PubMed] [Google Scholar]
- Blair B.G., Anderson K.L. Regulation of cellulose‐inducible structures of Clostridium cellulovorans. Can J Microbiol. 1999;45:242–249. doi: 10.1139/w99-004. [DOI] [PubMed] [Google Scholar]
- Chivian D., Brodie E.L., Alm E.J., Culley D.E., Dehal P.S., DeSantis T.Z. Environmental genomics reveals a single‐species ecosystem deep within earth. Science. 2008;322:755–758. doi: 10.1126/science.1155495. et al. [DOI] [PubMed] [Google Scholar]
- Coutinho P., Henrissat B. Carbohydrate‐active enzymes: an integrated database approach. In: Gilbert H.J., Davies G., Henrissat B., Svensson B., editors. Royal Society of Chemistry; 1999. pp. 3–14. [Google Scholar]
- Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4346–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R.H. Cellulases of mesophilic microorganisms: cellulosome and non‐cellulosome producers. Ann N Y Acad Sci. 2008;1125:267–279. doi: 10.1196/annals.1419.002. [DOI] [PubMed] [Google Scholar]
- Foong F., Hamamoto T., Shoseyov O., Doi R.H. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J Gen Microbiol. 1991;137:1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- Foong F.C.‐F., Doi R.H. Characterization and comparison of Clostridium cellulovorans endoglucanasesxylanases EngB and EngD expressed in Escherichia coli. J Bacteriol. 1992;174:1403–1409. doi: 10.1128/jb.174.4.1403-1409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerngross U.T., Romaniec M.P., Kobayashi T., Huskisson N.S., Demain A.L. Sequencing of a Clostridium thermocellum gene (CipA) encoding the cellulosomal SL‐protein reveals an usual degree of internal homology. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- Han S.‐O., Yukawa H., Inui M., Doi R.H. Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans. J Bacteriol. 2003;185:6067–6075. doi: 10.1128/JB.185.20.6067-6075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.‐O., Yukawa H., Inui M., Doi R.H. Isolation and expression of the xynB gene and its product, XynB, a consistent component of the Clostridium cellulovorans cellulosome. J Bacteriol. 2004a;186:8347–8355. doi: 10.1128/JB.186.24.8347-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.‐O., Yukawa H., Inui M., Doi R.H. Regulation of expression of cellulosomes and noncellulosomal (hemi) cellulolytic enzymes in Clostridium cellulovorans during growth on different carbon sources. J Bacteriol. 2004b;186:4218–4227. doi: 10.1128/JB.186.13.4218-4227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J.‐H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. Transfer of carbohydrate‐active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Himmel M.E., Ding S.Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W., Foust T.D. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Kahel‐Raifer H., Jindou S., Bahari L., Nataf Y., Shoham Y., Bayer E.A. The unique set of putative membrane‐associated anti‐sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate‐sensing mechanism involved in gene regulation. FEMS Microbiol Lett. 2010;308:84–93. doi: 10.1111/j.1574-6968.2010.01997.x. et al. [DOI] [PubMed] [Google Scholar]
- Kakiuchi M., Isui A., Suzuki K., Fujino T., Fujino E., Kimura T. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180:4303–4308. doi: 10.1128/jb.180.16.4303-4308.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi A., Murashima K., Doi R.H. Xylanase and acetyl xylan esterase activities of XynA, a key subunit of the Clostridium cellulovorans cellulosome for xylan degradation. Appl Environ Microbiol. 2002;68:6399–6402. doi: 10.1128/AEM.68.12.6399-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.‐C., Doi R.H. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene. 1998;211:39–47. doi: 10.1016/s0378-1119(98)00081-x. [DOI] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. tRNAscan‐SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1999;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd L.R., van Zyl W.H., McBride J.E., Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Lynd L.R., Laser M.S., Bransby D., Dale B.E., Davison B., Hamilton R. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–172. doi: 10.1038/nbt0208-169. et al. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A. Genome sequencing in microfabricated high‐density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D., Berka R.M., Henrissat B., Saloheimo M., Arvas M., Baker S.E. Genome sequencing and analysis of the biomass‐degrading fungus Trichoderma reesei (syn. Hypocrea jecorina. Nat Biotechnol. 2008;26:533–560. doi: 10.1038/nbt1403. et al. [DOI] [PubMed] [Google Scholar]
- Pagès S., Bélaïch A., Fierobe H.P., Tardif C., Gaudin C., Bélaïch J.P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin‐containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181:1801–1810. doi: 10.1128/jb.181.6.1801-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro B.A., Gilbert H.J., Sakka K., Sakka K., Fernandes V.O., Prates J.A. Functional insights into the role of novel type I cohesin and dockerin domains from Clostridium thermocellum. Biochem J. 2009;424:375–384. doi: 10.1042/BJ20091152. et al. [DOI] [PubMed] [Google Scholar]
- Rainey F.A., Stackebrandt E. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- Sabathe F., Bélaïch A., Soucaille P. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol Lett. 2002;217:15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x. [DOI] [PubMed] [Google Scholar]
- Salamitou S., Lemaire M., Fujino T., Ohayon H., Gounon P., Béguin P., Aubert J.P. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J Bacteriol. 1994;176:2828–2834. doi: 10.1128/jb.176.10.2828-2834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.J., Podkaminer K.K., Desai S.G., Bardsley J.S., Rogers S.R., Thorne P.G. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;105:13769–13774. doi: 10.1073/pnas.0801266105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O., Doi R.H. Essential 170 kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc Natl Acad Sci USA. 1990;87:2192–2195. doi: 10.1073/pnas.87.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O., Takagi M., Goldstein M., Doi R.H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A (CbpA) Proc Natl Acad Sci USA. 1992;89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat R., Mah R.A., Robinson R. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984;48:88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M.R., Blanc E., Coutinho P.M., Henrissat B. Evolutionary and mechanistic relationships between glycosidases acting on alpha‐ and beta‐bonds. Carbohydr Res. 2005;340:2728–2734. doi: 10.1016/j.carres.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Tamaru Y., Doi R.H. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J Bacteriol. 1999;181:3270–3276. doi: 10.1128/jb.181.10.3270-3276.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y., Doi R.H. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal ManA. J Bacteriol. 2000;182:244–247. doi: 10.1128/jb.182.1.244-247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y., Doi R.H. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc Natl Acad Sci USA. 2001;98:4125–4129. doi: 10.1073/pnas.071045598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y., Karita S., Ibrahim A., Chan H., Doi R.H. A large gene cluster for the Clostridium cellulovorans cellulosome. J Bacteriol. 2000;182:5906–5910. doi: 10.1128/jb.182.20.5906-5910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y., Ui S., Murashima K., Kosugi A., Chan H., Doi R.H., Liu B. Formation of protoplasts from cultured tobacco cells and Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl Environ Microbiol. 2002;68:2614–2618. doi: 10.1128/AEM.68.5.2614-2618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y., Miyake H., Kuroda K., Nakanishi A., Kawade Y., Yamamoto K. Genome sequence of the cellulosome‐producing mesophilic Clostridium cellulovorans 743B. J Bacteriol. 2010;192:901–902. doi: 10.1128/JB.01450-09. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


