Summary
The fate and persistence of chlorinated organics in the environment have been a concern for the past 50 years. Industrialization and extensive agricultural activities have led to the accumulation of these pollutants in the environment, while their adverse impact on various ecosystems and human health also became evident. This review provides an update on the current knowledge of specialized anaerobic bacteria, namely ‘Dehalococcoides’ spp., which are dedicated to the transformation of various chlorinated organic compounds via reductive dechlorination. Advances in microbiology and molecular techniques shed light into the diversity and functioning of Dehalococcoides spp. in several different locations. Recent genome sequencing projects revealed a large number of genes that are potentially involved in reductive dechlorination. Molecular approaches towards analysis of diversity and expression especially of reductive dehalogenase‐encoding genes are providing a growing body of knowledge on biodegradative pathways active in defined pure and mixed cultures as well as directly in the environment. Moreover, several successful field cases of bioremediation strengthen the notion of dedicated degraders such as Dehalococcoides spp. as key players in the restoration of contaminated environments.
Chlorine‐containing chemicals like hexachlorobenzene (HCB), tetra‐ and trichloroethenes (PCE and TCE), dichlorodiphenyltrichloroethane (DDT), dioxins, polychlorinated biphenyls (PCBs), chlorophenols (CPs) and chlorofluorocarbons (CFCs) are persistent pollutants in our environment. Recognition of the ability of microorganisms to degrade these hazardous compounds opened up a new vista for the microbially mediated remediation of polluted environments. In addition, it also triggered the scientific community to undertake continued efforts towards the discovery, isolation and characterization of new microbial species. Among these, ‘Dehalococcoides’ spp. represent dedicated degraders, which are specialized in the anaerobic transformation of chlorinated organic contaminants that may otherwise persist in the environment for decades. In 1997, Maymó‐Gatell and co‐authors isolated the first anaerobic bacterium, ‘Dehalococcoides ethenogenes’ strain 195 (Maymo‐Gatell et al., 1997), that can transform toxic PCE completely to non‐toxic ethene via the process of reductive dechlorination. Since then, Dehalococcoides spp. were found to be dechlorinating a variety of hazardous chlorinated pollutants like CPs, polychlorinated dibenzo‐p‐dioxins (Fennell et al., 2004), PCB congeners (Bedard et al., 2007), chloroethanes (Grostern and Edwards, 2006; Duhamel and Edwards, 2007) and chlorinated benzenes (Adrian et al., 2000; Fennell et al., 2004). Dehalococcoides is a taxon of many irregularities. Even though the genomes of several representatives of this genus are among the smallest found in free‐living bacteria (Kube et al., 2005; Seshadri et al., 2005), they also contain the highest number of putative reductive dehalogenase (rdh) genes that code for the key enzymes mediating reductive dechlorination, within all known dechlorinating genera. Regardless of their general specialization to reductive dechlorination, every strain isolated so far has its own choice of favourite chlorinated compound(s). The unusual dependence of Dehalococcoides spp. on chlorinated organic compounds for their growth made them interesting research subjects to study their application in bioremediation. Yet our knowledge about presence, activity and capabilities of members of this genus in the environment is rather limited, including their response to changes in environmental conditions. This review provides a summary of the present knowledge on the role of Dehalococcoides spp. in degradation of chlorinated organic contaminants and the traits of this interesting group of microorganisms.
Pollution of chlorinated compounds and their bioremediation
Chlorine‐containing organics (Table 1) are often believed to originate exclusively from industrial pollution. However, many living organisms (e.g. marine sponges or terrestrial antagonistic microorganisms as a part of their defence mechanisms) produce them naturally whereas chlorinated compounds are also released as a result of, for example, eruptions of volcanoes, forest fires and geothermal processes (Griebler et al., 2004; Bengtson et al., 2009). Nevertheless, it is their extensive industrial (e.g. solvent, metal degreasing, rubber production) and agricultural (e.g. pesticide component) application over the past 50 years that resulted in their deposition in various environments, especially in soils, groundwater aquifers and sediments (Bailey, 2001; Meijer et al., 2003; Barber et al., 2005; Hageman et al., 2006; Weber et al., 2008). Due to their physicochemical properties (Table 1), exposure to these compounds can have carcinogenic and lethal effects on biota. Therefore, the production and application of most of these compounds is no longer allowed in 90 countries since the Stockholm convention in 2001 (Decision No. 2455/2001/EC, 2001; UNEP, 2005). Finding the suitable clean‐up techniques for contaminated environments, however, remains challenging. Remediation of soils and groundwater can be achieved via physicochemical methods such as thermal cleaning, chemical oxidation or adsorption of pollutants on activated carbon (Lai et al., 2007), whereas there are no in situ remediation technologies for sediments other than complete removal of the contaminated sediment (Wenning et al., 2006). Moreover, the high ecological disturbance that these physicochemical treatment methods can cause in the environment makes them unsustainable solutions in the long term (Wenning et al., 2006). Other than harsh physicochemical treatments, a far more preferable option is bioremediation. During bioremediation chlorinated contaminants are largely transformed by microorganisms although degradation by higher organisms is also reported. Phytoremediation, where plants are employed to assimilate, degrade, metabolize or detoxify chlorinated compounds, is an effective bioremediation method (Susarla et al., 2002). For example, poplar trees were shown to assimilate and degrade TCE to 2,2,2‐trichloroethanol, trichloroacetic acid and dichloroacetic acid (Newman et al., 1997). Recently, it has also been shown that the presence of these trees may stimulate the transformation of PCE in the subsurface (James et al., 2009). In this study, in the test location populated with hybrid poplar trees PCE pollution was reduced by over 99%, in comparison with 2% removal in an unplanted control. Moreover, several plant species, especially varieties of Cucurbita pepo ssp. pepo (squash), were shown to extract milligrams of PCBs from soil in approximately 8 weeks time (Zeeb et al., 2006). Lately the generation of transgenic plants to improve the phytoremediation of these pollutants resulted in several promising demonstrations of TCE, 1,2‐dichloroethane (DCA) and chlorophenol removal in several laboratory scale tests (Wang et al., 2004; Dowling and Doty, 2009; James and Strand, 2009). In many ecosystems, fungi are among the major decomposers. Most fungi are robust organisms and are generally tolerant to high levels of pollution (Singh, 2006). Fungal lignocellulolytic enzymes have been related to the degradation of various pollutants when used in combination with mediators and reactive radicals. Being the most commonly studied example, white‐rot fungi are able to detoxify a wide range of pollutants including chlorinated organics, with lignin and manganese peroxidases (Tortella et al., 2005; Field and Sierra‐Alvarez, 2008).
Table 1.
Sources, biological impacts and physicochemical properties of chlorinated organic compounds that have been reported to be degraded by ‘Dehalococcoides’ spp.
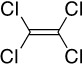
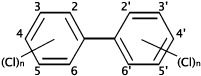
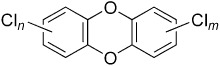
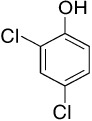
| HCBa,b | PCE/TCEc | PCBsa,d | Dioxinsa | CPse | |
|---|---|---|---|---|---|
 |
 gf gf
|
 |
 |
 |
|
| Natural sources | Volcanic activity, minerals | Volcanic activity, barley | Volcanic activity | Forest fires | Metabolites of microbes, sponges |
| Anthropogenic sources | Pesticide synthesis, waste incineration, dye production | Solvent (dry cleaning, metal cleansing), grain fumigation | Insulating fluid, microscope oil, stabilizing additive | Coal fired utilities, waste incineration, metal smelting, diesel truck, bleaching | Pesticides, bleaching wood pulp |
| Abiotic degradation | Photolysis | None | Ultrasound | Photolysis | Photolysis |
| Effects | Immune system and liver damage, cancer | Liver and kidney damage, neurotoxicity, possibly cancer | Skin rashes, dizziness, liver damage, reproductive damage, possibly cancer | Cancer, hepatotoxicity birth defects, endocrine disruption | Cancer, birth defects |
| Molecular weight | 285 | 165/131 | Various | Various (from 84–322) | Various (from 128–266) |
| Water solubility (mg l−1) | 0.005 | 150/1280 | 0.0027–0.42 × 10−3 | Insoluble | 10–905 |
| Vapour pressure (kPa)g | 0.1 × 10−3 | 1.9/7.8 | 1.1 × 10−3−1.3 × 10−7 (Criado et al., 2002) | NA | 1–12.7 × 10−3 |
Hexachlorobenzene [Agency for Toxic Substances and Disease Registry (ATSDR), 2002].
PCE: Tetrachloroethene and TCE: Trichloroethene (US EPA, 1985).
Polychlorinated biphenyls. There are theoretically 209 different PCB congeners, although only about 130 of these were found in commercial PCB mixtures (UNEP Chemicals, 1999).
Chlorophenols [Agency for Toxic Substances and Disease Registry (ATSDR), 1999].
Only PCE is illustrated.
At 20°C.
NA, not available.
The bacterial degradation of chlorinated pollutants can be a result of fortuitous co‐metabolic conversion, or it may contribute to the energy metabolism of the degrading organism. During the latter metabolic processes, chlorinated compounds are used either as carbon source or as electron acceptor (coupled to the oxidation of an electron donor), depending on the oxidation state of the compound. Although many chlorinated compounds may be transformed under aerobic conditions, the majority of polychlorinated compounds, such as those discussed in this review, are recalcitrant to aerobic degradation. Due to the electronegative nature of the chlorine atom, oxidation of the carbon backbone in the chlorinated compound becomes thermodynamically unfavourable (Wohlfarth and Diekert, 1997), especially in polychlorinated compounds. As a result they serve as energetically favourable electron acceptors in microbial metabolism in anoxic environments such as sediments, subsurface soils and groundwater aquifers. Consequently, anaerobic bacteria, which can use these compounds as electron acceptors in a process termed organohalide respiration, are good candidates for bioremediation (van Eekert and Schraa, 2001). Within the organohalide‐respiring bacteria, Dehalococcoides spp. and related isolates within the Chloroflexi represent a special case in the anaerobic detoxification of halogenated organic contaminants. It has been shown that several other bacteria belonging to the δ‐ and ε‐Proteobacteria (Anaeromyxobacter, Desulfuromonas, Desulfomonile, Desulfovibrio, Geobacter, Sulfurospirillum) or to the low‐GC Gram‐positive bacteria (Desulfitobacterium, Dehalobacter) are also able to degrade chlorinated organic contaminants through organohalide respiration (Fig. 1) (Smidt and de Vos, 2004). However, with the exception of Dehalobacter spp., none of these species are as specialized as Dehalococcoides, and they are reported to grow as well, for example, by metal reduction, denitrification or fermentation.
Figure 1.
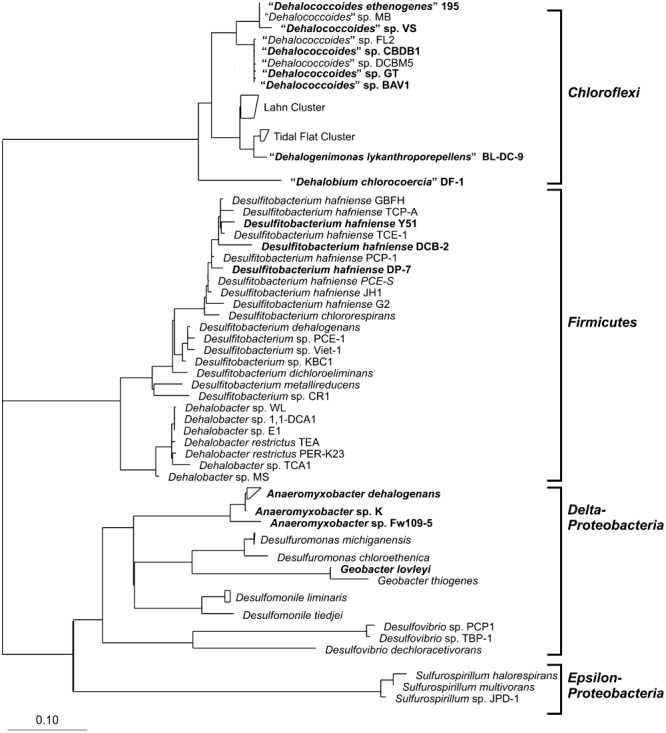
Phylogenetic tree of dechlorinating bacteria based on bacterial 16S rRNA sequences. Alignment and phylogenetic analysis were performed with the ARB software using the most recent release of the ARB‐SILVA project (SILVA 96) (Ludwig et al., 2004; Pruesse et al., 2007), and the tree was constructed using the neighbour joining method. The reference bar indicates the branch length that represents 10% sequence divergence. Boldface lettering indicates completed or ongoing genome sequencing.
The little bacteria that can: the genus Dehalococcoides
Dehalococcoides is a genus of strictly anaerobic Gram‐negative bacteria that to the best of our knowledge are restricted to gaining energy from the reduction of chlorinated compounds by organohalide respiration. Cultured Dehalococcoides spp. isolates have an irregular, spherical shape (approximately 0.5 µm) often referred to as coccoid. These mesophilic (25–40°C) bacteria prefer neutral pH environments. Their growth on alternative electron acceptors such as oxygen, nitrate or sulfate has never been reported (Kube et al., 2005; Seshadri et al., 2005). Reductive dechlorination by Dehalococcoides spp. occurs via the replacement of a chlorine atom in the chlorinated compound by hydrogen (reductive hydrogenolysis) and results in a net input of one proton and two electrons (Fig. 2) (Holliger et al., 1998). Gibbs free energy (ΔG0′) generated with reductive dechlorination of chlorinated compounds could range from −130 to −180 kJ mol−1 per chlorine removed. The redox potential thus generated is comparable to the redox potential of denitrification and higher than that generated by sulfate reduction. As a result it was suggested that reductively dechlorinating bacteria could out‐compete sulfate reducers and methanogens for reducing equivalents when the formation of reducing equivalents is rate limiting (Dolfing, 2003). Dehalococcoides spp. are also capable of degrading chlorinated aliphatic compounds, i.e. 1,2‐DCA, via so‐called dihaloelimination. In this process two neighbouring chlorine atoms are concurrently replaced via the formation of a double bond between the two carbon atoms. Dihaloelimination requires less H2 for the removal of chlorine atoms than reductive hydrogenolysis, thus its energy balance is more favourable under H2‐limited conditions (Smidt and de Vos, 2004).
Figure 2.
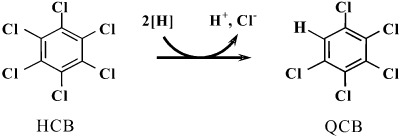
Reductive dechlorination of hexachlorobenzene (HCB) to pentachlorobenzene (QCB).
As the ecologists' quest prevails to delve Becking and Beijerinck's long running argument: ‘Everything is everywhere, but the environment selects’ (Beijerinck, 1913; Becking, 1934), the application of biomolecular tools, including the PCR amplification and sequencing of 16S ribosomal RNA (rRNA) genes from environmental samples, enables to study the full extent of microbial diversity and describe the biogeographical patterns exhibited by microorganisms at large spatial scales (Fierer and Jackson, 2006; Martiny et al., 2006). With the growing interest in Dehalococcoides' presence and functioning in the environment, several studies were conducted using Dehalococcoides‐specific 16S rRNA gene‐based approaches in uncontaminated and chlorinated ethene contaminated sediments, soils and groundwater aquifers (Löffler et al., 2000; Hendrickson et al., 2002; Kittelmann and Friedrich, 2008). Currently, more than 100 16S rRNA gene sequences of cultured and uncultured Dehalococcoides spp. have been deposited to the database of the National Center for Biotechnology Information (NCBI). The 16S rRNA gene in Dehalococcoides spp. is highly conserved throughout the entire genus (Fig. 1); however, various studies showed that this group is functionally very diverse (Maymo‐Gatell et al., 1997; Adrian et al., 2000; Hendrickson et al., 2002; He et al., 2003; Duhamel et al., 2004; Krajmalnik‐Brown et al., 2004). Eight Dehalococcoides strains have been isolated, mainly for their ability to degrade chlorinated ethenes (Table 2). Functional differences in these isolates can be observed in the chlorinated compound transformed and in the transformation end‐products. For example, the first isolate of the genusDehalococcoides ethenogenes strain 195 can completely dechlorinate PCE to ethene, although degradation of vinyl chloride (VC) to ethene is co‐metabolic (Maymo‐Gatell et al., 1997). Dehalococcoides ethenogenes strain 195 can also dechlorinate HCB to 1,3‐DCB (dichlorobenzene), 1,4‐DCB, 1,2‐DCB and 1,3,5‐TCB (trichlorobenzene). In contrast to D. ethenogenes strain 195, Dehalococcoides sp. CBDB1 dechlorinates HCB to 1,3‐DCB, 1,4‐DCB and 1,3,5‐TCB, and recently also transformation of PCE and TCE to trans‐DCE was observed (Adrian et al., 2007b). Dehalococcoides spp. are difficult to maintain in pure culture (Maymo‐Gatell et al., 1997; Adrian et al., 2000; He et al., 2003); they are more easily maintained in a microbial community, on which they depend for H2 supply, as long as ideal growth conditions are provided (Duhamel et al., 2004; Holmes et al., 2006). Examples include chlorinated ethene transforming Cornell (Maymo‐Gatell et al., 1997), Victoria (Hendrickson et al., 2002), Pinellas (Harkness et al., 1999), KB‐1 (Duhamel et al., 2002) and ANAS cultures (Richardson et al., 2002). In addition to Dehalococcoides spp., two other distantly related isolates within the Chloroflexi have recently been obtained (Fig. 2). The marine ‘Dehalobium chlorocoercia’ DF‐1 is able to dechlorinate a variety of PCBs (May et al., 2008). Most recently, ‘Dehalogenimonas lykanthroporepellens’ BL‐DC‐9 has been isolated from contaminated groundwater. This microorganism dechlorinates polychlorinated alkanes (Yan et al., 2009). Like Dehalococcoides spp., both isolates are strictly hydrogenotrophic.
Table 2.
Isolated strains of ‘Dehalococcoides’ spp. and the chlorinated substrates they transform.
| Chlorinated compound reduced | End‐products | References | |
|---|---|---|---|
| ‘Dehalococcoides ethenogenes’ strain 195 | PCE and TCE | Ethene | Maymo‐Gatell et al. (1997) |
| HCB | 1,3‐DCB, 1,4‐DCB, 1,2‐DCB and 1,3,5‐TCB | Fennell et al. (2004) | |
| 2,3‐DCP and 2,3,4‐TCP | 3‐MCP | Adrian et al. (2007b) | |
| 1,2 DCA | Ethene | Maymo‐Gatell et al. (1999) | |
| Dehalococcoides sp. BAV1 | VC | Ethene | He et al. (2003) |
| Dehalococcoides sp. CBDB1 | HCB | 1,3‐DCB, 1,4‐DCB and 1,3,5‐TCB | Adrian et al. (2000) |
| PCE and TCE | Trans‐1,2‐dichloroethene | Adrian et al. (2007b) | |
| 2,3‐DCP and 2,3,4‐TCP | 3‐MCP | Adrian et al. (2007b) | |
| Polychlorinated dioxins | Dichloro‐dioxins | Bunge et al. (2003) | |
| Polychlorinated biphenyls (Aroclor1260) | Various | Adrian et al. (2009) | |
| Dehalococcoides sp. VS | VC | Ethene | Cupples et al. (2003) |
| Dehalococcoides sp. FL2 | TCE | Cis‐1,2‐dichloroethene and trans‐1,2‐dichloroethene | He et al. (2005) |
| Dehalococcoides sp. GT | TCE | Ethene | Sung et al. (2006) |
| Dehalococcoides sp. DCMB5 | 1,2,4‐Trichlorodibenzo‐p‐dioxin | 2‐Monochlorodibenzo‐p‐dioxin | Bunge et al. (2008) |
| 1,2,3‐TCB | 1,3‐DCB | ||
| Dehalococcoides sp. Strain MB | PCE and TCE | Trans‐1, 2‐dichloroethene | Cheng and He (2009) |
Several enrichment studies showed the presence of Dehalococcoides spp. in different locations and environments in the Northern Hemisphere (mainly concentrated in North America, Europe and Japan). Dehalococcoides‐containing enrichment cultures originating from river sediments have been shown to dechlorinate PCB and dioxin congeners, PCE, TCE and a number of chlorinated benzenes (Ballerstedt et al., 2004; Yoshida et al., 2005; Bedard et al., 2007; Bunge et al., 2007; Futamata et al., 2007). Besides sediment enrichments, dechlorination by Dehalococcoides was also reported in groundwater aquifers (Bowman et al., 2006; Bürgmann et al., 2008; Imfeld et al., 2008; Lee et al., 2008; Himmelheber et al., 2009) and a denitrifying membrane‐biofilm reactor (Chung et al., 2008). Few studies have demonstrated that the bioaugmentation with reductively dehalogenating cultures can result in complete dechlorination of PCE and TCE to ethene (Ellis et al., 2000; Major et al., 2002; Lendvay et al., 2003; Scheutz et al., 2008). The maximum reported growth rates of Dehalococcoides spp. in pure and enrichment cultures are in the range of 0.2–0.4 day−1 under laboratory conditions (Maymo‐Gatell et al., 1997; Cupples et al., 2003; Adrian et al., 2007b; Duhamel and Edwards, 2007). Additionally, quantitative analyses of the Dehalococcoides spp. 16S rRNA gene at chlorinated ethene bioremediation sites (soil and groundwater) revealed abundances of 102–107 copies per gram material (Lendvay et al., 2003; Sleep et al., 2006). Recently, a groundwater bioremediation simulation study showed that growth rates obtained in laboratory conditions could also be replicated in large‐scale experiments, which resulted in up to 1012 16S rRNA gene copies l−1 (Vainberg et al., 2009). Hence, these pilot‐ as well as field‐scale bioremediation tests with Dehalococcoides‐containing cultures offer promising results for the further use of these microorganisms.
In spite of all the information obtained in physiological studies very little is known about the diversity, distribution and functioning of Dehalococcoides in different environments although they were detected at several contaminated locations. Hendrickson and co‐authors have demonstrated the presence of Dehalococcoides spp. in soil and groundwater samples from 24 sites scattered throughout North America and Europe (Hendrickson et al., 2002). Up to 200 µM PCE could be dechlorinated, and complete dechlorination to ethene could be correlated to the presence of Dehalococcoides spp. in the sampling locations. Recently, we conducted a large‐scale survey focusing on presence, activity and dechlorination potential of Dehalococcoides spp. in river sediments and floodplain soils from different polluted locations in Europe (Fig. 3) (Taş, 2009). Almost all of the tested sediment and soil samples showed the capacity to dechlorinate HCB and/or chlorinated ethenes irrespective of the in situ contaminant levels. Nevertheless, the HCB transformation rates observed in the laboratory‐scale microcosms and the number of 16S rRNA gene copies of Dehalococcoides spp. in the environmental samples did not show a strong correlation. In these river systems, Dehalococcoides spp. relative abundance was furthermore shown to change significantly along temporal and spatial gradients, but was also found to be influenced by other environmental factors such as water temperature (Taşet al., 2009).
Figure 3.
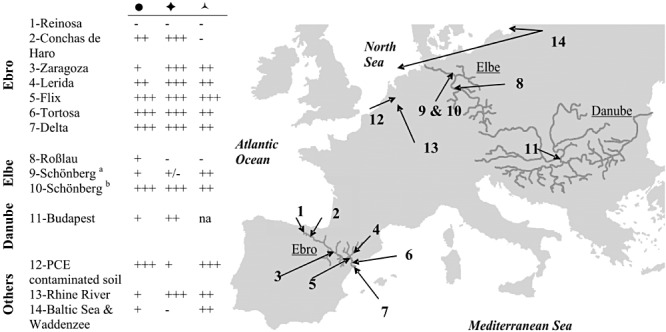
Summary of results from the locations studied byTaş (2009) with cultivation‐dependent and ‐independent molecular methods. ●: ‘Dehalococcoides’ spp. detection with 16S rRNA and/or 16S rRNA gene‐targeted methods;  : HCB transformation;
: HCB transformation;  : chlorinated ethene transformation; (−) no detection or no transformation; (+/++/+++) low to high rRNA copies or long to short lag phases in HCB and chlorinated ethene transformation; na: not available; (a) soil and (b) river sediment sample from Schönberg, Germany. Map was redrawn from OpenStreetMap (http://www.openstreetmap.org).
: chlorinated ethene transformation; (−) no detection or no transformation; (+/++/+++) low to high rRNA copies or long to short lag phases in HCB and chlorinated ethene transformation; na: not available; (a) soil and (b) river sediment sample from Schönberg, Germany. Map was redrawn from OpenStreetMap (http://www.openstreetmap.org).
As non‐fermentative microorganisms Dehalococcoides spp. and their organohalide‐respiring relatives Dehalobium chlorocoercia DF‐1 and Dehalogenimonas lykanthroporepellens DC‐9 depend on the H2 supply from other microorganisms for their growth (Smidt and de Vos, 2004; May et al., 2008; Yan et al., 2009). Recently, it has also been suggested that the activity of Dehalococcoides spp. in in situ conditions is linked to the performance of fermentative communities (Röling et al., 2007). Therefore, it is crucial to have insight in factors affecting nutrient fluxes and microbial communities involved in carbon, nitrogen and sulfur (C, N, S) cycling in the river basins to be able to understand the survival and functioning of Dehalococcoides spp. in different geographical locations. Because there are considerable differences between dechlorination capabilities of the known Dehalococcoides strains despite 16S rRNA identities of > 99%, their sole presence based on the detection of the 16S rRNA gene in an environment does not guarantee successful in situ dechlorination of a specific pollutant. Consequently, molecular tools that target metabolic activities of the entire microbial communities in the environment are needed to have a canonical assessment of the conditions.
Discoveries from Dehalococcoides spp. genomes
Our knowledge gap concerning the properties of Dehalococcoides spp. is closing rapidly with the developments in high‐throughput sequencing technologies. Full‐genome sequence analyses revealed that D. ethenogenes strain 195 (GenBank Accession No. NC_002936) and strain CBDB1 (NC_007356) genomes are approximately 1.47 and 1.39 million base pairs (Mbp) respectively. Both genomes comprise single circular chromosomes with 1591 predicted protein coding sequences (CDs) in strain 195 (Seshadri et al., 2005) and 1458 CDs in strain CBDB1 (Table 3). Up to 1217 of the CDs from strain CBDB1 have orthologous genes in D. ethenogenes strain 195 (83.5%) (Kube et al., 2005). Strain BAV1 (NC_009455) has a genome of 1.34 Mbp with 1385 CDs based on information provided in the Integrated Microbial Genomes (IMG) database, release March 2009 (Markowitz et al., 2008). All of these genomes are among the smallest for free‐living bacteria. Different Dehalococcoides spp. genomes share many common properties. For example, one copy of each rRNA gene is present in all Dehalococcoides genomes (Kube et al., 2005; Seshadri et al., 2005). In strains 195, CBDB1 and BAV1 the 16S rRNA gene is spatially separated from 5S and 23S rRNA genes. Comparative analysis of available Dehalococcoides genomes showed that 70% of all genes in these genomes have a high sequence and contextual conservation (McMurdie et al., 2008). Interestingly, D. ethenogenes strain 195 possesses a nitrogenase‐encoding operon, which is missing in strain CBDB1. Even though this finding suggests that D. ethenogenes strain 195 can fix nitrogen, diazotropic growth of the Dehalococcoides strains has not yet been reported.
Table 3.
Comparison of whole‐genome sequence statistics for reductively dechlorinating bacteria as presented in Integrated Microbial Genomes (IMG/M) database, March 2009 (Markowitz et al., 2008).
| Genome name | Phylum/genus | Bases (Mbp) | GC (%) | Genes | CDs | RNA | 16S | Orthologues | Paralogues | rdh genes |
|---|---|---|---|---|---|---|---|---|---|---|
| Anaeromyxobacter dehalogenans 2CP‐C | Proteobacteria Anaeromyxobacter | 5.01 | 0.75 | 4419 | 4361 | 58 | 2 | 4290 | 2468 | 2 |
| Geobacter lovleyi SZ | Proteobacteria Geobacter | 3.87 | 0.55 | 3514 | 3476 | 38 | 1 | 3287 | 1858 | 2 |
| Desulfitobacterium hafniense DCB‐2 | Firmicutes Desulfitobacterium | 5.28 | 0.48 | 4801 | 4712 | 89 | 5 | 4597 | 2921 | 7 |
| Desulfitobacterium hafniense Y51 | Firmicutes Desulfitobacterium | 5.73 | 0.47 | 5137 | 5060 | 77 | 6 | 4765 | 3200 | 4 |
| ‘Dehalococcoides ethenogenes’ strain 195 | Chloroflexi ‘Dehalococcoides’ | 1.47 | 0.49 | 1641 | 1591 | 51 | 1 | 1426 | 628 | 17 |
| ‘Dehalococcoides’ sp. BAV1 | Chloroflexi ‘Dehalococcoides’ | 1.34 | 0.47 | 1436 | 1385 | 51 | 1 | 1327 | 488 | 10 |
| ‘Dehalococcoides’ sp. CBDB1 | Chloroflexi ‘Dehalococcoides’ | 1.39 | 0.47 | 1516 | 1458 | 58 | 1 | 1378 | 541 | 32 |
| ‘Dehalococcoides’ sp. VS | Chloroflexi ‘Dehalococcoides’ | 2.39 | 0.55 | 2160 | 2096 | 64 | 1 | 2003 | 892 | 36 |
Genes: total gene count; CDs: coding sequences; RNA: number of rRNA, tRNA and other RNA genes; 16S: number of 16S rRNA gene copies; Orthologues: number of genes in orthologues; Paralogues: number of genes in paralogues; rdh genes: confirmed and predicted reductive dehalogenase‐encoding genes.
Different Dehalococcoides strains contain different numbers of rdh genes that encode protein, which have been proven or predicted to catalyse the dechlorination reaction. When compared with the genomes of other dechlorinating bacteria, Dehalococcoides have the highest number of rdh genes in their genomes (Table 3). Genomes of strains 195, CBDB1 and BAV1 have 17, 32 and 10 rdh genes, respectively, whereas only seven rdh genes were identified in the genome of Desulfitobacterium hafniense DCB‐2, four rdh genes in D. hafniense Y51 and two rdh genes in Geobacter lovleyi SZ and Anaeromyxobacter dehalogenans (Thomas et al., 2008). The draft genome of strain VS contains the highest number of rdh genes (36 full‐length genes) ever found in a single bacterial genome (McMurdie et al., 2008). Similarly, 14 and 19 rdh genes were detected via PCR amplification in Dehalococcoides sp. strains FL2 and DCMB5 respectively (Holscher et al., 2004; Bunge et al., 2008). Twelve rdh genes from strain CBDB1 have orthologues in D. ethenogenes strain 195 genome with 86.4–95.4% sequence identity. In D. ethenogenes strain 195 and strain CBDB1 genomes almost all of the rdh genes (except DET0079, TCE reductive dehalogenase tceA in D. ethenogenes strain 195 and cbdbA1583 in strain CBDB1) were found to be located in close proximity to genes for transcription regulators, and were predicted to be transcribed in the direction of DNA synthesis, which suggests tight regulation of rdh activity (Kube et al., 2005; Seshadri et al., 2005). However, the function of only a small number of these genes is known. Only two rdh genes from strain 195, DET0079 and DET0318, have been characterized as TCE (tceA) and PCE (pceA) reductive dehalogenases respectively (Fung et al., 2007). Another tceA gene was identified in Dehalococcoides sp. strain FL2 (GenBank Accession No. AY165309) (He et al., 2005). The cbdbA84 gene from strain CBDB1 was recently designated as a chlorobenzene reductive dehalogenase (cbrA), which is involved in dechlorination of 1,2,3,4‐TeCB and 1,2,3‐TCB (Adrian et al., 2007a). Additionally, two VC reductase genes were identified from strain BAV1 (bvcrA, DehaBAV1_0847) (Krajmalnik‐Brown et al., 2004) and strain VS (vcrA, GenBank Accession No. AY322364) (Muller et al., 2004). Since metabolic function cannot be inferred from Dehalococcoides phylogeny, detection methods based on process‐specific biomarkers are necessary to describe the bioremediation capacity and activity of Dehalococcoides in the environment. Therefore, genes like rdhs and the corresponding gene products that are specific to functions of interest can serve as useful biomarkers in monitoring of different Dehalococcoides activities. In the past years microarrays were shown to be useful tools for such monitoring activities and characterization of microbial communities (Zhou, 2003; Wang et al., 2009). Furthermore, functional gene arrays (FGAs), which target functional genes such as nitrogenases, cellulases etc., allow fast and comprehensive analysis of metabolic potential and activity of microbial communities in the environment by targeting a large number of genes or their transcripts in one single experiment (Wu et al., 2001; Taroncher‐Oldenburg et al., 2003; Steward et al., 2004; Zhou et al., 2008). Up to date the most extensive FGA platform is the GeoChip (He et al., 2007), which targets approximately 10 000 catabolic genes involved in major biogeochemical cycles, including those of carbon, nitrogen and sulfur, as well as organic pollutant degradation. Analysis of HCB‐contaminated sediments in the Ebro river basin (Spain) using the GeoChip amended with probes targeting 153 rdh genes showed that rdh gene diversity changed significantly between different sampling locations (Taşet al., 2009). More specifically, sediment samples taken at a site with high HCB pollution (Lacorte et al., 2006) were dominated by rdh genes of Dehalococcoides spp. strain CBDB1 and D. ethenogenes strain 195. In contrast samples, which were characterized by more diffuse pollution with a broader range of contaminants, a wide spectrum of rdh genes was detected including those from various other organohalide‐respiring microorganisms (Fig. 4). However, it should be noted that microarrays can only detect known sequences, which can cause an underestimation of functional gene diversity and abundance in environments for which limited sequence information is available. Application of FGAs in combination with newly developed techniques such as high‐throughput non‐gel‐based proteomics (Maron et al., 2007) and sequencing of the metatranscriptome offers a remarkable promise. Recent studies on D. ethenogenes strain 195 and Dehalococcoides spp. strain CBDB1 transcriptomes suggested continuous transcription of rdh genes such as tceA (Johnson et al., 2008) and cbrA (Wagner et al., 2009) during different growth phases. As a result gene transcripts of such genes can be studied using transcriptomic techniques with FGAs, in combination with proteomics methods (Morris et al., 2006; Morris et al., 2007) to identify the proteins with significant functional impact.
Figure 4.

Hierarchical cluster analysis of rdh gene profiles based on GeoChip functional gene array hybridization signals for samples from Flix and Rice Fields (RF, river delta) in the Ebro River. HCB is reported to be the dominant chlorinated contaminant in Ebro's basin where location Flix bares the highest HCB pollution (Lacorte et al., 2006). White represents no hybridization above background level and grey represents positive hybridization. The grey‐scale intensity indicates differences in hybridization signal intensity, with black representing the strongest signals. Samples are represented according to sampling month, year and sampling depth (i.e. F06D5: February 2006 depth 0–5 cm; J04D15: June 2004 depth 10–15 cm). For accession numbers of rdh gene targets, see Taş and colleagues (2009).
Future perspectives: reductive dechlorination, systems microbiology and microbial networks
The broad aim of systems microbiology is to define and understand the relationships between the individual components that build a cellular organism, a community and an ecological niche (Vieites et al., 2009). As a result, in the past, the focus of systems microbiology was on microbial isolates or enrichments (McHardy and Rigoutsos, 2007). To date, the majority of the research conducted in the field of reductive dechlorination has been predominantly focused on the identification of genes and proteins directly responsible for the dechlorination process (Cupples et al., 2003; Muller et al., 2004; Holmes et al., 2006; Adrian et al., 2007a; McMurdie et al., 2007; West et al., 2008; Wagner et al., 2009). These experimental studies, so far, allowed the analysis and characterization of several key genes. However, it is becoming evident that to understand microbial functions or functioning of microbial communities one must study the entire system (Vieites et al., 2009). The body of research summarized in this review also supports this idea and suggests that with biomolecular assays targeting ribosomal and process‐specific functional genes such as those encoding reductive dehalogenases, it will remain difficult to understand the full extent of the process, since the dechlorination process comprises an integral part of a complex web of metabolic and regulatory interactions (Rahm et al., 2006; West et al., 2008; Wagner et al., 2009). The application of novel, more comprehensive methods like whole genome shotgun (WGS) sequencing of environmental DNA and mRNA (functional metagenomics) (Tringe et al., 2005; Kalyuzhnaya et al., 2008), the establishment of large‐scale databases which contain metagenomic data from different environments (Seshadri et al., 2007; Pignatelli et al., 2009; Vogel et al., 2009) as well as the development of new computational resources for comparative (meta)genomic analyses (Peterson et al., 2001; Alm et al., 2005; Markowitz et al., 2006) enable us to develop and analyse data sets (and microbial networks) which so far are believed to be the closest to the actual environmental situations. Thus, today, it can be proposed to leave reductionist approaches that are limited to only one or a few selected biomarkers, and to study reductive dechlorination and the function of Dehalococcoides spp. in larger communities and in the environments in which they belong. As the functional properties of such communities are elucidated, we will be able to assess the true role and importance of Dehalococcoides spp. in the environment.
Acknowledgments
This work was supported by EU‐FP6 Project AquaTerra (Project no. GOCE 505248), NWO (2007/0144444/IB) and Ecogenomics (BSIK03011). Authors would like to thank Prof. J. Zhou and colleagues in IE6, Oklahoma for their cooperation in microarray experiments.
References
- Adrian L., Szewzyk U., Wecke J., Gorisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Adrian L., Rahnenfuhrer J., Gobom J., Holscher T. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2007a;73:7717–7724. doi: 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L., Hansen S.K., Fung J.M., Gorisch H., Zinder S.H. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ Sci Technol. 2007b;41:2318–2323. doi: 10.1021/es062076m. [DOI] [PubMed] [Google Scholar]
- Adrian L., Dudkova V., Demnerova K., Bedard D.L. Dehalococcoides’ sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2009;75:4516–4524. doi: 10.1128/AEM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR) 1999. ) Public health statement for chlorophenols [WWW document]. URL http://www.atsdr.cdc.gov/toxprofiles/phs107.html.
- Agency for Toxic Substances and Disease Registry (ATSDR) 2002. ) Toxicological profile for HCB [WWW document]. URL http://www.atsdr.cdc.gov/toxprofiles/tp90.html.
- Alm E.J., Huang K.H., Price M.N., Koche R.P., Keller K., Dubchak I.L., Arkin A.P. The MicrobesOnline web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey R.E. Global hexachlorobenzene emissions. Chemosphere. 2001;43:167–182. doi: 10.1016/s0045-6535(00)00186-7. [DOI] [PubMed] [Google Scholar]
- Ballerstedt H., Hantke J., Bunge M., Werner B., Gerritse J., Andreesen J.R., Lechner U. Properties of a trichlorodibenzo‐p‐dioxin‐dechlorinating mixed culture with a Dehalococcoides as putative dechlorinating species. FEMS Microbiol Ecol. 2004;47:223–234. doi: 10.1016/S0168-6496(03)00282-4. [DOI] [PubMed] [Google Scholar]
- Barber J.L., Sweetman A.J., Van Wijk D., Jones K.C. Hexachlorobenzene in the global environment: emissions, levels, distribution, trends and processes. Sci Total Environ. 2005;349:1–44. doi: 10.1016/j.scitotenv.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Becking B.L.G. W.P. Van Stockum & Zoon; 1934. [Google Scholar]
- Bedard D.L., Ritalahti K.M., Loffler F.E. The Dehalococcoides population in sediment‐free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2007;73:2513–2521. doi: 10.1128/AEM.02909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijerinck M. Jaarboek van de Koninklijke Akademie v. Wetenschappen; 1913. [Google Scholar]
- Bengtson P., Bastviken D., De Boer W., Öberg G. Possible role of reactive chlorine in microbial antagonism and organic matter chlorination in terrestrial environments. Environ Microbiol. 2009;11:1330–1339. doi: 10.1111/j.1462-2920.2009.01915.x. [DOI] [PubMed] [Google Scholar]
- Bowman K.S., Moe W.M., Rash B.A., Bae H.S., Rainey F.A. Bacterial diversity of an acidic Louisiana groundwater contaminated by dense nonaqueous‐phase liquid containing chloroethanes and other solvents. FEMS Microbiol Ecol. 2006;58:120–133. doi: 10.1111/j.1574-6941.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- Bunge M., Adrian L., Kraus A., Opel M., Lorenz W.G., Andreesen J.R. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature. 2003;421:357. doi: 10.1038/nature01237. et al. [DOI] [PubMed] [Google Scholar]
- Bunge M., Kähkönen M., Rämisch W., Opel M., Vogler S., Walkow F. Biological activity in a heavily organohalogen‐contaminated river sediment. Environ Sci Pollut Res. 2007;14:3–10. doi: 10.1065/espr2006.03.298. et al. [DOI] [PubMed] [Google Scholar]
- Bunge M., Wagner A., Fischer M., Andreesen J.R., Lechner U. Enrichment of a dioxin‐dehalogenating Dehalococcoides species in two‐liquid phase cultures. Environ Microbiol. 2008;10:2670–2683. doi: 10.1111/j.1462-2920.2008.01688.x. [DOI] [PubMed] [Google Scholar]
- Bürgmann H., Kleikemper J., Duc L., Bunge M., Schroth M., Zeyer J. Detection and quantification of Dehalococcoides‐related bacteria in a chlorinated ethene‐contaminated aquifer undergoing natural attenuation. Bioremediation J. 2008;12:193–209. [Google Scholar]
- Cheng D., He J. Isolation and characterization of ‘Dehalococcoides’ sp. strain MB that dechlorinates tetrachloroethene to trans‐1, 2‐dichloroethene. Appl Environ Microbiol. 2009 doi: 10.1128/AEM.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Krajmalnik‐Brown R., Rittmann B.E. Bioreduction of trichloroethene using a hydrogen‐based membrane biofilm reactor. Environ Sci Technol. 2008;42:477–483. doi: 10.1021/es702422d. [DOI] [PubMed] [Google Scholar]
- Criado M.R., Pereiro I.R., Torrijos R.C. Determination of polychlorinated biphenyl compounds in indoor air samples. J Chromatogr A. 2002;963:65–71. doi: 10.1016/s0021-9673(02)00643-x. [DOI] [PubMed] [Google Scholar]
- Cupples A.M., Spormann A.M., McCarty P.L. Growth of a Dehalococcoides‐like microorganism on vinyl chloride and cis‐dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decision No. 2455/2001/EC. Decision No. 2455/2001/EC of the European Parliament and of the Council of 20 November 2001. The list of priority substances in the field of water policy and amending Directive 2000/60/EC. OJ L. 2001;331:1–5. [Google Scholar]
- Dolfing J. Thermodynamic considerations for dehalogenation. In: Haggblom M.M., Bossert I.D., editors. Kluwer Academic Publishers; 2003. pp. 89–114. [Google Scholar]
- Dowling D.N., Doty S.L. Improving phytoremediation through biotechnology. Curr Opin Biotechnol. 2009;20:204–206. doi: 10.1016/j.copbio.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Edwards E.A. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2‐dichloroethane. Environ Sci Technol. 2007;41:2303–2310. doi: 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Wehr S.D., Yu L., Rizvi H., Seepersad D., Dworatzek S. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis‐dichloroethene and vinyl chloride. Water Res. 2002;36:4193–4202. doi: 10.1016/s0043-1354(02)00151-3. et al. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Mo K., Edwards E.A. Characterization of a highly enriched Dehalococcoides‐containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70:5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eekert M.H.A., Schraa G. The potential of anaerobic bacteria to degrade chlorinated compounds. Water Sci Technol. 2001;44:49–56. [PubMed] [Google Scholar]
- Ellis D.E., Lutz E.J., Odom J.M., Buchanan R.J., Bartlett C.L., Lee M.D. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ Sci Technol. 2000;34:2254–2260. et al. [Google Scholar]
- Fennell D.E., Nijenhuis I., Wilson S.F., Zinder S.H., Haggblom M.M. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol. 2004;38:2075–2081. doi: 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- Field J.A., Sierra‐Alvarez R. Microbial degradation of chlorinated dioxins. Chemosphere. 2008;71:1005–1018. doi: 10.1016/j.chemosphere.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Fierer N., Jackson R.B. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung J.M., Morris R.M., Adrian L., Zinder S.H. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3‐dichlorophenol. Appl Environ Microbiol. 2007;73:4439–4445. doi: 10.1128/AEM.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futamata H., Yoshida N., Kurogi T., Kaiya S., Hiraishi A. Reductive dechlorination of chloroethenes by Dehalococcoides‐containing cultures enriched from a polychlorinated‐dioxin‐contaminated microcosm. ISME J. 2007;1:471–479. doi: 10.1038/ismej.2007.42. [DOI] [PubMed] [Google Scholar]
- Gribble G.W. The diversity of naturally produced organohalogens. Chemosphere. 2003;52:289–297. doi: 10.1016/S0045-6535(03)00207-8. [DOI] [PubMed] [Google Scholar]
- Griebler C., Adrian L., Meckenstock R.U., Richnow H.H. Stable carbon isotope fractionation during aerobic and anaerobic transformation of trichlorobenzene. FEMS Microbiol Ecol. 2004;48:313. doi: 10.1016/j.femsec.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Grostern A., Edwards E.A. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl Environ Microbiol. 2006;72:428–436. doi: 10.1128/AEM.72.1.428-436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman K.J., Simonich S.L., Campbell D.H., Wilson G.R., Landers D.H. Atmospheric deposition of current‐use and historic‐use pesticides in snow at national parks in the western United States. Environ Sci Technol. 2006;40:3174–3180. doi: 10.1021/es060157c. [DOI] [PubMed] [Google Scholar]
- Harkness M.R., Bracco A.A., Brennan M.J., DeWeerd K.A., Spivack J.L. Use of bioaugmentation to stimulate complete reductive dechlorination of trichloroethene in Dover soil columns. Environ Sci Technol. 1999;33:1100–1109. [Google Scholar]
- He J., Ritalahti K.M., Yang K.‐L., Koenigsberg S.S., Loffler F.E. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- He J., Sung Y., Krajmalnik‐Brown R., Ritalahti K.M., Loffler F.E. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)‐ and 1,2‐dichloroethene‐respiring anaerobe. Environ Microbiol. 2005;7:1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- He Z., Gentry T.J., Schadt C.W., Wu L., Liebich J., Chong S.C. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1:67. doi: 10.1038/ismej.2007.2. et al. [DOI] [PubMed] [Google Scholar]
- Hendrickson E.R., Payne J.A., Young R.M., Starr M.G., Perry M.P., Fahnestock S. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene‐contaminated sites throughout North America and Europe. Appl Environ Microbiol. 2002;68:485–495. doi: 10.1128/AEM.68.2.485-495.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelheber D.W., Thomas S.H., Löffler F.E., Taillefert M., Hughes J.B. Microbial colonization of an in situ sediment cap and correlation to stratified redox zones. Environ Sci Technol. 2009;43:66–74. doi: 10.1021/es801834e. [DOI] [PubMed] [Google Scholar]
- Holliger C., Wohlfarth G., Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1998;22:383–398. [Google Scholar]
- Holmes V.F., He J., Lee P.K., Alvarez‐Cohen L. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol. 2006;72:5877–5883. doi: 10.1128/AEM.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holscher T., Krajmalnik‐Brown R., Ritalahti K.M., Von Wintzingerode F., Gorisch H., Loffler F.E., Adrian L. Multiple nonidentical reductive‐dehalogenase‐homologous genes are common in Dehalococcoides. Appl Environ Microbiol. 2004;70:5290–5297. doi: 10.1128/AEM.70.9.5290-5297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imfeld G., Nijenhuis I., Nikolausz M., Zeiger S., Paschke H., Drangmeister J. Assessment of in situ degradation of chlorinated ethenes and bacterial community structure in a complex contaminated groundwater system. Water Res. 2008;42:871–882. doi: 10.1016/j.watres.2007.08.035. et al. [DOI] [PubMed] [Google Scholar]
- James C.A., Strand S.E. Phytoremediation of small organic contaminants using transgenic plants. Curr Opin Biotechnol. 2009;20:237–241. doi: 10.1016/j.copbio.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C.A., Xin G., Doty S.L., Muiznieks I., Newman L., Strand S.E. A mass balance study of the phytoremediation of perchloroethylene‐contaminated groundwater. Environ Pollut. 2009 doi: 10.1016/j.envpol.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.R., Brodie E.L., Hubbard A.E., Andersen G.L., Zinder S.H., Alvarez‐Cohen L. Temporal transcriptomic microarray analysis of ‘Dehalococcoides ethenogenes’ strain 195 during the transition into stationary phase. Appl Environ Microbiol. 2008;74:2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya M.G., Lapidus A., Ivanova N., Copeland A.C., McHardy A.C., Szeto E. High‐resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. et al. [DOI] [PubMed] [Google Scholar]
- Kittelmann S., Friedrich M.W. Identification of novel perchloroethene‐respiring microorganisms in anoxic river sediment by RNA‐based stable isotope probing. Environ Microbiol. 2008;10:31–46. doi: 10.1111/j.1462-2920.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- Krajmalnik‐Brown R., Holscher T., Thomson I.N., Saunders F.M., Ritalahti K.M., Loffler F.E. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube M., Beck A., Zinder S.H., Kuhl H., Reinhardt R., Adrian L. Genome sequence of the chlorinated compound‐respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23:1269. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- Lacorte S., Raldua D., Martinez E., Navarro A., Diez S., Bayona J.M., Barcelo D. Pilot survey of a broad range of priority pollutants in sediment and fish from the Ebro river basin (NE Spain) Environ Pollut. 2006;140:471–482. doi: 10.1016/j.envpol.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Lai K.C.K., Surampalli R.Y., Tyagi R.D., Lo I.M.C., Yan S. Performance monitoring of remediation technologies for soil and groundwater contamination: review. Pract Periodical of Haz, Toxic, and Radioactive Waste Mgmt. 2007;11:132–157. [Google Scholar]
- Lee P.K.H., Macbeth T.W., Sorenson K.S., Jr, Deeb R.A., Alvarez‐Cohen L. Quantifying genes and transcripts to assess the in situ physiology of ‘Dehalococcoides’ spp. in a trichloroethene‐contaminated groundwater site. Appl Environ Microbiol. 2008;74:2728–2739. doi: 10.1128/AEM.02199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay J.M., Loffler F.E., Dollhopf M., Aiello M.R., Daniels G., Fathepure B.Z. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ Sci Technol. 2003;37:1422–1431. et al. [Google Scholar]
- Löffler F.E., Sun Q., Li J., Tiedje J.M. 16S rRNA gene‐based detection of tetrachloroethene‐dechlorinating Desulfuromonas and Dehalococcoides species. Appl Environ Microbiol. 2000;66:1369–1374. doi: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy A.C., Rigoutsos I. What's in the mix: phylogenetic classification of metagenome sequence samples. Curr Opin Microbiol. 2007;10:499–503. doi: 10.1016/j.mib.2007.08.004. [DOI] [PubMed] [Google Scholar]
- McMurdie P.J., Behrens S.F., Holmes S., Spormann A.M. Unusual codon bias in vinyl chloride reductase genes of Dehalococcoides species. Appl Environ Microbiol. 2007;73:2744–2747. doi: 10.1128/AEM.02768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Behrens S.F., Muller J., Goke J., Loffler F.E., Spormann A.M. 2008. , and ) Comparative genomics of two vinyl chloride respiring bacteria, Dehalococcoides sp. strains VS and BAV1. In The 12th International Symposium on Microbial Ecology ISME‐12. Cairns, Australia.
- Major D.W., McMaster M.L., Cox E.E., Edwards E.A., Dworatzek S.M., Hendrickson E.R. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol. 2002;36:5106–5116. doi: 10.1021/es0255711. et al. [DOI] [PubMed] [Google Scholar]
- Markowitz V.M., Ivanova N., Palaniappan K., Szeto E., Korzeniewski F., Lykidis A. An experimental metagenome data management and analysis system. Bioinformatics. 2006;22:e359–e367. doi: 10.1093/bioinformatics/btl217. et al. [DOI] [PubMed] [Google Scholar]
- Markowitz V.M., Ivanova N.N., Szeto E., Palaniappan K., Chu K., Dalevi D. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron P.‐A., Ranjard L., Mougel C., Lemanceau P. Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol. 2007;53:486–493. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- Martiny J.B.H., Bohannan B.J.M., Brown J.H., Colwell R.K., Fuhrman J.A., Green J.L. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. et al. [DOI] [PubMed] [Google Scholar]
- May H.D., Miller G.S., Kjellerup B.V., Sowers K.R. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl Environ Microbiol. 2008;74:2089–2094. doi: 10.1128/AEM.01450-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymo‐Gatell X., Chien Y.‐T., Gossett J.M., Zinder S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- Maymo‐Gatell X., Anguish T., Zinder S.H. Reductive dechlorination of chlorinated ethenes and 1, 2‐dichloroethane by ‘Dehalococcoides ethenogenes’ 195. Appl Environ Microbiol. 1999;65:3108–3113. doi: 10.1128/aem.65.7.3108-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer S.N., Ockenden W.A., Sweetman A., Breivik K., Grimalt J.O., Jones K.C. Global distribution and budget of PCBs and HCB in background surface soils: implications for sources and environmental processes. Environ Sci Technol. 2003;37:667–672. doi: 10.1021/es025809l. [DOI] [PubMed] [Google Scholar]
- Morris R.M., Sowell S., Barofsky D., Zinder S., Richardson R. Transcription and mass‐spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ Microbiol. 2006;8:1499–1509. doi: 10.1111/j.1462-2920.2006.01090.x. [DOI] [PubMed] [Google Scholar]
- Morris R.M., Fung J.M., Rahm B.G., Zhang S., Freedman D.L., Zinder S.H., Richardson R.E. Comparative proteomics of Dehalococcoides spp. reveals strain‐specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–326. doi: 10.1128/AEM.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J.A., Rosner B.M., Von Abendroth G., Meshulam‐Simon G., McCarty P.L., Spormann A.M. Molecular Identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman L.A., Strand S.E., Choe N., Duffy J., Ekuan G., Ruszaj M. Uptake and biotransformation of trichloroethylene by hybrid poplars. Environ Sci Technol. 1997;31:1062–1067. et al. [Google Scholar]
- Peterson J.D., Umayam L.A., Dickinson T., Hickey E.K., White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli M., Moya A., Tamames J. EnvDB, a database for describing the environmental distribution of prokaryotic taxa. Environ Microbiol Rep. 2009;1:191–197. doi: 10.1111/j.1758-2229.2009.00030.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glockner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahm B.G., Morris R.M., Richardson R.E. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl Environ Microbiol. 2006;72:5486–5491. doi: 10.1128/AEM.00855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R.E., Bhupathiraju V.K., Song D.L., Goulet T.A., Alvarez‐Cohen L. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ Sci Technol. 2002;36:2652–2662. doi: 10.1021/es0157797. [DOI] [PubMed] [Google Scholar]
- Röling W.F.M., Van Breukelen B.M., Bruggeman F.J., Westerhoff H.V. Ecological control analysis: being(s) in control of mass flux and metabolite concentrations in anaerobic degradation processes. Environ Microbiol. 2007;9:500–511. doi: 10.1111/j.1462-2920.2006.01167.x. [DOI] [PubMed] [Google Scholar]
- Scheutz C., Durant N.D., Dennis P., Hansen M.H., Jørgensen T., Jakobsen R. Concurrent ethene generation and growth of Dehalococcoides containing vinyl chloride reductive dehalogenase genes during an enhanced reductive dechlorination field demonstration. Environ Sci Technol. 2008;42:9302–9309. doi: 10.1021/es800764t. et al. [DOI] [PubMed] [Google Scholar]
- Seshadri R., Adrian L., Fouts D.E., Eisen J.A., Phillippy A.M., Methe B.A. Genome sequence of the PCE‐dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. et al. [DOI] [PubMed] [Google Scholar]
- Seshadri R., Kravitz S.A., Smarr L., Gilna P., Frazier M. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H. Wiley‐Interscience; 2006. [Google Scholar]
- Sleep B.E., Seepersad D.J., Mo K., Heidorn C.M., Hrapovic L., Morrill P.L. Biological enhancement of tetrachloroethene dissolution and associated microbial community changes. Environ Sci Technol. 2006;40:3623–3633. doi: 10.1021/es051493g. et al. [DOI] [PubMed] [Google Scholar]
- Smidt H., De Vos W.M. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- Steward G.F., Jenkins B.D., Ward B.B., Zehr J.P. Development and testing of a DNA macroarray to assess nitrogenase (nifH) gene diversity. Appl Environ Microbiol. 2004;70:1455–1465. doi: 10.1128/AEM.70.3.1455-1465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y., Ritalahti K.M., Apkarian R.P., Loffler F.E. Quantitative PCR confirms purity of strain GT, a novel trichloroethene‐to‐ethene‐respiring Dehalococcoides isolate. Appl Environ Microbiol. 2006;72:1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susarla S., Medina V.F., McCutcheon S.C. Phytoremediation: an ecological solution to organic chemical contamination. Ecol Eng. 2002;18:647–658. [Google Scholar]
- Taroncher‐Oldenburg G., Griner E.M., Francis C.A., Ward B.B. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl Environ Microbiol. 2003;69:1159–1171. doi: 10.1128/AEM.69.2.1159-1171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N. 2009. Dehalococcoides spp. in river sediments: insights in functional diversity and dechlorination activity. In Laboratory of Microbiology PhD Dissertations. Wageningen, the Netherlands: Wageningen University [WWW document]. URL http://edepot.wur.nl/5966.
- Taş N., Van Eekert M.H.A., Schraa G., Zhou J., De Vos W.M., Smidt H. Tracking functional guilds: ‘Dehalococcoides’ spp. in European river basins contaminated with hexachlorobenzene. Appl Environ Microbiol. 2009;75:4696–4704. doi: 10.1128/AEM.02829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.H., Wagner R.D., Arakaki A.K., Skolnick J., Kirby J.R., Shimkets L.J. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP‐C suggests an aerobic common ancestor to the delta‐proteobacteria. PLoS ONE. 2008;3:e2103. doi: 10.1371/journal.pone.0002103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortella G., Diez M., Durán N. Fungal diversity and use in decomposition of environmental pollutants. Crit Rev Microbiol. 2005;31:197–212. doi: 10.1080/10408410500304066. [DOI] [PubMed] [Google Scholar]
- Tringe S.G., Von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. et al. [DOI] [PubMed] [Google Scholar]
- US EPA. 1985. ) Health and environmental effects profile for chloroethene [WWW document]. URL http://www.epa.gov.
- UNEP. United Nations Environment Programme; 2005. [Google Scholar]
- UNEP Chemicals; U.N.E.P. United Nations Environment Programme; 1999. pp. 1–34. [Google Scholar]
- Vainberg S., Condee C., Steffan R. Large‐scale production of bacterial consortia for remediation of chlorinated solvent‐contaminated groundwater. J Ind Microbiol Biotechnol. 2009;36:1189–1197. doi: 10.1007/s10295-009-0600-5. [DOI] [PubMed] [Google Scholar]
- Vieites J.M., Guazzaroni M.E., Beloqui A., Golyshin P.N., Ferrer M. Metagenomics approaches in systems microbiology. FEMS Microbiol Rev. 2009;33:236–255. doi: 10.1111/j.1574-6976.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- Vogel T.M., Simonet P., Jansson J.K., Hirsch P.R., Tiedje J.M., Van Elsas J.D. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol. 2009;7:252–252. et al. [Google Scholar]
- Wagner A., Adrian L., Kleinsteuber S., Andreesen J.R., Lechner U. Transcription analysis of genes encoding homologues of reductive dehalogenases in Dehalococcoides sp. strain CBDB1 using terminal restriction fragment length polymorphism and quantitative PCR. Appl Environ Microbiol. 2009;75:1876–1884. doi: 10.1128/AEM.01042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhou H., Meng J., Peng X., Jiang L., Sun P. GeoChip‐based analysis of metabolic diversity of microbial communities at the Juan de Fuca Ridge hydrothermal vent. Proc Natl Acad Sci USA. 2009;106:4840–4845. doi: 10.1073/pnas.0810418106. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.‐D., Li Q.‐J., Luo B., Chen X.‐Y. Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat Biotechnol. 2004;22:893–897. doi: 10.1038/nbt982. [DOI] [PubMed] [Google Scholar]
- Weber R., Gaus C., Tysklind M., Johnston P., Forter M., Hollert H. Dioxin‐ and POP‐contaminated sites – contemporary and future relevance and challenges. Environ Sci Pollut Res Int. 2008;15:363–393. doi: 10.1007/s11356-008-0024-1. et al. [DOI] [PubMed] [Google Scholar]
- Wenning R.J., Sorensen M., Magar V.S. Importance of implementation and residual risk analyses in sediment remediation. Integr Environ Assess Manag. 2006;2:59–65. [PubMed] [Google Scholar]
- West K.A., Johnson D.R., Hu P., DeSantis T.Z., Brodie E.L., Lee P.K.H. Comparative genomics of ‘Dehalococcoides ethenogenes’ 195 and an enrichment culture containing unsequenced ‘Dehalococcoides’ strains. Appl Environ Microbiol. 2008;74:3533–3540. doi: 10.1128/AEM.01835-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfarth G., Diekert G. Anaerobic dehalogenases. Curr Opin Biotechnol. 1997;8:290–295. doi: 10.1016/s0958-1669(97)80006-7. [DOI] [PubMed] [Google Scholar]
- Wu L., Thompson D.K., Li G., Hurt R.A., Tiedje J.M., Zhou J. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl Environ Microbiol. 2001;67:5780–5790. doi: 10.1128/AEM.67.12.5780-5790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Rash B.A., Rainey F.A., Moe W.M. Isolation of novel bacteria within the Chloroflexi capable of reductive dechlorination of 1,2,3‐trichloropropane. Environ Microbiol. 2009;11:833–843. doi: 10.1111/j.1462-2920.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- Yoshida N., Takahashi N., Hiraishi A. Phylogenetic characterization of a polychlorinated‐dioxin‐ dechlorinating microbial community by use of microcosm studies. Appl Environ Microbiol. 2005;71:4325–4334. doi: 10.1128/AEM.71.8.4325-4334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeb B.A., Amphlett J.S., Rutter A., Reimer K.J. Potential for phytoremediation of polychlorinated biphenyl‐(PCB)‐contaminated soil. Int J Phytorem. 2006;8:199–221. doi: 10.1080/15226510600846749. [DOI] [PubMed] [Google Scholar]
- Zhou J. Microarrays for bacterial detection and microbial community analysis. Curr Opin Microbiol. 2003;6:288. doi: 10.1016/s1369-5274(03)00052-3. [DOI] [PubMed] [Google Scholar]
- Zhou J., Kang S., Schadt C.W., Garten C.T. Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA. 2008;105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]


