Summary
Ever since the introduction of the Salmonella typhimurium mammalian microsome mutagenicity assay (the ‘Ames test’) over three decades ago, there has been a constant development of additional genotoxicity assays based upon the use of genetically engineered microorganisms. Such assays rely either on reversion principles similar to those of the Ames test, or on promoter–reporter fusions that generate a quantifiable dose‐dependent signal in the presence of potential DNA damaging compounds and the induction of repair mechanisms; the latter group is the subject of the present review. Some of these assays were only briefly described in the scientific literature, whereas others have been developed all the way to commercial products. Out of these, only one, the umu‐test, has been fully validated and ISO‐ and OECD standardized. Here we review the main directions undertaken in the construction and testing of bacterial‐based genotoxicity bioassays, including the attempts to incorporate at least a partial metabolic activation capacity into the molecular design. We list the genetic modifications introduced into the tester strains, compare the performance of the different assays, and briefly describe the first attempts to incorporate such bacterial reporters into actual genotoxicity testing devices.
Introduction
The increasing need to assay and monitor the potential genotoxic effects of an ever‐growing number of chemicals and environmental samples is countered by the logistic, economical and ethical constraints imposed by the use of animal‐based test systems. Consequently, ever since the introduction of the revolutionary Salmonella typhimurium mammalian microsome mutagenicity assay (the ‘Ames test’) over three decades ago (Ames et al., 1973), continuous efforts have been directed towards the development, improvement and implementation of additional bacterial‐based genotoxicity assays. One group of these assays is based on the same principle of the Ames test, in that they quantify the reversion rate from a defined mutation back to the wild‐type (Biran et al., 2009; Reifferscheid and Buchinger, 2009). The present review will not discuss this group but rather concentrate on assays that employ genetically engineered microorganisms, ‘tailored’ to generate a quantifiable signal that reflects the genotoxic potency of the tested sample. Such assays possess several significant advantages including rapid response times, high reproducibility, facility of use and low operational cost. Yet, bacterial‐based assays cannot carry out the complex biochemical reactions collectively known as ‘metabolic activation’; these take place mainly in mammalian liver cells, in which xenobiotics may be transformed into genotoxic forms. Herein, we review the main directions undertaken in the construction and testing of bacterial‐based genotoxicity bioassays, including the attempts to incorporate at least a partial metabolic activation capacity into the molecular design.
The promoter–reporter concept
As will be discussed below, several genetic engineering approaches have been employed over the years in the construction of bacterial reporter strains that respond to the presence of genotoxic compounds. Many of these share the same basic principle: the fusion of a gene promoter, known to be activated by the presence of genotoxic chemicals, to a gene or a group of genes the activity of which can be monitored quantitatively, preferably in real time (Belkin, 2003). The gene promoter acts as the sensing element, which – upon activation – drives the transcription of the downstream reporter gene(s). Consequently, the gene promoter will dictate the response spectrum of the construct and, to some extent, its sensitivity. The reporter genes determine the nature of the generated signal (bioluminescence, fluorescence, etc.) and thus also the instrumentation required for its acquisition. The host cell, the third major component in the construction of a genotoxicity reporter strain, is selected for ease of genetic manipulation, for its relevance, and – most importantly – for its effects on detection sensitivity and threshold.
Sensing elements
In the selection of sensing elements to be used for the construction of genotoxicity reporter systems, the most promising candidates are promoters of genes involved in DNA repair. Such genes, induced in response to either actual DNA damage or to the presence of DNA damaging agents, are mainly part of one of two inducible systems: the recA‐dependent, lexA‐controlled SOS response and the recA‐independent, ada‐controlled adaptive system induced in response to alkylation damage of DNA. The latter system responds to the presence of methylated phosphotriesters generated by DNA alkylation that activate the ada gene product which, in turn, triggers the transcription of genes such as ada, alkA, alkB and aid (Volkert, 1988; Volkert et al., 1989).
The SOS response is under the control of the LexA protein that binds to the SOS box in the promoter region of the regulon genes, repressing their expression. De‐repression occurs when the RecA protein binds to single‐stranded DNA at replication forks that are blocked by DNA damage, forming RecA‐ssDNA nucleoprotein filaments (Courcelle and Hanawalt, 2003; Janion, 2008). Once bound to DNA, the RecA protein changes conformation and acts as a co‐protease in the cleavage of LexA, thus allowing transcription of the SOS genes (Little, 1991; Janion, 2001; Giese et al., 2008). Among these are genes such as uvrA, recA, recN or umuDC, responsible for DNA repair, and others such as sulA, that couple DNA damage to cell division (D'Ari, 1985; Janion, 2001). Expression of a given SOS gene depends on the specific LexA‐binding properties of its promoter, determined by the sequence of the LexA‐binding sites (SOS boxes), their number and their arrangement (Lewis et al., 1994; Fernández de Henestrosa et al., 2000; Norman et al., 2005).
SOS promoters
Several gene promoters from over 30 known SOS regulon genes induced upon DNA damage were used for the construction of genotoxicity sensors, as is briefly described below.
umuDC. The two proteins coded by this operon, UmuD and UmuC, are induced under DNA damage conditions by the LexA‐ and RecA‐dependent transcriptional upregulation of the SOS regulon. They first form a UmuD2C complex, which acts as a checkpoint inhibitor of cell division until repair can address the original DNA damage signal. After RecA/ssDNA‐mediated UmuD cleavage, these proteins form a (UmuD′)2C complex (DNA polymerase V), which carries out the error‐prone replication of damaged DNA (SOS mutagenesis; for review see Sutton et al., 2000).
The first description of a umuC′–lacZ fusion coded on plasmid pSK1002 in S. typhimurium for the detection of genotoxic agents was published by Oda and colleagues (1985), and has since been recognized as the ‘umu‐test’. The S. typhimurium strain carrying this fusion (TA1535) has undergone several modifications including excision repair deficiency (uvrB), an rfa deletion which increases permeability to many chemicals, and a deletion of the natural lac operon. The umu‐test was standardized according to the German Institute of Standardization (DIN 38415‐3) and the International Standardization Organization (ISO/CD 13829). It is now a part of the set of tools available to authorities and researchers for the investigation and monitoring of genotoxicity of environmental samples. The system was adapted to a 96‐well microtiter plate format (Reifferscheid et al., 1991) and has been used, for example, to detect a wide range of carcinogenic mutagens (Oda et al., 1985; Nakamura et al., 1987; Shimada and Nakamura, 1987; McDaniels et al., 1990; Reifferscheid and Heil, 1996), as well as genotoxic activity in disinfectants (Sakagami et al., 1988), complex mixtures (Whong et al., 1986; Hamer et al., 2000), environmental pollutants (Bihari et al., 1990), river waters and industrial wastewaters (Reifferscheid et al., 1991; Ehrlichmann et al., 2000; Dizer et al., 2002).
sulA (sfiA). The SulA protein, produced in large amounts during the SOS response, halts cell division in Escherichia coli by binding to the tubulin‐like GTPase, FtsZ (Higashitani et al., 1995). It has been used as a sensing element for genotoxicity detection in several cases, most notably in the colorimetric SOS chromotest (Quillardet et al., 1982), commercialized in 1984. The E. coli PQ37 tester strain used in the SOS chromotest harbours a sfiA′::lacZ fusion and carries a deletion of the normal lac region, so that β‐galactosidase activity is strictly dependent on sfiA expression. Similarly to the umu‐test bacterium it is mutated in the uvr‐system (uvrA) to hinder DNA repair, and in rfa to increase cell wall permeability (Quillardet and Hofnung, 1985). A different colorimetric assay based on a plasmid‐borne sulA′::lacZ fusion in S. typhimurium TA1538 was proposed by El Mzibri and colleagues (1996), who also described a procedure that includes metabolic activation based on S9‐mix.
recN. Another E. coli SOS gene promoter fusion that has been developed into a commercial product (VITOTOXTM) is based on the recN gene, coding for a protein that is involved in double stranded DNA break repair. The E. coli and several S. typhimurium strains (TA 98, TA 100 and TA104) are used as bacterial hosts. A multi‐copy plasmid harbouring a fusion of the recN promoter to the Vibrio fischeri luxCDABE genes drives the emission of light in response to the presence of DNA damaging agents (van der Lelie et al., 1997), allowing real‐time monitoring of the bacterial response. The VITOTOXTM test strains were evaluated with a variety of chemicals (van der Lelie et al., 1997; Verschaeve et al., 1999; Westerink et al., 2009) as well as river water (Vijayashree et al., 2005), ground water (Verschaeve, 2002) and air samples.
recA. The promoter of the RecA recombinase gene, which plays a key role in the SOS response by its co‐protease activity on the LexA repressor, has served as the basis of several genotoxicity sensors. Nunoshiba and Nishioka (1989; 1991) described the colorimetric E. coli‘Rec‐lac test’, based on the GE94 strain that carries the recA–lacZ fusion, and its DNA repair‐deficient derivative strains such as KY946 (uvrA), KY945 (recA) and KY943 (lexA). The system was tested against 4‐Nitroquinoline‐N‐oxid (4‐NQO), N‐methyl‐N′‐nitro‐N‐nitrosoguanidine (MNNG), mitomycin C (MMC) and UV radiation, as well as with hydrogen peroxide, formaldehyde, tert‐butyl hydroperoxide, cumene hydroperoxide and streptonigrin.
A different reporter system, the V. fischeri luxCDABE operon, was used by Vollmer and colleagues (1997) to generate several E. coli reporter strains, one of them (DPD2794) carrying a recA′::luxCDABE fusion on the multi‐copy plasmid pUCD615 (Vollmer et al., 1997). As in other constructs carrying this 5‐gene complement, these bioluminescent fusions allowed real‐time visualization of the transcriptional responses induced by DNA damage, without the need for cell‐free enzyme assays or the exogenous addition of luciferase substrates. To make full use of these advantages, Polyak and colleagues (2000) have alginate‐immobilized a similar recA′::lux harbouring strain to the tip of an optic fibre, the other end of which was connected to a photon counter. The instrument allowed a real‐time determination of genotoxicity by dipping the bacteria‐clad fibre end into a sample.
The recA gene promoter of the radiation resistant bacterium Deinococcus radiodurans, characterized by an extremely efficient DNA repair capabilities, was fused to the EGFP gene, generating a genotoxicity and radioactivity bioreporter that can persist in extremely genotoxic conditions (Gao et al., 2008). A real‐time concentration‐dependent fluorescent response to γ‐radiation and to MMC was demonstrated.
cda. The colicin D gene cda, a constituent of the ColD plasmid (Frey et al., 1986), also served as a basis for a bioluminescent genotoxicity sensor using Photobacterium leiognathi luxCDABE as a reporter. This ‘SOS lux’ test responded sensitively to diverse genotoxins such as MMC, MNNG, nalidixic acid (NA), dimethylsulfate (DMS), H2O2, CH2O, UV and γ‐radiation (Ptitsyn et al., 1997). This assay has later been combined with the GFPuv‐based Lac‐Fluoro test to generate a combined toxicity‐genotoxicity sensor (Baumstark‐Khan et al., 2001).
Four different SOS promoters (recA, umuCD, sulA and cda) were compared by Norman and colleagues (2005) using the same fluorescent reporter (gfpmut3*, in plasmid pANO1). The differences between the constructs were evaluated after exposure of cells harbouring the fusion plasmids (MG1655/pANO1::SOS promoter) to the known genotoxicant N‐methyl‐N′‐nitro‐N‐nitrosoguanidine (MNNG). A tolC mutation enhanced the sensitivity to this chemical, the only genotoxic agent tested in this study. Performance of the cda‐based sensor in response to MNNG clearly surpassed the other three with respect to the SOS‐induction factor, as a result of high rates of gene expression combined with a low background activity of the cda promoter. Thus, cda promoter was selected for the further development of the GenoTox test (Østergaard et al., 2007).
Non‐SOS promoters
alkA. Several bacterial DNA protection and repair systems that are independent of the SOS regulon have been described, one of which, most efficiently induced by alkylating agents, has been generally termed the ‘adaptive response’. Several genes of this system have been characterized, including alkA, which encodes a repair glycosylase (N3‐methyladenine DNA glycosylase II; Volkert, 1988). A promoter of this gene has been fused by Vollmer and colleagues (1997) to the V. fischeri luxCDABE genes. The construct displayed a very strong response to the alkylating agent MNNG, the magnitude of which was enhanced by very low background bioluminescence; the responses of an equivalent lacZ fusion were much more moderate.
nrdA. The expression of the E. coli nrdA gene, which encodes for a ribonucleoside diphosphate reductase, is strongly affected by DNA damage, induced, for example, by UV exposure, but is independent of LexA (Courcelle et al., 2001). The nrdA promoter was fused by Hwang and colleagues (2008) to Photorhabdus luminescens luxCDABE genes. The E. coli strain BBTNrdA carrying this plasmid‐borne fusion responded to the DNA damaging agents NA, MMC, MNNG, 4‐NQO and hydrogen peroxide, but not to other oxidants or phenolic compounds (Hwang et al., 2008).
Reporter systems
The spectrum of reporter systems available for monitoring gene expression by transcriptional fusions is continuously expanding, as is the instrumentation for signal detection and quantification. Colorimetric, fluorescent, bioluminescent and electrochemical detection of genotoxicity have been described, and are briefly outlined below.
Colorimetric and electrochemical (lacZ, phoA, uidA)
The β‐galactosidase gene, lacZ, has been used as a gene expression reporter for several decades. The most common substrates employed for assaying the activity of this enzyme are o‐nitrophenyl β‐d‐galactopyranoside (ONPG) and 5‐bromo‐4‐chloro‐3‐indolyl β‐d‐galactoside (X‐gal) for colorimetric detection, 4‐methylumbelliferyl‐β‐d‐galactopyranoside (MUG) for fluorimetry, 1,2‐dioxetane substrates for luminescence, and p‐aminophenyl‐β‐d‐galactopyranoside (pAPG) for electrochemical analysis. The advantages of colorimetric assays lie in their simplicity and rapidity, but the need for improved sensitivity, faster response times, a broader dynamic range and the capability of real‐time monitoring has led to a continuous search for alternatives (Jain and Magrath, 1991). The umu‐test (Oda et al., 1985), SOS chromotest (Quillardet et al., 1982), sulA‐test (El Mzibri et al., 1996) and Rec‐lac test (Nunoshiba and Nishioka, 1991) were all developed using lacZ as the reporter and ONPG as the substrate. Several remedies have been proposed to overcome interferences by colored samples, such as the inclusion of a washing step after the exposure of the bacteria to the samples (Nakamura et al., 1987; Pal et al., 1992), or a post‐treatment dilution and re‐incubation (McDaniels et al., 1990; Reifferscheid et al., 1991). The latter procedure was reported to enhance the sensitivity of the umu‐test to genotoxicants in environmental samples in a high‐throughput microtiter plate system (Hamer et al., 2000). Oda and colleagues (2004) achieved higher sensitivity by using a different substrate, chlorophenol red‐β‐D‐galactopyranoside (CRPG). The red reaction product following cleavage by the β‐galactosidase has a longer life time than o‐nitrophenol, the ONPG reaction product. Similar modifications were also introduced to the SOS chromotest (Ohta et al., 1984). The original colorimetric procedure of the assay (Quillardet et al., 1982) was successfully changed to a fluorimetric one by using the fluorescent substrate 4‐methylumbelliferyl‐β‐D‐galactopyranoside (MUG) (Fuentes et al., 2006).
A different approach was proposed by Matsui and colleagues (2006), who provided the umu‐test bacteria (TA1535/pSK1002) with the substrate pAPG, the end product of which (p‐aminophenol) can be monitored electrochemically. This approach, utilized earlier for other bacterial sensor systems (Biran et al., 1999; 2000; Paitan et al., 2003; Schwartz‐Mittelmann et al., 2003), requires the addition of an external substrate but does not involve lysis or permeabilization of the cells. Similarly to bioluminescence, it is thus suitable for continuous online measurement of enzymatic activity, even in turbid solutions and under anaerobic conditions (Badihi‐Mossberg et al., 2007). Matsui and colleagues (2006) have demonstrated this by scanning electrochemical microscopy (SECM) in a specialized glass biochip configuration, using 5 nl cell aliquots immobilized in collagen gel. Overall, lower limits of detection of 2‐aminoflouren (2‐AF), MMC and 2‐aminoanthracene (2‐AA; +S9‐mix) were obtained by the microbial chip as compared with the conventional umu‐test, but it should be noted that the definition of the detection limit was different and exposure times were longer. Buchinger and colleagues (2009) demonstrated the applicability of chrono‐amperometric detection using screen printed electrodes for assaying the activity of the umu‐test bacterial strain (TA1535/pSK1002). The effect of the S9‐mix on the measurement was evaluated and no interference was found. The response of the umu‐test strain to IQ, metabolically activated with an S9‐mix, was monitored both electrochemically and with the ISO‐standardized colorimetric detection, revealing a good correlation between the induction factors calculated for both methods.
Bioluminescence (lux, luc)
The reaction by which photons are released by a biological reaction is shared by numerous groups of organisms, including bacteria, protozoa, fungi, insects and fish. In all cases the reaction is catalysed by an enzyme generically referred to as luciferase, which oxidizes a substrate known as luciferin; however, the chemical and enzymatic nature of both entities vary greatly, depending on the organism from which the system is derived. The two bioluminescent systems most commonly used as reporters of gene activation are of bacterial and insect (firefly) origin. Firefly luciferase, coded by the luc gene, is a 62 kDa monomeric protein, and its activity is oxygen‐ and ATP‐dependent. Its luciferin, benzothiazoyl‐thiazole, has to be added externally when luc is used as a reporter gene. Bacterial luciferase catalyses the oxidation of a reduced flavin mononucleotide (FMNH2) by a long‐chain fatty aldehyde to FMN and the corresponding fatty acid, in the presence of molecular oxygen. All bacterial luciferases are heterodimeric proteins composed of two subunits, α (40 kDa) and β (37 kDa), encoded by the luxA and luxB genes of the lux operon. Three other genes in this operon (luxCDE) encode the synthesis and recycling enzymes of the fatty acid aldehyde (Meighen, 1993). Constructs carrying just luxAB are sufficient to generate a bioluminescent signal, but necessitate the external addition of a long‐chain aldehyde. The commonly used luciferases of V. fischeri and Vibrio harveyi have limited upper temperatures of 30°C or 37°C respectively. In recent years, Photorhabdus luminescens lux genes are thus often used due to the higher upper temperature limit (45°C) of their gene products (Meighen and Szittner, 1992).
The non‐invasive protocol using lux fusions allows real‐time reporting of the transcriptional activation of the monitored gene promoters. As described above, the VITOTOXTM test uses the V. fischeri luxCDABE operon under the control of recN promoter (van der Lelie et al., 1997; Verschaeve et al., 1999), and the SOS lux test employs the luxCDABFE operon of P. leiognathi under control of the cda gene promoter of the plasmid ColD (Ptitsyn et al., 1997). Vollmer and colleagues (1997) have fused the E. coli recA, uvrA and alkA gene promoters to V. fischeri luxCDABE. Further modifications to the same system (Davidov et al., 2000; Rosen et al., 2000) included integration of the recA′::lux fusion into the E. coli chromosome, a change of the reporter system to P. luminescens lux, and the use of either S. typhimurium or a tolC E. coli mutant as alternative hosts. Application of the P. luminescens reporter, which allowed a working temperature of 37°C, resulted in a more rapid response to various genotoxic chemicals and UV.
The luxCDABE genes of V. fischeri were also fused to the recA promoter of Pseudomonas aeruginosa (Elasri and Miller, 1998). As a soil and freshwater bacterium, P. aeruginosa was presented as a good candidate to serve as a sensor for the state of natural bacterial communities of both pristine and polluted habitats. Light production in response to UV exposure was monitored in this strain as part of a study of UV effects on natural bacterial populations.
To increase the sensitivity of the umu‐test and to expand its detection capabilities, two groups independently replaced its β‐galactosidase reporting gene by either bacterial (Justus and Thomas, 1998) or insect (Schmid et al., 1997) luciferase. In both cases, improvements in performance were reported, including enhanced sensitivity, improved signal to noise ratios, stronger signals and a better neutralization of color interferences.
In an attempt to compare between bioluminescence, colorimetric and electrochemical detection, a single gene promoter, sulA, was fused to either P. luminescens luxCDABE (Yagur‐Kroll et al., 2009) or to the E. coli alkaline phosphatase gene phoA, the activity of which was monitored both colorimetrically (with p‐nitrophenyl phosphate as a substrate) or electrochemically with p‐aminophenyl phosphate. Figure 1 displays the correlation between the three assays, comparing colorimetric detection to either bioluminescence (Fig. 1A) or electrochemical detection (Fig. 1B). While the correlations demonstrate the validity of all three reporters, the higher dynamic range of the bioluminescence and electrochemical measurements provide a more reliable data over a much broader range of genotoxicant concentrations.
Figure 1.
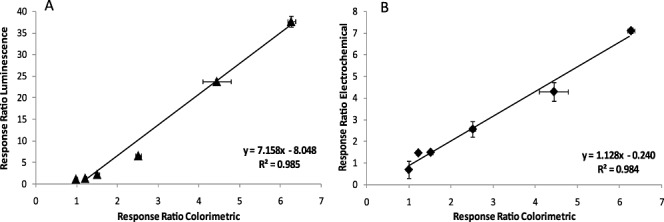
Correlations between the activation of the sulA promoter fused to different reporter systems, comparing colorimetric (sulA′::phoA) detection with either (A) bioluminescent (sulA′::luxCDABE) detection or (B) electrochemical (sulA′::phoA) detection. Response ratio – fold induction over the non‐induced control.
Fluorescent protein genes
The highly stable green fluorescent protein, GFP, of the jellyfish Aequorea victoria was the first fluorescent protein the gene of which was utilized as a molecular reporter (Chalfie et al., 1994). It was soon followed by additional fluorescent protein genes isolated from various marine organisms as well as by mutated forms with improved performance (Crameri et al., 1996; Tsien, 1998; Matz et al., 1999; Fradkov et al., 2000; Nagai et al., 2002; Wiedenmann et al., 2004; Shaner et al., 2007). The GFP protein has a high quantum yield and can be expressed in both prokaryotic and eukaryotic systems with no need for a substrate or cofactor (Kain and Kitts, 1997). To increase the sensitivity of assays based on the GFP reporter system, several green fluorescent protein mutants were constructed (Cormack et al., 1996; Crameri et al., 1996; Heim and Tsien, 1996; Welsh and Kay, 1997).
Arai and colleagues (2001) have modified the umu‐test by replacing the lacZ gene with a DNA fragment encoding for EGFP (enhanced green fluorescent protein). This construct was tested in E. coli strain KY706 with 3 µg ml−1 4‐NQO, a concentration that strongly induced β‐galactosidase activity in the umu‐test. Detection sensitivity of the GFP reporter system became similar to that of β‐galactosidase only after the introduction of additional modifications to the plasmid. These included utilization of tandem lacUV5 and chimeric trp/umu promoters, and coexpression of the E. coli recA5327 mutant. An additional construct that harbours the fusion of the umuDC promoter to the gfp gene was generated by Justus and Thomas (1999), who reported an overall performance inferior to that of the lacZ‐based assay.
Other fluorescent genotoxicity sensors were constructed by fusing the gfp and gfpmut3 reporting genes to the recA promoter (Kostrzynska et al., 2002). GFPmut3 is a mutant that is approximately 20 times more fluorescent than wild‐type GFP and only weakly excited by UV light (Cormack et al., 1996). The use of the wild‐type gfp yielded dose‐dependent but weak responses, while with gfpmut3, the detection thresholds for MMC, MNNG, NA, hydrogen peroxide and formaldehyde were comparable to the SOS chromotest (Quillardet et al., 1982), the umu‐test (Nakamura et al., 1987) and the SOS lux test (Ptitsyn et al., 1997).
The three fluorescent protein genes coding for EGFP, GFPuv and DsRed [a red fluorescent protein derived from the sea anemone Discosoma sp. (Matz et al., 1999)] were similarly fused to the recA promoter (Sagi et al., 2003), and the responses to nalidixic acid were compared with a luminescent recA′::lux strain. Performance was usually poorer compared with bioluminescent recA‐based reporters: lag times were longer and detection thresholds were higher, unless incubation times were very long. The recA′::DsRed plasmid, hosted in E. coli UTL2, was used in order to monitor antigenotoxic activity of plant extracts that exhibited some protection against MMC, NA and hydrogen peroxide (Bartolome et al., 2006).
The use of fluorescent proteins as reporters has been characterized as superior in terms of stability but inferior to enzyme‐based reporters in terms of sensitivity and response kinetics (Hakkila et al., 2002; Sagi et al., 2003). Norman and colleagues (2006) have demonstrated that these drawbacks can be circumvented by the use of flow cytometry, displaying a response threshold of strain cda′::gfpmut3 to MNNG of 5 nM, 10‐fold lower than the minimal detectable concentration (MDC) of the umu‐test (Reifferscheid et al., 1991). Moreover, the experimental procedure enabled the detection of MMC in spiked soil.
Cytotoxicity controls
As samples or chemicals of suspected DNA damaging activity are also likely to be cytotoxic, genotoxicity assays often incorporate suitable controls to neutralize or correct for the effects that cell damage or cell death may exert on assay results. One simple measure is an optical determination of cell growth in parallel to assaying reporter gene activity (Baun et al., 1999). However, this solution is limited, as optical density does not necessarily reflect the viability of a cell suspension. A different approach is based on the inclusion of an additional, constitutive reporting strain or enzyme which serves as a ‘light off’ sensor: a decrease in its signal indicates a toxic effect of the sample. This approach, for example, was adopted in the VITOTOXTM test that introduced a constitutive light‐producing strain with a lux operon under the control of the strong promoter, pr1 (Verschaeve et al., 1999).
The SOS lux test similarly incorporated a cytotoxic reporting strain harbouring a constitutive lac‐GFPuv plasmid in the same S. typhimurium host strain (Baumstark‐Khan et al., 2001). In a further development of this system a SWITCH plasmid was added, combining the SOS lux plasmid pPLS‐1 and the LAC‐Fluoro plasmid pGFPuv (Baumstark‐Khan et al., 2005).
Using a different approach, the tester strain in the SOS chromotest was made constitutive for alkaline phosphatase synthesis (Torriani and Rothman, 1961). This enzyme, non‐inducible by DNA‐damaging agents, is assayed in parallel to β‐galactosidase and the ratio of the two activities is taken as a measure of the specific activity of β‐galactosidase (Quillardet et al., 1982).
The toxicity of a sample can also be evaluated with promoters that are induced by a broad spectrum of environmental insults and are thus good indicators of toxic cellular stress, such as the promoter of the grpE gene, a component of the chaperone network in E. coli (Van Dyk et al., 1994; de Marco et al., 2007). The use of two strains, one harbouring the plasmid recA′::GFPuv and the other grpE′::lux allowed an assessment of the toxicity of the sample along with its genotoxicity (Sagi et al., 2003). A dual‐function toxicity/genotoxicity bioreporter system was reported by Hever and Belkin (2006) who described a plasmid containing both recA′::EGFP and grpE′::DsRed fusions. A somewhat different double reporter concept was demonstrated by Mitchell and Gu (2004), who presented a strain containing a fluorescent genotoxicity reporter fusion (recA′::GFPuv4) and a bioluminescent oxidative stress reporter (katG′::luxCDABE).
Detection performance
As described in detail above, numerous genotoxicity bioassays based on genetically engineered bacteria have been presented over the years. While some of them, such as the umu‐test and the SOS chromotest, have undergone intensive validation and were tested against hundreds of compounds, others have only been briefly described along with their responses to a very limited range of chemicals. Quite clearly therefore pending further validation of the latter group, the validated tests are of a much higher value for routine testing and their results merit higher credibility. Detailed reports of an extensive testing of these assays and their comparison to the Ames test can be found in Nakamura and colleagues (1987), Reifferscheid and Heil (1996), and Quillardet and Hofnung (1993). The umu‐test has been standardized and accepted for wastewater quality testing (ISO/CD 13829, DIN 38423‐5).
Table 1 summarizes reported detection thresholds of selected genotoxicants exhibited by many of the assays described in the present review. Even a brief glance at Table 1 reveals that of all the tested systems, only two, the umu‐test and the SOS chromotest, were challenged with the required spectrum of genotoxic chemicals necessary to demonstrate their applicability to environmental testing. All others were only preliminarily challenged with a very limited number of compounds. In fact, Table 1 only lists chemicals that have been tested by at least one bioassay in addition to the umu‐test and the SOS chromotest; it thus does not contain the detection thresholds for hundreds of other compounds that have been reported for these two tests (Quillardet et al., 1982; 1985; Ohta et al., 1984; Oda et al., 1985; Nakamura et al., 1987; von der Hude et al., 1988; Quillardet and Hofnung, 1993; Reifferscheid and Heil, 1996). Based on this limited comparison it may also be observed that detection thresholds vary greatly between the different assays, sometimes by several orders of magnitude. Other factors such as response times, detection spectra or facility of use that have not been compared in Table 1 confer additional advantages to some of the reporter strains.
Table 1.
Published detection thresholds of selected chemicals by genetically engineered bacterial genotoxicity reporters.
| umu‐test | SOS Chromotest | SOS lux test (2), (3), (18), (21) | VITOTOXTM (24), (26), (29) | sulA‐test (4) | recA′::lux (10), (28) | nrdA′::lux (6) | recA′::gfp (7) | cda′::gfp (13) | recA′::DsRed (1) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Promoter | umuCD | sulA (sfiA) | Cda (ColD) | recN | sulA | recA | nrdA | recA | Cda (ColD) | recA |
| Reporter gene(s) | lacZ | lacZ | luxCDABFE P. leiognathi | luxCDABE V. fischeri | lacZ | luxCDABE V. fischeri | luxCDABE P. luminescens | gfpmut3 | gfpmut3 | DsRed2 |
| Host strain | S. typhimurium TA1535 | E. coli PQ37 | S. typhimurium TA1535 | S. typhimurium TA104 | S. typhimurium TA1535 | E. coli RFM443 | E. coli RFM443 | E. coli C600 | S. typhimurium TA1535 | E. coli UTL2 |
| Compound | MDC (µM) | |||||||||
| Fungal toxins and antibiotics | ||||||||||
| MMC | 0.01 (12) | 0.016 (19) | 4.3 × 10−3 | 0.046 | 5.3 × 10−3 | 2.9 × 10−4 | 0.93 | 0.012 | 9.1 × 10−3 | 0.011 |
| Doxorubicin | 1.1 (12) | 0.41 (11) | 0.9 | 0.43 | ||||||
| NA | 2.4 (12) | 4.6 | 0.69 | 10.77 | 3.57 | 1.02 | 3.01 | |||
| Bleomycin | 0.04 (12) | 22.5 (11) | 0.35 | |||||||
| Esters | ||||||||||
| MMS | 150 (22) | 63 (19) | 10 | 117 | ||||||
| EMS | 1.8 × 103 (12) | 104 (27) | 2061 | 3414 | ||||||
| DMS | 300 (12) | 6.7 (19) | 7.5 | |||||||
| Nitroso‐, nitro‐ | ||||||||||
| DEN | 5 × 104 (12) | 199 (19) | 2349 | 1.5 × 104 | ||||||
| MNNG | 3.0 (5) | 0.45 (27) | 0.6 | 0.40 | 1 | 0.33 | 1.06 | 0.763 | 0.16 | |
| NPAHs | ||||||||||
| 4‐Nopd | 32 (14) | ND (16) | 10.4 | |||||||
| 3‐NFA | 0.04 (15) | 0.25 (8) | 0.06 | |||||||
| 4‐NQO | 0.1 (12) | 0.02 (19) | 0.042 | 3.16 | 0.018 | 13.1 | ||||
| Furazolidone | 0.22 (17) | < 0.44 (16) | 0.002 | |||||||
| HAs | ||||||||||
| 2‐AA | 1 (22) | 3 (27) | 1.08 | 0.037 | 0.08 | |||||
| 2‐AF | 0.4 (22) | < 274 (27) | 1.1 | |||||||
| PAHs | ||||||||||
| B[a]P | 4 (12) | 2.33 (19) | 3.6 | 0.58 | 5 × 10−5 | |||||
| Fluoranthene | ND (12) | ND (9) | 15.3 | |||||||
| Chrysene | 65 (12) | < 87 (16) | 21.9 | |||||||
| Acridine | ||||||||||
| Acridine orange | 12 (12) | 75 (19) | 0.83 | |||||||
| ICI 191 | 0.5 (12) | 0.7 (27) | 0.68 | 0.3 | ||||||
| 9‐aminoacridine | 46 (12) | ND (16) | 5 | |||||||
| Miscellaneous | ||||||||||
| H2O2 | 1322 (12) | 1000 (27) | 50 | 59 | 84 | 23.5 | 1470 | 286 | 24.7 | 588 |
| CH2O | 623 (12) | > 1000 (27) | 6500 | 305 | 141 | |||||
| Epi | 649 (12) | 3300 (27) | 1383 | |||||||
| EtBr | 127 (12) | 1165 (20) | 0.32 | |||||||
| Metals | ||||||||||
| K2Cr2O7 | 258 (12) | 68 (25) | 0.68 | 14 | ||||||
| CdCl2 | ND (23) | ND (20) | ND | 0.99 | ||||||
MDC, the lowest concentration at which the response is systematically over twice the background; ND, not detected; MMC, mitomycin C; NA, nalidixic acid; MMS, methyl methanesulfonate; EMS, ethyl methanesulfonate; DMS, dimethylsulfate; DEN, diethylnitrosamine; MNNG, N‐methyl‐N′‐nitro‐N‐nitrosoguanidine; 4‐Nopd, 4‐nitro‐o‐phenylenediamine; 3‐NFA, 3‐nitrofluoranthene; 4‐NQO, 4‐nitroquinoline‐N‐oxid; 2‐AA, 2‐aminoanthracene; 2‐AF, 2‐aminoflouren; B[a]P, benzo[a]pyrene; H2O2, hydrogen peroxide; CH2O, formaldehyde; Epi, epichlorohydrin; EtBr, ethidium bromide.
Numbers in parenthesis indicate the following references: 1 –Bartolome et al. (2006), 2 –Baumstark‐Khan et al. (2001), 3 –Baumstark‐Khan et al. (2007), 4 –El Mzibri et al. (1996), 5 –Hamer et al. (2000), 6 –Hwang et al. (2008), 7 –Kostrzynska et al. (2002), 8 –Mersch‐Sundermann et al. (1991), 9 –Mersch‐Sundermann et al. (1992), 10 –Min et al. (1999), 11 –Muller and Janz (1992), 12 –Nakamura et al. (1987), 13 –Norman et al. (2005), 14 –Oda et al. (1985), 15 –Oda et al. (1992), 16 –Ohta et al. (1984), 17 –Pal et al. (1992), 18 –Ptitsyn et al. (1997), 19 –Quillardet et al. (1982), 20 –Quillardet and Hofnung (1985), 21 –Rabbow et al. (2002), 22 –Reifferscheid et al. (1991), 23 –Reifferscheid and Heil (1996), 24 –van der Lelie et al. (1997), 25 –Venier et al. (1989), 26 –Verschaeve et al. (1999), 27 –von der Hude et al. (1988), 28 –Vollmer et al. (1997) 29 –Westerink et al. (2009).
Further comparison of the performance of some of these bioassays has been performed in several hands‐on workshops conducted in Belgium (Mol TECHNOTOX; Corbisier et al., 2000; Baumstark‐Khan et al., 2007) and in the USA (Eilatox‐Oregon; Hakkila et al., 2004; Meriläinen and Lampinen, 2004; Pancrazio et al., 2004; Pedahzur et al., 2004). In addition to highlighting differences in response characteristics, such workshops help to emphasize the difficulties encountered when taking a newly developed test out of the lab and into the field and emphasized the merits of utilizing standard testing protocols for performance validation.
Sensitivity enhancement and expansion of response spectrum
Very early in the short history of genetically engineered bacterial reporters, it became apparent that simple promoter–reporter fusions may be sufficient to demonstrate the applicability of the concept for genotoxicity testing, but that additional molecular manipulations are required in order to turn them into efficient tools for routine use. Such manipulations have taken several forms, including modification of the sensing elements, introduction of mutations into the reporter strains for enhanced sensitivity and permeability, and the incorporation of metabolic activation capabilities.
Table 2 lists some of the genetic manipulations introduced into the E. coli or S. typhimurium host strains and their reported effects. The modifications can be divided into two classes: deficiencies that reduce the ability of the cells to defend against DNA damaging agents, and new or enhanced capabilities of bacterial cells to metabolically activate pre‐genotoxic compounds, thus at least partially mimicking the metabolic pathways such compounds may undergo in mammalian systems.
Table 2.
Molecular modifications introduced into genotoxicity reporter strains to enhance sensitivity, expand the response spectrum and incorporate metabolic activation capabilities.
| Manipulation | Test | Strain | Modified capabilities | Effect | Reference |
|---|---|---|---|---|---|
| uvrAB mutation | SOS Chromotest | PQ37/sfiA::Mud(Ap lac) cts | Deficiency in nucleotide excision repair | Increased sensitivity toward certain genotoxicants | Ames et al. (1973), Baumstark‐Khan et al. (2001), Nunoshiba and Nishioka (1991), El Mzibri et al. (1996), Oda et al. (1985), Quillardet et al. (1982), Østergaard et al. (2007) |
| umu‐test | TA1535/umuDC′::lacZ | ||||
| GenoTox | TA1535/cda′::gfp | ||||
| SOS lux | TA1538/cda′::lux | ||||
| VITOTOXTM | TA104/recN2‐4′::lux | ||||
| sulA‐test | TA1538/sulA′::lacZ | ||||
| Rec‐lac test | KY946 ϕ(recA–lacZ) | ||||
| tag mutation | SOS‐Chromotest | PQ243/sfiA::Mud(Ap lac) cts | Inactivation of the constitutive 3‐methyl‐adenine DNA glycosylase I | Response to lower concentrations of alkylating agent as MNNG, MMS, etc. | Costa de Oliveira et al. (1987), Quillardet and Hofnung (1993) |
| oxyR mutation | SOS Chromotest | PQ300/sfiA::Mud(Ap lac) cts | Depletes the oxidative stress responses under the control of OxyR transcription regulator | More sensitive to various classes of peroxides and compounds generating peroxides | Muller and Janz (1992), Quillardet and Hofnung (1993) |
| rfa mutation | SOS Chromotest | PQ37/sfiA::Mud(Ap lac) cts | Mutation in the core enzymes of lypopolysaccharide (LPS) biosynthesis. Incomplete LPS composed of the ketodeoxyoctanoate‐lipid core. | Higher permeability to substances, especially important with larger hydrophobic genotoxins. | Ames et al. (1973), El Mzibri et al. (1996), Oda et al. (1985), Østergaard et al. (2007), Verschaeve et al. (1999), Quillardet and Hofnung (1985), Rettberg et al. (2001) |
| umu‐test | TA1535/umuDC′::lacZ | ||||
| GenoTox | TA1535/cda′::gfp | ||||
| SOS lux | TA1538/cda′::lux | ||||
| VITOTOXTM | TA104/recN2‐4′::lux | ||||
| sulA‐test | TA1538/sulA′::lacZ | ||||
| tolC mutation | GenoTox | N43/cda′::gfpmut3 | Inactivation of the efflux, outer membrane transporter‐TolC | Limited efflux capability, increases sensitivity to genotoxins | Davidov et al. (2000), Norman et al. (2005), Rettberg et al. (2001), Maehana et al. (2004) |
| SOS lux | PB3/cda′::lux | ||||
| recA′::lux | DE112/recA′::lux | ||||
| SOSluc | KT1008/umuD′::luc | ||||
| S. typhimurium NR overexpression | umu‐test | NM1011/umuDC′::lacZ | High nitroreductase activity | Highly sensitive towards many nitroarenes as 2‐NF, 1‐NP, etc. | Oda et al. (1992) |
| S. typhimurium O‐AT overexpression | umu‐ test | NM2009/umuD′::lacZ | Thirteen‐fold higher isoniazid‐N‐acetyltransferase activity | High sensitivity toward nitro‐ and dinitro‐containing compounds, as wells as arylamins, aminoanzo and HAs. | Oda et al. (1993), Oda et al. (1995) |
| S. typhimurium O‐AT and NR overexpression | umu‐test | NM3009/umuDC′::lacZ | High O‐AT and NR activity | Increased sensitivity to aromatic amines and nitroarens with/without external MA (S‐9) | Oda et al. (1995), Oda et al. (2004), Østergaard et al. (2007) |
| GenoTox | TGO2/cda′::gfp | ||||
| Human N‐ATs (NAT1 and NAT2) expression in S. typhimurium | umu‐test | NM6001/umuDC′::lacZ | N‐acetyltransferase 1 or N‐acetyltransferase 2 activity | Increased sensitivity to aromatic amines and heterocyclic aromatic amines | Oda et al. (1999) |
| NM6002/umuDC′::lacZ | |||||
| Human CYP1A2 and NADPH–P450 reductase expression in S. typhimurium | umu‐test | OY1001/umuDC′::lacZ | 7‐Ethoxyresorufin O‐de‐ethylation and NADPH–cytochrome c reductase activity | Detection of some carcinogenic HAs, without the addition of metabolic activation system (S‐9) | Aryal et al. (1999) |
| Human CYP1A2 and NADPH–P450 reductase with O‐AT expression in S. typhimurium | umu‐test | OY1002/umuDC′::lacZ | 7‐Ethoxyresorufin O‐de‐ethylation and NADPH–cytochrome c reductase activity, joint with high O‐AT activity | More sensitive to HAs than the previous strain, with ought external MA. Detects the mutagens APNH and APH. | Aryal et al. (2000), Oda et al. (2004) |
CYP1A2, cytochrome P450 1A2; O‐AT, O‐acetyltransferase; NR, nitroreductase; N‐AT, N‐acetyltransferase; HA, hetrocyclic amines; MA, metabolic activation; APNH, aminophenylnorharman; APH, aminophenylharman.
Introduction of host strain mutations for enhanced sensitivity
As listed in Table 2, several mutations have been introduced into genotoxicity reporter strains to enhance their sensitivity and thus lower their detection thresholds. While some mutations, such as tag or oxyR, have only been reported once (for the SOS chromotest; Quillardet and Hofnung, 1993), others have become almost a pre‐requisite in microbial genotoxicity reporters. Most notable in the latter group are uvrAB mutants deficient in excision repair, and rfa mutations that, by increasing membrane permeability, allow the build‐up of higher intracellular concentrations of the tested chemicals (Makela et al., 1974; Ames et al., 1975).
Altering the sensing element: manipulation of regulatory sequences
Molecular manipulations of the DNA fragment harbouring the sensing promoter element in order to improve the bacterial response to target chemicals have also been described. One of the first examples is the VITOTOXTM strain (van der Lelie et al., 1997). In addition to the wild‐type recN promoter, two different mutant promoters were constructed and tested alone and in combination: a deletion of one of the LexA binding sites, and a ‘promoter up’ mutation where a consensus nucleotide was introduced in the −35 region. Each of the single mutants was superior to the wild‐type in at least one respect, but the double mutation resulted in poorer performance. The influence of addition/subtraction of lexA binding sites on reporter gene expression in the SOS promoter::uidA fusion under genotoxic stress was examined with several SOS promoters (umuD, sulA, recA and recN) (Dreier et al., 2002). Highest signals were produced by constructs that either harboured an additional lexA binding site in the sulA promoter that overlapped the −35 promoter region, or that lacked one of the two recN LexA binding sites. Another successful effort was reported by Arai and colleagues (2001), who improved the performance of a umuDC′::gfp construct by replacing the wild‐type −35 promoter sequence with the −35 sequence of the highly active trp gene promoter.
Further exploration of different approaches for enhancing the sensing performance of a sulA′::luxCDABE fusion by manipulation of the promoter region was carried out by Yagur‐Kroll and colleagues (2009). Four independent strategies were used: modifying the length of the DNA segment containing the promoter region, introduction of random mutations by a directed evolution process, insertion of site‐directed mutations into the −35 and −10 regions, and promoter duplication (Fig. 2). Two manipulations had the most dramatic effect: extending the promoter‐containing fragment into the sulA open reading frame and a modification of the −35 consensus site. A mutation near the lexA binding site and adding an additional recA promoter improved performance to a lower extent. Such manipulations, however, may be promoter‐specific and thus not necessarily universal in nature.
Figure 2.
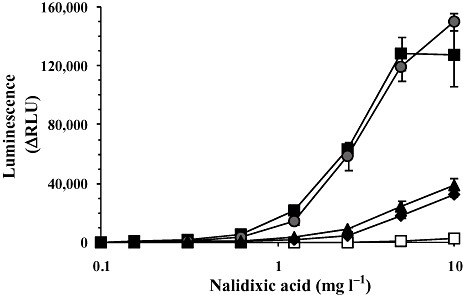
Effects on reporter performance of several manipulations of the promoter region of a sulA′::lux construct in E. coli. Data represent the increase in bioluminescence (in relative light units) following a 60 min exposure to different nalidixic acid concentrations. (□) wild‐type; (◆) a mutation near the lexA binding site; ( ) a tandem sulA‐recA promoter; (●) a modification of the −35 consensus sequence; (
) a tandem sulA‐recA promoter; (●) a modification of the −35 consensus sequence; ( ) a promoter‐containing fragment extending into the sulA ORF. Activity is reported as the difference in luminescence intensity (in arbitrary relative light units, RLU) between the induced and non‐induced reactions.
) a promoter‐containing fragment extending into the sulA ORF. Activity is reported as the difference in luminescence intensity (in arbitrary relative light units, RLU) between the induced and non‐induced reactions.
Introduction of metabolic activation enzymes
To at least partially alleviate the restriction of metabolic activation potential in prokaryotes and correspondingly reduce the dependency on external metabolic activation by rodent‐derived cytochrome P450 (S9) preparations (Malling, 1971; Ames et al., 1973), several attempts have been made to genetically engineer bacterial cells to incorporate some of the enzymatic activities involved in the activation process of xenobiotics. Some of these efforts are summarized in Table 2, clearly demonstrating the viability and potential of this approach. Nevertheless, while the study of single enzymes is a necessary step in improving our understanding of the activation of promutagens to mutagens (Oda et al., 1999; Muckel et al., 2002), it should be remembered that the CYP P‐450 is composed of a diverse group of enzymes that are not very likely to be engineered together into a single reporter strain in the near future.
Devices incorporating bacterial genotoxicity reporters
Several reports describe the incorporation of genetically engineered bacteria into specially designed hardware to generate dedicated genotoxicity biosensors. Polyak and colleagues (2000) have immobilized the E. coli strain DPD1718 (Davidov et al., 2000; Rosen et al., 2000) that contains a chromosomally integrated recA′::lux fusion in sodium alginate onto the tip of an optical fibre. The luminescent signal induced in the bacteria by the presence of genotoxicants was collected by the fibre and electronically amplified. Sensor strains embedded in alginate retained their sensitivity following a 2‐month incubation, but at the cost of a significantly delayed response (Davidov et al., 2000). A biosensor composed of a high‐density living bacterial cell array was fabricated by depositing single E. coli cells carrying a recA′::gfpmut2 into a microwell array formed on one end of an imaging fibre bundle (Kuang et al., 2004). Each fibre in the array had its own distinct light pathway, allowing thousands of individual cell responses to be monitored simultaneously with both spatial and temporal resolution. An active sensing lifetime of more than 6 h and a shelf‐life of 2 weeks were demonstrated. An on‐chip whole‐cell genotoxicity bioassay was developed (Tani et al., 2004), using a three‐dimensional microfluidic network system composed of one perforated microwell chip bound in two microchannel chips, in which the sensor cells were immobilized in agarose. The bioluminescent responses to MMC of the sensor strain, SOSluc, in a wild‐type E. coli or its tolC deficient derivative were measured by a charge‐coupled camera (Maehana et al., 2004). The immobilized cells were stable for at least 1 week at 4°C. Following further optimization, the detection limits of model hydrophobic and hydrophilic genotoxicants were comparable to those obtained by the reference method used (Maehana et al., 2006)
Electrochemical mutagen screening based on the umu‐test was performed on a microbial chip combined with a SECM device (Matsui et al., 2006). The microbial chip was fabricated by embedding 5 nl of collagen‐immobilized genetically engineered S. typhimurium strain (TA1535/pSK1002 in a microcavity on a glass substrate. β‐Galactosidase activity was monitored by electrochemical determination of the concentration of p‐aminophenol (pAP), the hydrolysis product of pAPG. Recently, a novel microelectro‐mechanical system (MEMS)‐based micro‐chip incorporated in a micro‐fluidic system was constructed and characterized by Ben‐Yoav and colleagues (2009) (Fig. 3). The activity of minute volumes (down to 2.5 nl) of a S. typhimurium strain carrying a umuC′::lacZ fusion and an E. coli strain harbouring a sulA′::phoA fusion was measured after exposure to IQ and NA respectively. The presence of genotoxic materials was detected by chrono‐amperometry using the substrates pAPP (for the phoA reporter) and pAPG (for the lacZ reporter), generating a significant signal after 3 min with MDCs of 0.31 and 42 µM for IQ after metabolic activation and NA respectively.
Figure 3.
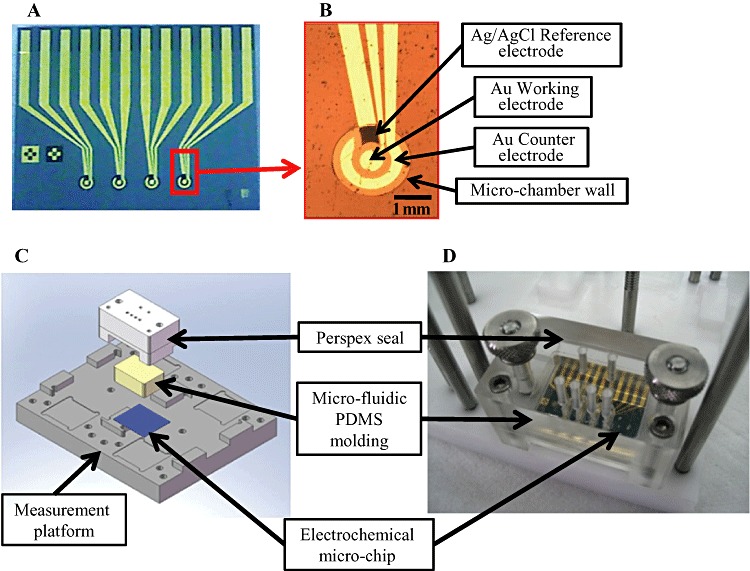
A. A silicon‐based micro‐chip comprised of four electrochemical micro‐chambers (2 mm in diameter). B. Close‐up view of a single three‐electrode electrochemical micro‐chamber. C. Layout of the measurement platform comprised of a Perspex seal, micro‐fluidic PDMS moulding, and an electrochemical micro‐chip. D. The assembled micro‐fluidic electrochemical chip system.
Although such biosensors open potential horizons for field applications and high‐throughput screening systems, their current status requires additional development before they become available for routine use. Such future development should clearly take into account the necessity of including metabolic activation as an integral part of the process, as well the need for significant improvement in long‐term maintenance of cell viability and activity.
Summary and outlook
Genetically engineered bacterial sensor systems play a central role in effect‐directed analysis of environmental contaminants and the assessment of the DNA damaging potential of chemicals. The basis for a successful construction of bacterial sensors consists of three elements: (i) a bacterial tester strain that offers an appropriate genetic background facilitating high permeability for chemicals, an appropriate DNA‐repair capacity and negligible background activity of the reporter gene; (ii) a sensitive promoter that offers well‐balanced repressor binding properties for sensitive induction of the reporter gene; and (iii) a sensitive, fast responding reporter system with a broad dynamic range and preferably the capability of real‐time monitoring.
The discovery and the understanding of the bacterial SOS‐system, a regulatory pathway that is mainly responsible for inducible DNA repair and induced mutagenesis in bacteria, opened diverse possibilities for genetically tailoring bacteria for the specific, sensitive and fast detection of genotoxic contaminants. The fusion of SOS gene promoters to reporter genes that generate quantifiable signals allow to easily detect the genotoxic potency of a sample. This review describes the current stage of development of the sensing systems by discussing the molecular, biochemical and physico‐chemical characteristics of the different promoters and suitable reporter genes based on colorimetric, luminometric, fluorimetric and electrochemical detection.
The potential of future sensor developments with enhanced external and/or internal metabolic competence is immense. Against the background of a worldwide increasing freshwater demand, and the need for reclamation of process water as drinking water resource, bacterial sensors can play a crucial role in risk minimization. To fully reach this objective, additional progress needs to be made in several directions including enhancement of sensitivity, expansion of the response spectrum, stabilization of the more sensitive reagents and, most importantly, the introduction of broad‐spectrum metabolic activation capabilities into the sensor strains.
Acknowledgments
The authors are grateful for funding provided by the German BMBF and the Israeli MOST in the framework of project WT601 (‘DipChip’) of the German/Israeli binational Water Technology Program. [Correction added on 26 January 2010, after first online publication: the preceding sentence was added after first online publication.]
References
- Ames B.N., Durston W.E., Yamasaki E., Lee F.D. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA. 1973;70:2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B.N., McCann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the salmonella/mammalian‐microsome mutagenicity test. Mutat Res Environ Mutagen Relat Subj. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Arai R., Makita Y., Oda Y., Nagamune T. Construction of green fluorescent protein reporter genes for genotoxicity test (SOS/umu‐test) and improvement of mutagen‐sensitivity. J Biosci Bioeng. 2001;92:301–304. doi: 10.1263/jbb.92.301. [DOI] [PubMed] [Google Scholar]
- Aryal P., Yoshikawa K., Terashita T., Guengerich F.P., Shimada T., Oda Y. Development of a new genotoxicity test system with Salmonella typhimurium OY1001/1A2 expressing human CYP1A2 and NADPH‐P450 reductase. Mutat Res Genet Toxicol Environ Mutagen. 1999;442:113–120. doi: 10.1016/s1383-5718(99)00070-4. [DOI] [PubMed] [Google Scholar]
- Aryal P., Terashita T., Guengerich F.P., Shimada T., Oda Y. Use of genetically engineered Salmonella typhimurium OY1002/1A2 strain coexpressing human cytochrome P450 1A2 and NADPH‐cytochrome P450 reductase and bacterial O‐acetyltransferase in SOS/umu assay. Environ Mol Mutagen. 2000;36:121–126. doi: 10.1002/1098-2280(2000)36:2<121::aid-em6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Badihi‐Mossberg M., Buchner V., Rishpon J. Electrochemical biosensors for pollutants in the environment. Electroanalysis. 2007;19:2015–2028. [Google Scholar]
- Bartolome A., Mandap K., David K.J., Sevilla Iii F., Villanueva J. SOS‐red fluorescent protein (RFP) bioassay system for monitoring of antigenotoxic activity in plant extracts. Biosens Bioelectron. 2006;21:2114–2120. doi: 10.1016/j.bios.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Baumstark‐Khan C., Rode A., Rettberg P., Horneck G. Application of the Lux‐Fluoro test as bioassay for combined genotoxicity and cytotoxicity measurements by means of recombinant Salmonella typhimurium TA1535 cells. Anal Chim Acta. 2001;437:23–30. [Google Scholar]
- Baumstark‐Khan C., Cioara K., Rettberg P., Horneck G. Determination of geno‐ and cytotoxicity of groundwater and sediments using the recombinant SWITCH Test*. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2005;40:245–263. doi: 10.1081/ese-200045529. [DOI] [PubMed] [Google Scholar]
- Baumstark‐Khan C., Rabbow E., Rettberg P., Horneck G. The combined bacterial Lux‐Fluoro test for the detection and quantification of genotoxic and cytotoxic agents in surface water: results from the ‘Technical Workshop on Genotoxicity Biosensing’. Aquat Toxicol. 2007;85:209–218. doi: 10.1016/j.aquatox.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Baun A., Andersen J.S., Nyholm N. Correcting for toxic inhibition in quantification of genotoxic response in the umuC test. Mutat Res Genet Toxicol Environ Mutagen. 1999;441:171–180. doi: 10.1016/s1383-5718(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Belkin S. Microbial whole‐cell sensing systems of environmental pollutants. Curr Opin Microbiol. 2003;6:206–212. doi: 10.1016/s1369-5274(03)00059-6. [DOI] [PubMed] [Google Scholar]
- Ben‐Yoav H., Biran A., Pedahzur R., Belkin S., Buchinger S., Reifferscheid G., Shacham‐Diamand Y. A whole cell electrochemical biosensor for water genotoxicity bio‐detection. Electrochim Acta. 2009;54:6113–6118. [Google Scholar]
- Bihari N., Vukmirovic M., Batel R., Zahn R. Application of the SOS umu‐test in detection of pollution using fish liver S9 fraction. Comp Biochem Physiol. 1990;95:15–18. doi: 10.1016/0742-8413(90)90075-k. [DOI] [PubMed] [Google Scholar]
- Biran I., Klimentiy L., Hengge‐Aronis R., Ron E.Z., Rishpon J. On‐line monitoring of gene expression. Microbiology. 1999;145:2129–2133. doi: 10.1099/13500872-145-8-2129. [DOI] [PubMed] [Google Scholar]
- Biran I., Babai R., Levcov K., Rishpon J., Ron E.Z. Online and in situ monitoring of environmental pollutants: electrochemical biosensing of cadmium. Environ Microbiol. 2000;2:285–290. doi: 10.1046/j.1462-2920.2000.00103.x. [DOI] [PubMed] [Google Scholar]
- Biran A., Pedahzur R., Buchinger S., Reifferscheid G., Belkin S. Genetically engineered bacteria for genotoxicity assessment. In: Barcelo D., Hansen P.‐D., editors. Springer; 2009. pp. 161–186. [Google Scholar]
- Buchinger S., Grill P., Morosow V., Ben‐Yoav H., Shacham‐Diamand Y., Biran A. Evaluation of chrono‐amperometric signal detection for the analysis of genotoxicity by a whole cell biosensor. Analytica Chimica Acta. 2009 doi: 10.1016/j.aca.2009.11.027. et al (in press): doi: 10.1016/j.aca.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu G., Euskirchen G., Ward W.W., Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–804. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Corbisier P., Hansen P.‐D., Barcelo D. 2000. , and ) Proceedings of the BIOSET technical workshop on genotoxicity biosensing TECHNOTOX. [WWW document]. URL http://wwwa.vito.be/english/environment/pdf/technotox/Proceedings4_00.PDF.
- Cormack B.P., Valdivia R.H., Falkow S. FACS‐optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Costa de Oliveira R., Laval J., Boiteux S. Induction of SOS and adaptive responses by alkylating agents in Escherichia coli mutants deficient in 3‐methyladenine‐DNA glycosylase activities. Mutat Res DNA Rep. 1987;183:11–20. doi: 10.1016/0167-8817(87)90040-x. [DOI] [PubMed] [Google Scholar]
- Courcelle J., Hanawalt P.C. RecA‐dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- Courcelle J., Khodursky A., Peter B., Brown P.O., Hanawalt P.C. Comparative gene expression profiles following UV exposure in wild‐type and SOS‐deficient Escherichia coli. Genet. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri A., Whitehorn E.A., Tate E., Stemmer W.P.C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotech. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- D'Ari R. The SOS system. Biochimie. 1985;67:343–347. doi: 10.1016/s0300-9084(85)80077-8. [DOI] [PubMed] [Google Scholar]
- Davidov Y., Rosen R., Smulski D.R., Van Dyk T.K., Vollmer A.C., Elsemore D.A. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat Res Genet Toxicol Environ Mutagen. 2000;466:97–107. doi: 10.1016/s1383-5718(99)00233-8. et al. [DOI] [PubMed] [Google Scholar]
- Dizer H., Wittekindt E., Fischer B., Hansen P. The cytotoxic and genotoxic potential of surface water and wastewater effluents as determined by bioluminescence, umu‐assays and selected biomarkers. Chemosphere. 2002;46:225–233. doi: 10.1016/s0045-6535(01)00062-5. [DOI] [PubMed] [Google Scholar]
- Dreier J., Breitmaier E.B., Gocke E., Apfel C.M., Page M.G.P. Direct influence of S9 liver homogenate on fluorescence signals: impact on practical applications in a bacterial genotoxicity assay. Mutat Res Genet Toxicol Environ Mutagen. 2002;513:169–182. doi: 10.1016/s1383-5718(01)00309-6. [DOI] [PubMed] [Google Scholar]
- Ehrlichmann H., Dott W., Eisentraeger A. Assessment of the water‐extractable genotoxic potential of soil samples from contaminated sites. Ecotoxicol Environ Saf. 2000;46:73–80. doi: 10.1006/eesa.1999.1875. [DOI] [PubMed] [Google Scholar]
- El Mzibri M., De Méo M.P., Laget M., Guiraud H., Séree E., Barra Y., Duménil G. The Salmonella sulA‐test: a new in vitro system to detect genotoxins. Mutat Res Genet Toxicol. 1996;369:195–208. doi: 10.1016/s0165-1218(96)00052-3. [DOI] [PubMed] [Google Scholar]
- Elasri M., Miller R. A Pseudomonas aeruginosa biosensor responds to exposure to ultraviolet radiation. Appl Microbiol Biotechnol. 1998;50:455–458. doi: 10.1007/s002530051320. [DOI] [PubMed] [Google Scholar]
- Fernández de Henestrosa A.R., Ogi T., Aoyagi S., Chafin D., Hayes J.J., Ohmori H., Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Fradkov A.F., Chen Y., Ding L., Barsova E.V., Matz M.V., Lukyanov S.A. Novel fluorescent protein from Discosoma coral and its mutants possesses a unique far‐red fluorescence. FEBS Lett. 2000;479:127–130. doi: 10.1016/s0014-5793(00)01895-0. [DOI] [PubMed] [Google Scholar]
- Frey J., Ghersa P., Palacios P.G., Belet M. Physical and genetic analysis of the ColD plasmid. J Bacteriol. 1986;166:15–19. doi: 10.1128/jb.166.1.15-19.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes J.L., Alonso A., Cuétara E., Vernhe M., Alvarez N., Sánchez‐Lamar A., Llagostera M. Usefulness of the SOS Chromotest in the study of medicinal plants as radioprotectors. Int J Radiat Biol. 2006;82:323–329. doi: 10.1080/09553000600733168. [DOI] [PubMed] [Google Scholar]
- Gao G., Fan L., Lu H., Hua Y. Engineering Deinococcus radiodurans into biosensor to monitor radioactivity and genotoxicity in environment. Chin Sci Bull. 2008;53:1675–1681. [Google Scholar]
- German Institute of Standardization. German Institute of Standardization; 1996. [Google Scholar]
- Giese K.C., Michalowski C.B., Little J.W. RecA‐dependent cleavage of LexA dimers. J Mol Biol. 2008;377:148–161. doi: 10.1016/j.jmb.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkila K., Maksimow M., Karp M., Virta M. Reporter genes lucFFluxCDABEgfp, and dsred have different characteristics in whole‐cell bacterial sensors. Anal Biochem. 2002;301:235–242. doi: 10.1006/abio.2001.5517. [DOI] [PubMed] [Google Scholar]
- Hakkila K., Lappalainen J., Virta M. Toxicity detection from EILATox‐Oregon Workshop samples by using kinetic photobacteria measurement: the flash method. J Appl Toxicol. 2004;24:349–353. doi: 10.1002/jat.1021. [DOI] [PubMed] [Google Scholar]
- Hamer B., Bihari N., Reifferscheid G., Zahn R.K., Müller W.E.G., Batel R. Evaluation of the SOS/umu‐test post‐treatment assay for the detection of genotoxic activities of pure compounds and complex environmental mixtures. Mutat Res Genet Toxicol Environ Mutagen. 2000;466:161–171. doi: 10.1016/s1383-5718(00)00016-4. [DOI] [PubMed] [Google Scholar]
- Heim R., Tsien R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hever N., Belkin S. A dual‐color bacterial reporter strain for the detection of toxic and genotoxic effects. Eng Life Sci. 2006;6:319–323. [Google Scholar]
- Higashitani A., Higashitani N., Horiuchi K. A cell division inhibitor SulA of Escherichia coli directly interacts with FtsZ through GTP hydrolysis. Biochem Biophys Res Comm. 1995;209:198–204. doi: 10.1006/bbrc.1995.1489. [DOI] [PubMed] [Google Scholar]
- Von Der Hude W., Behm C., Gurtler R., Basler A. Evaluation of the SOS chromotest. Mutat Res Environ Mutagen Relat Subj. 1988;203:81–94. doi: 10.1016/0165-1161(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Hwang E.T., Ahn J.‐M., Kim B.C., Gu M.B. Construction of a nrdAluxCDABE fusion and its use in Escherichia coli as a DNA damage biosensor. Sensors. 2008;8:1297–1307. doi: 10.3390/s8021297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Standardization Organization. International Standardization Organization; 2000. [Google Scholar]
- Jain V.K., Magrath I.T. A chemiluminescent assay for quantitation of β‐galactosidase in the femtogram range: application to quantitation of β‐galactosidase in lacZ‐transfected cells. Anal Biochem. 1991;199:119–124. doi: 10.1016/0003-2697(91)90278-2. [DOI] [PubMed] [Google Scholar]
- Janion C. Some aspects of the SOS response system – A critical survey. Acta Biochim Pol. 2001;48:599–610. [PubMed] [Google Scholar]
- Janion C. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci. 2008;4:338–344. doi: 10.7150/ijbs.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justus T., Thomas S.M. Construction of a umuC′‐luxAB plasmid for the detection of mutagenic DNA repair via luminescence. Mutat Res Fund Mol Mech Mutagen. 1998;398:131–141. doi: 10.1016/s0027-5107(97)00215-7. [DOI] [PubMed] [Google Scholar]
- Justus T., Thomas S.M. Evaluation of transcriptional fusions with green fluorescent protein versus luciferase as reporters in bacterial mutagenicity tests. Mutagenesis. 1999;14:351–356. doi: 10.1093/mutage/14.4.351. [DOI] [PubMed] [Google Scholar]
- Kain S.R., Kitts P. Expression and detection of green fluorescent protein (GFP) In: Tuan R.S., editor. Humana Press; 1997. pp. 305–324. [DOI] [PubMed] [Google Scholar]
- Kostrzynska M., Leung K.T., Lee H., Trevors J.T. Green fluorescent protein‐based biosensor for detecting SOS‐inducing activity of genotoxic compounds. J Microbiol Methods. 2002;48:43–51. doi: 10.1016/s0167-7012(01)00335-9. [DOI] [PubMed] [Google Scholar]
- Kuang Y., Biran I., Walt D.R. Living bacterial cell array for genotoxin monitoring. Anal Chem. 2004;76:2902–2909. doi: 10.1021/ac0354589. [DOI] [PubMed] [Google Scholar]
- Van Der Lelie D., Regniers L., Borremans B., Provoost A., Verschaeve L. The VITOTOX® test, an SOS bioluminescence Salmonella typhimurium test to measure genotoxicity kinetics. Mutat Res Genet Toxicol Environ Mutagen. 1997;389:279–290. doi: 10.1016/s1383-5718(96)00158-1. [DOI] [PubMed] [Google Scholar]
- Lewis L.K., Harlow G.R., Gregg‐Jolly L.A., Mount D.W. Identification of high affinity binding sites for LexA which define new DNA damage‐inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Little J.W. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- Maehana K., Tani H., Shiba T., Kamidate T. Effects of using a low‐copy plasmid and controlling membrane permeability in SOS‐based genotoxic bioassay. Anal Chim Acta. 2004;522:189–195. [Google Scholar]
- Maehana K., Tani H., Kamidate T. On‐chip genotoxic bioassay based on bioluminescence reporter system using three‐dimensional microfluidic network. Anal Chim Acta. 2006;560:24–29. [Google Scholar]
- Makela P.H., Mayer H., Whang H.Y., Neter E. Participation of lipopolysaccharide genes in the determination of the enterobacterial common antigen: analysis of R mutants of Salmonella minnesota. J Bacteriol. 1974;119:760–764. doi: 10.1128/jb.119.3.760-764.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malling H.V. Dimethylnitrosamine: formation of mutagenic compounds by interaction with mouse liver microsomes. Mutat Res Fund Mol Mech Mutagen. 1971;13:425–429. doi: 10.1016/0027-5107(71)90054-6. [DOI] [PubMed] [Google Scholar]
- De Marco A., Deuerling E., Mogk A., Tomoyasu T., Bukau B. Chaperone‐based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;7:32. doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui N., Kaya T., Nagamine K., Yasukawa T., Shiku H., Matsue T. Electrochemical mutagen screening using microbial chip. Biosens Bioelectron. 2006;21:1202–1209. doi: 10.1016/j.bios.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Matz M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., Lukyanov S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotech. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- McDaniels A.E., Reyes A.L., Wymer L.J., Rankin C.C., Stelma G.N., Jr Comparison of the Salmonella (Ames) test, Umu tests, and the sos chromotests for detecting genotoxins. Environ Mol Mutagen. 1990;16:204–215. doi: 10.1002/em.2850160308. [DOI] [PubMed] [Google Scholar]
- Meighen E.A. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- Meighen E.A., Szittner R.B. Multiple repetitive elements and organization of the lux operons of luminescent terrestrial bacteria. J Bacteriol. 1992;174:5371–5381. doi: 10.1128/jb.174.16.5371-5381.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriläinen J., Lampinen J. EILATox‐Oregon Workshop: blind study evaluation of vitotox test with genotoxic and cytotoxic sample library. J Appl Toxicol. 2004;24:327–332. doi: 10.1002/jat.1019. [DOI] [PubMed] [Google Scholar]
- Mersch‐Sundermann V., Kern S., Wintermann F. Genotoxicity of nitrated polycyclic aromatic hydrocarbons and related structures on Escherichia coli PQ37 (SOS chromotest) Environ Mol Mutagen. 1991;18:41–50. doi: 10.1002/em.2850180108. [DOI] [PubMed] [Google Scholar]
- Mersch‐Sundermann V., Mochayedi S., Kevekordes S. Genotoxicity of polycyclic aromatic hydrocarbons in Escherichia coli PQ37. Mutat Res Genet Toxicol. 1992;278:1–9. doi: 10.1016/0165-1218(92)90279-9. [DOI] [PubMed] [Google Scholar]
- Min J., Kim E.J., LaRossa R.A., Gu M.B. Distinct responses of a recAluxCDABE Escherichia coli strain to direct and indirect DNA damaging agents. Mutat Res Genet Toxicol Environ Mutagen. 1999;442:61–68. doi: 10.1016/s1383-5718(99)00059-5. [DOI] [PubMed] [Google Scholar]
- Mitchell R.J., Gu M.B. Construction and characterization of novel dual stress‐responsive bacterial biosensors. Biosens Bioelectron. 2004;19:977–985. doi: 10.1016/j.bios.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Muckel E., Frandsen H., Glatt H.R. Heterologous expression of human N‐acetyltransferases 1 and 2 and sulfotransferase 1A1 in Salmonella typhimurium for mutagenicity testing of heterocyclic amines. Food Chem Toxicol. 2002;40:1063–1068. doi: 10.1016/s0278-6915(02)00032-7. [DOI] [PubMed] [Google Scholar]
- Muller J., Janz S. Assessment of oxidative DNA damage in the oxyR‐deficient sos chromotest strain escherichia coli PQ300. Environ Mol Mutagen. 1992;20:297–306. doi: 10.1002/em.2850200408. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell‐biological applications. Nat Biotech. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakamura S.‐i., Oda Y., Shimada T., Oki I., Sugimoto K. SOS‐inducing activity of chemical carcinogens and mutagens in Salmonella typhimurium TA1535/pSK1002: examination with 151 chemicals. Mutat Res Lett. 1987;192:239–246. doi: 10.1016/0165-7992(87)90063-7. [DOI] [PubMed] [Google Scholar]
- Norman A., Hansen L.H., Sørensen S.J. Construction of a ColD cda promoter‐based SOS‐green fluorescent protein whole‐cell biosensor with higher sensitivity toward genotoxic compounds than constructs based on recAumuDC, or sulA promoters. Appl Environ Microbiol. 2005;71:2338–2346. doi: 10.1128/AEM.71.5.2338-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman A., Hansen L.H., Sørensen S.J. A flow cytometry‐optimized assay using an SOS‐green fluorescent protein (SOS‐GFP) whole‐cell biosensor for the detection of genotoxins in complex environments. Mutat Res Genet Toxicol Environ Mutagen. 2006;603:164–172. doi: 10.1016/j.mrgentox.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T., Nishioka H. Genotoxicity of quinoxaline 1,4‐dioxide derivatives in Escherichia coli and Salmonella typhimurium. Mutat Res DNA Repair. 1989;217:203–209. doi: 10.1016/0921-8777(89)90072-4. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T., Nishioka H. ‘Rec‐lac test’ for detecting SOS‐inducing activity of environmental genotoxic substances. Mutat Res DNA Repair. 1991;254:71–77. doi: 10.1016/0921-8777(91)90042-n. [DOI] [PubMed] [Google Scholar]
- Oda Y., Nakamura S.‐i., Oki I., Kato T., Shinagawa H. Evaluation of the new system (umu‐test) for the detection of environmental mutagens and carcinogens. Mutat Res Environ Mutagen Relat Subj. 1985;147:219–229. doi: 10.1016/0165-1161(85)90062-7. [DOI] [PubMed] [Google Scholar]
- Oda Y., Shimada T., Watanabe M., Ishidate M., Nohmi T. A sensitive umu‐test system for the detection of mutagenic nitroarenes in Salmonella typhimurium NM1011 having a high nitroreductase activity. Mutat Res Environ Mutagen Relat Subj. 1992;272:91–99. doi: 10.1016/0165-1161(92)90037-m. [DOI] [PubMed] [Google Scholar]
- Oda Y., Yamazaki H., Watanabe M., Nohmi T., Shimada T. Highly sensitive umu test system for the detection of mutagenic nitroarenes in Salmonella typhimurium NM3009 having high O‐acetyltransferase and nitroreductase activities. Environ Mol Mutagen. 1993;21:357–364. doi: 10.1002/em.2850210407. [DOI] [PubMed] [Google Scholar]
- Oda Y., Yamazaki H., Watanabe M., Nohmi T., Shimada T. Development of high sensitive umu‐test system: rapid detection of genotoxicity of promutagenic aromatic amines by Salmonella typhimurium strain NM2009 possessing high O‐acetyltransferase activity. Mutat Res Environ Mutagen Relat Subj. 1995;334:145–156. doi: 10.1016/0165-1161(95)90005-5. [DOI] [PubMed] [Google Scholar]
- Oda Y., Yamazaki H., Shimada T. Role of human N‐acetyltransferases, NAT1 or NAT2, in genotoxicity of nitroarenes and aromatic amines in Salmonella typhimurium NM6001 and NM6002. Carcinogenesis. 1999;20:1079–1083. doi: 10.1093/carcin/20.6.1079. [DOI] [PubMed] [Google Scholar]
- Oda Y., Kunihiro F., Masaaki K., Akihiko N., Taro Y. Use of a high‐throughput umu‐microplate test system for rapid detection of genotoxicity produced by mutagenic carcinogens and airborne particulate matter. Environ Mol Mutagen. 2004;43:10–19. doi: 10.1002/em.10209. [DOI] [PubMed] [Google Scholar]
- Ohta T., Nakamura N., Moriya M., Shirasu Y., Kada T. The SOS‐function‐inducing activity of chemical mutagens in Escherichia coli. Mutat Res DNA Repair Rep. 1984;131:101–109. doi: 10.1016/0167-8817(84)90048-8. [DOI] [PubMed] [Google Scholar]
- Østergaard T.G., Hansen L.H., Binderup M.‐L., Norman A., Sørensen S.J. The cda GenoTox assay: a new and sensitive method for detection of environmental genotoxins, including nitroarenes and aromatic amines. Mutat Res Genet Toxicol Environ Mutagen. 2007;631:77–84. doi: 10.1016/j.mrgentox.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Paitan Y., Biran D., Biran I., Shechter N., Babai R., Rishpon J., Ron E.Z. On‐line and in situ biosensors for monitoring environmental pollution. Biotechnol Adv. 2003;22:27–33. doi: 10.1016/j.biotechadv.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Pal A.K., Rahman M.S., Chatterjee S.N. On the induction of Umu gene expression in Salmonella typhimurium strain TA1535/pSK1002 by some nitrofurans. Mutat Res Genet Toxicol. 1992;280:67–71. doi: 10.1016/0165-1218(92)90019-v. [DOI] [PubMed] [Google Scholar]
- Pancrazio J.J., McFadden P.N., Belkin S., Marks R.S. EILATox‐Oregon Biomonitoring Workshop: summary and observations. J Appl Toxicol. 2004;24:317–321. doi: 10.1002/jat.1032. [DOI] [PubMed] [Google Scholar]
- Pedahzur R., Polyak B., Marks R.S., Belkin S. Water toxicity detection by a panel of stress‐responsive luminescent bacteria. J Appl Toxicol. 2004;24:343–348. doi: 10.1002/jat.1023. [DOI] [PubMed] [Google Scholar]
- Polyak B., Bassis E., Novodvorets A., Belkin S., Marks R.S. Optical fiber bioluminescent whole‐cell microbial biosensors to genotoxicants. Water Sci Technol. 2000;42:305–311. [Google Scholar]
- Ptitsyn L.R., Horneck G., Komova O., Kozubek S., Krasavin E.A., Bonev M., Rettberg P. A biosensor for environmental genotoxin screening based on an SOS lux assay in recombinant Escherichia coli cells. Appl Environ Microbiol. 1997;63:4377–4384. doi: 10.1128/aem.63.11.4377-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillardet P., Hofnung M. The SOS chromotest, a colorimetric bacterial assay for genotoxins: procedures. Mutat Res Environ Mutagen Relat Subj. 1985;147:65–78. doi: 10.1016/0165-1161(85)90020-2. [DOI] [PubMed] [Google Scholar]
- Quillardet P., Hofnung M. The SOS chromotest: a review. Mutat Res Rev Genet Toxicol. 1993;297:235–279. doi: 10.1016/0165-1110(93)90019-j. [DOI] [PubMed] [Google Scholar]
- Quillardet P., Huisman O., D'Ari R., Hofnung M. SOS chromotest, a direct assay of induction of an SOS function in Escherichia coli K‐12 to measure genotoxicity. Proc Natl Acad Sci USA. 1982;79:5971–5975. doi: 10.1073/pnas.79.19.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillardet P., De Bellecombe C., Hofnung M. The SOS chromotest, a colorimetric bacterial assay for genotoxins: validation study with 83 compounds. Mutat Res Environ Mutagen Relat Subj. 1985;147:79–95. doi: 10.1016/0165-1161(85)90021-4. [DOI] [PubMed] [Google Scholar]
- Rabbow E., Rettberg P., Baumstark‐Khan C., Horneck G. SOS‐LUX‐ and LAC‐FLUORO‐TEST for the quantification of genotoxic and/or cytotoxic effects of heavy metal salts. Anal Chim Acta. 2002;456:31–39. [Google Scholar]
- Reifferscheid G., Buchinger S. Cell‐based genotoxicity testing: genetically modified and genetically engineered bacteria in environmental genotoxicology. Adv Biochem Eng Biotechnol. 2009 doi: 10.1007/10_2009_8. [DOI] [PubMed] [Google Scholar]
- Reifferscheid G., Heil J. Validation of the SOS/umu‐test using test results of 486 chemicals and comparison with the Ames test and carcinogenicity data. Mutat Res Genet Toxicol. 1996;369:129–145. doi: 10.1016/s0165-1218(96)90021-x. [DOI] [PubMed] [Google Scholar]
- Reifferscheid G., Heil J., Oda Y., Zahn R.K. A microplate version of the SOS/umu‐test for rapid detection of genotoxins and genotoxic potentials of environmental samples. Mutat Res Environ Mutagen Relat Subj. 1991;253:215–222. doi: 10.1016/0165-1161(91)90134-t. [DOI] [PubMed] [Google Scholar]
- Rettberg P., Bandel K., Baumstark‐Khan C., Horneck G. Increased sensitivity of the SOS‐LUX‐Test for the detection of hydrophobic genotoxic substances with Salmonella typhimurium TA1535 as host strain. Anal Chim Acta. 2001;426:167–173. [Google Scholar]
- Rosen R., Davidov Y., LaRossa R.A., Belkin S. Microbial sensors of ultraviolet radiation based on recA′lux fusions. Appl Biochem Biotechnol. 2000;89:151–160. doi: 10.1385/abab:89:2-3:151. [DOI] [PubMed] [Google Scholar]
- Sagi E., Hever N., Rosen R., Bartolome A.J., Premkumar J.R., Ulber R. Fluorescence and bioluminescence reporter functions in genetically modified bacterial sensor strains. Sensor Actuator B Chem. 2003;90:2–8. et al. [Google Scholar]
- Sakagami Y., Yamazaki H., Ogasawara N., Yokoyama H., Ose Y., Sato T. The evaluation of genotoxic activities of disinfectants and their metabolites by umu‐test. Mutat Res Lett. 1988;209:155–160. doi: 10.1016/0165-7992(88)90034-6. [DOI] [PubMed] [Google Scholar]
- Schmid C., Reifferscheid G., Zahn R.K., Bachmann M. Increase of sensitivity and validity of the SOS/umu‐test after replacement of the β‐galactosidase reporter gene with luciferase. Mutat Res Genet Toxicol Environ Mutagen. 1997;394:9–16. doi: 10.1016/s1383-5718(97)00118-6. [DOI] [PubMed] [Google Scholar]
- Schwartz‐Mittelmann A., Neufeld T., Biran D., Rishpon J. Electrochemical detection of protein‐protein interactions using a yeast two hybrid: 17‐β‐estradiol as a model. Anal Biochem. 2003;317:34–39. doi: 10.1016/s0003-2697(03)00110-6. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Patterson G.H., Davidson M.W. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Shimada T., Nakamura S.‐i. Cytochrome P‐450‐mediated activation of pro‐carcinogens and promutagens to DNA‐damaging products by measuring expression of umu gene in Salmonella typhimurium TA1535/pSK1002. Biochem Pharmacol. 1987;36:1979–1987. doi: 10.1016/0006-2952(87)90497-7. [DOI] [PubMed] [Google Scholar]
- Sutton M.D., Smith B.T., Godoy V.G., Walker G.C. THE SOS RESPONSE: recent insights into umuDC‐dependent mutagenesis and DNA damage tolerance. Annu Rev Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- Tani H., Maehana K., Kamidate T. Chip‐based bioassay using bacterial sensor strains immobilized in three‐dimensional microfluidic network. Anal Chem. 2004;76:6693–6697. doi: 10.1021/ac049401d. [DOI] [PubMed] [Google Scholar]
- Torriani A., Rothman F. Mutants of Escherichia coli constitutive for alkaline phosphatase. J Bacteriol. 1961;81:835–836. doi: 10.1128/jb.81.5.835-836.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y. The green flourescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Van Dyk T.K., Majarian W.R., Konstantinov K.B., Young R.M., Dhurjati P.S., LaRossa R.A. Rapid and sensitive pollutant detection by induction of heat shock gene‐bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venier P., Montini R., Zordan M., Clonfero E., Paleologo M., Levis A.G. Induction of SOS response in Escherichia coli strain PQ37 by 16 chemical compounds and human urine extracts. Mutagenesis. 1989;4:51–57. doi: 10.1093/mutage/4.1.51. [DOI] [PubMed] [Google Scholar]
- Verschaeve L. Genotoxicity studies in groundwater, surface waters, and contaminated soil. Scientific World J. 2002;2:1247–1253. doi: 10.1100/tsw.2002.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschaeve L., Van Gompel J., Thilemans L., Regniers L., Vanparys P., Van Der Lelie D. VITOTOX® bacterial genotoxicity and toxicity test for the rapid screening of chemicals. Environ Mol Mutagen. 1999;33:240–248. [PubMed] [Google Scholar]
- Vijayashree B., Ahuja Y., Regniers L., Rao V., Verschaeve L. Genotoxicity of the Musi River (Hyderabad, India) investigated with the VITOTOX test. Folia Biol (Praha) 2005;51:133–139. [PubMed] [Google Scholar]
- Volkert M.R. Adaptive response of Escherichia coli to alkylation damage. Environ Mol Mutagen. 1988;11:241–255. doi: 10.1002/em.2850110210. [DOI] [PubMed] [Google Scholar]
- Volkert M.R., Gately F.H., Hajec L.I. Expression of DNA damage‐inducible genes of Escherichia coli upon treatment with methylating, ethylating and propylating agents. Mutat Res DNA Repair. 1989;217:109–115. doi: 10.1016/0921-8777(89)90062-1. [DOI] [PubMed] [Google Scholar]
- Vollmer A.C., Belkin S., Smulski D.R., Van Dyk T.K., LaRossa R.A. Detection of DNA damage by use of Escherichia coli carrying recA′::luxuvrA′::lux, or alkA′::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh S., Kay S.A. Reporter gene expression for monitoring gene transfer. Curr Opin Biotechnol. 1997;8:617–622. doi: 10.1016/s0958-1669(97)80038-9. [DOI] [PubMed] [Google Scholar]
- Westerink W.M., Stevenson J.C.R., Lauwers A., Griffioen G., Horbach G.J., Schoonen W.G.E.J. Evaluation of the VitotoxTM and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat Res: Genet Toxicol Environ Mutagen. 2009;676:113–130. doi: 10.1016/j.mrgentox.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Whong W.‐Z., Wen Y.‐F., Stewart J., Ong T.‐m. Validation of the SOS/Umu test with mutagenic complex mixtures. Mutat Res Lett. 1986;175:139–144. doi: 10.1016/0165-7992(86)90112-0. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J., Ivanchenko S., Oswald F., Schmitt F., Röcker C., Salih A. EosFP, a fluorescent marker protein with UV‐inducible green‐to‐red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagur‐Kroll S., Bilic B., Belkin S. Strategies for enhancing bioluminescent bacterial sensor performance by promoter region manipulation. Microb Biotechnol. 2009 doi: 10.1111/j.1751-7915.2009.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


