Summary
Monitoring of pathogenic strains of Fusarium oxysporum (Fox), which cause wilt and rots on agricultural and ornamental plants, is important for predicting disease outbreaks. Since both pathogenic and non‐pathogenic strains of Fox are ubiquitous and are able to colonize plant roots, detection of Fox DNA in plant material is not the ultimate proof of an ongoing infection which would cause damage to the plant. We followed the colonization of tomato plants by strains Fox f. sp. radicis‐lycopersici ZUM2407 (a tomato foot and root rot pathogen), Fox f. sp. radicis‐cucumerinum V03‐2g (a cucumber root rot pathogen) and Fox Fo47 (a well‐known non‐pathogenic biocontrol strain). We determined fungal DNA concentrations in tomato plantlets by quantitative PCR (qPCR) with primers complementary to the intergenic spacer region (IGS) of these three Fox strains. Two weeks after inoculation of tomato seedlings with these Fox strains, the DNA concentration of Forl ZUM2407 was five times higher than that of the non‐compatible pathogen Forc V03‐2g and 10 times higher than that of Fo47. In 3‐week‐old plantlets the concentration of Forl ZUM2407 DNA was at least 10 times higher than those of the other strains. The fungal DNA concentration, as determined by qPCR, appeared to be in good agreement with data of the score of visible symptoms of tomato foot and root rot obtained 3 weeks after inoculation of tomato with Forl ZUM2407. Our results show that targeting of the multicopy ribosomal operon results in a highly sensitive qPCR reaction for the detection of Fox DNA. Since formae speciales of Fox cannot be distinguished by comparison of ribosomal operons, detection of Fox DNA is not evidence of plant infection by a compatible pathogen. Nevertheless, the observed difference in levels of plant colonization between pathogenic and non‐pathogenic strains strongly suggests that a concentration of Fox DNA in plant material above the threshold level of 0.005% is due to proliferation of pathogenic Fox.
Introduction
Fusarium oxysporum (Fox) is a well‐known pathogen of agricultural and ornamental crops (Nelson et al., 1981). Phytopathogenic strains of Fox are responsible for yield loss of many economically important crops worldwide. For example, wilt of tomato caused by Fusarium oxysporum f.sp. lycopersici (Fol) and foot and root rot of tomato caused by Fusarium oxysporum f.sp. radicis‐lycopersici (Forl) have been reported in at least 32 countries (Jones et al., 1991). These diseases occur both in greenhouse and field and result in significant crop losses (Hahn, 2002; Cai et al., 2003). Root and stem rot of cucumber, caused by Fusarium oxysporum f. sp. radicis‐cucumerinum (Forc), also significantly reduce yield in greenhouses in many countries (Vakalounakis et al., 2004). Besides yield decreases, many Fusarium sp. strains produce toxins which can accumulate in the end products and therefore may become dangerous for human and animal health (Pitt, 2000).
Fox is a cosmopolitan species whose representatives can survive as saprophytes in soil (Burgess, 1981). Due to their ability to utilize a large variety of nutrients, both pathogenic and non‐pathogenic Fox strains can colonize the rhizospheres of various plants and, moreover, enter into the endophytic stage (Garret, 1970). Some non‐pathogenic strains of Fox have been shown to control tomato foot and root rot (TFRR) caused by Forl (Olivain and Alabouvette, 1999; Bolwerk et al., 2005). Moreover ‘BioFox C’, a product based on a non‐pathogenic strain of Fox, is used for the protection of basil, carnation, cyclamen and tomato against pathogenic Fox and Fusarium moniliforme (Jones and Burges, 1998).
Monitoring of plant pathogens is crucial for disease management. Early detection, identification and quantification of the infestation level can help to choose appropriate defence measures. Monitoring of a phytopathogenic microorganism can be done indirectly by following disease symptoms appearing on the plants or by analysing volatiles excreted during pathogen multiplication (Prithiviraj et al., 2004). Direct approaches, such a dilution plating of infested plant or soil on selective media (Vujanovic et al., 2002), detection of fungal spores in plant material (Hahn, 2002), immunological and molecular detection of the causal agent of the disease, give more precise information about the pathogens (Paulitz, 2000).
Development of real‐time PCR (RT‐PCR) has provided a powerful tool for pathogen monitoring. RT‐PCR technique is highly sensitive for the detection of fungal strains (Zhang et al., 2005; Pasquali et al., 2006). It allows to detect the pathogen earlier than symptoms of the disease appear on the plants (Pasquali et al., 2004). With the use of real‐time PCR it is possible to perform a semi‐quantification of fungal pathogens such as F. oxysporum, Fusarium solani, Pythium ultimum and Rhizoctonia solani in a single assay (Lievens et al., 2005).
Since plants can be colonized by pathogenic and non‐pathogenic Fox strains, detection of Fox in planta is not necessarily evidence of attack of a pathogen. The patterns of tomato root penetration by pathogenic and non‐pathogenic Fox appear to be quite similar and the differences are mainly quantitative (Olivain and Alabouvette, 1999). In the case of non‐pathogenic Fox, flax plants appeared to be able to stop invasion of the fungus by building barriers in the cortex, whereas pathogenic strains appeared to avoid the defence system of the host plant (Olivain et al., 2003). Microscopical observations showed that, due to the reaction of the plant, the non‐pathogenic strain Fo47 is restricted in multiplication in tomato and flax (Olivain and Alabouvette, 1999; Olivain et al., 2003).
If differences in proliferation of Fox strains in planta exist, one should be able to detect them by quantitative PCR. To test this idea we compared colonization of tomato plants by different Fox strains: (i) the tomato foot and root rot pathogen Forl ZUM2407 strain, (ii) the cucumber root rot pathogen Forc V03‐2g and (iii) the non‐pathogenic biocontrol strain Fox Fo47 using qPCR. The results are reported in this paper.
Results
Infection of tomato with Fox strains from different formae speciales
Tomato seeds were inoculated with strains Forl ZUM2407, Forc V03‐2g and non‐pathogenic Fox Fo47 in concentrations of 105 spores per litre of PNS. No difference in rate and level of germination was observed between untreated seeds and seeds inoculated with any of these three Fox strains. Tomato plants harvested 1 week after sowing had no symptoms of TFRR. Two‐ and three‐week‐old tomato plants sometimes had brownish lesion and some of the plants were dead. The results of the disease evaluation in which no differences in the extension of the lesions were taken into account are shown in Fig. 2A. Among the inoculated 2‐week‐old plants, no statistical difference in disease incidence was evident (Fig. 1A), but the disease was more severe on plants treated with Forl ZUM2407 (Fig. 1B). Actually tomato plants inoculated with Forc V03‐2g and Fox Fo47 strains had mainly light lesions. Three weeks post inoculation both disease incidence and severity were statistically higher on tomato plants inoculated with Forl ZUM2407 (Fig. 1A and B)
Figure 2.
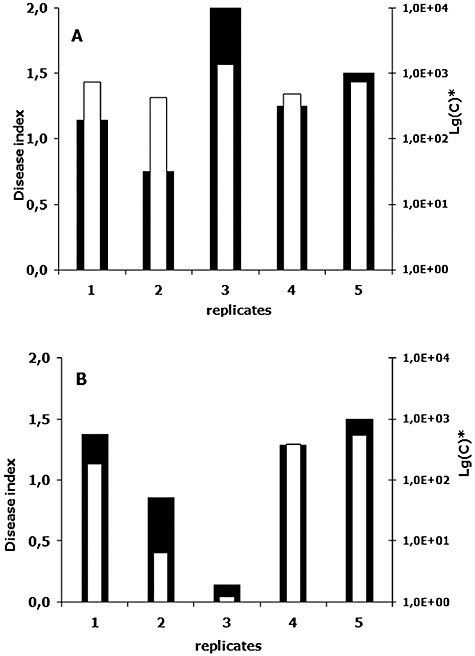
Comparison of disease severity index and fungal DNA concentration. (A) and (B) represent results of two independent experiments on tomato infection with Forl ZUM2407. Plants were scored 3 weeks after inoculation of the seeds with fungal spores. Black bars correspond to disease index (left y‐axis), white ones show fungal DNA concentration in a logarithmic scale (right y‐axis). Note that in Lg(C) ‘C’ is the fungal DNA concentration in fg per ng of plant DNA.
Figure 1.
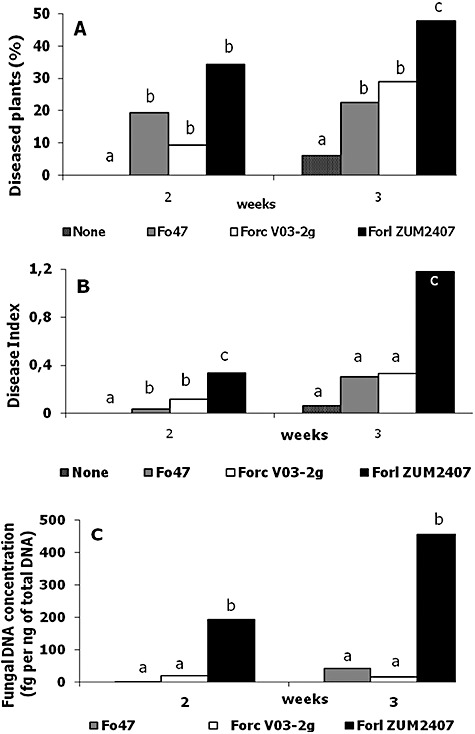
Quantification of tomato infection by three Fox strains based on the results of two independent experiments. A. Disease level. B. Disease severity index. C. Fungal DNA concentration in tomato plantlets. For statistics, a variance analysis followed by Fisher's least‐significant‐difference test (a = 0.05) was used. Statistically different values are labelled with different letters (a, b and c).
Quantification of fungal DNA in planta
Equal efficacy of templates for DNA fragment amplification is important for comparison of different strains. DNA samples isolated from Forl ZUM2407, Forc V03‐2g and Fox Fo47 strains were used to check the efficiency of the fragment amplification with primers OMP1049 and OMP1050. No significant difference was observed when DNA dilutions, ranging from 10 ng to 1 fg, from each individual strain were compared using qPCR.
To follow proliferation of Fox strains, total DNA was isolated from harvested and assessed groups (replicates) of tomato plants and, subsequently, used for qPCR. Comparisons of the disease severity indexes and DNA quantifications are shown on Fig. 2. In both independent trials the changes in DNA concentrations and disease indexes reveal the same trend. Results of the quantification show that the concentration of Forl ZUM2407 DNA is 5–10 times higher than those observed for the other Fox strains (Fig. 1C).
Two weeks after the inoculation, the DNA concentrations of the Forl ZUM2407 in tomato tissue were 5 and 10 times higher than that of the other Fox strains, in the first and second experiment, respectively, whereas no statistical differences were detected between Forc V03‐2g and Fox Fo47 DNA concentration (Fig. 1C).The DNA concentration of strains Forc V03‐2g and Fox Fo47 did not exceed 100 fg per 1 ng of total DNA (Fig. 3).
Figure 3.
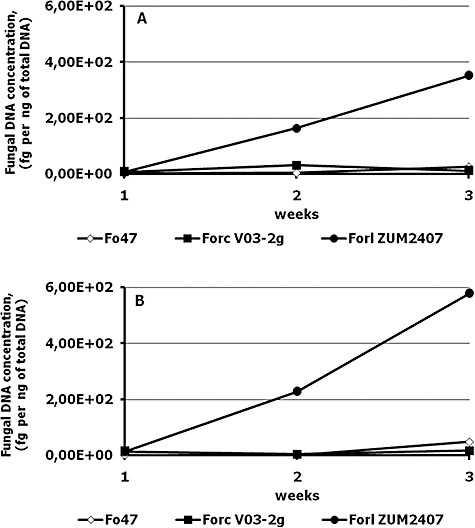
Changes in fungal DNA concentrations during growth of inoculated tomato plants. (A) and (B) represent two independent experiments.
Discussion
The ribosomal operon is present in 200 copies per haploid Fox genome, which offers a high sensitivity for the qPCR reaction. The space between the 18S and 28S rRNA genes of the ribosomal operon is designated as the intergenic spacer region (IGS) region. We used this IGS region as the target for qPCR. It was possible to detect 20 fg of DNA from the three Fox strains in 1 ng of tomato plant DNA, using primer pair OMP1049–OMP1050, which amplifies a 150 bp fragment within the IGS. Similar results were obtained when the IGS region was used for the detection of Fox f. sp. vasinfectum in cotton seedlings (Abd‐Elsalam et al., 2006). The lower detection level of 100 fg of pure fungal DNA reported in their article might be explained by the bigger size of the fragment used for amplification (438 bp).
We followed three different strains in plant infection/colonization and compared the results of two scoring systems with those obtained with PCR quantification. Results of scores without differentiation of lesion extension did not show a statistically significant difference between Fox strains in the second week after inoculation (Fig. 1A). In contrast, when the disease severity is taken into account, a statistically significant difference was observed between Forl ZUM2407 and the other two Fox strains, since Fox Fo47 and Forc V03‐2g strains did produce only small lesions on tomato plantlets (Fig. 1B).
Fungal DNA concentrations determined by qPCR were in a good agreement with the results of the disease indexes. Replicates containing more plants with larger lesions were given a higher disease index and they also showed a higher DNA concentration of Forl strain ZUM2407 (Fig. 2). A similar correlation between biomass of Alternaria brassicola and Botrytis cinerea was obtained when disease progression of the pathogens on Arabidopsis thaliana was quantified (Brouwer et al., 2003).
PCR‐based methods are very sensitive. Therefore, they can reduce the detection time (Lievens et al., 2003). Choice of the target DNA fragment is pivotal for the monitoring. Two types of targets, namely fragments specific to a certain group of Fox strains (supposedly forma specialis) and orthologous sequences (ribosomal operon, tubulin gene, etc.) can be used for the detection of the fungal strains in various substrates. For example, anonymous fragments generated with RAPD methods were used for the detection of specific pathogenic Fox in Paris daisy and basil (Pasquali et al., 2004; 2006). The target fragments used for amplification gave a high sensitivity for the detection of specific pathogens of Paris daisy and basil. The authors do not discuss whether all Fox that are pathogenic to these crops can be detected using primers specific for these anonymous fragments. Similarly to many formae speciales, such as Fox. f. sp. cubense and Fox. f. sp. melonis, Forl is comprised of strains which have a polyphyletic evolutionary origin (Jacobson and Gordon, 1990; Kistler, 1997). This means that some of the unrelated Fox strains that are pathogenic to the same plant species can miss an anonymous fragment, which is the target for qPCR. Also, vice versa, detection of DNA fragments in the samples cannot fully guarantee that the detected Fox is a specific pathogen of the given plant, except when the genes are involved in host‐plant infection (Rep et al., 2004).
Orthologuous sequences cannot be used for distinguishing heterogenic formae specialis or for discrimination of pathogenic Fox strains from non‐pathogenic ones. On the other hand, multicopy orthologuous sequences are convenient targets for qPCR due to the wide range of strains, which can be detected due to the high sensitivity of the reaction.
The fungal DNA concentration, as determined in plants inoculated with different Fox strains, correlated with disease severity and showed a statistically significant difference between Forl strain ZUM2407 and the other two Fox strains (Fig. 1C).
The level of colonization of tomato plants by the three Fusarium strains was followed using the fungal DNA concentration in isolated DNA of the plant as a criterion. The DNA concentration of Forl strain ZUM2407 has increased from week 1 to week 3, whereas the concentration of DNA from strains Fox Fo47 and Forc V03‐2g hardly changed and never exceeded 100 fg per 1 ng of total DNA (Fig. 3).
According to Olivain and Alabouvette (1999), the difference in colonization level of tomato by pathogenic and non‐pathogenic strains of Fox is mainly quantitative. Perhaps this difference can be explained by an observation made during flax root colonization by pathogenic and non‐pathogenic strains of Fox. Strain Fox Fo47 triggers formation of barriers, which apparently stop further invasion by the fungus, whereas the pathogenic strain avoids the plant's defence system (Olivain et al., 2003). In this scenario non‐pathogenic strains are doomed to stay outside the inner root parts and have to compete with rhizosphere microorganisms for the limited amounts of nutrients from root exudates and/or remain restricted in proliferation by defence mechanisms of the host within the root cortex. In contrast, pathogenic strains proliferate on the abundant level of nutrients present in the plant's cortex and root stele.
Our results show that the non‐pathogenic Fox strain Fo47 and the non‐compatible pathogen Forc V03‐2g could not exceed the level of 40 fg per 1 ng of total DNA, neither in week two nor in week three post inoculation. This concentration might reflect the level of colonization by the non‐pathogenic strain and by the non‐compatible pathogenic Fusarium, and this colonization level apparently is not dangerous for tomato plant health. In contrast, the phytopathogenic strain Forl ZUM2407, using nutrients of plant cortex and vascular system, could proliferate considerably. This proliferation resulted in disease symptom development. Differences in fungal biomass of non‐pathogenic/non‐compatible pathogenic and compatible phytopathogenic strains can be detected using RT‐PCR. Therefore, exploitation of a conserved multicopy region, such as an IGS fragment, allows the highly sensitive detection of Fox strains and the quantification of Fox DNA in plant material. It can be an option for distinguishing the disease progress of Fox pathogens from root colonization by non‐pathogenic Fox strains.
Experimental procedures
Strains and growth conditions
Strains used in this study are listed in Table 1. Strains were kept frozen at −80°C. When needed, cultures of the strains were plated on Czapek‐Dox agar (CDA, Difco Laboratories, Detroit, MI, USA) and grown at 28°C for 5–10 days. To avoid bacterial contamination CDA was amended with kanamycin (Duchefa, Haarlem, the Netherlands) and tetracyclin (Duchefa, Haarlem, the Netherlands) in final concentrations of 50 and 40 µg ml−1 respectively.
Table 1.
Strains used in the study.
| Strain | Host | GenBank Accession No. of the IGS region | Reference/source |
|---|---|---|---|
| Fox f. sp. radicis‐lycopersici ZUM2407 | Tomato | EF437260 | Syngenta, The Netherlands |
| Fox f. sp. radicis‐cucumerinum V03‐2g | Cucumber | EF437279 | ARRIAM, Russiaa |
| Fox Fo47 | None | EF437222 | Olivain and Alabouvette (1999) |
All‐Russian Research Institute of Agricultural Microbiology, Saint‐Petersburg‐Pushkin, Russia.
Plant inoculation with Fox strains and scoring of disease symptoms
To obtain spores, 1 l Erlenmeyer flasks containing 200 ml of Czapek‐Dox broth (CDB, Difco Laboratories, Detroit, MI, USA) were inoculated with one‐third of a 10‐day‐old CDA Petri dish culture of the Fox strains. The cultures were grown at 28°C for 72 h under aeration (110 r.p.m.). Spores were separated from the mycelium and the agar pieces by filtering of the cultures through miracloth (Omnilabo International BV, Breda, the Netherlands). The microspore concentration was determined using light microscopy. The filtrate was adjusted to concentrations of 105 spores per litre of Plant Nutrient Solution (PNS; PPO, Naaldwijk, the Netherlands).
Tomato (Solanum lycopersicum L.) seeds (cv. Carmello) were sown in stonewool plugs (Grodan BV, Roermond, the Netherlands). Plastic trays with the stonewool plugs were soaked in PNS supplemented with Fox. Seeds were allowed to germinate for 3 days in the dark at 23–27°C and subsequently the plantlets were grown in a greenhouse at 21–24°C, 70% relative humidity and 16 h daylight.
For each treatment, 144 plants were tested. Forty‐eight plants were harvested after 1, 2 and 3 weeks and the disease level was assessed using six replicates of 8 plants each. To evaluate the disease, plants were removed from the stonewool, washed, and the plant roots were examined for TFRR (tomato foot and root rot) symptoms as indicated by browning and lesions. Only roots without any disease symptoms were classified as healthy. The disease level, in this case, was calculated as the percentage of plants with a lesion. Alternatively, the disease was assessed by indexation of the disease severity: healthy plants were given a value of 0, plants with small lesions (< 2 mm) were given a value of 1, plants with developed lesions received a value of 2, and plants with large lesions (rotten foot, vast root rot) a value of 3. The value for dead plants was 4. The disease index (DI) was calculated using following formula:
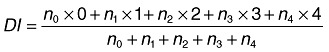 |
in which n0, n1, n2, n3 and n4 are the numbers of plants with indexes of 0, 1, 2, 3 and 4 respectively.
Differences in disease level among treatments were determined by analysis of variance (anova) and mean comparisons were performed by Fisher's least‐significant difference test (a = 0.05), using spss software (SPSS, Chicago, IL, USA). The experiment was performed twice.
Sample collection and DNA isolation
Each replicate containing eight whole plants was pulverized in liquid nitrogen. One gram of ground plant material was mixed with 1 ml of extraction buffer consisting of 2% hexadecatrimethylammonium bromide (CTAB; Sigma‐Aldrich Chemie BV, Zwijndrecht, the Netherlands) – 100 mM Tris‐HCl (pH 8.0) – 1.4 M NaCl – 20 mM EDTA (MP Biochemicals, Amsterdam, the Netherlands). The mixture was incubated at 55°C for 20 min and then centrifuged at 14 500 r.p.m. The supernatant was extracted with one volume of chloroform. The upper phase was transferred to a new Eppendorf tube and the DNA was precipitated by adding 0.6 volume of isopropanol followed by centrifugation. The pellets were dissolved in 500 µl of TE buffer (pH 8.0). To remove RNA from the preparations, RNase (Sigma‐Aldrich Chemie BV, Zwijndrecht, the Netherlands) was added at a final concentration 2 µg ml−1. DNA preparations were incubated at 60°C for 30 min and subjected to phenol‐chloroform extraction. DNA was precipitated by adding 50 µl of 3 M sodium acetate and 350 µl of isopropanol followed by centrifugation at 14 500 r.p.m. for 5 min. DNA pellets were washed twice with 70% ethanol and dried. DNA was dissolved in 50 µl of milliQ water, quantified and adjusted to a concentration of 5 ng µl−1. To isolate the DNA from Fox strains, the fungi were cultured for 5 days at 28°C on sterile filter paper placed on plate with CDA. The filter papers with fungal hyphea of the strains were removed from plate and ground in liquid nitrogen; further isolation was performed as described for the isolation of DNA from tomato plants.
Quantitative PCR reaction
Primers OMP1049 (5′‐TGCGATTTGGACGAGATATGTG‐3′) and OMP1050 (5′‐ATTTGCCTACCCTGTACCTACC‐3′) for quantitative PCR reaction were designed using Beacon Designer 5.0 (Bio‐Rad Laboratories BV, Veenendaal, the Netherlands) on the basis of the IGS sequence of Forl ZUM2407 strain. Real‐time PCR was performed in Chromo4 Multicolor Real‐Time PCR Detection System (Bio‐Rad Laboratories BV, Veenendaal, the Netherlands) with the following thermal profile: initial DNA denaturation and polymerase activation at 95°C for 10 min, followed by 40 cycles each containing denaturation and annealing steps at 95°C and at 58°C, respectively, both for 15 s. Amplification cycles were followed by a melting curve built from 50°C to 90°C, with measurements made every 0.2°C. The PCR mixture was prepared using qPCR Core kit for SYBR Green I No ROX (Eurogentec, Seraing, Belgium) according to the recommendations of the manufacturer (see reference number RT‐0000‐06, at http://www.eurogentec.com). A standard curve for quantification was generated by plotting the log of the concentrations (from 28 fg to 5 ng) of total DNA isolated from Forl strain ZUM2407 in the presence of 5 ng of tomato plant DNA.
Acknowledgments
The research described here was supported by the Technology Foundation Stichting voor de Technische Wetenschappen, Applied Science Division of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek, and the Technology Programme of the Ministry of Economic Affairs (LBI.5884) and by the Dutch Programme EET (Economy, Ecology, Technology) a joint initiative of the Ministries of Economic Affairs, Education, Culture and Sciences and of Housing, Spatial Planning and the Environment. We thank Dr Bernadette Kroon (Syngenta B.V., Enkhuizen, the Netherlands) for providing us with tomato seeds.
References
- Abd‐Elsalam K.A., Asran‐Amal A., Schnieder F., Migheli Q., Vetreet J.A. Molecular detection of Fusarium oxysporum f. sp vasinfectum in cotton roots by PCR and real‐time PCR assay. J Plant Dis Prot. 2006;113:14–19. [Google Scholar]
- Bolwerk A., Lagopodi A.L., Lugtenberg B.J.J., Bloemberg G.V. Visualization of interactions between a pathogenic and a beneficial Fusarium strain during biocontrol of tomato foot and root rot. Mol Plant Microbe Interact. 2005;18:710–721. doi: 10.1094/MPMI-18-0710. [DOI] [PubMed] [Google Scholar]
- Brouwer M., Lievens B., Van Hemelrijck W., Van den A.G., Cammue B.P., Thomma B.P. Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol Lett. 2003;228:241–248. doi: 10.1016/S0378-1097(03)00759-6. [DOI] [PubMed] [Google Scholar]
- Burgess L.W. General ecology of the fusaria. In: Nelson P.E., Toussoun T.A., Cook R.J., editors. The Pennsylvania State University Press; 1981. pp. 225–235. [Google Scholar]
- Cai G., Rosewich L., Schneider R.W., Kistler H.C., Davis R.M., Elias K.S., Miyao E.M. Origin of race 3 of Fusarium oxysporum f.sp. lycopersici at a single site in California. Ecol Popul Biol. 2003;93:1014–1022. doi: 10.1094/PHYTO.2003.93.8.1014. [DOI] [PubMed] [Google Scholar]
- Garret S.D. Cambridge University Press; 1970. [Google Scholar]
- Hahn F. Fungal spore detection on tomatoes using spectral Fourier signatures. Biosyst Eng. 2002;81:249–259. [Google Scholar]
- Jacobson D.J., Gordon T.R. Variability of mitochondrial‐Dna as an indicator of relationships between populations of Fusarium oxysporum f.sp. melonis. Mycol Res. 1990;94:734–744. [Google Scholar]
- Jones J., Jones J.P., Stall R.E., Zitter T.A. American Phytophathological Society; 1991. [Google Scholar]
- Jones K.A., Burges H.D. Technology of formulation and application. In: Burges H.D., editor. Kluwer Academic Publishers; 1998. pp. 7–30. [Google Scholar]
- Kistler H.C. Genetic diversity in the plant‐pathogenic fungus Fusarium oxysporum. Phytopathology. 1997;87:474–479. doi: 10.1094/PHYTO.1997.87.4.474. [DOI] [PubMed] [Google Scholar]
- Lievens B., Brouwer M., Vanachter A.C.R.C., Levesque C.A., Cammue B.P.A., Thomma B.P.H.J. Design and development of a DNA array for rapid detection and identification of multiple tomato vascular wilt pathogens. FEMS Microbiol Lett. 2003;223:113–122. doi: 10.1016/S0378-1097(03)00352-5. [DOI] [PubMed] [Google Scholar]
- Lievens B., Brouwer M., Vanachter A.C.R.C., Levesque C.A., Cammue B.P.A., Thomma B.P.H.J. Quantitative assessment of phytopathogenic fungi in various substrates using a DNA macroarray. Environ Microbiol. 2005;7:1698–1710. doi: 10.1111/j.1462-2920.2005.00816.x. [DOI] [PubMed] [Google Scholar]
- Nelson P.E., Toussoun T.A., Cook R.J. Pennsylvania State University Press; 1981. , and Fusarium: Diseases, Biology and Taxonomy. University Park, PA, USA: [Google Scholar]
- Olivain C., Alabouvette C. Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with a non‐pathogenic strain. New Phytol. 1999;141:497–510. doi: 10.1046/j.1469-8137.1997.00855.x. [DOI] [PubMed] [Google Scholar]
- Olivain C., Trouvelot S., Binet M.N., Cordier C., Pugin A., Alabouvette C. Colonization of flax roots and early physiological responses of flax cells inoculated with pathogenic and nonpathogenic strains of Fusarium oxysporum. Appl Environ Microbiol. 2003;69:5453–5462. doi: 10.1128/AEM.69.9.5453-5462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali M., Acquadro A., Balmas V., Migheli Q., Gullino M.L., Garibaldi A. Development of PCR primers for a new Fusarium oxysporum pathogenic on Paris daisy (Argyranthemum frutescens L.) Eur J Plant Pathol. 2004;110:7–11. [Google Scholar]
- Pasquali M., Piatti P., Gullino M.L., Garibaldi A. Development of a real‐time polymerase chain reaction for the detection of Fusarium oxysporum f. sp basilici from basil seed and roots. J Phytopathol. 2006;154:632–636. [Google Scholar]
- Paulitz T.C. Population dynamics of biocontrol agents and pathogens in soils and rhizospheres. Eur J Plant Pathol. 2000;106:401–413. [Google Scholar]
- Pitt J.I. Toxigenic fungi: which are important? Med Mycol. 2000;38(1):17–22. [PubMed] [Google Scholar]
- Prithiviraj B., Vikram A., Kushalappa A.C., Yaylayan V. Volatile metabolite profiling for the discrimination of onion bulbs infected by Erwinia carotovora ssp carotovora, Fusarium oxysporum and Botrytis allii. Eur J Plant Pathol. 2004;110:371–377. [Google Scholar]
- Rep M., van der Does H.C., Meijer M., van Wijk R., Houterman P.M., Dekker H.L. A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. et al. [DOI] [PubMed] [Google Scholar]
- Vakalounakis D.J., Wang Z., Fragkiadakis G.A., Skaracis G.N., Li D.‐B. Characterization of Fusarium oxysporum isolates obtained from cucumber (Cucumis sativus) in China by pathogenicity, VCGs and RAPD. Plant Dis. 2004;88:645–649. doi: 10.1094/PDIS.2004.88.6.645. [DOI] [PubMed] [Google Scholar]
- Vujanovic V., Hamel C., Jabaji‐Hare S., St Arnaud M. Development of a selective myclobutanil agar (MBA) medium for the isolation of Fusarium species from asparagus fields. Can J Microbiol. 2002;48:841–847. doi: 10.1139/w02-082. [DOI] [PubMed] [Google Scholar]
- Zhang Z.G., Zhang J.Y., Wang Y.C., Zheng X.B. Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues and soil (vol 249, pg 39, 2005) FEMS Microbiol Lett. 2005;251:357. doi: 10.1016/j.femsle.2005.05.057. [DOI] [PubMed] [Google Scholar]


