Summary
Shewanella are renowned for their ability to utilize a wide range of electron acceptors (EA) for respiration, which has been partially accredited to the presence of a large number of the c‐type cytochromes. To investigate the involvement of c‐type cytochrome proteins in aerobic and anaerobic respiration of Shewanella oneidensis Mr ‐1, 36 in‐frame deletion mutants, among possible 41 predicted, c‐type cytochrome genes were obtained. The potential involvement of each individual c‐type cytochrome in the reduction of a variety of EAs was assessed individually as well as in competition experiments. While results on the well‐studied c‐type cytochromes CymA(SO4591) and MtrC(SO1778) were consistent with previous findings, collective observations were very interesting: the responses of S. oneidensis Mr ‐1 to low and highly toxic metals appeared to be significantly different; CcoO, CcoP and PetC, proteins involved in aerobic respiration in various organisms, played critical roles in both aerobic and anaerobic respiration with highly toxic metals as EA. In addition, these studies also suggested that an uncharacterized c‐type cytochrome (SO4047) may be important to both aerobiosis and anaerobiosis.
Introduction
Cytochromes are proteins carrying haem as a prosthetic group. The haem is not only the functional unit to realize electron transfer via the valence change of haem iron but also used for the classification of cytochromes. Cytochromes of the c‐type exhibit a unique characteristic that the haem cofactor is attached to the protein covalently in contrast to the noncovalent attachment in other cytochromes (a‐, b‐, d‐ and o‐type). Thioether bonds are formed between the haem and the cysteines of the haem binding motif CXXCH, which is well reserved across species (Thöny‐Meyer, 1997). Both soluble periplasmic and membrane‐bound c‐type cytochromes are identified in bacterial respiratory chains, mainly passing electrons from the bc1 complex to terminal oxidoreductases (Thöny‐Meyer, 1997).
Shewanella oneidensis Mr ‐1, a Gram‐negative facultative anaerobe, is renowned for its remarkable anaerobic respiration ability. In recent years, studies aiming at harnessing the bacterial ability for the bioremediation of metal/radionuclide contaminants in the environment have been carried out intensively (Fredrickson et al., 2008). The profile of c‐type cytochromes in S. oneidensis changes with time. Based on the first genome annotation, the microorganism possessed 44 genes for c‐type cytochromes (Heidelberg et al., 2002). By screening for the cytochrome c haem‐binding site, Meyer et al. identified 42 possible cytochrome c proteins (Meyer et al., 2004). The most recent annotation reduced the number of c‐type cytochromes to 41 after eliminating truncated or disrupted genes (Romine et al., 2008). Nevertheless, compared with Escherichia coli which hosts only five to seven c‐type cytochromes, S. oneidensis has a large number of such proteins, which may be responsible for its diverse respiratory capability (Blattner et al., 1997; Heidelberg et al., 2002; Fredrickson et al., 2008).
While some of these c‐type cytochromes have been extensively studied, such as MtrA(SO1777), MtrC(SO1778), OmcA(SO1779) and CymA(SO4591) (Myers and Myers, 1997; 2001; 2002; Beliaev et al., 2001, 2005; Tsapin et al., 2001; Schwalb et al., 2002; 2003; Pitts et al., 2003; Shi et al., 2006; Bretschger et al., 2007; Donald et al., 2008), little has been done towards understanding of the rest. The objective of this study is to systematically characterize c‐type cytochromes for their involvement in energy metabolisms. A total of 37 in‐frame deletion c‐type cytochrome mutants were generated and evaluated for their ability to reduce a variety of electron acceptors (EAs). With integrated unique barcodes and competition assays, we were able to identify in this study c‐type cytochromes which play a minor role in aerobic and anaerobic respiration.
Results
Barcoding mutagenesis of c‐type cytochrome genes
The annotation of c‐type cytochrome genes in the S. oneidensis genome has changed over time (Heidelberg et al., 2002; Meyer et al., 2004; Romine et al., 2008). In the latest annotation, 41 genes are predicted to encode intact c‐type cytochromes in the S. oneidensis Mr ‐1 genome (Romine et al., 2008) (Table 1). The primary aim of this study was to examine the effects of c‐type cytochromes on respiration of S. oneidensis Mr ‐1 by mutational analysis of each individual predicted c‐type cytochrome gene in the Mr ‐1 genome. To this end, mutagenesis was carried out using two different approaches: fusion PCR (Gao et al., 2006a) and cre‐lox (Marx and Lidstrom, 2002; Gao et al., 2006b) (Table 1). After three attempts, the cre‐lox approach failed to produce deletion mutants for scyA(SO0264), tocC(SO1233), ccpA(SO2178) and SO3056 genes whereas the fusion PCR was unable to generate a mutant devoid of SO1748. A bioinformatics analysis of these genes failed to reveal any common characteristics in sequences and possible secondary structures, implicating a gene‐specific phenomenon. In total, 36 out of 41 genes were successfully deleted.
Table 1.
Strains and plasmids used in this study.a
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strain | ||
| WM3064 | Host for pir‐dependent plasmids and donor strain for conjugation; ΔdapA | Lab stock |
| S. oneidensis strains | ||
| MR‐1 | Wild‐type | Lab stock |
| JZ0479 (Δso0479) | so0479 deletion mutant derived from MR‐1; Δso0479::loxP | This study |
| JZ0610 (ΔpetC) | petC deletion mutant derived from MR‐1; ΔpetC::loxP | This study |
| MR0714 (Δso0714) | so0714 deletion mutant derived from MR‐1; Δso0714 | This study |
| MR0716 (ΔsorB) | sorB deletion mutant derived from MR‐1; ΔsorB | This study |
| MR0717 (Δso0717) | so0717 deletion mutant derived from MR‐1; Δso0717 | This study |
| JZ0845 (ΔnapB) | napB deletion mutant derived from MR‐1; ΔnapB::loxP | Gao et al., 2009 |
| JZ0939 (Δso0939) | so0939 deletion mutant derived from MR‐1; Δso0939::loxP | This study |
| MR0970 (ΔfccA) | fccA deletion mutant derived from MR‐1; ΔfccA | This study |
| JZ1413 (Δso1413) | so1413 deletion mutant derived from MR‐1; Δso1413::loxP | This study |
| JZ1421 (Δso1421) | so1421 deletion mutant derived from MR‐1; Δso1421::loxP | This study |
| JZ1427 (ΔdmsC) | dmsC deletion mutant derived from MR‐1; ΔdmsC::loxP | This study |
| JZ1659 (Δso1659) | so1659 deletion mutant derived from MR‐1; Δso1659::loxP | This study |
| MR1777 (ΔmtrA) | mtrA deletion mutant derived from MR‐1; ΔmtrA | This study |
| MR1778 (ΔmtrC) | mtrC deletion mutant derived from MR‐1; ΔmtrC | This study |
| MR1779 (ΔomcA) | omcA deletion mutant derived from MR‐1; ΔomcA | This study |
| MR1780 (ΔmtrF) | mtrF deletion mutant derived from MR‐1; ΔmtrF | This study |
| MR1782 (ΔmtrD) | mtrD deletion mutant derived from MR‐1; ΔmtrD | This study |
| MR2361 (ΔccoP) | ccoPdeletion mutant derived from MR‐1; ΔccoP | This study |
| MR2363 (ΔccoO) | ccoO deletion mutant derived from MR‐1; ΔccoO | This study |
| JZ2727 (ΔcctA) | cctA deletion mutant derived from MR‐1; ΔcctA::loxP | This study |
| MR2930 (Δso2930) | so2930 deletion mutant derived from MR‐1; Δso2930 | This study |
| JZ2931 (Δso2931) | so2931 deletion mutant derived from MR‐1; Δso2931::loxP | This study |
| MR3300 (Δso3300) | so3300 deletion mutant derived from MR‐1; Δso3300 | This study |
| JZ3420 (Δso3420) | so3420 deletion mutant derived from MR‐1; Δso3420::loxP | This study |
| MR3980 (ΔnrfA) | nrfA deletion mutant derived from MR‐1; ΔnrfA | Gao et al., 2009 |
| MR4047 (Δso4047) | so4047 deletion mutant derived from MR‐1; Δso4047 | This study |
| MR4048 (Δso4048) | so4048 deletion mutant derived from MR‐1; Δso4048 | This study |
| MR4142 (Δso4142) | so4142deletion mutant derived from MR‐1; Δso4142 | This study |
| JZ4144 (Δso4144) | so4144 deletion mutant derived from MR‐1; Δso4144::loxP | This study |
| MR4360 (Δso4360) | so4360 deletion mutant derived from MR‐1; Δso4360 | This study |
| JZ4484 (Δshp) | shp deletion mutant derived from MR‐1; Δshp::loxP | This study |
| JZ4485 (Δso4485) | so4485 deletion mutant derived from MR‐1; Δso4485::loxP | This study |
| JZ4572 (Δso4572) | so4572 deletion mutant derived from MR‐1; Δso4572::loxP | This study |
| MR4591 (ΔcymA) | cymA deletion mutant derived from MR‐1; ΔcymA | Gao et al., 2009 |
| JZ4606 (ΔcyoA) | cyoA deletion mutant derived from MR‐1; ΔcyoA::loxP | This study |
| JZ4666 (ΔcytB) | cytB deletion mutant derived from MR‐1; ΔcytB::loxP | This study |
| Plasmids | ||
| pDS3.0 | Apr, Gmr, derivative from suicide vector pCVD442 | Lab stock |
| pJK100 | Allelic exchange vector | Lab stock |
| pCM157 | cre expression vector | Lab stock |
| pBBRMCS‐5 | Complementation vector | Lab stock |
Plasmids containing mutational structures were constructed as described in the text and not included in the table.
To confer a way to identify each individual mutant from a pool of mutants by either PCR or specifically designed microarrays, a unique barcode(s) was included in the primers such that a signature tag(s) can be inserted at the deletion location in each mutant. A single barcode was used with the fusion PCR method whereas two barcodes were included in the cre‐lox approach. The barcode‐tagged PCR products containing mutagenesis construct were cloned into the mutagenesis vectors, which eventually promoted deletion of the targeted gene and integration of the barcode(s).
In order to rule out polarity issues introduced by mutagenesis, two approaches were adopted. First, each mutation was created by an in‐frame deletion strategy, preventing a frameshift mutation in the mutated gene consisting of the remaining coding sequence and the inserted gene‐specific barcode(s). All resulting in‐frame mutations were verified by DNA sequencing the mutated genes. Second, complementation experiments were performed to validate that the observed phenotype was specific to the mutation. In total, nine of the mutants which showed an apparent phenotype were applied to complementation. In all cases, physiological differences were insignificant between the mutation strain containing the plasmid‐borne corresponding gene and the wild‐type containing the empty vector (Table S1). These results indicate that mutations exert no polar effect, at least in mutants with apparent phenotypes.
Growth of the c‐type cytochrome mutants in the presence and absence of oxygen
Throughout the entire characterization, the defined medium M1‐L was used with oxygen or one of following chemical agents as the sole EA: DMSO, fumarate, TMAO, NO3‐, NO2‐, Fe(III), Mn(IV) and Cr(VI) under anaerobic conditions respectively.
Aerobic respiration. Shewanella oneidensis is among the most diverse respiratory organisms described so far, which is believed to be in part due to the large number of c‐type cytochromes (Fredrickson et al., 2008). To gain insights into the roles of c‐type cytochromes in aerobic respiration, a growth assay of all obtained mutants under aerobic conditions was performed. As presented in Fig. 1, 10 of these mutants exhibited a growth defect when grown with oxygen as the sole EA. These included strains carrying deletion in petC(SO0610), SO0939, mtrA(SO1777), mtrF(SO1780), ccoP(SO2361), ccoO(SO2363), SO4047, SO4144 and cyoA(SO4606). It is not surprising that the petC(SO0610), ccoP(SO2361), ccoO(SO2363) and cyoA(SO4606) mutants showed reduced growth rates given that these genes encode proteins active in aerobic respiration (Berry et al., 2000; Pinchuk et al., 2009). In contrast, the observation that mutations in genes mtrA(SO1777), mtrF(SO1780) and nrfA(SO3980) resulted in altered growth rates was intriguing. All of these proteins play roles in the metal and nitrite reduction under anaerobic conditions and the underlying mechanism is worth exploring (Gao et al., 2009; C. Reardon and J. Fredrickson, unpublished results).
Figure 1.
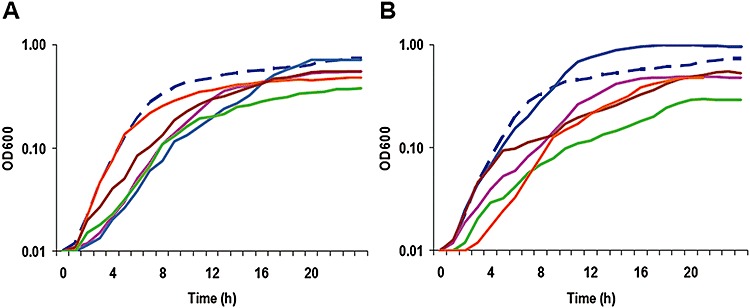
Aerobic growth of S. oneidensis c‐type cytochrome mutants and their parental strain Mr ‐1 in M1‐L medium. In both panels, Mr ‐1 was represented by the dash line in dark blue.
A. ΔpetC(SO0610) ( ), Δso0939 (
), Δso0939 ( ), ΔmtrA(SO1777) (
), ΔmtrA(SO1777) ( ), ΔmtrF(SO1780) (
), ΔmtrF(SO1780) ( ), ΔccoP(SO2361) (
), ΔccoP(SO2361) ( ).
B. ΔccoO(SO2363) (
).
B. ΔccoO(SO2363) ( ), ΔnrfA(SO3980) (
), ΔnrfA(SO3980) ( ), Δso4047 (
), Δso4047 ( ), Δso4144 (
), Δso4144 ( ), ΔcyoA(SO4606) (
), ΔcyoA(SO4606) ( ).
The data are averages from three independent cultures. Error bars representing ± 1 standard deviation (less than 5% of presented data) from the mean were omitted for clarity. Only cultures differed from Mr ‐1 significantly were shown.
).
The data are averages from three independent cultures. Error bars representing ± 1 standard deviation (less than 5% of presented data) from the mean were omitted for clarity. Only cultures differed from Mr ‐1 significantly were shown.
Mn(IV), Fe (III) and Cr (VI) reduction. After more than a decade of intensive study on reduction of Fe(III) and Mn(IV) in S. oneidensis, a number of proteins important for this process have been identified and characterized. Among them, CymA(SO4591), MtrA(SO1777), MtrC(SO1778), OmcA(SO1779), MtrF(SO1780) and MtrD(SO1782) are c‐type cytochromes. Recently, the ability of all obtained S. oneidensis c‐type cytochrome mutants to reduce Fe(III) and Mn(IV) has been assessed (Bretschger et al., 2007). While the stress responses to toxic heavy metals such as chromium, strontium and vanadium have been investigated (Brown et al., 2006; Chourey et al., 2006), the role of c‐type cytochromes in reduction of this type of metals remains undefined. In this study, we first examined the ability of each mutation strain to reduce chromate in 24 h. As shown in Fig. 2, 13 mutants exhibited Cr(VI) reduction capacities significantly lower than the parental strain, including ΔmtrA(SO1777), ΔmtrC(SO1778), ΔomcA(SO1779), ΔmtrF(SO1780), ΔmtrD(SO1782), ΔcctA(SO2727), ΔcymA(SO4591), ΔpetC(SO0610), ΔccoP(SO2361), ΔccoO(SO2363), Δso0970, Δso4047 and Δso4360. Consistent with previous reports, the mutants (ΔmtrA, ΔmtrC, ΔomcA, ΔmtrF, ΔmtrD, ΔcctA and ΔcymA) devoid of one of the well‐defined metal reducing proteins and ΔpetC were defective in Cr(VI) reduction. The result revealed that five other cytochromes were important for the process. To validate the above observation, we quantitatively tested Cr(VI) reduction of these 13 mutants in a time‐course manner. Responses of these strains to Cr(VI) were not uniform (Fig. 3A and B). Two strains, ΔpetC and ΔcymA, lost their ability to reduce Cr(VI) almost completely. While some strains, such as ΔmtrC, ΔmtrD, ΔccoP, ΔccoO and ΔcctA, displayed a slow start and less than 50% of reduction in the period of 30 h, most of the mutants responded to the Cr(VI) quickly and then continued reduction at a relatively stable rate. This observation implicates that these c‐type cytochromes may be involved in Cr(VI) reduction/response through different mechanisms.
Figure 2.
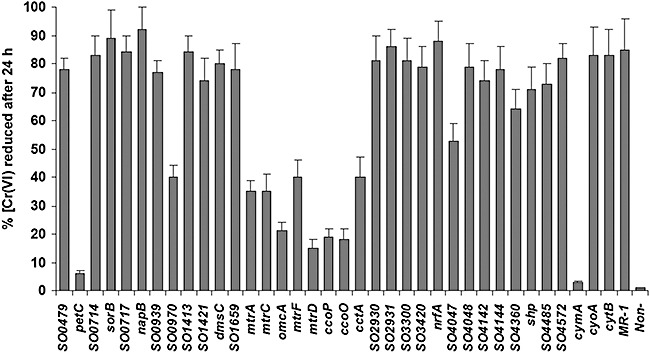
Reduction of Cr(VI) by S. oneidensis Mr ‐1 and c‐type cytochrome mutants. The percentages of reduced Cr(VI) were measured 24 h after the initiation of growth. The concentration of reduced Cr(VI) was determined using the DPC method as described in the Experimental procedures. The values were the means ± standard deviations (error bars) of at least three measurements.
Figure 3.
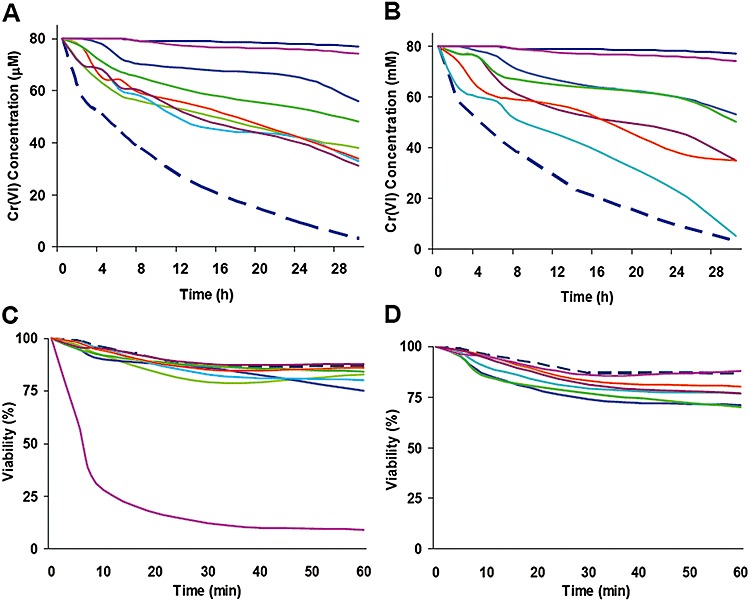
Cr(VI) reduction and survival of S. oneidensis Mr ‐1 and the c‐type cytochrome mutants. Cr(VI) reduction (A and B) and survival assay (C and D) were conducted independently. For each experiment, all tested strains were assayed at the same time but results were presented in two separate panels for clarity. Error bars representing ± 1 standard deviation (less than 5% of presented data) from the mean were omitted for clarity. In all panels, Mr ‐1 and non‐cell control was represented by the dash line in dark blue. Non‐cell control represented by solid lines in dark blue was also included in (A) and (B). In (A) and (C): ΔpetC(SO0610) ( ), Δso0970 (
), Δso0970 ( ), ΔmtrA(SO1777) (
), ΔmtrA(SO1777) ( ), ΔmtrC(SO1778) (
), ΔmtrC(SO1778) ( ), ΔomcA(SO1779) (
), ΔomcA(SO1779) ( ), ΔmtrF(SO1780) (
), ΔmtrF(SO1780) ( ), ΔmtrD(SO1782) (
), ΔmtrD(SO1782) ( ). In (B) and (D): ΔccoP(SO2361) (
). In (B) and (D): ΔccoP(SO2361) ( ), ΔccoO(SO2363) (
), ΔccoO(SO2363) ( ), ΔcctA(SO2727) (
), ΔcctA(SO2727) ( ),Δso4047 (
),Δso4047 ( ),Δso4360 (
),Δso4360 ( ), ΔcymA(SO4591) (
), ΔcymA(SO4591) ( ).
).
Given that the Cr(VI) reductase activity of S. oneidensis was identified to be associated with the cytoplasmic membrane and the Mn(IV) and Fe(III) reducing proteins (Myers et al., 2000), it is expected that mutants lacking well‐defined metal reduction proteins were defective in Cr(VI) reduction. However, interesting results were observed. For example, ΔomcA(SO1779) and ΔmtrD(SO1782) exhibited more severe defect thanΔmtrC(SO1778) which lacks the most critical protein in the Mn(IV) and Fe(III), suggesting that MtrC could not be the major terminal reductase for Cr(IV) reduction. For strains carrying mutation in non‐metal reduction genes, ΔpetC(SO0610) was found to be defective to a similar extent that ΔcymA(SO4591) displayed. In addition, the ability to reduce Cr(VI) of the strains devoid of CcoP(SO2361) or CcoO(SO2363) was greatly impaired. On the basis that CcoP(SO2361), CcoO(SO2363) and PetC(SO0610) are reductase/oxidase functioning during aerobiosis, these proteins may be especially active in response to toxic heavy metals. The quantitative assay also validated the defect of the mutants Δso0970, Δso4047 and Δso4360 in Cr(VI) reduction (Fig. 3A and B). While SO0970 was identified to be abundant in insoluble fraction by mass spectrometry in response to Fe(III) (14), little is known about cytochromes SO4047 or SO4360.
It is reasonable to speculate that the impaired ability of these 13 mutants to reduce Cr(VI) may not be exclusively due to the defect in the reduction pathway. To test this, we carried out the survival assay with these strains. The results presented in Fig. 3C and D showed clearly that ΔpetC(SO0610) and ΔcymA responded to 1 mM Cr(VI) substantially different in terms of survival although neither of these two strains was able to reduce Cr(VI). While ΔcymA displayed a comparable survival rate to Mr ‐1, only a quarter of ΔpetC(SO0610) cells were still viable 10 min after the treatment. It was also evident that mutation in mtrA(SO1777), mtrC(SO1778), omcA(SO1779), mtrF(SO1780) or mtrD(SO1782) did not negatively affect its resistance to Cr(VI) whereas ΔccoP and ΔccoO were significantly more sensitive than Mr ‐1. These observations suggest two explanations to the defect of the c‐type cytochrome mutants in Cr(VI) reduction. On one hand, it is due to the loss of reducing components as either terminal reductases or electron transporters. On the other hand, it may be resulted from the damaged cellular detoxification system but not the reduction pathway per se.
DMSO, fumarate, nitrate and nitrite utilization. Based on the annotation by TIGR, S. oneidensis possesses two operons that encode DMSO reductases and their accessory proteins: so1427‐30 and so4360‐57 (amino acid sequence identity: SO1427/SO4360, 47%; SO1428/SO4359, 28%; SO1429/SO4328, 35%; SO1430/SO4327, 58%). The essentiality of so1427‐30 has been previously established (Gralnick et al., 2005; 2006). In addition to verifying previous findings, our analysis demonstrated that so4360 was not required for DMSO respiration. Only two of obtained mutants ΔfccA(SO0970), soluble fumarate reductase and ΔcymA(SO4591) showed significant defect when grown on fumarate (Table 2). Both of these genes encode the well‐known proteins involved in fumarate reduction (Gordon et al., 1998; Meyer et al., 2004). Three c‐type cytochromes, CymA(SO4591), NapB(SO0845) and NrfA(SO3980), are involved in nitrate/nitrite reduction and their roles in the process have been examined comprehensively and reported recently (Gao et al., 2009).
Table 2.
Mutants defective in respiration on at least one electron acceptor under anaerobic conditions.
| Mutant | Possible function of deleted gene | DMSO | Fumarate | TMAO | NO3‐ | NO2‐ | Fe‐Citrate | MnO2 | Cr(VI) |
|---|---|---|---|---|---|---|---|---|---|
| JZ0479 | Tetrathionate reductase complex | + | + | + | + | + | + | + | + |
| JZ0610 | Ubiquinol‐monoheme cytochrome c5 reductase (petC) | + | + | ‐‐ | + | + | + | ‐‐ | — |
| MR0716 | Sulfite reduction (sorB) | + | + | ‐‐ | + | + | + | + | + |
| JZ0845 | Nitrate reduction napB) | + | + | + | + | + | + | + | + |
| JZ0939 | Unkown | + | + | + | + | + | + | + | + |
| MR0970 | Fumarate reduction (fccA) | + | — | + | + | + | + | + | ‐‐ |
| JZ1421 | Flavocytochrome ifcA | + | + | + | + | + | + | + | + |
| JZ1427 | DMSO reduction (dmsE) | — | + | + | + | + | + | + | + |
| MR1777 | Metal oxide reduction (mtrA) | + | + | + | + | + | ‐‐ | ‐‐ | ‐‐ |
| MR1778 | Metal oxide reduction (mtrC/omcB) | + | + | + | + | + | ‐‐ | ‐‐ | ‐‐ |
| MR1779 | Metal oxide reduction (omcA) | + | + | + | + | + | + | ‐‐ | ‐‐ |
| MR1780 | Metal oxide reduction (mtrF), presumably like omcA | + | + | + | + | + | + | ‐‐ | ‐‐ |
| MR1782 | Metal oxide reduction (mtrD), donor to OM cytochromes | + | + | + | + | + | ‐‐ | ‐‐ | ‐‐ |
| MR2361 | cbb3‐type cytochrome c oxidase (ccoP) Subunit III | + | + | + | + | + | + | + | ‐‐ |
| MR2363 | cbb3‐type cytochrome c oxidase (ccoO). Subunit II | + | + | + | + | + | + | + | ‐‐ |
| JZ3980 | Nitrite reduction (NrfA) | + | + | + | + | — | + | + | + |
| JZ2727 | Anaerobic electron shuttle (cctA) | + | + | + | + | + | + | + | ‐‐ |
| MR4047 | Sulfur cycle, soxA‐like protein | + | + | + | + | + | + | + | ‐‐ |
| MR4360 | Unknown | + | + | + | + | + | + | + | ‐‐ |
| MR4591 | Electron transporter (cymA) | — | — | + | — | — | ‐‐ | ‐‐ | — |
+: Growth of the mutant was comparable with Mr ‐1.
‐‐: Growth of the mutant was significantly impaired (growth rate and/or maximum cell density < 80% of Mr ‐1).
—: Growth of the mutant was not observed.
Relative fitness of c‐type cytochrome mutants during aerobiosis and anaerobiosis
Although valuable information about these c‐type cytochrome mutants was obtained from the conventional physiological characterization of the individual mutants, subtle differences in the influence of the individual mutations on growth remain undefined. In S. oneidensis, there may exist a significant amount of functional redundancy among the various cytochromes because of the inherent branching of most electron transport systems. Therefore, a potentially more sensitive method for characterizing the obtained c‐type cytochrome mutants was designed and utilized. In this study, q‐PCR‐based competition experiments were taken to estimate the influence of each mutation on fitness under various growth conditions. The feasibility of these experiments was made possible by the presence of the unique tags in each mutant integrated during mutagenesis, which allow mutant‐specific fragments (98–102 bp) generation by PCR. After multiple rounds of primer optimization, single clear bands from all mutants but not ΔfccA(SO0970), Δso1413 and Δso4048 were obtained, which qualified the corresponding mutants for q‐PCR analysis (data not shown).
The relative fitness of each c‐type cytochrome mutant under aerobic conditions was determined and shown in Fig. 4A. Populations of two mutants ΔccoP(SO2361) and ΔccoO(SO2363) were substantially reduced (∼102–103) after 5 days and fell below the detectable level from the mixed cultures after 10 days, suggesting that these two genes are critical for aerobic growth. Four other mutants [ΔpetC(SO0610), ΔmtrC(SO1777), ΔmtrD(SO1782) and ΔSO4360] exhibited significant reduction in fitness (at least 103‐fold) during the 10 day incubation. These data were generally in agreement with the observation from single cultures that mutations in all these genes but SO4360 resulted in substantially reduced growth rates. For other mutants displaying an evident phenotype in single cultures, results from two assays were less consistent, suggesting that two cultural conditions are remarkably different. In total, there were only six mutants whose aerobic growth benefits from the loss of the corresponding c‐type cytochrome. Among them, however, ΔnrfA(SO3980) is the only strain devoid of a gene which is well defined.
Figure 4.
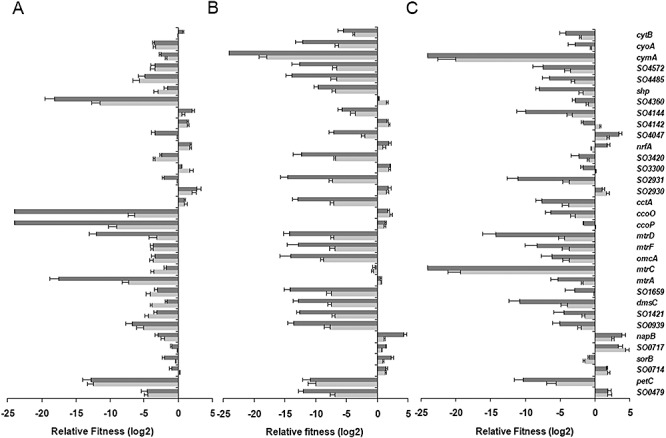
Group competition assays among c‐type cytochrome mutants using qPCR. A same number of cells from overnight culture of each mutant strain were mixed and incubated under tested conditions for competition. The experiments lasted for 10 days and sampled on the first, fifth and tenth days, and results presented were from samples on days 5 (light grey) and 10 (dark grey) respectively. A. Group competition assays under aerobic conditions. B. Group competition assays under anaerobic conditions with fumarate as the sole EA. C. Group competition assays under anaerobic conditions with Fe‐citrate as the sole EA.
The group competition experiments with fumarate as the sole EA under anaerobic conditions revealed a relatively higher number of genes impacting the fitness of S. oneidensis under this specific condition (Fig. 4B). Up to 15 mutants showed a significant decrease in fitness as defined by at least a 103‐fold decrease in population after 10 transfers. Among them, ΔcymA(SO4591), a key branch point in anaerobic respiration, was not detectable at all. This result suggests that multiple c‐type cytochromes may contribute to fitness under anaerobic conditions. Similarly, the number of mutants showing significant decrease in population in Fe‐citrate competition cultures was larger compared with that in aerobic competition profiles (Fig. 4C). While it is not surprising that ΔmtrC(SO1778) was unable to grow over the detectable level in mixed cultures, other mutants devoid of well‐defined metal reduction proteins, such as ΔmtrA(SO1777), ΔomcA(SO1779), ΔmtrF(SO1780) and ΔmtrD(SO1782), displayed a moderate loss in fitness, suggesting that these proteins are not essential to iron reduction. Only five mutants [ΔSO2931, ΔpetC(SO0610), ΔmtrD(SO1782), ΔmtrC(SO1778), ΔdmsC(SO1427)] showed more than 103 times reduction in population when grown on fumarate and Fe‐citrate.
While the group competition experiments revealed a number of c‐type cytochromes impacting fitness under aerobic and/or anaerobic growth condition, they have one indisputable drawback: the wild type was excluded because of the lack of a unique tag. To further evaluate the fitness of selected mutants, we performed two pairwise competition experiments between the wild type and either of three mutants ΔcytcB(SO4666),ΔccoP(SO2361) or ΔnrfA(SO3980). These three mutants were chosen because they represent three different categories: no significant change, increase and decrease in fitness through the group competition experiments. The results of pairwise competition experiments were presented in Table 3. In T0 samples, the average numbers of colonies of were 227, 286 and 245, of which 48.7%, 50.4% and 51.5% were identified by colony PCR (100 colonies examined per plate) to be the wild‐type in experiments paired with ΔcytcB(SO4666),ΔccoP(SO2361) and ΔnrfA(SO3980) respectively. Results from the 1 day and 5 day competition experiments agreed well with each other. After 5 day competition, the percentages of the wild‐type were changed to approximately 49.1%, 78.2% and 39.8%, representing relative fitness values 1.007, 1.058 and 0.98 over ΔcytcB(SO4666),ΔccoP(SO2361) and ΔnrfA(SO3980) respectively. These results indicate that mutation in the cytcB gene does not affect the fitness of S. oneidensis, while mutations in the ccoP and nrfA significantly altered the ability of S. oneidensis to grow under the tested conditions.
Table 3.
Pairwise competition experiments.
| Gene | Samples | No. of colonies | Percentage of MR‐1a | Percentage of the mutanta | Relative fitnessb |
|---|---|---|---|---|---|
| Δso4666 | Day 0 | 227 ± 21 | 48.7 | 51.3 | |
| Day 1 | 332 ± 27 | 48.3 | 51.7 | 0.984 ± 0.023 | |
| Day 5 | 318 ± 19 | 49.1 | 50.9 | 1.007 ± 0.019 | |
| Δso2361 | Day 0 | 286 ± 16 | 50.4 | 49.6 | |
| Day 1 | 312 ± 23 | 56.3 | 43.7 | 1.053 ± 0.032 | |
| Day 5 | 289 ± 15 | 78.2 | 21.8 | 1.058 ± 0.029 | |
| Δso3980 | Day 0 | 245 ± 18 | 51.5 | 48.5 | |
| Day 1 | 331 ± 32 | 48.7 | 51.3 | 0.976 ± 0.025 | |
| Day 5 | 273 ± 19 | 39.8 | 60.2 | 0.980 ± 0.014 |
The averaged percentage of either the wild‐type or mutant colonies identified by PCR (100 colonies per plate).
The relative fitness was given as the sampled day vs. the day 0.
Discussion
The presented mutational analysis enables us to assess the importance of each individual c‐type cytochrome in respiration of a variety of EAs not only independently but also in competition experiments either pairwise or in a group. While the group competition assays revealed subtle differences in the roles of c‐type cytochromes in regards to the fitness of the cell under specific growth conditions, the pairwise competition assays between the wild‐type and certain mutants validate the results from the former. Most of the results from the competition experiments are consistent with previous findings on roles of individual c‐type cytochromes in reduction of Fe(III) and Mn(IV) (Bretschger et al., 2007), indicating that the competition assay is a solid approach.
A combination of growth profiling and competition assay revealed a number of new findings about c‐type cytochromes in S. oneidensis, the most prominent of which is that the response of S. oneidensis to highly toxic metals such as Cr(VI) is evidently different from that to low toxic metals (Fe(III) and Mn(IV)). Although Cr(VI) at 1 mM repressed transcription of mtrA, mtrC and omcA substantially (Brown et al., 2006), these genes were required for Cr(VI) reduction. Surprisingly, the proteins showing the strongest impact on Cr(VI) reduction were OmcA and MtrD, both of which played minor roles in reduction of Fe(III) and Mn(IV). Shewanella oneidensis possesses a single OmcA between two sets of metal reduction complexes: the MtrB‐MtrA‐MtrC and the MtrF‐MtrE‐MtrD (Fredrickson et al., 2008). It is possible that the MtrF‐MtrE‐MtrD metal reductase complex may be responsible for highly toxic metals and OmcA may function as a component in both complexes.
In addition to the well‐defined c‐type cytochromes involved in metal reduction, our analysis demonstrated that CcoP, CcoO, PetC and three other c‐type cytochromes were critical to anaerobic Cr(VI) reduction. The Cco complex, a member of the cbb3‐type cytochrome oxidase family, consists of four subunits, of which c‐type cytochromes CcoO and CcoP are subunits II and III respectively (Hemp et al., 2005; Ducluzeau et al., 2008; Peters et al., 2008; Pinchuk et al., 2009). These two subunits transfer electrons from the donor to the catalytic binuclear centre within the subunit I (Shi et al., 2007; Ducluzeau et al., 2008). It has been firmly established that Cco complex is necessary for aerobic respiration and may play a critical role in removal of reactive oxygen species during respiration (Pinchuk et al., 2009). Therefore, it is reasonable to assume that these proteins may be required for detoxification rather than reduction per se in response to Cr(VI). It is striking that the removal of PetC resulted in the most severe defect in Cr(VI) reduction by S. oneidensis. As a subunit of bacterial cytochrome bc1‐complex, the protein is required for coupling electron transfer to proton translocation across membrane (Berry et al., 2000; Schneider et al., 2004; Ouchane et al., 2005; Baniulis et al., 2008). The findings presented here implicate that the protein may have a more profound role in bacterial physiology.
Involvement of c‐type cytochromes in aerobiosis was assessed and results were intriguing. Although further exploration on the role of CcoO, CcoP and PetC in aerobic respiration is needed, the observation that mutants devoid of these proteins were defective is anticipated. In contrast, the fact that mutation in mtrA, mtrF or nrfA resulted in an altered growth rate compared with the wild type was not readily acceptable. NrfA, recently confirmed to be the only nitrite reductase converting nitrite to ammonium in S. oneidensis (Gao et al., 2009), is the only c‐type cytochrome whose absence promotes aerobic growth. It is worth mentioning that SO4047, a SoxA‐like diheme c‐type cytochrome, was found to be important in both aerobic growth and Cr(VI) reduction. However, little is known about this protein except that expression of both so4047 and so4048 (encoding a c‐type cytochrome in the same operon) was not altered when exposed to different EAs, including sulfur (Beliaev et al., 2005). Nevertheless, the protein may have a more general role in bacterial respiration and demands a further analysis.
Functional redundancy has been regarded to be common in S. oneidensis, that is, multiple genes present in the genome can carry out similar physiological functions although they may be different in efficiencies and under different regulation (Pitcher and Watmough, 2004; Bretschger et al., 2007; Shi et al., 2007). The c‐type cytochromes are no exception. The data presented here are particularly valuable in resolving functional redundancy among c‐type cytochromes sharing a high level sequence similarity. Similar to bacteria with the TCA cycle, S. oneidensis Mr ‐1 hosts fumarate reductases for aerobic respiration, which are membrane bound with both covalently bound flavin adenine dinucleotide (FAD) and iron‐sulfur centres as cofactors (Tsapin et al., 2001). In addition, S. oneidensis Mr ‐1 contains another version of fumarate reductases, which are soluble periplasmic tetraheme flavocytochrome c (Gao et al., 2009). A total of six genes in the Mr ‐1 genome are annotated to encode the soluble fumarate reductase‐like proteins (Tsapin et al., 2001). However, fumarate reduction is abolished only in the fccA deletion mutant, implicating that FccA is the fumarate reductase functioning under anaerobic conditions. In the case of DMSO reduction, both SO1427‐1430 and SO4357‐4360 display high sequence similarities to the well‐studied DMSO reductase and accessory proteins. Our data demonstrated that DmsC(SO1427) rather than SO4360 is required for DMSO reduction. SO4357‐4360 may be involved in the aerobic respiration, evidenced by the reduced fitness of the SO4360 deletion strain in the group competition assay.
It is reasonable to assume that functional redundancy of c‐type cytochromes is not limited to terminal reductases. Multiple c‐type cytochromes may contribute to the same biological process as electron transport proteins. As a result, the abilities of S. oneidensis to respire most of EAs could not be abolished by mutation in a single c‐type cytochrome gene. For example, mutation in mtrC can only reduce the bacterial capacity of respiring Fe(III) and MnO2 to some extent even in the absence of omcA (Myers and Myers, 2001; Shi et al., 2006). In contrast to functional redundancy, S. oneidensis displays an interesting economy by sharing CymA in anaerobic respiration of a variety of EAs (Myers and Myers, 1997; Schwalb et al., 2002; Schwalb et al., 2003). The membrane‐bound proteins are in place of otherwise specific electron transport proteins to deliver electrons to metal, thiosulfate, DMSO, fumarate, nitrate and nitrite terminal reductases.
Results presented in this study also suggest a possibility that some c‐type cytochromes may evolve new functions. In this case, PetC appears to be a good example. Although the specific physiological function of PetC remains unknown, the protein is proposed to function as subunits of multiple enzymes involved in aerobic or photosynthetic bioenergetic electron transport chain (Schneider et al., 2004). In S. oneidensis, PetC appeared to be critical during either aerobiosis or anaerobiosis based on the findings from the group competitive exclusion. Given that many S. oneidensis proteins (i.e. ArcA, EtrA, Crp) deviate from their conventional function significantly (Maier and Myers, 2001; Saffarini et al. 2003; Gao et al., 2008), it is not surprising that PetC extends its role into anaerobic respiration.
It is important to note that variation in genes encoding c‐type cytochromes among the sequenced Shewanella strains (http://www.jgi.doe.gov) is substantial (Table S2), correlated with the extensive physiological diversity (Fredrickson et al., 2008). The data presented here only addresses whether the predicted S. oneidensis genes for c‐type cytochromes present in other genomes because most of other sequenced Shewanella genomes have not been annotated. Surprisingly, only five and 15 out of the 41 genes are present in all and just one less of Shewanella species, respectively, indicating that the core of c‐type cytochrome genes (∼36.6% in S. oneidensis) across the genus is rather small. It is conceivable that the core will be even smaller with more Shewanella genomes sequenced if the same criterion applied. This systematic study provides insights into a full set of c‐type cytochromes, thus represents a primary step towards understanding of these proteins at the omics level.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. For genetic manipulation of E. coli WM3064 and S. oneidensis strains were grown in Lysogeny broth (LB, Difco, Detroit, MI) medium maintained at 37°C and the room temperature respectively. Where required, the growth medium was supplemented with chemical agents at the following concentrations: 2, 6‐diaminopimelic acid (DAP) at 30 µM, ampicillin at 50 µg ml−1, kanamycin at 50 µg ml−1 and gentamicin at 15 µg ml−1.
Barcode‐tagged in‐frame mutagenesis
In‐frame targeted gene deletion mutants were created by either cre‐lox recombination or fusion PCR method as previously described (Marx and Lidstrom, 2002; Gao et al., 2006a,b). To facilitate identification of each mutant within mixed cultures, the primers used for construction of the mutants (Table S3) were designed such that unique barcode(s) of approximately 20 bp was inserted in place of the gene deleted. Two barcodes were used in the cre‐lox method whereas one barcode was used in the fusion PCR method as the linker. All primers used and plasmids containing mutagenesis structure constructed in this study were given in Table S3. For each mutant, the desired deletion was verified by PCR sequence analysis of the mutated region of the chromosomal DNA. To rule out polarity issues introduced by the mutations, complementation experiments were carried out in mutants with phenotypes as described previously (Gao et al., 2008).
Physiological characterization of c‐type cytochrome mutants
Growth of the mutants and wild‐type Mr ‐1 in M1‐L supplemented with one of EAs was performed in microtitre plates, in triplicate and measured using a Bioscreen C microbiology reader (Labsystems Oy, Helsinki, Finland) as previously described (Gao et al., 2008). M1‐L was derived from M1 defined medium (Myers and Myers, 2002) by adding 0.02% (w/v) of vitamin‐free Casamino Acids and 15 mM lactate as the electron donor. Non‐metal EAs tested in this study included fumarate (20 mM), nitrate (2 mM), nitrite (1 mM), thiosulfate (3 mM), TMAO (20 mM) and DMSO (20 mM). For mutants showing an altered growth curve, additional verification of aerobic growth of these mutants was conducted in flasks with vigorous shaking. To evaluate utilization of MnO2 (5 mM), and ferric citrate (10 mM) as EAs, reduction was monitored qualitatively by observing a change in colour of the cultures and growth measured by cell counting under a microscope (Nikon Optiphot, Nikon, Japan). Chromium reduction was estimated quantitatively as follows. Cells were grown in an anaerobic chamber in M1‐L medium amended with 20 mM fumarate to 0.2 of OD600, pelleted by centrifugation, suspended in M1‐L medium supplemented with 0.1 mM potassium chromate (K2CrO4, Sigma‐Aldrich, St. Louis, MO) to 0.15 of OD600, and sampled every 3 h. Soluble Cr(VI) concentrations in the supernatant fraction of each sample were quantified spectrophotometrically at a wavelength of 540 nm using the 1,5‐diphenyl‐carbazide (DPC) in a sulfuric acid solution following the procedure described previously (Park et al., 2000; Chourey et al., 2006).
Survival assays by viable‐cell counting were performed to further characterize Mr ‐1 and the mutants defective in Cr(VI) reduction. Cultures were grown to an optical density of ∼0.5 at 600 nm and aliquoted for the assay. Potassium chromate was added to aliquoted cultures to final concentrations of 1 mM and the cultures were serially diluted and plated onto LB plates 5, 10, 30 and 60 min after the treatment. Plates from dilutions that gave 100–250 colony forming units (CFU) per plate were used to for calculation. Experiments were done in triplicate.
Growth competition assays in liquid media
A single starter culture (ST0) was prepared for both aerobic and anaerobic competition assays. It was prepared by mixing approximately 1 × 107 cells of each strain, grown independently to the stationary phase in M1‐L under aerobic condition, centrifugation of the mixture at 4000 r.p.m., and then resuspending the cells in 1 ml of M1‐L. For aerobic assays, an aliquot of ST0 was adjusted to 2 × 106 cells·per millilitre in a volume of 0.1 ml (∼6 × 104 cells per mutant) to inoculate 9.9 ml of fresh M1‐L, and grown for 24 h until the stationary phase. For anaerobic competition experiments, an aliquot of ST0 was adjusted to 2 × 105 cells·per millilitre in a volume of 0.1 ml, purged with nitrogen gas, and used to inoculate 9.9 ml of fresh M1‐L supplemented with 20 mM fumarate and 10 mM ferric citrate, and grown for 24 and 48 h until stationary phase respectively. After a round of the incubation, 0.1 ml of the mixed culture was inoculated to fresh 9.9 ml of the same medium and the rest was taken as the sample of ST1. The experiment was repeated to collect the sample on the next day as ST2. In total, the procedure was repeated for 10 consecutive rounds. Strain frequencies were estimated by quantitative PCR.
For pairwise competition between wild type and certain mutant strains, the same procedure was used except that the initial inoculation was prepared with approximately ∼1 × 106 cells per strain. To determine the relative fitness, samples were series diluted with fresh LB and aliquots of 0.1 ml of appropriate diluted samples were plated onto LB plates. A total of 100 colonies from plates containing 150–300 colonies were randomly picked and tested by colony PCR with the same pair of primers used for the targeted gene (Table S3). Wild type and mutant cells were differentiated from each other by PCR product size (either the wild type copy or the deleted copy). Relative fitness, W, was calculated according to the method described previously (Lenski et al., 1991; Gao et al., 2009). In both the group competition and pairwise competition assays, data were collected from three independent experiments.
Quantitative PCR
Strain frequencies in group competition experiments were assessed by quantitative PCR using iQ5 Multicolor Real‐time PCR detection system with the bundled software (Bio‐Rad). Using the unique ‘barcode’ incorporated during construction of the mutant, specific primers were designed for each mutant (Table S3). For one‐tag and two‐tag mutants, the tags (5′→3′) and C‐terminal tags (5′→3′) were used as mutant‐specific primers (forward) respectively. The reverse primers began with the nucleotide approximately 98–102 bp downstream. The annealing temperature of these primers was kept in the range of from 58 to 62°C by adjusting the length of primers. The tags (3′→5′) and N‐terminal tags (3′→5′) were used instead as mutant‐specific primers if the original design failed to generate clear and strong bands. All primers were given in `Table S3. For each experiment (the same set of samples), all reactions were performed simultaneously with the iQ SYBR Green Supermix using the genomic DNA from collected samples as the template and a pair of verified primers for one of the mutants. The standard curve was constructed as described previously (Gao et al., 2004). Assessments were performed in triplicate and were averaged to give the final value.
Acknowledgments
This research was supported by The U.S. Department of Energy under the Genomics: GTL Program through the Shewanella Federation, Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee‐Battelle LLC for the Department of Energy under contract DOE‐AC05‐00OR22725. This research was also supported by National Natural Science Foundation of China (30870032) to H. Gao.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Mutants subjected to complementation assays.
Distribution of analogues to S. oneidensis cytochrome c genes cross Shewanella.
Primers used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Baniulis D., Yamashita E., Zhang H., Hasan S.S., Cramer W.A. Structure‐function of the cytochrome b6f complex. Photochem Photobiol. 2008;84:1349–1358. doi: 10.1111/j.1751-1097.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- Beliaev A.S., Saffarini D.A., McLaughlin J.L., Hunnicutt D. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR‐1. Mol Microbiol. 2001;39:722–730. doi: 10.1046/j.1365-2958.2001.02257.x. [DOI] [PubMed] [Google Scholar]
- Beliaev A.S., Klingeman D.M., Klappenbach J.A., Wu L., Romine M.F., Tiedje J.M. Global transcriptome analysis of Shewanella oneidensis MR‐1 exposed to different terminal electron acceptors. J Bacteriol. 2005;187:7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry E.A., Guergova‐Kuras M., Huang L.‐S., Crofts A.R. Structure and function of cytochrome bc complexes. Ann Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M. The complete genome sequence of Escherichia coli K‐12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. et al. [DOI] [PubMed] [Google Scholar]
- Bretschger O., Obraztsova A., Sturm C.A., Chang I.S., Gorby Y.A., Reed S.B. Current production and metal oxide reduction by Shewanella oneidensis MR‐1 wild type and mutants. Appl Environ Microbiol. 2007;73:7003–7012. doi: 10.1128/AEM.01087-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.D., Thompson M.R., VerBerkmoes N.C., Chourey K., Shah M., Zhou J. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics. 2006;5:1054–1071. doi: 10.1074/mcp.M500394-MCP200. et al. [DOI] [PubMed] [Google Scholar]
- Chourey K., Thompson M.R., Morrell‐Falvey J., VerBerkmoes N.C., Brown S.D., Shah M. Global molecular and morphological effects of 24‐hour chromium(VI) exposure on Shewanella oneidensis MR‐1. Appl Environ Microbiol. 2006;72:6331–6344. doi: 10.1128/AEM.00813-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald J.W., Hicks M.G., Richardson D.J., Palmer T. The c‐type cytochrome OmcA localizes to the outer membrane upon heterologous expression in Escherichia coli. J Bacteriol. 2008;190:5127–5131. doi: 10.1128/JB.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau A.‐L., Ouchane S., Nitschke W. The cbb3 Oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- Fredrickson J.K., Romine M.F., Beliaev A.S., Auchtung J.M., Driscoll M.E., Gardner T.S. Towards environmental systems biology of Shewanella. Nat Rev Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. et al. [DOI] [PubMed] [Google Scholar]
- Gao H., Wang Y., Liu X., Yan T., Wu L., Alm E. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Yang Z.K., Wu L., Thompson D.K., Zhou J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR‐1 and mutational analysis of its classical cold shock proteins. J Bacteriol. 2006b;188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Wang X., Yang Z.K., Palzkill T., Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR‐I by integrated genomic analyses. BMC Genomics. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Yang Z.K., Barua S., Reed S., Romine M., Fredrickson J. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems. ISME J. 2009;3:966–976. doi: 10.1038/ismej.2009.40. et al. [DOI] [PubMed] [Google Scholar]
- Gao W., Liu Y., Giometti C., Tollaksen S., Khare T., Wu L. Knock‐out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR‐1. BMC Genomics. 2006a;7:76. doi: 10.1186/1471-2164-7-76. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E.H.J., Pealing S.L., Chapman S.K., Ward F.B., Reid G.A. Physiological function and regulation of flavocytochrome c(3), the soluble fumarate reductase from Shewanella putrefaciens NCIMB 400. Microbiology. 1998;144:937–945. doi: 10.1099/00221287-144-4-937. [DOI] [PubMed] [Google Scholar]
- Gralnick J.A., Brown C.T., Newman D.K. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56:1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- Gralnick J.A., Vali H., Lies D.P., Newman D.K. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR‐1. Proc Nat Acad Sci USA. 2006;103:4669–4674. doi: 10.1073/pnas.0505959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg J.F., Paulsen I.T., Nelson K.E., Gaidos E.J., Nelson W.C., Read T.D. Genome sequence of the dissimilatory metal ion‐reducing bacterium Shewanella oneidensis. Nat Biotech. 2002;20:1118–1123. doi: 10.1038/nbt749. et al. [DOI] [PubMed] [Google Scholar]
- Hemp J., Christian C., Barquera B., Gennis R.B., Martinez T.J. Helix switching of a key active site residue in the cytochrome cbb3 oxidases. Biochem. 2005;44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- Lenski R.E., Rose M.R., Simpson S.C., Tadler S.C. Long‐term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Maier T.M., Myers C.R. Isolation and characterization of a Shewanella putrefaciens MR‐1 electron transport regulator etrA mutant: Reassessment of the role of EtrA. J Bacteriol. 2001;183:4918–4926. doi: 10.1128/JB.183.16.4918-4926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C.J., Lidstrom M.E. Broad‐host‐range cre‐lox system for antibiotic marker recycling in Gram‐negative bacteria. Biotechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- Meyer T.E., Tsapin A.I., Vandenberghe I., De Smet L., Frishman D., Nealson K.H. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS. 2004;8:57–77. doi: 10.1089/153623104773547499. et al. [DOI] [PubMed] [Google Scholar]
- Myers C.R., Myers J.M. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR‐1. J Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C.R., Myers J.M. MtrB Is required for proper incorporation of the cytochromes OmcA and OmcB into the outer membrane of Shewanella putrefaciens MR‐1. Appl Environ Microbiol. 2002;68:5585–5594. doi: 10.1128/AEM.68.11.5585-5594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C.R., Carstens B.P., Antholine W.E., Myers J.M. Chromium(VI) reductase activity is associated with the cytoplasmic membrane of anaerobically grown Shewanella putrefaciens MR‐1. J Appl Microbiol. 2000;88:98–106. doi: 10.1046/j.1365-2672.2000.00910.x. [DOI] [PubMed] [Google Scholar]
- Myers J.M., Myers C.R. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR‐1 in reduction of manganese dioxide. Appl Environ Microbiol. 2001;67:260–269. doi: 10.1128/AEM.67.1.260-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchane S., Nitschke W., Bianco P., Vermeglio A., Astier C. Multiple Rieske genes in prokaryotes: exchangeable Rieske subunits in the cytochrome bc1 complex of Rubrivivax gelatinosus. Mol Microbiol. 2005;57:261–275. doi: 10.1111/j.1365-2958.2005.04685.x. [DOI] [PubMed] [Google Scholar]
- Park C.H., Keyhan M., Wielinga B., Fendorf S., Matin A. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol. 2000;66:1788–1795. doi: 10.1128/aem.66.5.1788-1795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Kulajta C., Pawlik G., Daldal F., Koch H.‐G. Stability of the cbb3‐type cytochrome oxidase requires specific CcoQ‐CcoP interactions. J Bacteriol. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk G.E., Rodionov D.A., Yang C., Li X., Osterman A.L., Dervyn E. Genomic reconstruction of Shewanella oneidensis MR‐1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc Nat Acad Sci USA. 2009;106:2874–2879. doi: 10.1073/pnas.0806798106. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher R.S., Watmough N.J. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Pitts K.E., Dobbin P.S., Reyes‐Ramirez F., Thomson A.J., Richardson D.J., Seward H.E. Characterization of the Shewanella oneidensis MR‐1 decaheme cytochrome MtrA: expression in Escherichia coli confers the ability to reduce soluble Fe(III) chelates. J Biol Chem. 2003;278:27758–27765. doi: 10.1074/jbc.M302582200. [DOI] [PubMed] [Google Scholar]
- Romine M.F., Carlson T.S., Norbeck A.D., McCue L.A., Lipton M.S. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR‐1 genome. Appl Environ Microbiol. 2008;74:3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarini D.A., Schultz R., Beliaev A. Involvement of cyclic AMP (CAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D., Berry S., Volkmer T., Seidler A., Rogner M. PetC1 is the major Rieske iron‐sulfur protein in the cytochrome b(6)f complex of Synechocystis sp PCC 6803. J Biol Chem. 2004;279:39383–39388. doi: 10.1074/jbc.M406288200. [DOI] [PubMed] [Google Scholar]
- Schwalb C., Chapman S.K., Reid G.A. The membrane‐bound tetrahaem c‐type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella. Biochem Soc Trans. 2002;30:658–662. doi: 10.1042/bst0300658. [DOI] [PubMed] [Google Scholar]
- Schwalb C., Chapman S.K., Reid G.A. The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochem. 2003;42:9491–9497. doi: 10.1021/bi034456f. [DOI] [PubMed] [Google Scholar]
- Shi L., Chen B., Wang Z., Elias D.A., Mayer M.U., Gorby Y.A. Isolation of a high‐affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c‐type cytochromes of Shewanella oneidensis MR‐1. J Bacteriol. 2006;188:4705–4714. doi: 10.1128/JB.01966-05. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Squier T.C., Zachara J.M., Fredrickson J.K. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c‐type cytochromes. Mol Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny‐Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapin A.I., Vandenberghe I., Nealson K.H., Scott J.H., Meyer T.E., Cusanovich M.A. Identification of a small tetraheme cytochrome c and a flavocytochrome c as two of the principal soluble cytochromes c in Shewanella oneidensis strain MR1. Appl Environ Microbiol. 2001;67:3236–3244. doi: 10.1128/AEM.67.7.3236-3244.2001. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutants subjected to complementation assays.
Distribution of analogues to S. oneidensis cytochrome c genes cross Shewanella.
Primers used in this study.


