Summary
Atrazine is an herbicide of the s‐triazine family that is used primarily as a nitrogen source by degrading microorganisms. While many catabolic pathways for xenobiotics are subjected to catabolic repression by preferential carbon sources, atrazine utilization is repressed in the presence of preferential nitrogen sources. This phenomenon appears to restrict atrazine elimination in nitrogen‐fertilized soils by indigenous organisms or in bioaugmentation approaches. The mechanisms of nitrogen control have been investigated in the model strain Pseudomonas sp. ADP. Expression of atzA, atzB ad atzC, involved in the conversion of atrazine in cyanuric acid, is constitutive. The atzDEF operon, encoding the enzymes responsible for cyanuric acid mineralization, is a target for general nitrogen control. Regulation of atzDEF involves a complex interplay between the global regulatory elements of general nitrogen control and the pathway‐specific LysR‐type regulator AtzR. In addition, indirect evidence suggests that atrazine transport may also be a target for nitrogen regulation in this strain. The knowledge about regulatory mechanisms may allow the design of rational bioremediation strategies such as biostimulation using carbon sources or the use of mutant strains impaired in the assimilation of nitrogen sources for bioaugmentation.
Introduction
Atrazine (2‐chloro‐4‐ethylamino‐6‐isopropylamino‐1,3,5‐triazine) is an effective herbicide used worldwide for broad‐leaf weed control. Because of its high mobility in soil and its massive application, atrazine has often been detected in surface and ground waters at concentrations well above the legal limits (Kolpin and Kalkhoff, 1993; Richards and Baker, 1993; Biradar and Rayburn, 1995; Hayes et al., 2002; 2003; Tappe et al., 2002). The high incidence of atrazine contamination, along with an increasing concern about the toxicological properties of atrazine, has prompted researchers to seek bioremediation options for atrazine‐polluted sites (Allran and Karasov, 2001). Unlike most xenobiotics, atrazine and other s‐triazines are used preferentially, and often exclusively, as nitrogen sources. The enzymology of the atrazine degradative pathways is now well understood (Ellis et al., 2006). However, gene regulation has scarcely been studied and is emerging as a main factor that limits atrazine bioremediation in the field.
Atrazine‐degrading microorganisms and pathways
Early atrazine‐utilizing isolates partially metabolized the herbicide by means of dealkylation and deamination reactions that release the side‐chains, but were unable to dehalogenate atrazine or cleave the triazine ring (Cook et al., 1984; Mulbry, 1994; Nagy et al., 1995; Shao et al., 1995). However, since the early 1990s, an increasing number of bacteria capable of mineralizing atrazine have been isolated and characterized (Yanze‐Kontchou and Gschwind, 1994; Mandelbaum et al., 1995; Struthers et al., 1998). Although atrazine is used primarily as a nitrogen source due to the fully oxidized state of the s‐triazine ring carbon atoms (Mandelbaum et al., 1995; Radosevich et al., 1995; Struthers et al., 1998; Topp et al., 2000a), several organisms can metabolize the N‐alkyl substituents as the sole carbon source (Shapir et al., 2007).
Atrazine mineralization is achieved in two stages. Initially, the chlorine and N‐alkyl side‐chains are removed to yield cyanuric acid (2,4,6‐trihydroxy‐1,3,5‐triazine). The enzymes involved in this stage often have broad substrate specificity and may perform a variety of reactions on different substrates (Shapir et al., 2007). The s‐triazine ring is then cleaved and subsequently converted to ammonium and carbon dioxide by a set of enzymes that have a much narrower substrate range. Based on the enzymatic activities compiled at the University of Minnesotta Biocatalysis and Biodegradation Database (http://umbbd.msi.umn.edu/) (Ellis et al., 2006), conversion of atrazine to cyanuric acid can be achieved via three distinct pathways, one of which is purely hydrolytic, while the other two are mixed oxidative‐hydrolytic (Fig. 1). The hydrolytic pathway has been extensively characterized in Pseudomonas sp. ADP, and consists of three enzymatic steps catalysed by the gene products of atzA, atzB and atzC (Martinez et al., 2001). AtzA is responsible for dechlorination of atrazine (de Souza et al., 1996), and AtzB and AtzC catalyse the elimination of N‐alkyl substituents from the s‐triazine ring to yield cyanuric acid (Boundy‐Mills et al., 1997; Sadowsky et al., 1998). The atzA, atzB and atzC genes have been shown to be widespread and plasmid‐borne in multiple atrazine‐degrading isolates from all continents (de Souza et al., 1998a,b; Topp et al., 2000a; Rousseaux et al., 2001). In a few Gram‐positive strains, trzN substitutes for atzA. TrzN is a broad substrate range hydrolase that performs dechlorination of atrazine and can remove multiple functional groups from a wide variety of s‐triazines (Topp et al., 2000b; Shapir et al., 2005a). Oxidative‐hydrolytic pathways involve the initial oxidative N‐dealkylation of atrazine to deisopropylatrazine or deethylatrazine. The products of the above reactions may be dealkylated again to deisopropyldeethylatrazine, or may be subjected to hydrolytic dechlorination, deamination and/or dealkylation to yield cyanuric acid. Enzymes catalysing the oxidative reactions include Rhodococcus sp. N186/21 cytochrome P450 (Nagy et al., 1995) and AtrA from Rhodococcus sp. TE1 (Shao et al., 1995). The enzyme TriA displays a 98% identity with AtzA, but is active in deamination of atrazine derivatives rather than dehalogenation (Seffernick et al., 2001). On the contrary, TrzA from Rhodococcus corallinus NRRLB‐15444R and the broad‐specificity hydrolase AtzB catalyse the dechlorination of dealkylated atrazine (Cook et al., 1984; Mulbry, 1994; Seffernick et al., 2002). Although a number of bacteria harbouring the complete set of genes encoding for the hydrolytic pathway have been identified, no single organism studied to date accomplishes all the enzymatic reactions required for atrazine conversion to cyanuric acid via an oxidative‐hydrolytic pathway. Degradation may nevertheless be achieved by a consortium of organisms harbouring the appropriate combination of enzymes, as shown for deisopropylatrazine with a mixed culture of R. corallinus NRRLB‐15444R and Pseudomonas huttiensis NRRLB‐12228 (Cook et al., 1984).
Figure 1.
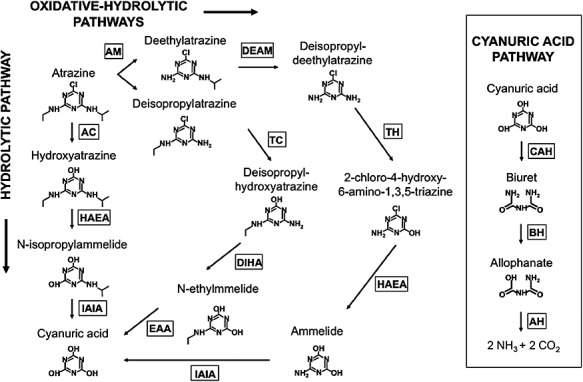
Atrazine degradative pathways. Left: enzymatic steps catalysing the conversion of atrazine to the common intermediate cyanuric acid. Right: cyanuric acid degradative pathway. AC, atrazine chlorohydrolase; HAEA, hidroxyatrazine ethylaminohydrolase; IAIA, N‐isopropylammelide isopropylamidohydrolase; TC, s‐triazine chlorohydrolase; AM, atrazine monooxygenase; DEAM, deethylatrazine monooxygenase; DIHA, deisopropyhidroxylatrazine amidohydrolase; EAA, N‐ethylammelide amidohydrolase, TH, s‐triazine hydrolase; CAH, cyanuric acid hydrolase; BH, biuret hydrolase; AH, allophanate hydrolase.
Cyanuric acid is a common intermediate of many s‐triazine biodegradative pathways (Cook et al., 1985) and its metabolization invariably occurs via hydrolytic cleavage of the s‐triazine ring and consecutive hydrolysis of biuret and allophanate to produce ammonium and carbon dioxide (Cheng et al., 2005). In the case of Pseudomonas sp. ADP and several other organisms (Martinez et al., 2001; Cheng et al., 2005), the enzymes responsible for these reactions are encoded by the atzDEF operon. Homologues to AtzD (TrzD) and AtzF (TrzF) with small differences in substrate affinity and specificity have been shown to perform the equivalent reactions in other bacteria (Karns, 1999; Shapir et al., 2002; Fruchey et al., 2003; Shapir et al., 2005b). Some of the best‐characterized atrazine‐degrading bacterial strains, along with their enzymatic activities and genes, are listed in Table 1.
Table 1.
Bacterial strains harbouring enzymatic activities involved in atrazine catabolism.
| Strain | Enzymatic activities and genes | References |
|---|---|---|
| Agrobacterium radiobacter J14a | AC (atzA), HAEA (atzB), IAIA (atzC), CAH (atzD), BH (atzE), AH (atzF) | De Souza et al. (1998a); Struthers et al. (1998); Cheng et al. (2005) |
| Alcaligenes sp. SG1 | AC (atzA), HAEA (atzB), IAIA (atzC) CAH (trzD), BH,a AHa | K.L. Boundy‐Mills (unpublished); Cheng et al. (2005) |
| Arthrobacter aurescens TC1 | AC (trzN), HAEA (atzB), IAIA (atzC) | Strong et al. (2002); Shapir et al. (2005a) |
| Clavibacter michiganese ATZ1 | AC (atzA), HAEA (atzB) | de Souza et al. (1998a) |
| Enterobacter cloacae 99 | CAH (trzD), BH,a AH (trzF) | Cook and Hutter (1981); Cook et al. (1985); Eaton and Karns (1991b); Cheng et al. (2005) |
| Nocardioides sp. C190 | AC (trzN) | Topp et al. (2000b) |
| Nocardia sp. | AM, DEAM | Giardina et al. (1982); Giardi et al. (1985) |
| Pseudoaminobacter sp | AC (atzA), HAEA (atzB), IAIA (atzC), CAH,a BH,a AHa | Topp et al. (2000a) |
| Pseudomonas sp. ADP | AC (atzA), HAEA (atzB), IAIA (atzC), CAH (atzD), BH (atzE), AH (atzF) | Mandelbaum et al. (1995); de Souza et al. (1996); Boundy‐Mills et al. (1997); Martinez et al. (2001); Shapir et al. (2002); Fruchey et al. (2003); Shapir et al. (2005b) |
| Pseudomonas sp. CN1 | IAIA (atzC), CAH,a BH,a AHa | de Souza et al. (1998a) |
| Acidovorax avenae ssp. citrulli (Pseudomonas sp. NRRLB‐12227) | IAIA (trzC), CAH (trzD), BH,a AH,a TH (triA) | Cook and Hutter (1981); Cook et al. (1985); Eaton and Karns (1991a); Karns (1999); Seffernick et al. (2001); Seffernick et al. (2002) |
| Pseudomonas huttiensis NRRLB‐12228 | EAA, IAIA (trzC), CAH (trzD), BH,a AHa | Cook and Hutter (1981); Cook et al. (1984); Cook et al. (1985); Eaton and Karns (1991b) |
| Ralstonia brasilensis M91‐3 | AC (atzA), HAEA (atzB), IAIA (atzC), CAH (trzD), BH,a AHa | Radosevich et al. (1995); de Souza et al. (1998a); Cheng et al. (2005) |
| Ralstonia picketii D | CAH (atzD), BH (atzE), AH (atzF) | Cheng et al. (2005) |
| Rhizobium sp. PATR | AC (atzA) | Bouquard et al. (1997) |
| Rhodococcus corallinus NRRLB‐15444R | TC (trzA), DIHA | Cook et al. (1984); Mulbry (1994) |
| Rhodococcus sp. NI86/21 | AM (thcB) | Nagy et al. (1995) |
| Rhodococcus sp. TE1 | AM (atrA) | Behki et al. (1993); Shao and Behki (1995) |
Enzymatic activities are assumed based on experimental evidence of cyanuric acid mineralization by this strain.
When identified, the genes encoding the enzymatic activities are included in parentheses. Abbreviations used are the same as in Fig. 1.
Effect of environmental nitrogen on atrazine degradation
The influence of nitrogen compounds on the efficiency of atrazine elimination has been a focus of research, since most degrading strains use atrazine as a nitrogen source and agricultural soils are often rich in nitrogen due to routine fertilization. Numerous studies have established that nitrogen amendments have a negative effect on atrazine biodegradation by indigenous microbial populations in soils (Entry et al., 1993; Alvey and Crowley, 1995; Abdelhafid et al., 2000a,b; Guillén Garcés et al., 2007). The effect of nitrogen sources on atrazine degradation has also been tested in pure cultures of several degrading strains (Bichat et al., 1999; Gebendinger and Radosevich, 1999; García‐González et al., 2003). Atrazine degradation by Agrobacterium radiobacter J14a is not influenced by the simultaneous presence of ammonium, nitrate, urea or glycine in the growth medium (Bichat et al., 1999). However, this may be exceptional, as this is one of the few bacterial strains known to use atrazine as a carbon and nitrogen source (Struthers et al., 1998). In contrast, atrazine degradation by Ralstonia sp. M91‐3 is inhibited by the presence of preferential nitrogen sources, such as ammonium, nitrate or urea, both in growing cells (Bichat et al., 1999) and in washed cell suspensions previously grown in the presence of the indicated nitrogen sources (Radosevich et al., 1995; Struthers et al., 1998; Gebendinger and Radosevich, 1999).
Nitrogen control of atrazine degradation has been characterized extensively in Pseudomonas sp. ADP. Bichat and colleagues (1999) showed that this bacterial strain metabolizes atrazine rapidly in the presence of glycine, but significantly slower in the presence of ammonium, nitrate or urea. A later study (García‐González et al., 2003) demonstrated that atrazine degradation is inhibited in resting cell suspensions previously cultured in the presence of nitrogen sources that support fast growth of Pseudomonas sp. ADP, such as ammonium, urea, proline or nitrate. In contrast, cells acclimated in media containing a growth‐limiting nitrogen source, such as serine, or a pathway metabolite, such as cyanuric acid, biuret or atrazine, efficiently eliminated atrazine in the resting cell assay. The presence of atrazine in growth media containing other nitrogen sources did not stimulate degradation, indicating that the pathway is not induced by its substrate. Nitrogen control of atrazine degradation requires assimilation of the repressing nitrogen source, since the inhibitory effect is suppressed in the presence of a glutamine synthetase inhibitor (García‐González et al., 2003).
The relevance of the negative effect of preferential nitrogen sources on atrazine degradation in cultures or cell suspensions to the bioremediation of contaminated soils has been addressed in Pseudomonas sp. ADP; García‐González and colleagues (2003) showed that nitrate amendment inhibited atrazine mineralization in non‐sterile soil microcosms bioaugmented with Pseudomonas sp. ADP. This effect is the result of nitrogen control, as evidenced by the fact that a mutant defective in nitrate assimilation effectively mineralized atrazine in the presence of nitrate in the soil microcosm assay. Inhibition of atrazine utilization by Pseudomonas sp. ADP mimics the results obtained with indigenous populations (Entry et al., 1993; Alvey and Crowley, 1995; Abdelhafid et al., 2000a,b; Guillén Garcés et al., 2007), indicating that nitrogen control is likely a prevalent trait among atrazine‐degrading strains that may limit degradation in soils with high nitrogen content. Indeed, the success of biostimulation strategies using carbon sources as amendments (Abdelhafid et al., 2000a,b; Silva et al., 2004) may reflect the shift in carbon/nitrogen balance caused by the added carbon, as this is the signal sensed by general nitrogen control (Ninfa and Atkinson, 2000; Commichau et al., 2006).
Organization and regulation of the Pseudomonas sp. ADP atrazine catabolic genes
The Pseudomonas sp. ADP atz genes are harboured on the catabolic plasmid pADP‐1. Two sets of genes can be distinguished with significant differences in organization and regulation. The genes atzA, atzB and atzC, encoding broad‐specificity enzymes, are dispersed in an unstable region of the catabolic plasmid and flanked by insertion sequence elements (Martinez et al., 2001). Northern blot hybridization and real‐time RT‐PCR experiments showed that their transcription is constitutive (Martinez et al., 2001; Devers et al., 2004), and atrazine chlorohydrolase assays in crude extracts failed to reveal any post‐transcriptional regulation (V. García‐González, PhD thesis). New catabolic pathways normally exhibit unregulated or suboptimally regulated expression profiles, while in further evolved pathways, catabolic genes are assembled in operons whose expression responds to both substrate‐specific and general physiological signals (Cases and de Lorenzo, 2001). Since atrazine was introduced in the environment about 50 years ago, it has been proposed that acquisition and evolution of atzA, atzB and atzC have taken place in recent times (Martinez et al., 2001; Shapir et al., 2007). In contrast, the three genes encoding narrow‐specificity enzymes involved in the utilization of cyanuric acid are clustered in the atzDEF operon, which is located in a stable region of pADP‐1 (Martinez et al., 2001). Cyanuric acid is not only a common metabolite of multiple s‐triazine degradation pathways, but also a compound that is naturally present in soils (Fruchey et al., 2003). Cyanuric acid is used as a nitrogen source by multiple bacteria and supports fast growth of Pseudomonas sp. ADP (Neumann et al., 2004). Due to the frequent occurrence of cyanuric acid in nature, the atzDEF genes may have assembled and evolved to achieve high specificity prior to the acquisition of atzA, atzB and atzC via transposition or recombination events.
Consistent with its early assembly and evolution, the atzDEF operon displays a complex and sophisticated regulation that integrates a specific response to the substrate of the pathway with a general response to the physiological status of the cell (Fig. 2). Expression of atzDEF is positively regulated in response to nitrogen limitation and cyanuric acid (García‐González et al., 2005). Transcription from the atzDEF promoter is strictly dependent on the product of the divergently transcribed gene, atzR (García‐González et al., 2005). AtzR is a LysR‐type transcriptional regulator (LTTR) and, as most members of this family of proteins, simultaneously activates the expression of the catabolic genes and represses its own synthesis by binding to a single site with conserved structure located at the divergent promoter region (Schell, 1993; Porrúa et al., 2007). Interestingly, AtzR can activate atzDEF expression in response to either nitrogen limitation or cyanuric acid, but also in response to both signals concurrently in a synergistic manner, which is an unusual feature for this type of regulators. Similarly to other LTTRs (Wang and Winans, 1995; Van Keulen et al., 2003), the interaction of AtzR with its effector molecule, cyanuric acid, triggers the repositioning of AtzR at its binding site to achieve a conformation of the AtzR–DNA complexes that is competent for activation (Porrúa et al., 2007). The nitrogen status modulates the activity of AtzR by two different mechanisms. First, transcription of atzR is a target for nitrogen regulation. In this case, the global regulator NtrC is responsible for activation of PatzR in response to nitrogen limitation (García‐González et al., 2005). NtrC is an archetypical activator of σ54‐RNA polymerase (Reitzer et al., 1989; Su et al., 1990; Rippe et al., 1997), and atzR is expressed from a σ54‐dependent promoter, but unlike typical NtrC‐regulated promoters, transcription activation does not require specific DNA binding by NtrC (Porrúa et al., 2007; O. Porrúa, unpubl. results). Second, the ability of AtzR to activate the PatzDEF promoter is promoted in nitrogen‐limiting conditions. Recent studies suggest that the signal transduction protein GlnK is the element that completes the regulatory circuit and communicates the nitrogen limitation signal to AtzR via protein–protein interaction (V. García‐González, PhD thesis). Expression of GlnK is also subjected to positive control by NtrC in enterobacteria (Zimmer et al., 2000) and in Pseudomonas putida KT2440 (Hervás et al., 2008). Since atzDEF is transcribed from a σ70‐dependent promoter (García‐González et al., 2005), this circuit allows the general nitrogen control system to simultaneously regulate the expression of both the σ54‐dependent PatzR and the σ70‐dependent PatzDEF.
Figure 2.
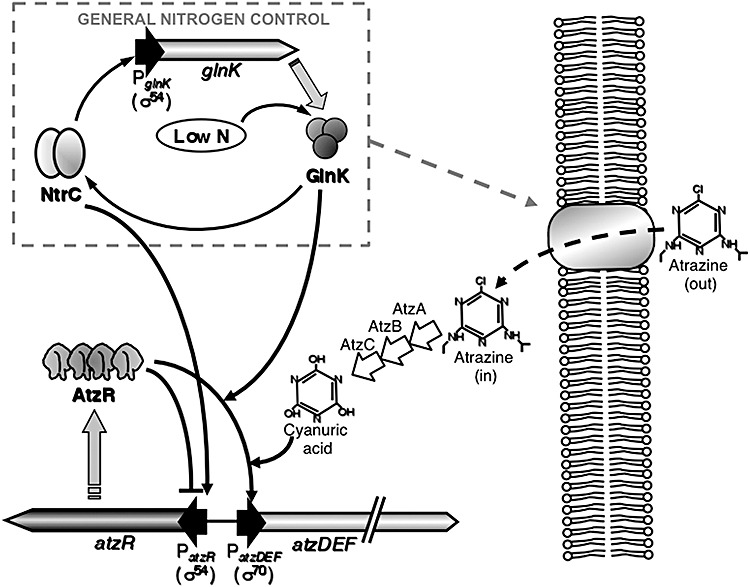
Proposed regulatory circuit for the Pseudomonas sp. ADP atrazine degradative genes. Some elements of the general nitrogen control circuit, such as GlnD and NtrB, have been omitted for simplicity. Dashed lines denote hypothetical elements in the circuit for which experimental evidence is insufficient.
The shape of regulatory circuit for the atzDEF operon is a typical example of a feed‐forward loop, in which a global regulatory element (NtrC) simultaneously controls the synthesis of the specific regulator (AtzR) and expression of the structural genes downstream (atzDEF). Feed‐forward loop circuits are often found in nature because they permit not only the integration of the general and specific regulation, but also a response of the regulated genes that is proportional to the intensity of the stimuli (Cases and de Lorenzo, 2005). It should be mentioned that a very similar arrangement is found in the cascade regulation of the Klebsiella pneumoniae nitrogen fixation genes, where NtrC activates transcription of both nifLA and glnK, and GlnK abolishes formation of the inhibitory NifL–NifA complex, allowing nifHDKTY activation (Stips et al., 2004).
Is atrazine transport a target for nitrogen control?
Nitrogen control of the cyanuric acid utilization operon atzDEF does not suffice to explain the inhibition of atrazine degradation by the presence of preferential nitrogen sources. Since expression of atzA, atzB and atzC is constitutive (Martinez et al., 2001; Devers et al., 2004), conversion of atrazine to cyanuric acid is expected to occur in any condition. Accordingly, growth in medium containing ammonium did not affect atrazine elimination in cell‐free extracts or toluene‐permeabilized Pseudomonas sp. ADP cells (V. García‐González, unpubl. results). This is in sharp contrast with the inhibitory effect of ammonium on atrazine utilization by whole cells (García‐González et al., 2003). Since disruption of the cell membrane eliminates nitrogen regulation, we believe that atrazine transport may also be a target for the nitrogen control system (Fig. 2). The existence of a specific transport system for atrazine was proposed first by Gebendinger and Radosevich (1999)Ralstonia brasilensis M91‐3, based on a major discrepancy between the atrazine degradation rates in cell extracts and whole cells. Transcriptomic studies have shown that transport systems for nitrogen sources are highly represented among the functions of the genes subjected to general nitrogen control in Escherichia coli and P. putida (Zimmer et al., 2000; Hervás et al., 2008). An atrazine transporter may have evolved from a pyrimidine transport system, due to the structural similarity between the s‐triazine ring and the diazine ring in pyrimidines (Wackett et al., 2002). Interestingly, synthesis of the cytosine permease CodB is subjected to general nitrogen control in both P. putida and E. coli (Zimmer et al., 2000; Hervás et al., 2008). Finally, the pADP‐1 plasmid harbours genes encoding transport systems that may be suitable candidates for an atrazine transporter, such as orf46, annotated as a putative uracil/xanthine permease, or orf94‐95‐96‐97‐98, encoding a putative ABC‐type transporter, which are located immediately downstream from and may be co‐transcribed with atzR (Martinez et al., 2001). A similar arrangement of genes encoding a transport system, clustered with s‐triazine utilization genes, is observed at the melamine degradative plasmid pPDL12 of Enterobacter chloacae 99 (GenBank sequence Accession No. AF342826), and the atrazine utilization genes of Arthrobacter aurescens TC1 (Sajjaphan et al., 2004).
Final remarks and future perspectives
While the catabolic potential of any microbial strain is dictated by its enzymatic complement, it is well known that most strains that perform well in a lab setting fail to behave similarly in natural conditions (Young and Burns, 1993). The reasons often mentioned are the suboptimal growth environmental conditions and competition with well‐adapted indigenous organisms. A third factor may be the presence of environmental traits that repress the genes responsible for the catabolic activities. Recent findings point out that atrazine degradation in agricultural soils may be limited by the effect of nitrogen added as fertilizer on catabolic or transport gene expression (García‐González et al., 2003), and a complex regulatory circuit that may involve nitrogen control of atrazine utilization at three different levels has been described (Fig. 2). Based on the new knowledge on nitrogen control of atrazine biodegradation, rational strategies to overcome this limitation may be designed. Some of these strategies may include biostimulation using carbon sources in order to increase the carbon/nitrogen balance (Abdelhafid et al., 2000a,b; Silva et al., 2004), the application of inhibitors of nitrogen assimilation (García‐González et al., 2003), or the use of mutant strains impaired in the assimilation of nitrogen sources for bioaugmentation (García‐González et al., 2003).
Acknowledgments
The work on atrazine degradation in our laboratory has been supported by Grants QLK3‐CT‐1999‐00041 (European Union), BIO2004‐01354 and BIO2007‐63754 (Ministerio de Educación y Ciencia, Spain), and fellowships from the I3P (CSIC/Ministerio de Educación y Ciencia, Spain) and FPU (Ministerio de Educación y Cultura, Spain) programmes, awarded to O.P. and V.G.‐G. respectively.
References
- Abdelhafid R., Houot S., Barriuso E. Dependence of atrazine degradation on C and N availability in adapted and non‐adapted soils. Soil Biol Biochem. 2000a;32:389–401. [Google Scholar]
- Abdelhafid R., Houot S., Barriuso E. How increasing availabilities of carbon and nitrogen affect atrazine behaviour in soils. Biol Fertil Soils. 2000b;30:333–340. [Google Scholar]
- Allran J.W., Karasov W.H. Effects of atrazine on embryos, larvae, and adults of anuran amphibians. Environ Toxicol Chem. 2001;20:769–775. doi: 10.1897/1551-5028(2001)020<0769:eoaoel>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Alvey S., Crowley D.E. Influence of organic amendments on biodegradation of atrazine as a nitrogen source. J Environ Qual. 1995;24:1156–1162. [Google Scholar]
- Behki R., Topp E., Dick W., Germon P. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol. 1993;59:1955–1959. doi: 10.1128/aem.59.6.1955-1959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichat F., Sims G.K., Mulvaney R.L. Microbial utilization of heterocyclic nitrogen from atrazine. Soil Sci Soc Am J. 1999;63:100–110. [Google Scholar]
- Biradar D.P., Rayburn A.L. Chromosomal damage induced by herbicide contamination at concentrations observed in public water supplies. J Environ Qual. 1995;24:1222–1225. [Google Scholar]
- Boundy‐Mills K.L., De Souza M.L., Mandelbaum R.T., Wackett L.P., Sadowsky M.J. The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.916-923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquard C., Ouazzani J., Prom J.‐C., Michel‐Briand Y., Plésiat P. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl Environ Microbiol. 1997;63:916–923. doi: 10.1128/aem.63.3.862-866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I., De Lorenzo V. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 2001;20:1–11. doi: 10.1093/emboj/20.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I., De Lorenzo V. Promoters in the environment: transcriptional regulation in its natural context. Nat Rev Microbiol. 2005;3:105–118. doi: 10.1038/nrmicro1084. [DOI] [PubMed] [Google Scholar]
- Cheng G., Shapir N., Sadowsky M.J., Wackett L.P. Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl Environ Microbiol. 2005;71:4437–4445. doi: 10.1128/AEM.71.8.4437-4445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau F.M., Forchhammer K., Stulke J. Regulatory links between carbon and nitrogen metabolism. Curr Opin Microbiol. 2006;9:167–172. doi: 10.1016/j.mib.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Cook A.M., Hutter R. s‐Triazines as nitrogen sources for bacteria. J Agric Food Chem. 1981;29:1135–1143. [Google Scholar]
- Cook A.M., Grossenbacher H., Hutter R. Deethylsimazine: bacterial dechlorination, deamination and complete degradation. J Agric Food Chem. 1984;32:581–585. [Google Scholar]
- Cook A.M., Beilstein P., Grossenbacher H., Hutter R. Ring cleavage and degradative pathway of cyanuric acid in bacteria. Biochem J. 1985;231:25–30. doi: 10.1042/bj2310025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers M., Soulas G., Martin‐Laurent F. Real‐time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Methods. 2004;56:3–15. doi: 10.1016/j.mimet.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Eaton R.W., Karns J.S. Cloning and analysis of s‐triazine catabolic genes from Pseudomonas sp. strain NRRLB‐12227. J Bacteriol. 1991a;173:1215–1222. doi: 10.1128/jb.173.3.1215-1222.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton R.W., Karns J.S. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s‐triazine‐degrading bacterial strains. J Bacteriol. 1991b;173:1363–1366. doi: 10.1128/jb.173.3.1363-1366.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L.B.M., Roe D., Wackett L.P. The University of Minnesota biocatalysis/biodegradation database: the first decade. Nucleic Acids Res. 2006;34:D517–D521. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entry J.A., Mattson K.G., Emmingham W.H. The influence of nitrogen on atrazine and 2,4‐dichlorophenoxyacetic acid mineralization in grassland soils. Biol Fertil Soils. 1993;16:179–182. [Google Scholar]
- Fruchey I., Shapir N., Sadowsky M.J., Wackett L.P. On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl Environ Microbiol. 2003;69:3653–3657. doi: 10.1128/AEM.69.6.3653-3657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐González V., Govantes F., Shaw L.J., Burns R.G., Santero E. Nitrogen control of atrazine utilization in Pseudomonas sp. strain ADP. Appl Environ Microbiol. 2003;69:6987–6993. doi: 10.1128/AEM.69.12.6987-6993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐González V., Govantes F., Porrúa O., Santero E. Regulation of the Pseudomonas sp. strain ADP cyanuric acid degradation operon. J Bacteriol. 2005;187:155–167. doi: 10.1128/JB.187.1.155-167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebendinger N., Radosevich M. Inhibition of atrazine degradation by cyanazine and exogenous nitrogen in bacterial isolate M91‐3. Appl Microbiol Biotechnol. 1999;51:375–381. doi: 10.1007/s002530051405. [DOI] [PubMed] [Google Scholar]
- Giardi M.T., Giardina M.C., Filacchioni G. Chemical and biological degradation of primary metabolites of atrazine by a Nocardia strain. Agric Biol Chem. 1985;49:1551–1558. [Google Scholar]
- Giardina M.C., Giardi M.T., Filacchioni G. Atrazine metabolism by Nocardia: elucidation of initial pathway and synthesis of potential metabolites. Agric Biol Chem. 1982;46:1439–1445. [Google Scholar]
- Guillén Garcés R.A., Hansen A.M., Van Afferden M. Mineralization of atrazine in agricultural soil: inhibition by nitrogen. Environ Toxicol Chem. 2007;26:844–850. doi: 10.1897/06-328r.1. [DOI] [PubMed] [Google Scholar]
- Hayes T., Haston K., Tsui M., Hoang A., Haeffele C., Vonk A. Herbicides: feminization of male frogs in the wild. Nature. 2002;419:895–896. doi: 10.1038/419895a. [DOI] [PubMed] [Google Scholar]
- Hayes T., Haston K., Tsui M., Hoang A., Haeffele C., Vonk A. Atrazine‐induced hermaphroditism at 0.1 ppb in American leopard frogs (Rana pipiens): laboratory and field evidence. Environ Health Perspect. 2003;111:568–575. doi: 10.1289/ehp.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás A.B., Canosa I., Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008;190:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karns J.S. Gene sequence and properties of an s‐triazine ring‐cleavage enzyme from Pseudomonas sp. strain NRRLB‐12227. Appl Environ Microbiol. 1999;65:3512–3517. doi: 10.1128/aem.65.8.3512-3517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpin D.W., Kalkhoff S.J. Atrazine degradation in a small stream in Iowa. Environ Sci Technol. 1993;27:134–139. [Google Scholar]
- Mandelbaum R.T., Wackett L.P., Allan D.L. Isolation and characterization of a Pseudomonas sp. that mineralizes the s‐triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez B., Tomkins J., Wackett L.P., Wing R., Sadowsky M.J. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP‐1 from Pseudomonas sp. strain ADP. J Bacteriol. 2001;183:5684–5697. doi: 10.1128/JB.183.19.5684-5697.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulbry W.W. Purification and characterization of an inducible s‐triazine hydrolase from Rhodococcus corallinus NRRL B‐15444R. Appl Environ Microbiol. 1994;60:613–618. doi: 10.1128/aem.60.2.613-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I., Compernolle F., Ghys K., Vanderleyden J., De Mot R. A single cytochrome P‐450 system is involved in degradation of the herbicides EPTC (S‐ethyl dipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl Environ Microbiol. 1995;61:2056–2060. doi: 10.1128/aem.61.5.2056-2060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Teras R., Monson L., Kivisaar M., Schauer F., Heipieper H.J. Simultaneous degradation of atrazine and phenol by Pseudomonas sp. strain ADP: effects of toxicity and adaptation. Appl Environ Microbiol. 2004;70:1907–1912. doi: 10.1128/AEM.70.4.1907-1912.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A.J., Atkinson M.R. PII signal transduction proteins. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- Porrúa O., Garcia‐Jaramillo M., Santero E., Govantes F. The LysR‐type regulator AtzR binding site: DNA sequences involved in activation, repression and cyanuric acid‐dependent repositioning. Mol Microbiol. 2007;66:410–427. doi: 10.1111/j.1365-2958.2007.05927.x. [DOI] [PubMed] [Google Scholar]
- Radosevich M., Traina S.J., Hao Y.L., Tuovinen O.H. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol. 1995;61:297–302. doi: 10.1128/aem.61.1.297-302.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L.J., Movsas B., Magasanik B. Activation of glnA transcription by nitrogen regulator I (NRI)‐phosphate in Escherichia coli: evidence for a long‐range physical interaction between NRI‐phosphate and RNA polymerase. J Bacteriol. 1989;171:5512–5522. doi: 10.1128/jb.171.10.5512-5522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R.P., Baker D.B. Pesticide concentration patterns in agricultural drainage networks in the Lake Erie basin. Environ Toxicol Chem. 1993;12:13–36. [Google Scholar]
- Rippe K., Guthold M., Von Hippel P.H., Bustamante C. Transcriptional activation via DNA‐looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase x sigma 54 holoenzyme by scanning force microscopy. J Mol Biol. 1997;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- Rousseaux S., Hartmann A., Soulas G. Isolation and characterisation of new Gram‐negative and Gram‐positive atrazine degrading bacteria from different French soils. FEMS Microbiol Ecol. 2001;36:211–222. doi: 10.1111/j.1574-6941.2001.tb00842.x. [DOI] [PubMed] [Google Scholar]
- Sadowsky M.J., Tong Z., De Souza M., Wackett L.P. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine‐metabolizing enzymes. J Bacteriol. 1998;180:152–158. doi: 10.1128/jb.180.1.152-158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjaphan K., Shapir N., Wackett L.P., Palmer M., Blackmon B., Tomkins J., Sadowsky M.J. Arthrobacter aurescens TC1 atrazine catabolism genes trzNatzB, and atzC are linked on a 160‐kilobase region and are functional in Escherichia coli. Appl Environ Microbiol. 2004;70:4402–4407. doi: 10.1128/AEM.70.7.4402-4407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M.A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- Seffernick J.L., De Souza M.L., Sadowsky M.J., Wackett L.P. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J Bacteriol. 2001;183:2405–2410. doi: 10.1128/JB.183.8.2405-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seffernick J.L., Shapir N., Schoeb M., Johnson G., Sadowsky M.J., Wackett L.P. Enzymatic degradation of chlorodiamino‐s‐triazine. Appl Environ Microbiol. 2002;68:4672–4675. doi: 10.1128/AEM.68.9.4672-4675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z.Q., Behki R. Cloning of the genes for degradation of the herbicides EPTC (S‐ethyl dipropylthiocarbamate) and atrazine from Rhodococcus sp. strain TE1. Appl Environ Microbiol. 1995;61:2061–2065. doi: 10.1128/aem.61.5.2061-2065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z.Q., Seffens W., Mulbry W., Behki R.M. Cloning and expression of the s‐triazine hydrolase gene (trzA) from Rhodococcus corallinus and development of Rhodococcus recombinant strains capable of dealkylating and dechlorinating the herbicide atrazine. J Bacteriol. 1995;177:5748–5755. doi: 10.1128/jb.177.20.5748-5755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapir N., Osborne J.P., Johnson G., Sadowsky M.J., Wackett L.P. Purification, substrate range, and metal center of AtzC: the N‐isopropylammelide aminohydrolase involved in bacterial atrazine metabolism. J Bacteriol. 2002;184:5376–5384. doi: 10.1128/JB.184.19.5376-5384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapir N., Rosendahl C., Johnson G., Andreina M., Sadowsky M.J., Wackett L.P. Substrate specificity and colorimetric assay for recombinant TrzN derived from Arthrobacter aurescens TC1. Appl Environ Microbiol. 2005a;71:2214–2220. doi: 10.1128/AEM.71.5.2214-2220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapir N., Sadowsky M.J., Wackett L.P. Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J Bacteriol. 2005b;187:3731–3738. doi: 10.1128/JB.187.11.3731-3738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapir N., Mongodin E.F., Sadowsky M.J., Daugherty S.C., Nelson K.E., Wackett L.P. Evolution of catabolic pathways: genomic insights into microbial s‐triazine metabolism. J Bacteriol. 2007;189:674–682. doi: 10.1128/JB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E., Fialho A.M., Sa‐Correia I., Burns R.G., Shaw L.J. Combined bioaugmentation and biostimulation to cleanup soil contaminated with high concentrations of atrazine. Environ Sci Technol. 2004;38:632–637. doi: 10.1021/es0300822. [DOI] [PubMed] [Google Scholar]
- De Souza M.L., Sadowsky M.J., Wackett L.P. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification, and protein characterization. J Bacteriol. 1996;178:4894–4900. doi: 10.1128/jb.178.16.4894-4900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza M.L., Seffernick J., Martinez B., Sadowsky M.J., Wackett L.P. The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol. 1998a;180:1951–1954. doi: 10.1128/jb.180.7.1951-1954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza M.L., Wackett L.P., Sadowsky M.J. The atzABC genes encoding atrazine catabolism are located on a self‐transmissible plasmid in Pseudomonas sp. strain ADP. Appl Environ Microbiol. 1998b;64:2323–2326. doi: 10.1128/aem.64.6.2323-2326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stips J., Thummer R., Neumann M., Schmitz R.A. GlnK effects complex formation between NifA and NifL in Klebsiella pneumoniae. Eur J Biochem. 2004;271:3379–3388. doi: 10.1111/j.1432-1033.2004.04272.x. [DOI] [PubMed] [Google Scholar]
- Strong L.C., Rosendahl C., Johnson G., Sadowsky M.J., Wackett L.P. Arthrobacter aurescens TC1 metabolizes diverse s‐triazine ring compounds. Appl Environ Microbiol. 2002;68:5973–5980. doi: 10.1128/AEM.68.12.5973-5980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struthers J.K., Jayachandran K., Moorman T.B. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl Environ Microbiol. 1998;64:3368–3375. doi: 10.1128/aem.64.9.3368-3375.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA‐looping and enhancer activity: association between DNA‐bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappe W., Groeneweg J., Jantsch B. Diffuse atrazine pollution in German aquifers. Biodegradation. 2002;13:3–10. doi: 10.1023/a:1016325527709. [DOI] [PubMed] [Google Scholar]
- Topp E., Zhu H., Nour S.M., Houot S., Lewis M., Cuppels D. Characterization of an atrazine‐degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl Environ Microbiol. 2000a;66:2773–2782. doi: 10.1128/aem.66.7.2773-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp E., Mulbry W.M., Zhu H., Nour S.M., Cuppels D. Characterization of S‐triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl Environ Microbiol. 2000b;66:3134–3141. doi: 10.1128/aem.66.8.3134-3141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keulen G., Ridder A.N., Dijkhuizen L., Meijer W.G. Analysis of DNA binding and transcriptional activation by the LysR‐type transcriptional regulator CbbR of Xanthobacter flavus. J Bacteriol. 2003;185:1245–1252. doi: 10.1128/JB.185.4.1245-1252.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L.P., Sadowsky M.J., Martinez B., Shapir N. Biodegradation of atrazine and related s‐triazine compounds: from enzymes to field studies. Appl Microbiol Biotechnol. 2002;58:39–45. doi: 10.1007/s00253-001-0862-y. [DOI] [PubMed] [Google Scholar]
- Wang L., Winans S.C. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligand‐responsive DNA bending. J Mol Biol. 1995;253:691–702. doi: 10.1006/jmbi.1995.0583. [DOI] [PubMed] [Google Scholar]
- Yanze‐Kontchou C., Gschwind N. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl Environ Microbiol. 1994;60:4297–4302. doi: 10.1128/aem.60.12.4297-4302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C.S., Burns R.G. Detection, survival and activity of bacteria added to soil. In: Bollag J.‐M., Stotzky G., editors. Marcel Dekker; 1993. pp. 1–63. [Google Scholar]
- Zimmer D.P., Soupene E., Lee H.L., Wendisch V.F., Khodursky A.B., Peter B.J. Nitrogen regulatory protein C‐controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA. 2000;97:14674–14679. doi: 10.1073/pnas.97.26.14674. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


