Summary
Nisin A is the most thoroughly investigated member of the lantibiotic family of antimicrobial peptides. In addition to a long history of safe use as a food antimicrobial, its activity against multi‐drug resistant pathogens has resulted in a renewed interest in applying nisin as a chemotherapeutic to treat bacterial infections. The wealth of Nisin‐related information that has been generated has also led to the development of the biotechnological capacity to engineer novel Nisin variants with a view to improving the function and physicochemical properties of this already potent peptide. However, the identification of bioengineered Nisin derivatives with enhanced antimicrobial activity against Gram‐positive targets is a recent event. In this study, we created stable producers of the most promising derivatives of Nisin A generated to date [M21V (hereafter Nisin V) and K22T (hereafter Nisin T)] and assessed their potency against a range of drug‐resistant clinical, veterinary and food pathogens. Nisin T exhibited increased activity against all veterinary isolates, including streptococci and staphylococci, and against a number of multi‐drug resistant clinical isolates including MRSA, but not vancomycin‐resistant enterococci. In contrast, Nisin V displayed increased potency against all targets tested including hVISA strains and the hyper‐virulent Clostridium difficile ribotype 027 and against important food pathogens such as Listeria monocytogenes and Bacillus cereus. Significantly, this enhanced activity was validated in a model food system against L. monocytogenes. We conclude that Nisin V possesses significant potential as a novel preservative or chemotherapeutic compound.
Introduction
The lantibiotics are an ever‐expanding family of antimicrobial peptides that are produced by a diverse range of bacteria (McAuliffe et al., 2001; Twomey et al., 2002; Chatterjee et al., 2005; Bierbaum and Sahl, 2009). These gene‐encoded, ribosomally synthesized peptides are distinguished by the presence of post‐translationally modified amino acids such as dehydroalanine (Dha), dehydrobutyrine (Dhb) and the eponymous lanthionine (Lan) and β‐methyllanthionine (MeLan) formed by thioether linkages between dehydrated residues and neighbouring cysteines. As a result of their highly potent biological activities, lantibiotics have the potential to be employed as novel antimicrobials to combat medically significant bacteria and their multi‐drug resistant forms (Cotter et al., 2005a; Lawton et al., 2007; Piper et al., 2009a,b). The prototypical and most thoroughly investigated lantibiotic is Nisin A, a 34 amino acid polycyclic peptide that exhibits antibacterial activity against a wide range of clinical and food‐borne pathogens, including staphylococci, bacilli, clostridia and Listeria. Nisin exerts its antimicrobial activity both by pore formation and by inhibition of cell wall synthesis through specific binding to lipid II, an essential precursor of the bacterial cell wall (Breukink et al., 1999; Wiedemann et al., 2001; Bonelli et al., 2006). As a consequence of these two distinct and cooperative mechanisms, microbes have been unable to develop any significant resistance to Nisin A despite its widespread use in the food industry (Breukink and de Kruijff, 1999). Nisin A has a long record of safe use and has been approved as a natural biopreservative by the US FDA (United States Food and Drug Administration), the WHO (World Health Organization, 1969) and by the EU as additive E234 (Delves‐Broughton, 1990; Chen and Hoover, 2003; Guinane et al., 2005) for use in a wide variety of foods including processed cheese, dairy products and canned foods (for reviews see Chen and Hoover, 2003; Deegan et al., 2006; Sobrino‐Lopez and Martin‐Belloso, 2008). Furthermore, the efficacy of both Nisin A and its natural variant Nisin Z against the Gram‐positive pathogens responsible for bovine mastitis has resulted in its incorporation into a number of products dedicated to controlling or treating such infections (Sears et al., 1992; Cotter et al., 2005a; Cao et al., 2007; Wu et al., 2007). The in vivo effectiveness of Nisin A has been tested and it was shown to be more effective than vancomycin when treating mice infected with Streptococcus pneumoniae (Goldstein et al., 1998) while multi‐drug resistant bacteria such as methicillin‐resistant Staphylococcus aureus (MRSA), vancomycin‐intermediate S. aureus (VISA), heterogeneous vancomycin‐intermediate S. aureus (hVISA) and vancomycin‐resistant enterococci (VRE) have all been shown to be susceptible to Nisin A (Severina et al., 1998; Brumfitt et al., 2002; Piper et al., 2009b). Nisin F, another natural variant, was also found to successfully control S. aureus infection in rats (De Kwaadsteniet et al., 2009). Nisin A also effectively inhibits the microorganisms responsible for periodontal disease (Howell et al., 1993; Turner et al., 2004). Other applications have been suggested for Nisin A arising from its strong spermicidal (Aranha et al., 2004; Reddy et al., 2004; Gupta et al., 2009) and anti‐fungal properties (Akerey et al., 2009).
It has been suggested that, due to its gene‐encoded nature, the efficacy of Nisin A as an antimicrobial could be further improved through bioengineering‐based approaches. Although such efforts were first made in the early 1990s, the results were disappointing in that, while bioengineered derivatives were generated that facilitated the elucidation of structure‐function analyses or resulted in increased activity against Gram‐negative or non‐pathogenic targets (Kuipers et al., 1996; Yuan et al., 2004; Rink et al., 2007), no Nisin A or Z derivatives with enhanced potency against Gram‐positive pathogens had been identified. The recent identification, following the utilization of random and site saturation mutagenesis approaches, of a number of Nisin A derivatives with enhanced bioactivity against Gram‐positive pathogens was thus a significant development (Field et al., 2008). Specific activity studies with three of these Nisin A derivatives (M21V, K22T and N20P) revealed that M21V possesses enhanced specific activity against S. aureus DPC 5245 and Listeria monocytogenes EGDe and 10403S, that K22T possesses enhanced specific activity against Streptococcus agalactiae ATCC 13813 and S. aureus ST528 (MRSA) and that N20P possesses enhanced specific activity against S. aureus ST528 but reduced activity against S. agalactiae 13813 (Field et al., 2008). Here we create stable producers, and carry out a more detailed analysis, of the two bioengineered Nisin derivatives with enhanced broad spectrum activity relative to Nisin A [M21V (hereafter Nisin V) and K22T (hereafter Nisin T)]. This analysis incorporates a wider selection of strains and species of medically significant pathogens, including hVISA, VRE and additional MRSA strains, Clostridium difficile ribotype 027, S. agalactiae and L. monocytogenes isolates, including a mutant that exhibits enhanced Nisin A resistance. Notably, we demonstrate that the enhanced specific activity of Nisin V over Nisin A against L. monocytogenes F2365 is retained in food matrices.
Results
Creation of stable Nisin V‐ and Nisin T‐producers
To facilitate ongoing Nisin V‐ and T‐related research, we created producers, which are likely to be more genetically stable, through double cross‐over homologous recombination. This yields strains which are useful for peptide purification, but also is a strategy which is less likely to impinge on the food‐grade status of the producers as no heterologous DNA is present in the final constructs. The nisV and nisT genes (generated by PCR‐based mutagenesis) were each inserted at the appropriate location in the Lactococcus lactis NZ9800 chromosome via double cross‐over recombination to generate L. lactis NZ9800::nisV and L. lactis NZ9800::nisT. Lactococcus lactis NZ9800 is a derivative of NZ9700 that has a four‐base pair deletion in the nisA gene (Kuipers et al., 1993). Results of deferred antagonism assays with S. agalactiae ATCC 13813 and L. monocytogenes EGDe indicated that gene replacement had successfully occurred and that the bioactivity profiles of the newly constructed strains differed from that of L. lactis NZ9700 (Fig. 1A and B). Mass spectrometry analysis confirmed the production of peptides with masses corresponding to Nisin V (3321 amu) and Nisin T (3326 amu) (Fig. 1C). We also confirmed the absence of the pORI280 shuttle vector employed to facilitate the recombination process (data not shown). It was noted that these newly created Nisin V and T producers produced levels of peptide that corresponded closely with those produced when the corresponding structural genes were expressed on a multicopy vector (data not shown).
Figure 1.
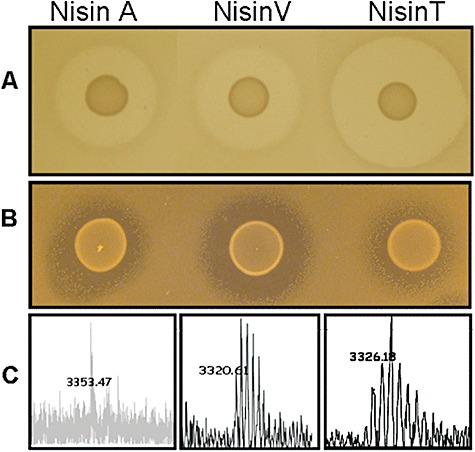
A. Deferred antagonism assays of the Nisin A producing strain L. lactis NZ9700 and the stable nisin derivative producing strains L. lactis NZ9800::nisV and L. lactis NZ9800::nisT against the sensitive indicator S. agalactiae ATCC 13813. B. L. monocytogenes EGDe. C. Colony Mass Spectrometry analysis of the nisin A (3353 amu), Nisin V (3321 amu) and Nisin T (3326 amu) producing strains utilized in this study.
MIC‐based assessment of Nisin A, V and T activity against human pathogens
While bioactivity studies with producing strains give a valuable insight into the net consequences of bioengineering lantibiotic genes, they do not differentiate between changes that alter production and those that impact on specific activity. Importantly, previous studies have established that purified Nisin V and Nisin T both exhibit a twofold enhanced specific activity, relative to Nisin A, against S. aureus DPC 5245, that Nisin T exhibits twofold enhanced potency against S. agalactiae ATCC 13813 and that Nisin V is twofold more potent against L. monocytogenes EGDe (Field et al., 2008). These data were generated by means of agar diffusion assays using equimolar concentrations of the purified peptides. To ensure that this enhanced activity was not as a consequence of altered diffusion rates in agar, the specific activity of the peptides was assessed against a wide range of targets using classical broth‐based minimum inhibitory concentration (MIC) determination assays. Targets included S. aureus strains ST 528 (MRSA), ST 530 (MRSA), ST 534 (MRSA), hVISA 32679, hVISA 32652 and DPC5247, VRE strains VRE Ec538, VRE Ec725, VRE Ec533 and VRE Ec748, C. difficile ribotype 027, S. agalactiae ATCC 13813 and Group B, S. mutans, L. lactis spp. cremoris HP, Bacillus cereus as well as L. monocytogenes EGDe and 10403S, L. innocua FH1848 and the Nisin A resistant mutant L. monocytogenes LO28ΔlisK. MRSA strains were included given their notoriety in nosocomial and community‐acquired infections throughout the world (Zinn et al., 2004; Grundmann et al., 2006). Investigations established that the MIC of Nisin A with respect to the MRSA strains ST 528, ST 530 and ST 534 was 0.5, 0.5 and 1 mg l−1, respectively (Table 1), which was in close agreement with previous results (Piper et al., 2009b). Nisin T was twofold more active than Nisin A against ST 528 and ST 530 (0.26 mg l−1 in each case) but was of equal activity against ST 534. In contrast, Nisin V was consistently twofold more potent against all three MRSA indicator strains tested (0.26, 0.26 and 0.52 mg l−1 respectively) (Table 1). There was a particular desire to include clinical hVISA and VRE strains as indicators in this study in light of the different means via which these pathogens protect themselves against the lipid II binding activity of vancomycin. hVISA and VISA strains are notable in that they possess a thickened cell wall, which is thought to obstruct access of vancomycin to its molecular target, the D‐Ala‐D‐Ala motif in the lipid II pentapeptide (Cui et al., 2006). hVISA have a vancomycin MIC of ≤ 2 mg l−1 but contain a resistant subpopulation able to grow at higher vancomycin concentrations (14 mg l−1) (Howden et al., 2006). VRE differ from VISA in that their resistance is not attributed to cell wall thickening, but results from the incorporation of D‐Ala‐D‐Lac in place of D‐Ala‐D‐Ala, resulting in a markedly lower binding affinity for vancomycin (Bugg et al., 1991). Studies with purified Nisin A established that the MIC against both hVISA 32679 and hVISA 32652 was 2.51 mg l−1[Table 1; in close agreement with previous data generated with a commercial Nisin A preparation, Nisaplin (Piper et al., 2009b)]. These strains were found to be more sensitive to at least one of the bioengineered peptides. While Nisin T was twice as active against hVISA 32679 only (1.25 mg l−1; Table 1), Nisin V exhibited enhanced activity against both strains (1.25 mg l−1; Table 1). Nisin V also outperformed Nisin A and Nisin T when the four VRE isolates (EC 538, EC 533, EC 725 and EC 748) were targeted. The MIC for Nisin V against EC 538, EC 533, EC 748 and EC 725 were 2.09, 1, 1 and 0.52 mg l−1, respectively, which in each case was twofold more active than either Nisin A or Nisin T (Table 1). Another multi‐drug resistant clinical pathogen C. difficile was also included in our investigations. Clostridium difficile‐associated diarrhoea (CDAD) is the most frequent hospital‐acquired diarrhoea in economically developed countries (Arvand et al., 2009). Recently, a hyper‐virulent strain designated ribotype (RT) 027 has been implicated in C. difficile outbreaks associated with increased morbidity and mortality over the last decade (Arvand et al., 2009; Hookman and Barkin, 2009). In MIC experiments conducted with this strain, Nisin A was found to be inhibitory at 8.38 g l−1. However, both Nisin V and Nisin T were twice as active against this target (MIC 4.19 g l−1 in both cases). Another, although less virulent, human pathogen is S. mutans, which is generally considered to be one of the main etiological agents of dental caries in humans (Hamada and Slade, 1980; Loesche, 1986) and begins to colonize in infants as young as 6 months of age (Wan et al., 2003). While this pathogen has been shown to be effectively inhibited by Nisin A (Badaoui Najjar et al., 2009), we demonstrate that this activity can be further improved upon in that both Nisin V and Nisin T were twofold more active than Nisin A (4.19, 4.19 and 8.38 mg l−1 respectively).
Table 1.
Activity of Nisin A, V and T against a range of indicator organisms.
| Strain | Nisin A | Nisin V | Nisin T |
|---|---|---|---|
| mg l−1 (µM) | mg l−1 (µM) | mg l−1 (µM) | |
| S. agalactiae ATCC13813 | 0.13 (0.039) | 0.06 (0.019) | 0.06 (0.019) |
| S. agalactiae Group B | 0.26 (0.078) | 0.13 (0.039) | 0.13 (0.039) |
| S. mutans | 8.38 (2.50) | 4.19 (1.25) | 4.19 (1.25) |
| C. difficile ribotype 027 | 8.38 (2.50) | 4.19 (1.25) | 4.19 (1.25) |
| L. monocytogenes EGDe | 12.57 (3.75) | 6.28 (1.875) | 12.57 (3.75) |
| L. monocytogenes 10403S | 12.57 (3.75) | 6.28 (1.875) | 12.57 (3.75) |
| L. monocytogenes LO28 | 6.28 (1.875) | ND | ND |
| L. monocytogenes LO28ΔlisK | 12.57 (3.75) | 6.28 (1.875) | 12.57 (3.75) |
| L. innocua FH1848 | 12.57 (3.75) | 6.28 (1.875) | 12.57 (3.75) |
| S. aureus ST 528 (MRSA) | 0.52 (0.156) | 0.26 (0.078) | 0.26 (0.078) |
| S. aureus ST 530 (MRSA) | 0.52 (0.156) | 0.26 (0.078) | 0.26 (0.078) |
| S. aureus ST 534 (MRSA) | 1 (0.312) | 0.52 (0.156) | 1 (0.312) |
| hVISA 32679a | 2.51 (0.75) | 1.25 (0.375) | 1.25 (0.375) |
| hVISA 32652a | 2.51 (0.75) | 1.25 (0.375) | 2.51 (0.75) |
| S. aureus DPC 5247 | 0.2 (0.0625) | 0.1 (0.0312) | 0.1 (0.0312) |
| E. faecium VRE Ec538b | 4.19 (1.25) | 2.09 (0.625) | 4.19 (1.25) |
| E. faecium VRE Ec725b | 1.04 (0.312) | 0.52 (0.156) | 1.04 (0.312) |
| E. faecium VRE Ec533b | 2 (0.625) | 1 (0.312) | 2 (0.625) |
| E. faecium VRE Ec748b | 2 (0.625) | 1 (0.312) | 2 (0.625) |
| Bacillus cereus | 4.19 (1.25) | 2.09 (0.625) | 4.19 (1.25) |
| L. lactis spp cremoris HP | 0.2 (0.0625) | 0.1 (0.0312) | 0.05 (0.0156) |
Heterogenous Vancomycin‐intermediate S. aureus.
Vancomycin‐resistant enterococci.
Results given are mean values of three independent determinations.
ND, not determined.
Activity of Nisin derivatives against bovine mastitic pathogens
Because Nisin A has been incorporated into products to prevent or treat bovine mastitis, we chose three representative mastitic isolates for MIC determination studies, including two S. agalactiae and one S. aureus. This is of particular commercial relevance as mastitis is the most common infectious disease among dairy herds and is estimated to cost US dairy farmers US$1.7 billion annually (Viguier et al., 2009), with S. aureus being responsible for 15–30% of the infections (Zadoks and Fitzpatrick, 2009). Streptococcus agalactiae is also of relevance as it is commonly found in the human gastrointestinal, reproductive and urinary tracts and is the causative agent of Group B streptococcal septicemia, which can be fatal for newborn infants. In line with previous agar‐based investigations, Nisin T was found to be twofold more active against S. agalactiae ATCC 13813 (0.06 mg l−1) than Nisin A (0.13 mg l−1). Nisin T was also found to be more active against a second representative strain, S. agalactiae Group B, with MICs of 0.13 and 0.26 mg l−1 for Nisin T and Nisin A respectively. Notably, although agar‐based assays had previously indicated that Nisin V and Nisin A possessed similar potency against S. agalactiae ATCC13813 (Field et al., 2008), broth‐based MIC data obtained here established that Nisin V is twofold more active than Nisin A against both S. agalactiae targets. This discrepancy could possibly be as a result of the difference between broth based assays compared with solid agar based tests which are known to be affected by several parameters (Wolf and Gibbons, 1996). Having previously established (again through agar‐based approaches) that Nisin V and Nisin T exhibit enhanced activity against the bovine mastitis‐associated strain S. aureus DPC5245 (Field et al., 2008), another such isolate, S. aureus DPC 5247 (Fitzgerald et al., 1997), was employed here. Significantly, several studies indicate that the majority of bovine mastitis cases are caused by only a few specialized clones of S. aureus that have a broad distribution (Fitzgerald et al., 1997; Zadoks et al., 2000; Rabello et al., 2005) and thus establishing the sensitivity of another representative of these to the bioengineered Nisin A derivatives is important. MIC analysis revealed that both Nisin V and Nisin T were twofold more active (0.1 mg l−1) than Nisin A (0.2 mg l−1) against S. aureus DPC 5247. These results are in agreement with deferred antagonism assays using the L. lactis Nisin producing strains (L. lactis NZ9800::nisV and L. lactis NZ9800::nisT and L. lactis NZ9700) against both mastitis‐associated S. aureus strains (data not shown).
Activity of Nisin derivatives against food‐associated strains
In addition to clinical and veterinary pathogens, a decision was made to investigate the sensitivity of two representative food pathogens. Nisin A is effective against L. monocytogenes but it is not very potent (Vignolo et al., 2000; Chi‐Zhang et al., 2004; von Staszewski and Jagus, 2008). Indeed, we also observed that the Listeria strains tested here exhibited the greatest natural resistance to Nisin A of all targets tested. This is particularly relevant as this pathogen is the causative agent of listeriosis, one of the most significant foodborne diseases in industrialized countries. Although agar‐based analysis had suggested that Nisin V possesses enhanced activity, relative to Nisin A, against two strains of L. monocytogenes (EGDe and 10403S) (Field et al., 2008), here the activity of both Nisin V and Nisin T were investigated using a broth‐based approach and a larger selection of L. monocytogenes isolates (Table 2). These investigations revealed that Nisin V is indeed twofold more active (6.28 mg l−1) than Nisin A (12.57 mg l−1) against both L. monocytogenes EGDe and 10403S, whereas the activity of Nisin T is equal to that of Nisin A (12.57 mg l−1). This trend was also apparent against a food (fish paste/smoked haddock) isolate, L. innocua FH 1848, with the MICs of Nisin V, Nisin T and Nisin A being 6.28, 12.57 and 12.57 mg l−1 respectively. Finally, although the development of enhanced resistance by food pathogens to Nisin A has not been reported as a major issue, it remains a concern. Mutants have been generated in the laboratory which exhibit enhanced resistance to Nisin A. One such mutant is L. monocytogenes LO28ΔlisK (Cotter et al., 2002). This mutant lacks the LisK histidine kinase component of the LisRK two‐component signal transduction system (Cotter et al., 1999; Cotter et al., 2002). Here we quantify this resistance and establish that is corresponds to a twofold increase in MIC (LO28, 6.28 mg l−1; LO28ΔlisK 12.57 mg l−1). MIC determination studies revealed that while Nisin T again did not exhibit enhanced potency against this L. monocytogenes target (12.57 mg l−1), Nisin V is twofold more active (6.28 mg l−1). We can conclude that the mechanism of enhanced resistance mediated by the deletion of lisK is overcome by Nisin V. A second food pathogen, B. cereus, was also selected for investigation. Bacillus cereus is ubiquitous in the environment but is also commonly found in food production settings due to its ability to form biofilms and highly adhesive endospores, allowing it to survive food processing treatments and spread to a variety of foods (Lotte et al., 2008). Notably, previous studies have demonstrated that Nisin (in the form of Nisaplin) was able to inhibit the growth of vegetative B. cereus cells in beef gravy at a concentration of 5 mg l−1 at 15°C (Beuchat et al., 1997). Results from MIC determinations with a B. cereus isolate indicate that while the activity of Nisin A and Nisin T are comparable with one another (4.19 mg l−1), Nisin V is twice as potent (2.09 mg l−1). Lastly, due to its high sensitivity to lantibiotics, the cheesemaking strain L. lactis spp. cremoris HP has been routinely used as an indicator organism in several studies (Cotter et al., 2006a,b; Wiedemann et al., 2006; Field et al., 2007). The MIC of Nisin A against L. lactis HP was 0.2 mg l−1 (62.5 nM) which is in close agreement to that obtained for Nisin Z (48 nM) (Wiedemann et al., 2006). Further analysis with the bioengineered peptide revealed that Nisin V is twofold more active than Nisin A (MIC value of 0.1 mg l−1). In contrast, the Nisin T peptide exhibited a fourfold increase in specific activity compared with Nisin A (0.05 mg l−1).
Table 2.
Strains and plasmids utilized in this study.
| Strains/plasmids | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| L. lactis NZ9700 | Wild‐type Nisin producer | Kuipers et al., 1993; 1998 |
| L. lactis NZ9800 | L. lactis NZ9700ΔnisA | Kuipers et al., 1993; 1998 |
| L. lactis NZ9800 pVE6007 | L. lactis NZ9800 harbouring pVE6007 | Field et al., 2008 |
| L. lactis NZ9800::nisV | This study | |
| L. lactis NZ9800::nisT | This study | |
| E. coli EC101 | E. coli host for pORI280 | Law et al., 1995 |
| Indicator organisms | ||
| Strep. agalactiae | Nisin‐sensitive indicator | UCC culture collection |
| Strep. agalactiae ATCC13813 | Nisin‐sensitive indicator | American Type Culture Collection |
| S. aureus DPC5245 | Nisin‐sensitive indicator | DPC Collection |
| ST528 (MRSA) | Nisin‐sensitive indicator | BSAC |
| ST530(MRSA) | Nisin‐sensitive indicator | BSAC |
| ST534(MRSA) | Nisin‐sensitive indicator | BSAC |
| hVISA32679 | Nisin‐sensitive indicator | BSAC |
| hVISA32652 | Nisin‐sensitive indicator | BSAC |
| VRE Ec538 | Nisin‐sensitive indicator | BSAC |
| VRE Ec725 | Nisin‐sensitive indicator | BSAC |
| VRE Ec533 | Nisin‐sensitive indicator | BSAC |
| VRE Ec748 | Nisin‐sensitive indicator | BSAC |
| C. difficile ribotype 027 | Nisin‐sensitive indicator | UCC culture collection |
| L. monocytogenes 10403S | Nisin‐sensitive indicator | UCC culture collection |
| L. monocytogenes EGDe | Nisin‐sensitive indicator | UCC culture collection |
| L. monocytogenes LO28 | Nisin‐sensitive indicator | UCC culture collection |
| L. monocytogenes LO28ΔlisK | Nisin‐sensitive indicator | Cotter et al., 1999 |
| L. monocytogenes F2365lux | Nisin‐sensitive indicator | Riedel et al., 2007 |
| L. innocua FH1848 | Nisin‐sensitive indicator | UCC culture collection |
| B. cereus | Nisin‐sensitive indicator | UCC culture collection |
| L. lactis spp cremoris HP | Nisin‐sensitive indicator | UCC culture collection |
| Plasmids | ||
| pORI280 | RepA‐, LacZ+ | Leenhouts et al., 1996 |
| pDF06 | pORI280‐nisA | Field et al., 2008 |
| pDF08 | pORI280‐nisV | This study |
| pDF09 | pORI280‐nisT | This study |
| pVE6007 | CmR; temp sensitive | Maguin et al., 1992 |
BSAC, British Society for Antimicrobial Chemotherapy; DPC, Dairy Products Research Centre.
Growth and kill curve‐based comparisons of the activity of Nisin A, T and V
While MIC analysis can illustrate the increased specific activity of the bioengineered peptides compared with Nisin A, they are end‐point assays and cannot reveal the more subtle details of the impact of an antimicrobial on bacterial viability that are apparent when growth curve analysis is performed. Furthermore, because such assays are based on the ability of the antimicrobial to retard growth, they do not provide an accurate insight into its ability to kill the pathogen. To address this issue, two of the indicators employed above, S. agalactiae ATCC 13813 and L. monocytogenes EGDe, were selected for further growth and kill analysis with a view to comparing the results with MIC values. For S. agalactiae ATCC 13813, at the concentration of peptide employed (0.04 mg l−1; Fig. 2A), Nisin A caused a slight delay in growth relative to the non‐Nisin‐containing control. Identical concentrations of the Nisin V and Nisin T resulted in a greater lag time, with the lag time being greatest when Nisin T was employed. Thus while the twofold dilutions of peptide employed for MIC determination did not reveal a difference in the specific activity of Nisin V and Nisin T against S. agalactiae ATCC13813, it is apparent from growth curve assays with equimolar concentrations of peptide that Nisin T is, in fact, slightly more potent. Thus while Nisin V is typically more potent than Nisin T, this result coupled with MIC data for L. lactis HP, reveals that in some select instances Nisin T is the more active of the two. An investigation of the growth of L. monocytogenes EGDe in the presence of sublethal quantities (4.19 mg l−1) of the Nisin peptides (as previously employed by Begley et al., 2006), and a non‐Nisin‐containing control, highlighted the greater potency of Nisin V as evident from a greatly extended lag. As expected, on the basis of MIC analysis, Nisin A and Nisin T did not differ dramatically (Fig. 2B).
Figure 2.
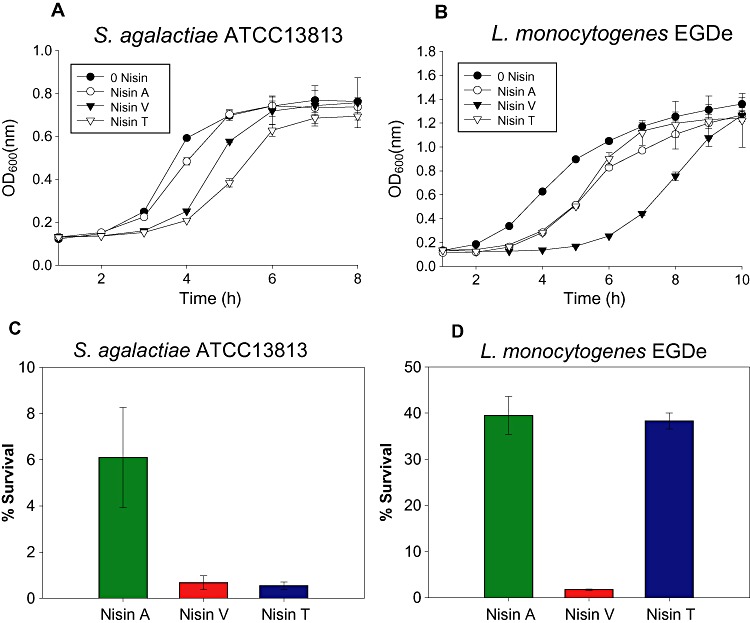
Growth curve analysis of strains (A) S. agalactiae ATCC 13813 and (B) L. monocytogenes EGDe in 0.04 mg l−1 and 4.19 mg l−1, respectively, of Nisin A, V and T peptides and no peptide (control), and Kill curve analysis of strains (C) S. agalactiae ATCC 13813 and (D) L. monocytogenes EGDe in 0.1 and 7.5 mg l−1 respectively of Nisin A, V and T.
In order to compare the bactericidal activity of Nisin A, Nisin V and Nisin T over a defined period of time, S. agalactiae ATCC 13813 and L. monocytogenes EGDe were exposed to 0.1 and 7.5 mg l−1 concentrations, respectively, of peptide for 1 h. While Nisin A reduced S. agalactiae numbers by > 90% after 1 h (6% survival), Nisin V and Nisin T brought about reductions of > 99% (Fig. 2C). Survival rates of 0.675% and 0.543% for Nisin V and Nisin T, respectively, correspond to respective nine‐ and 11‐fold reductions in cell numbers relative to Nisin A. Similar assays with L. monocytogenes EGDe reveal that while exposure to Nisin A or Nisin T results in similar cell survival rates (39% and 38%, respectively; Fig. 2D), the same concentration of the Nisin V peptide results in a reduction of > 98% in L. monocytogenes EGDe (1.76% survival) i.e. a greater than 20‐fold difference in cell counts relative to Nisin A.
Investigation of the anti‐Listeria activity of Nisin variants in a food matrix
Having established the potency of Nisin V against L. monocytogenes using a variety of laboratory‐based assays, we sought to determine whether this enhanced effectiveness could be translated to a food matrix. This was particularly important given that the effectiveness of Nisin A in food can be influenced by a wide range of factors including fat content (Jung et al., 1992; Davies et al., 1999), proteolytic degradation (Murray and Richard, 1997), partitioning into polar or nonpolar food components (Murray and Richard, 1997) and sodium chloride concentrations (Chollet et al., 2008). Thus, to evaluate the efficacy of Nisin V in a situation where Nisin A is traditionally used, the efficacy of Nisin A, Nisin V and Nisin T was compared using frankfurters, a food frequently associated with L. monocytogenes contamination, spiked with a strain L. monocytogenes F2365 associated with an epidemic outbreak of listeriosis (Linnan et al., 1988; Mascola et al., 1988). Previously, L. monocytogenes F2365 was tagged with a luciferase‐based reporter system that allows for real‐time monitoring in food as well as in vivo locations (Riedel et al., 2007). The resultant strain was named F2365lux. For food assays, a commercially available frankfurter was homogenized and placed into sterile containers to which L. monocytogenes F2365lux was added to a concentration of 1 × 107 cfu l−1. Each homogenate of 0.2 ml was transferred to multiwell plates and bioluminescence was quantified using a Xenogen IVIS 100 imager (Time T0). Purified Nisin A, Nisin V or Nisin T peptide was then added to reach a final concentration of 7.5 mg l−1. Following incubation at 37°C for 1 h, bacterial growth was monitored by both bioluminescence imaging (RLU) and plate counts. In the presence of either Nisin A or Nisin T L. monocytogenes F2365lux numbers increased as indicated by increased bioluminescence from 2.54+E05 relative light units (RLU) and 2.64+E05 RLU to 3.16+E05 RLU and 3.64+E05 RLU, respectively) after 1 h (Fig. 3) whereas in the corresponding Nisin V‐treated samples, there was a marked decrease in bioluminescence from 2.53+E05 RLU to 1.71+E05 (Fig. 3). CFU counts after 1 h established that this difference in bioluminescence corresponded to the presence of almost 1 log fewer F2365lux cells in the Nisin V‐treated frankfurter (9.45 ± 2.7%) relative to the numbers present in the Nisin A‐ and Nisin T‐treated samples [100% and 89.16 ± 20.8% respectively (Fig. 3)]. While these results are in close agreement with the broth‐based kill curve experiments described above, they are important in their own right in that they demonstrate that the enhanced potency of Nisin V is maintained even within a complex and high‐fat food matrix (total fat content 31.5%). This is reassuring given the problems associated with the inactivity of Nisin in certain foods.
Figure 3.
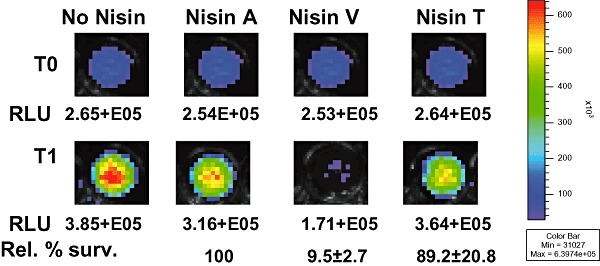
IVIS imaging of the kill effect of Nisin peptides (7.5 mg l−1) against L. monocytogenes F2365lux in frankfurter meat over a 1 h exposure period. The top row depicits the image at time 0 (T0). The bottom row depicts the survival of L. monocytogenes F2365lux after 1 h (T1). The data points represent the corresponding RLU values for images. Rel % Surv figures refer to percentage survival of F2365lux as determined by CFU counts after 1 h (T1) where WT Nisin A = 100%.
Discussion
The diminished capacity of currently available antibiotics to control unwanted bacteria is a major cause for concern. Due to their many unique properties, the lantibiotic class of bacteriocins would seem to have the potential to breach the gap between effective antibiosis and increasingly drug‐resistant clinical and veterinary microbes. Due to its existing broad spectrum activity against a wide range of targets, and its gene‐encoded nature, we regard Nisin A as an excellent target for bioengineering‐based development to generate more potent microbial inhibitors. The ability to generate these variants in a food‐grade manner, as employed here, also provides for the alternative application of these peptides as enhanced food preservatives. Such an application is particularly important in light of the fact that Nisin A has been approved for use, and is widely employed, in over 50 countries worldwide (Chen and Hoover, 2003). Significantly, it has been anticipated that the use of Nisin A is likely to increase in coming years due to the increased consumer demand for minimally processed foods lacking chemical preservatives and the fact that Nisin A is one of only two natural preservatives (the other being the antifungal, natamycin) to have been added to the European Union food additive list. The fact that Nisin V and T differ from Nisin A with respect to only one amino acid suggests that one of these peptides is more likely to be successfully added to this food additive list than any other compound. Such minimal changes also reduce the likelihood of negative consequences such as the peptides becoming cytolytic/hemolytic activity. Significantly Nisin V and T are similar to Nisin A in that no hemolytic activity is apparent even when assessed at very high levels (i.e. 500 mg l−1; data not shown).
Reflecting the multiple ways in which enhanced Nisin derivatives could be applied, the potency of purified Nisin A, V and T was compared against three targets groups; drug‐resistant clinical, veterinary and food pathogens. As representative clinical pathogens, four of the most notorious categories of drug resistant pathogens were selected; VRE (four strains), hVISA (two strains), MRSA (three strains) and the fluoroquinolone‐resistant C. difficile RT 027. The enhanced activity of Nisin V against all 10 of these targets is impressive and not only validates bioengineering strategies for peptide improvement and design, but also serves to highlight the potential of this lantibiotic as an antimicrobial for clinical use. The enhanced efficacy of Nisin V against these targets reveals that the mechanisms via which several of these pathogens have developed resistance to vancomycin do not negate the beneficial consequences of the M21V change, thus establishing that enhanced antimicrobial activity is through some as yet unknown mechanism. The second bioengineered peptide, Nisin T, exhibited enhanced activity against only four of these targets and thus the further investigation of this peptide as a novel anti‐VRE, hVISA or MRSA compound is not merited at this point. The contrasting, generally enhanced sensitivity of S. agalactiae and mastitis infection‐associated S. aureus to Nisin T provides further evidence that some bioengineered Nisin derivatives exhibit strain‐specific enhanced potency. Indeed, given the potency of Nisin V against these mastitis‐associated agents it would thus seem that both peptides merit further attention as novel anti‐mastitis antimicrobials. Bovine mastitis is the cause of a significant economic cost to dairy enterprises through premature culling, extensive antibiotic treatments, prophylactic antibiotic use, reduction in milk yields and milk wastage. Nisin A is already employed commercially as an anti‐mastitis product in the form of Wipe Out ® and recently use of a Nisin A‐containing therapeutic, Mast Out ®, was shown to result in highly statistically significant cure‐rates in 300 cows with subclinical mastitis (http://www.immucell.com). It has also been suggested that this product may qualify for use in the USA without a requirement to discard milk or withhold meat from human consumption as a consequence of treatment. Thus the existence of bioengineered Nisin derivatives that consistently exhibit enhanced activity against mastitis associated pathogens is noteworthy. However, further animal‐trial‐based investigations are required to confirm the efficacy of these peptides. From a human medicine perspective, the potential of Nisin in treating infectious mastitis in lactating mothers has already been demonstrated (Fernandez et al., 2008). Significantly, Nisin prepared from the Nis+ strain L. lactis ESI 515 effectively reduced staphylococcal numbers in breast milk and led to the complete disappearance of clinical signs of mastitis after 2 weeks of treatment. More importantly, Nisin was successful where therapy using traditional antibiotics had failed to provide any improvement in symptoms (Fernandez et al., 2008).
The enhanced nature of the activity of Nisin V and Nisin T against the hyper‐virulent strain C. difficile RT 027 is notable. Alternative and novel strategies are urgently needed to treat this increasingly problematic pathogen, especially as reports of resistance to vancomycin (Pelaez et al., 2002) and metronidazole (Brazier et al., 2001) have emerged. Previous examinations of the antimicrobial efficacy of nisin against five strains of C. difficile isolated from diseased patients recorded MIC values of between 0.125 mg l−1 and 2 mg l−1 (Kerr et al., 1997). In this study, the recorded MIC value for Nisin A against C. difficile RT 027 was 8.38 mg l−1, which could be due to differences in the methodologies employed or be a reflection of the enhanced stress resistance of the 027 ribotype.
Finally, the growing consumer desire for food that is minimally processed and chemical‐free presents new and difficult challenges for the production of fresher and safer food. In this regard Nisin A remains the most commercially important bacteriocin as the only such antimicrobial with GRAS status and WHO, EU and FDA approval. Listeria monocytogenes is of particular concern given its widespread distribution in the environment, robustness and ability to grow at refrigeration temperatures. Although the incidence of listeriosis is low, mortality rates associated with outbreaks are high (Schlech, 2000) and consequently several countries worldwide have adopted a zero tolerance in particular foods (Warriner and Namvar, 2009). Therefore, any new technologies or means to enhance the control of L. monocytogenes in foods are particularly desirable. While Nisin A has performed this role for decades, Nisin A does not possess very potent anti‐Listeria activity and thus enhanced derivatives could be employed to increase manufacturer and consumer confidence while still maintaining a ‘natural’ status. It is thus significant that the Nisin V peptide consistently exhibits superior activity against a range of L. monocytogenes isolates. This activity is not diminished in the face of more resistant strains of Listeria such as the LO28ΔlisK mutant utilized in this study. Moreover, its enhanced efficacy against L. monocytogenes F2365lux in frankfurter meat indicates it maintains this advantage over wild‐type Nisin even within complex food matrices.
From a commercial perspective, it is notable that neither Nisin A nor any other lantibiotic is currently employed commercially as a clinical antimicrobial. However, clinical trials involving duramycin have been reported (Hancock and Sahl, 2006) while mersacidin, among others, are in pre‐clinical development (Boakes and Wadman, 2009). In addition, the increasing number of in vivo studies using planosporicin, actagardine and microbisporicin demonstrate the potential of lantibiotics for chemotherapeutic exploitation. In addition to the added potency, the use of bioengineered Nisin derivatives as clinical therapeutic agents has additional advantages. One of the major drawbacks associated with novel peptide drugs is the cost of manufacture which can be as high as $100–600 per gram (Hancock and Sahl, 2006). Indeed, although Nisin A was shown to be as effective as penicillin in curing mice infected with S. pyogenes and S. aureus (Bavin et al., 1952) as early as the 1950s, a high cost of production and its rapid clearance in vivo were perceived as major obstacles for potential clinical use (Breukink and de Kruijff, 2006). However, due to the use of Nisin A as a food biopreservative, modern large scale fermentation and purification procedures are already in place representing an established base for large scale production of Nisin‐like compounds. It is anticipated that these technologies, coupled with the existing awareness of those in the food and veterinary industries of the value of Nisin A as an antimicrobial will be advantageous should novel bioengineered Nisin derivatives be targeted for commercial development.
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Lactococcus lactis strains were grown in M17 broth supplemented with 0.5% glucose (GM17) or GM17 agar at 30°C. Escherichia coli was grown in Luria–Bertani broth with vigorous shaking or agar at 37°C. Staphylococcus aureus strains were grown in Mueller–Hinton broth (Oxoid) or MH agar at 37°C, streptococci were grown in Tryptic Soy Broth (TSB) or TSB agar at 37°C, Listeria strains were grown in Brain Heart Infusion (BHI) or BHI agar at 37°C. VRE were cultured in cation‐adjusted Mueller–Hinton broth in accordance with the CLSI microbroth method at 37°C without aeration. Antibiotics were used where indicated at the following concentrations: Chloramphenicol at 10 and 20 µg ml−1, respectively, for L. lactis and E. coli. Erythromycin was used at 150 µg ml−1 and 5 µg ml−1 for E. coli and L. lactis respectively. Xgal (5‐bromo‐4‐chloro‐3‐indolyl‐β‐D‐galactopyranoside) was used at a concentration of 40 µg ml−1.
Creation of stable Nisin‐producing derivatives
Mutagenesis of the nisA gene was achieved using a combination of the Quickchange site‐directed mutagenesis strategy (Stratagene) and double cross‐over mutagenesis with pORI280 (RepA‐, LacZ+) as described previously (Cotter et al., 2003; 2005b; 2006a; Field et al., 2008) using the Quickchange protocol as per manufacturer's guidelines and using E. coli EC101 (RepA+) as host. To introduce the desired mutations within the hinge‐region of the nisA gene, the plasmid pDF06 (a 774 bp product encompassing approx. 300 bp either side of the nisA gene cloned into the vector pORI280) was amplified with the QuickChange system (Stratagene) using the primers nisinVFor 5′GAGCTCTGATGGGTTGTAACGTTAAAACAGCAACTTGTCATT3′ and nisinVRev 5′CAATGACAAGTTGCTGTTTTAACGTTACAACCCATCAGAGCT3′ or nisinTFor 5′CTCTGATGGGTTGTAACATGACAACAGCAACTTGTCATTGTA3′ and nisinTRev 5′CTACAATGACAAGTTGCTGTTGTCATGTTACAACCCATCAGA3′ (codon changes underlined). The resulting PCR products were transformed into E. coli EC101 (RepA+). To detect altered pORI280‐nisA transformants, candidates were screened by PCR using a specific ‘check’ primer i.e. nisinTcheck 5′TGATGGGTTGTAACATGAC and nisinV check 5′GCTCTGATGGGTTGTAACG designed to amplify mutated plasmid template only and a reverse primer oDF106 5′TAGAATTCAACAGACCAGCATTA3′. Plasmids from positive candidates were sequenced (MWG Biotech, Germany) using the primers pORI280FOR 5′CTCGTTCATTATAACCCTC3′ and pORI280REV 5′CGCTTCCTTTCCCCCCAT3′ to verify the deliberate mutation in each case and to confirm no other changes had been introduced. pDF08 (pORI280‐nisM21V) and pDF09 (pORI280‐nisK22T) were then introduced separately into NZ9800 pVe6007 by electroporation (Holo and Nes, 1995) and transformants were selected by growth on GM17‐Ery‐Xgal plates at 30°C. Integration of pDF08 and pDF09 by single cross‐over recombination and curing of the temperature sensitive plasmid pVe6007 was achieved by growth at 37°C in GM17‐Ery broth and plating on GM17‐Ery‐Xgal agar at the same temperature. Selected colonies were checked for their inability to grow on GM17‐Cm agar at 30°C and then subcultured in GM17 at 37°C. Each subculture was spread on GM17‐Xgal plates to identify candidates where pORI280 had excised and was lost (LacZ‐) due to a second cross‐over event. Mutant and wild‐type revertants were distinguished by PCR using the specific check primer in each case and oDF106 and also by deferred antagonism assay as candidate mutants exhibited a Bac+ phenotype and wild‐type revertants a Bac‐ phenotype. Bac+ candidates were analysed by Mass Spectrometry to verify production of the mutant Nisin peptide.
Nisin purification
Lactococcus lactis NZ9700 or the mutant Nisin strain of interest was subcultured twice in GM17 broth at 1% at 30°C before use. Two litres of modified TY broth were inoculated with the culture at 0.5% and incubated at 30°C overnight. The culture was centrifuged at 7000 r.p.m. for 15 min. The cell pellet was resuspended in 300 ml of 70% isopropanol 0.1% TFA and stirred at room temperature for approximately 3 h. The cell debris was removed by centrifugation at 7000 r.p.m. for 15 min and the supernatant retained. The isopropanol was evaporated using a rotary evaporator (Buchi) and the sample pH adjusted to 4 before applying to 10 g (60 ml) of Varian C‐18 Bond Elut Column (Varian, Harbor City, CA) pre‐equilibrated with methanol and water. The columns were washed with 100 ml of 20% ethanol and the inhibitory activity was eluted in 100 ml of 70% IPA 0.1% TFA. Fifteen millilitres of aliquots was concentrated to 2 ml through the removal of propan‐2‐ol by rotary evaporation. Aliquots (1.5 ml) were applied to a Phenomenex (Phenomenex, Cheshire, UK) C12 reverse phase (RP)‐HPLC column (Jupiter 4u proteo 90 Å, 250 × 10.0 mm, 4 µm) previously equilibrated with 25% propan‐2‐ol, 0.1% triflouroacetic acid TFA. The column was subsequently developed in a gradient of 30% propan‐2‐ol containing 0.1% TFA to 60% propan‐2‐ol containing 0.1% TFA from 10 to 45 min at a flow rate of 1.2 ml min−1.
Mass Spectrometry
For Colony Mass Spectrometry (CMS) bacteria were collected with sterile plastic loops and mixed with 50 µl of 70% isopropanol adjusted to pH 2 with HCl. The suspension was vortexed, the cells spun down in a benchtop centrifuge at 14 000 r.p.m. for 2 min, and the supernatant was removed for analysis. Mass Spectrometry in all cases was performed with an Axima CFR plus MALDI TOF mass spectrometer (Shimadzu Biotech, Manchester, UK). A 0.5 µl aliquot of matrix solution [alpha‐cyano‐4‐hydroxy cinnamic acid (CHCA), 10 mg ml−1 in 50% acetonitrile‐0.1% (v/v) trifluoroacetic acid] was placed onto the target and left for 1–2 min before being removed. The residual solution was then air‐dried and the sample solution (resuspended lyophilized powder or CMS supernatant) was positioned onto the precoated sample spot. Matrix solution (0.5 µl) was added to the sample and allowed to air‐dry. The sample was subsequently analysed in positive‐ion reflectron mode.
Minimum inhibitory concentration assays
Minimum inhibitory concentration determinations were carried out in triplicate in 96‐well microtitre plates. The 96‐well microtitre plates were pre‐treated with bovine serum albumin (BSA) prior to addition of the peptides. Briefly, to each well of the microtitre plate 200 µl of phosphate buffered saline (PBS) containing 1% (w/v) bovine serum albumin (PBS/BSA) was added and incubated at 37°C for 30 min. The wells were washed with 200 µl of PBS and allowed to dry. Target strains were grown overnight in the appropriate conditions and medium, subcultured into fresh broth and allowed to grow to an OD600 of ∼0.5, diluted to a final concentration of 105 cfu ml−1 in a volume of 0.2 ml. The lyophilized peptides were resuspended in 0.005% acetic acid to a stock concentration of 30 µM. Wild‐type Nisin and Nisin mutant peptides were adjusted to a 2.5 µM, 7.5 µM (Listeria) or 10 µM (C. difficile) starting concentration and twofold serial dilutions of each peptide were made in 96‐well plates for a total of 12 dilutions. The target strain was then added and after incubation for 16 h at 30°C or 37°C the MIC was read as the lowest peptide concentration causing inhibition of visible growth.
Clostridium difficile RT 027 was grown anaerobically at 37°C overnight on Fastidious Anaerobic Agar (LabM). A fresh isolated colony was transferred into freshly boiled Reinforced Clostridium Medium (RCM) (Merck) and incubated for ∼6 h anaerobically at 37°C. A 1:100 dilution was made into double strength RCM broth, and a 0.1 ml inoculum was added into each well containing the serially diluted nisin peptides (in sterile 50 mM phosphate buffer pH 6.5). The plates were incubated for 20 h in an anaerobic chamber at 37°C and again the MIC read as the lowest peptide concentration causing inhibition of visible growth.
Growth/kill experiments
For peptide Kill assays, fresh overnight cultures were transferred (107 cfu ml−1 in a volume of 1.0 ml) into TSB‐YE or BHI broth containing the relevant concentration of wild‐type or mutant Nisin and incubated for 60 min at 37°C. Cell growth was measured by performing viable cell counts by diluting cultures in one‐quarter‐strength Ringer solution and enumeration on TSB‐YE or BHI agar plates. For growth experiments, overnight cultures were transferred (107 cfu ml−1 in a volume of 1.0 ml) into TSB‐YE or BHI supplemented with the relevant concentration of wild‐type and mutant Nisins, and subsequently 0.2 ml was transferred to 96‐well microtitre plates (Sarstedt). Cell growth was measured spectrophotometrically over 24 h periods by using a Spectra Max 340 spectrophotometer (Molecular Devices, Sunnyvale, CA).
Bioassays for antimicrobial activity
Deferred antagonism assays were performed by replicating strains on GM17 or GM17 Xgal agar plates and allowing them to grow overnight before overlaying with either GM17/BHI/TS/MH agar (0.75% w/v agar) seeded with the appropriate indicator strain.
Frankfurter meat trial
Thirty‐five grams of frankfurter meat (78% pork meat and 12% pork fat) was weighed and placed into a sterile blender and homogenized on full for 30 s with 35 ml of PBS. Meat and meat juices were extracted from the homogenate and placed into sterile 1.5 ml of eppendorf tubes. The samples were then inoculated with approximately 1 × 107 cfu ml−1L. monocytogenes F2365lux. Samples were then treated with 7.5 µg ml−1 of purified Nisin V, Nisin T and Nisin A peptide to achieve final volumes of 1 ml. The kill effect of Nisin against Listeria was examined by two techniques: (i) 200 µl of treated homogenate was transferred to 96‐well plates and kill of Listeria monitored by bioluminescence using the Xenogen IVIS 100 system (Xenogen, ALmeda, CA) with a 2 min exposure time and (ii) 200 µl of treated homogenate was transferred to a sterile eppendorf tube and incubated at 37°C, the number of surviving Listeria cells was monitored by serial dilution and plate count technique using Listeria Selective Agar (Oxoid). The effect of Nisin against Listeria was monitored at time 0 and after 1 h. The test was conducted in triplicate.
Acknowledgments
This work was supported by the Irish Government under the National Development Plan, through a Science Foundation Ireland Investigator award to C. H., R. P. R and P. D. C. (06/IN.1/B98), an Irish Research Council for Science Engineering and Technology (IRCSET) EMBARK postgraduate fellowship to D. F and by an Enterprise Ireland Proof of Concept award.
References
- Akerey B., Le‐Lay C., Fliss I., Subirade M., Rouabhia M. In vitro efficacy of nisin Z against Candida albicans adhesion and transition following contact with normal human gingival cells. J Appl Microbiol. 2009;107:1298–1307. doi: 10.1111/j.1365-2672.2009.04312.x. [DOI] [PubMed] [Google Scholar]
- Aranha C., Gupta S., Reddy K.V. Contraceptive efficacy of antimicrobial peptide Nisin: in vitro and in vivo studies. Contraception. 2004;69:333–338. doi: 10.1016/j.contraception.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Arvand M., Hauri A.M., Zaiss N.H., Witte W., Bettge‐Weller G. Clostridium difficile ribotypes 001, 017, and 027 are associated with lethal C. difficile infection in Hesse, Germany. Euro Surveill. 2009;14:19403. doi: 10.2807/ese.14.45.19403-en. [DOI] [PubMed] [Google Scholar]
- Badaoui Najjar M., Kashtanov D., Chikindas M. Natural antimicrobials ε‐poly‐l‐lysine and Nisin A for control of oral microflora. Probiotics Antimicrob Proteins. 2009;1:143–147. doi: 10.1007/s12602-009-9020-0. [DOI] [PubMed] [Google Scholar]
- Bavin E.M., Beach A.S., Falconer R., Friedmann R. Nisin in experimental tuberculosis. Lancet. 1952;1:127–129. doi: 10.1016/s0140-6736(52)92429-x. [DOI] [PubMed] [Google Scholar]
- Begley M., Hill C., Ross R.P. Tolerance of listeria monocytogenes to cell envelope‐acting antimicrobial agents is dependent on SigB. Appl Environ Microbiol. 2006;72:2231–2234. doi: 10.1128/AEM.72.3.2231-2234.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchat L.R., Clavero M.R., Jaquette C.B. Effects of nisin and temperature on survival, growth, and enterotoxin production characteristics of psychrotrophic Bacillus cereus in beef gravy. Appl Environ Microbiol. 1997;63:1953–1958. doi: 10.1128/aem.63.5.1953-1958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbaum G., Sahl H.G. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- Boakes S., Wadman S. The therapeutic potential of lantibiotics. Innov Pharm Technol. 2009:22–25. [Google Scholar]
- Bonelli R.R., Schneider T., Sahl H.G., Wiedemann I. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode‐of‐action studies. Antimicrob Agents Chemother. 2006;50:1449–1457. doi: 10.1128/AAC.50.4.1449-1457.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier J.S., Fawley W., Freeman J., Wilcox M.H. Reduced susceptibility of Clostridium difficile to metronidazole. J Antimicrob Chemother. 2001;48:741–742. doi: 10.1093/jac/48.5.741. [DOI] [PubMed] [Google Scholar]
- Breukink E., De Kruijff B. The lantibiotic nisin, a special case or not? Biochim Biophys Acta. 1999;1462:223–234. doi: 10.1016/s0005-2736(99)00208-4. [DOI] [PubMed] [Google Scholar]
- Breukink E., De Kruijff B. Lipid II as a target for antibiotics. Nat Rev Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- Breukink E., Wiedemann I., Van Kraaij C., Kuipers O.P., Sahl H., De Kruijff B. Use of the cell wall precursor lipid II by a pore‐forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- Brumfitt W., Salton M.R., Hamilton‐Miller J.M. Nisin, alone and combined with peptidoglycan‐modulating antibiotics: activity against methicillin‐resistant Staphylococcus aureus and vancomycin‐resistant enterococci. J Antimicrob Chemother. 2002;50:731–734. doi: 10.1093/jac/dkf190. [DOI] [PubMed] [Google Scholar]
- Bugg T.D., Wright G.D., Dutka‐Malen S., Arthur M., Courvalin P., Walsh C.T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- Cao L.T., Wu J.Q., Xie F., Hu S.H., Mo Y. Efficacy of nisin in treatment of clinical mastitis in lactating dairy cows. J Dairy Sci. 2007;90:3980–3985. doi: 10.3168/jds.2007-0153. [DOI] [PubMed] [Google Scholar]
- Chatterjee C., Paul M., Xie L., Van Der Donk W.A. Biosynthesis and mode of action of lantibiotics. Chem Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Chen H., Hoover D.G. Bacteriocins and their food applications. Compr Rev Food Sci Food Saf. 2003;2:82–100. doi: 10.1111/j.1541-4337.2003.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Chi‐Zhang Y., Yam K.L., Chikindas M.L. Effective control of Listeria monocytogenes by combination of nisin formulated and slowly released into a broth system. Int J Food Microbiol. 2004;90:15–22. doi: 10.1016/s0168-1605(03)00168-5. [DOI] [PubMed] [Google Scholar]
- Chollet E., Sebti I., Martial‐Gros A., Degraeve P. Nisin preliminary study as a potential preservative for sliced ripened cheese: NaCl, fat and enzymes influence on nisin concentration and its antimicrobial activity. Food Control. 2008;19:982–989. [Google Scholar]
- Cotter P.D., Emerson N., Gahan C.G.M., Hill C. Identification and disruption of lisRK, a genetic locus encoding a two‐component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J Bacteriol. 1999;181:6840–6843. doi: 10.1128/jb.181.21.6840-6843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Guinane C.M., Hill C. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob Agents Chemother. 2002;46:2784–2790. doi: 10.1128/AAC.46.9.2784-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Hill C., Ross R.P. A food‐grade approach for functional analysis and modification of native plasmids in Lactococcus lactis. Appl Environ Microbiol. 2003;69:702–706. doi: 10.1128/AEM.69.1.702-706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Hill C., Ross R.P. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005a;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Connor P.M.O., Draper L.A., Lawton E.M., Deegan L.H., Hill C., Ross R.P. Posttranslational conversion of L‐serines to d‐alanines is vital for optimal production and activity of the lantibiotic lacticin 3147. Proc Natl Acad Sci USA. 2005b;102:18584–18589. doi: 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Deegan L.H., Lawton E.M., Draper L.A., O'Connor P.M., Hill C., Ross R.P. Complete alanine scanning of the two‐component lantibiotic lacticin 3147: generating a blueprint for rational drug design. Mol Microbiol. 2006a;62:735–747. doi: 10.1111/j.1365-2958.2006.05398.x. [DOI] [PubMed] [Google Scholar]
- Cotter P.D., Draper L.A., Lawton E.M., McAuliffe O., Hill C., Ross R.P. Overproduction of wild‐type and bioengineered derivatives of the lantibiotic lacticin 3147. Appl Environ Microbiol. 2006b;72:4492–4496. doi: 10.1128/AEM.02543-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Iwamoto A., Lian J.‐Q., Neoh H.‐m., Maruyama T., Horikawa Y., Hiramatsu K. Novel mechanism of antibiotic resistance originating in vancomycin‐intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E.A., Milne C.F., Bevis H.E., Potter R.W., Harris J.M., Williams G.C. Effective use of nisin to control lactic acid bacterial spoilage in vacuum‐packed bologna‐type sausage. J Food Prot. 1999;62:1004–1010. doi: 10.4315/0362-028x-62.9.1004. et al. [DOI] [PubMed] [Google Scholar]
- De Kwaadsteniet M., Doeschate K.T., Dicks L.M. Nisin F in the treatment of respiratory tract infections caused by Staphylococcus aureus. Lett Appl Microbiol. 2009;48:65–70. doi: 10.1111/j.1472-765X.2008.02488.x. [DOI] [PubMed] [Google Scholar]
- Deegan L.H., Cotter P.D., Hill C., Ross P. Bacteriocins: biological tools for bio‐preservation and shelf‐life extension. Int Dairy J. 2006;16:1058–1071. [Google Scholar]
- Delves‐Broughton J. Nisin and its uses as a food preservative. Food Technol. 1990;44:100–117. [Google Scholar]
- Fernandez L., Delgado S., Herrero H., Maldonado A., Rodriguez J.M. The bacteriocin nisin, an effective agent for the treatment of staphylococcal mastitis during lactation. J Hum Lact. 2008;24:311–316. doi: 10.1177/0890334408317435. [DOI] [PubMed] [Google Scholar]
- Field D., Collins B., Cotter P.D., Hill C., Ross R.P. A system for the random mutagenesis of the two‐peptide lantibiotic lacticin 3147: analysis of mutants producing reduced antibacterial activities. J Mol Microbiol Biotechnol. 2007;13:226–234. doi: 10.1159/000104747. [DOI] [PubMed] [Google Scholar]
- Field D., Connor P.M., Cotter P.D., Hill C., Ross R.P. The generation of nisin variants with enhanced activity against specific gram‐positive pathogens. Mol Microbiol. 2008;69:218–230. doi: 10.1111/j.1365-2958.2008.06279.x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J.R., Meaney W.J., Hartigan P.J., Smyth C.J., Kapur V. Fine‐structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119:261–269. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B.P., Wei J., Greenberg K., Novick R. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J Antimicrob Chemother. 1998;42:277–278. [PubMed] [Google Scholar]
- Grundmann H., Aires‐de‐Sousa M., Boyce J., Tiemersma E. Emergence and resurgence of meticillin‐resistant Staphylococcus aureus as a public‐health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- Guinane C.M., Cotter P.D., Hill C., Ross R.P. Microbial solutions to microbial problems; lactococcal bacteriocins for the control of undesirable biota in food. J Appl Microbiol. 2005;98:1316–1325. doi: 10.1111/j.1365-2672.2005.02552.x. [DOI] [PubMed] [Google Scholar]
- Gupta S.M., Aranha C.C., Bellare J.R., Reddy K.V.R. Interaction of contraceptive antimicrobial peptide nisin with target cell membranes: implications for use as vaginal microbicide. Contraception. 2009;80:299–307. doi: 10.1016/j.contraception.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Hamada S., Slade H.D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Mol Biol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E.W., Sahl H.‐G. Antimicrobial and host‐defense peptides as new anti‐infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Holo H., Nes I.F. Transformation of Lactococcus by electroporation. Methods Mol Biol. 1995;47:195–199. doi: 10.1385/0-89603-310-4:195. [DOI] [PubMed] [Google Scholar]
- Hookman P., Barkin J.S. Clostridium difficile associated infection, diarrhea and colitis. World J Gastroenterol. 2009;15:1554–1580. doi: 10.3748/wjg.15.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden B.P., Johnson P.D.R., Ward P.B., Stinear T.P., Davies J.K. Isolates with Low‐Level Vancomycin Resistance Associated with Persistent Methicillin‐Resistant Staphylococcus aureus Bacteremia. Antimicrob Agents Chemother. 2006;50:3039–3047. doi: 10.1128/AAC.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell T.H., Fiorellini J.P., Blackburn P., Projan S.J., De La Harpe J., Williams R.C. The effect of a mouthrinse based on nisin, a bacteriocin, on developing plaque and gingivitis in beagle dogs. J Clin Periodontol. 1993;20:335–339. doi: 10.1111/j.1600-051x.1993.tb00369.x. [DOI] [PubMed] [Google Scholar]
- Jung D.‐S., Bodyfelt F.W., Daeschel M.A. Influence of fat and emulsifiers on the efficacy of nisin in inhibiting Listeria monocytogenes in fluid milk. J Dairy Sci. 1992;75:387–393. doi: 10.3168/jds.S0022-0302(92)77773-X. [DOI] [PubMed] [Google Scholar]
- Kerr K.G., Copley R.M., Wilcoy M.H. Activity of nisin against Clostridium difficile. Lancet. 1997;349:1026–1027. doi: 10.1016/s0140-6736(05)62927-3. [DOI] [PubMed] [Google Scholar]
- Kuipers O.P., Beerthuyzen M.M., Siezen R.J., De Vos W.M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- Kuipers O.P., Bierbaum G., Ottenwalder B., Dodd H.M., Horn N., Metzger J. Protein engineering of lantibiotics. Antonie Van Leeuwenhoek. 1996;69:161–169. doi: 10.1007/BF00399421. et al. [DOI] [PubMed] [Google Scholar]
- Kuipers O.P., De Ruyter P.G., Kleerebezem M., De Vos W.M. Quorum sensing‐controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- Law J., Buist G., Haandrikman A., Kok J., Venema G., Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. Appl Environ Microbiol. 1995;177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton E.M., Ross R.P., Hill C., Cotter P.D. Two‐peptide lantibiotics: a medical perspective. Mini Rev Med Chem. 2007;7:1236–1247. doi: 10.2174/138955707782795638. [DOI] [PubMed] [Google Scholar]
- Leenhouts K., Buist G., Bolhuis A., Ten Berge A., Kiel J., Mierau I. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet. 1996;253:217–224. doi: 10.1007/s004380050315. et al. [DOI] [PubMed] [Google Scholar]
- Linnan M.J., Mascola L., Lou X.D., Goulet V., May S., Salminen C. Epidemic listeriosis associated with Mexican‐style cheese. N Engl J Med. 1988;319:823–828. doi: 10.1056/NEJM198809293191303. et al. [DOI] [PubMed] [Google Scholar]
- Loesche W.J. Role of Streptococcus mutans in human dental decay. Microbiol Mol Biol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotte P.S.A., Annette F., Per Einar G. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- McAuliffe O., Ross R.P., Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 2001;25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- Maguin E., Duwat P., Hege T., Ehrlich D., Gruss A. New thermosensitive plasmid for gram‐positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola L., Lieb L., Fannin S.L., Chiu J., Linnan M.J. Listeriosis: an uncommon opportunistic infection in patients with acquired immunodeficiency syndrome: a report of five cases and a review of the literature. Am J Med. 1988;84:162–164. doi: 10.1016/0002-9343(88)90027-7. [DOI] [PubMed] [Google Scholar]
- Murray M., Richard J.A. Comparative study of the antilisterial activity of nisin A and pediocin AcH in fresh ground pork stored aerobically at 5°C. J Food Prot. 1997;60:1534–1540. doi: 10.4315/0362-028X-60.12.1534. [DOI] [PubMed] [Google Scholar]
- Pelaez T., Alcala L., Alonso R., Rodriguez‐Creixems M., Garcia‐Lechuz J.M., Bouza E. Reassessment of Clostridium difficile susceptibility to Metronidazole and Vancomycin. Antimicrob Agents Chemother. 2002;46:1647–1650. doi: 10.1128/AAC.46.6.1647-1650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper C., Cotter P.D., Ross R.P., Hill C. Discovery of medically significant lantibiotics. Curr Drug Discov Technol. 2009a;6:1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- Piper C., Draper L.A., Cotter P.D., Ross R.P., Hill C. A comparison of the activities of lacticin 3147 and nisin against drug‐resistant Staphylococcus aureus and Enterococcus species. J Antimicrob Chemother. 2009b;63:546–551. doi: 10.1093/jac/dkp221. [DOI] [PubMed] [Google Scholar]
- Rabello R.F., Souza C.R.V.M., Duarte R.S., Lopes R.M.M., Teixeira L.M., Castro A.C.D. Characterization of Staphylococcus aureus isolates recovered from bovine mastitis in Rio de Janeiro, Brazil. J Dairy Sci. 2005;88:3211–3219. doi: 10.3168/jds.S0022-0302(05)73004-6. [DOI] [PubMed] [Google Scholar]
- Reddy K.V., Aranha C., Gupta S.M., Yedery R.D. Evaluation of antimicrobial peptide nisin as a safe vaginal contraceptive agent in rabbits: in vitro and in vivo studies. Reproduction. 2004;128:117–126. doi: 10.1530/rep.1.00028. [DOI] [PubMed] [Google Scholar]
- Riedel C.U., Monk I.R., Casey P.G., Morrissey D., O'Sullivan G.C., Tangney M. Improved luciferase tagging system for Listeria monocytogenes allows real‐time monitoring in vivo and in vitro. Appl Environ Microbiol. 2007;73:3091–3094. doi: 10.1128/AEM.02940-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink R., Wierenga J., Kuipers A., Kluskens L.D., Driessen A.J., Kuipers O.P., Moll G.N. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C‐terminal truncation. Appl Environ Microbiol. 2007;73:5809–5816. doi: 10.1128/AEM.01104-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlech W.F., III Foodborne listeriosis. Clin Infect Dis. 2000;31:770–775. doi: 10.1086/314008. [DOI] [PubMed] [Google Scholar]
- Sears P.M., Smith B.S., Stewart W.K., Gonzalez R.N., Rubino S.D., Gusik S.A. Evaluation of a nisin‐based germicidal formulation on teat skin of live cows. J Dairy Sci. 1992;75:3185–3190. doi: 10.3168/jds.S0022-0302(92)78083-7. et al. [DOI] [PubMed] [Google Scholar]
- Severina E., Severin A., Tomasz A. Antibacterial efficacy of nisin against multidrug‐resistant Gram‐positive pathogens. J Antimicrob Chemother. 1998;41:341–347. doi: 10.1093/jac/41.3.341. [DOI] [PubMed] [Google Scholar]
- Sobrino‐Lopez A., Martin‐Belloso O. Use of nisin and other bacteriocins for preservation of dairy products. Int Dairy J. 2008;18:329–343. [Google Scholar]
- Von Staszewski M., Jagus R.J. Natural antimicrobials: effect of microgard(TM) and nisin against Listeria innocua in liquid cheese whey. Int Dairy J. 2008;18:255–259. [Google Scholar]
- Turner S.R., Love R.M., Lyons K.M. An in‐vitro investigation of the antibacterial effect of nisin in root canals and canal wall radicular dentine. Int Endod J. 2004;37:664–671. doi: 10.1111/j.1365-2591.2004.00846.x. [DOI] [PubMed] [Google Scholar]
- Twomey D., Ross R.P., Ryan M., Meaney B., Hill C. Lantibiotics produced by lactic acid bacteria: structure, function and applications. Antonie Van Leeuwenhoek. 2002;82:165–185. [PubMed] [Google Scholar]
- Vignolo G., Palacios J., Farias M.E., Sesma F., Schillinger U., Holzapfel W., Oliver G. Combined effect of bacteriocins on the survival of various Listeria species in broth and meat system. Curr Microbiol. 2000;41:410–416. doi: 10.1007/s002840010159. [DOI] [PubMed] [Google Scholar]
- Viguier C., Arora S., Gilmartin N., Welbeck K., O'Kennedy R. Mastitis detection: current trends and future perspectives. Trends Biotechnol. 2009;27:486–493. doi: 10.1016/j.tibtech.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Wan A.K.L., Seow W.K., Purdie D.M., Bird P.S., Walsh L.J., Tudehope D.I. A longitudinal study of Streptococcus mutans colonization in infants after tooth eruption. J Dent Res. 2003;82:504–508. doi: 10.1177/154405910308200703. [DOI] [PubMed] [Google Scholar]
- Warriner K., Namvar A. What is the hysteria with Listeria? Trends Food Sci Technol. 2009;20:245–254. [Google Scholar]
- Wiedemann I., Breukink E., Van Kraaij C., Kuipers O.P., Bierbaum G., De Kruijff B., Sahl H.G. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem. 2001;276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- Wiedemann I., Bottiger T., Bonelli R.R., Wiese A., Hagge S.O., Gutsmann T. The mode of action of the lantibiotic lacticin 3147 – a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol. 2006;61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. et al. [DOI] [PubMed] [Google Scholar]
- Wolf C.E., Gibbons W.R. Improved method for quantification of the bacteriocin nisin. J Appl Bacteriol. 1996;80:453–457. doi: 10.1111/j.1365-2672.1996.tb03242.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Specifications for identity and purity of some antibiotics. World Health Organization/Food Add. 1969;69:53–67. [Google Scholar]
- Wu J., Hu S., Cao L. Therapeutic effect of nisin Z on subclinical mastitis in lactating cows. Antimicrob Agents Chemother. 2007;51:3131–3135. doi: 10.1128/AAC.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Zhang Z.Z., Chen X.Z., Yang W., Huan L.D. Site‐directed mutagenesis of the hinge region of nisinZ and properties of nisinZ mutants. Appl Microbiol Biotechnol. 2004;64:806–815. doi: 10.1007/s00253-004-1599-1. [DOI] [PubMed] [Google Scholar]
- Zadoks R., Van Leeuwen W., Barkema H., Sampimon O., Verbrugh H., Schukken Y.H., Van Belkum A. Application of pulsed‐field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J Clin Microbiol. 2000;38:1931–1939. doi: 10.1128/jcm.38.5.1931-1939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks R.N., Fitzpatrick J.L. Changing trends in mastitis. Ir Vet J. 2009;62:59–70. doi: 10.1186/2046-0481-62-S4-S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn C.S., Westh H., Rosdahl V.T., Group S.S. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb Drug Resist. 2004;10:160–168. doi: 10.1089/1076629041310055. [DOI] [PubMed] [Google Scholar]


