Summary
Bacterial membranes constitute the first physical barrier against different environmental stresses. Pseudomonas putida DOT‐T1E accumulates cyclopropane fatty acids (CFAs) in the stationary phase of growth. In this strain the cfaB gene encodes the main cyclopropane synthase responsible of the synthesis of CFAs, and its expression is mediated by RNA polymerase with sigma factor σ38. We generated a cfaB mutant of P. putida DOT‐T1E and studied its response to solvents, acid pH and other stress conditions such as temperature changes, high osmolarity and the presence of antibiotics or heavy metals in the culture medium. A CfaB knockout mutant was more sensitive to solvent stress than the wild‐type strain, but in contrast to Escherichia coli and Salmonella enterica, the P. putida cfaB mutant was as tolerant to acid shock as the wild‐type strain. The cfaB mutant was also as tolerant as the parental strain to a number of drugs, antibiotics and other damaging agents.
Introduction
Bacterial life in the environment is threatened by predators, and by many abiotic factors such as temperature changes, presence of toxic and chaotropic compounds, exposure to ultraviolet radiation, desiccation conditions and many other physical and chemical agents (Ramos et al., 2002). Cellular membranes constitute the first physical barrier against physicochemical challenges, and are provided with a series of sensors that trigger a succession of responses to overcome damage caused by noxious agents. One such response is the alteration of membrane lipid composition, an observation made more than 30 years ago by Sinensky (1974), who proposed the term ‘homeoviscous adaptation’ to refer to these changes.
A number of environment protection agencies have declared organic solvents as hazardous pollutants (http://www.epa.gov; European Union's Guideline 1999/13/EG). These chemicals are inherently toxic, and when they reach soils and waters, they affect the biodiversity of specific niches (Huertas et al., 2000). A limited number of microbes survive exposure to solvents, and the molecular mechanisms of solvent resistance have been partially elucidated in Gram‐negative bacteria (Sikkema et al., 1995; Segura et al., 1999; Ramos et al., 2002). Some bacterial strains belonging to the genus Pseudomonas (as P. putida DOT‐T1E or P. putida S12) are extremely tolerant to organic solvents and extrusion of toxic compounds via a series of efflux pumps has been proposed to be the main mechanism of solvent tolerance (Isken and de Bont, 1996; Kieboom et al., 1998; Kim et al., 1998; Ramos et al., 1998; Rojas et al., 2001; Rodriguez‐Herváet al., 2007); however, membrane modifications play an important role in the response to toluene and other solvents as well as the response to other environmental stresses such as acid stress or temperature shifts (Pinkart et al., 1996; Chang and Cronan, 1999; Junker and Ramos, 1999; Loffhagen et al., 2001; Kim et al., 2005; Bernal et al., 2007a).
Changes in the overall degree of fatty acid saturation or in the cis–trans isomerization of unsaturated fatty acids have been directly implicated in the cellular responses to fluidizing agents such as temperature or organic solvents (Heipieper et al., 1992; Sikkema et al., 1995; Weber and de Bont, 1996; Ramos et al., 1997; Junker and Ramos, 1999; Härtig et al., 2005). The amount of trans unsaturated fatty acids in the P. putida membrane increased up to 10‐fold just 5 min after toluene addition, enhancing membrane rigidity (Junker and Ramos, 1999; Bernal et al., 2007b). However, less is known about the role in the response to these environmental challenges of other membrane phenomena such as the modification of phospholipid head groups or the formation of cyclopropane fatty acids (CFA) (Ramos et al., 1997; Bernal et al., 2007a). Cyclopropane fatty acids have been proposed to be involved in the resistance of some γ‐proteobacteria to certain stresses such as low pH (Chang and Cronan, 1999; Kim et al., 2005) or freeze‐drying conditions (Muñoz‐Rojas et al., 2006). However, little is known about the effect of changes in cyclopropane levels in response to organic solvents. The CFAs are produced by a postsynthetic modification of cis unsaturated fatty acids, which involves the addition of a methylene group across the double bound of these unsaturated fatty acids (Grogan and Cronan, 1997). This reaction is catalysed by the enzyme cyclopropane synthase (Fig. 1A). In Escherichia coli and P. putida, CFAs accumulated during the late‐exponential and stationary phases of bacterial growth (Grogan and Cronan, 1997; Muñoz‐Rojas et al., 2006). In the solvent‐sensitive P. putida KT2440 genome, two different putative cyclopropane synthase genes (cfaA and cfaB) have been annotated (Nelson et al., 2002). A P. putida KT2440 cfaB mutant was shown to be unable to synthesize CFA and that a cfaA mutant had only slightly lower levels of CFAs than the parental strain (Muñoz‐Rojas et al., 2006).
Figure 1.
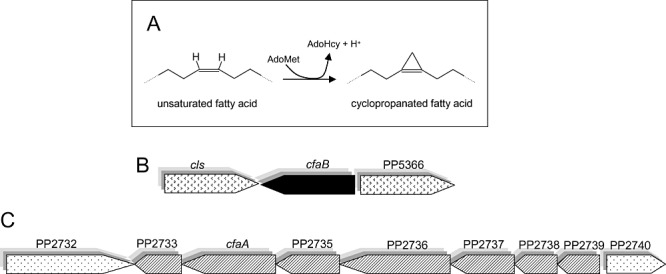
A. Reaction catalysed by the cyclopropane synthase. B. Schematic representation of the ORFs surrounding the cfaB gene in P. putida DOT‐T1E. cls stands for the cardiolipin synthase gene, PP5366 encoded for a putative dihydrolipoamide dehydrogenase 3. C. Schematic representation of the ORFs surrounding the cfaA gene in P. putida DOT‐T1E. PP2732, PP2733, PP2735 and PP2736 are hypothetical proteins; PP2737 encode for a putative oxidoreductase (short‐chain dehydrogenase/reductase family), PP2738 for a putative transcriptional regulator, PP2739 for a putative sensory box protein and PP2740 for a putative transcriptional regulator of the MerR family.
In this study, we identified the cfaA and cfaB genes in P. putida strain DOT‐T1E. Pseudomonas putida DOT‐T1E is a highly solvent‐tolerant strain that was isolated from a waste water treatment plant in Granada (Ramos et al., 1995). This strain is able to grow in the presence of high concentrations of toluene (up to 90% toluene [v/v]) and it is also able to degrade toluene via the toluene dioxygenase pathway (Mosqueda et al., 1999). This is a model strain regarding solvent tolerance not only because of its high tolerance but because of the accumulated data about the molecular mechanisms of solvent tolerance (Ramos et al., 2002; Rodriguez‐Herváet al., 2007). The cfaB gene product seems to be the only one responsible for the synthesis of CFA. Expression of the cfaB gene was shown to be RpoS‐dependent. We generated a cfaB mutant of P. putida DOT‐T1E and studied its response to solvents, low pH and other stresses such as temperature shifts, high osmolarity and the presence of antibiotics or heavy metals in the medium. The mutant was more sensitive to toluene shocks than the wild‐type strain, but in contrast to E. coli and Salmonella enterica, the P. putida cfaB mutant was as tolerant to acid shock as the wild‐type strain.
Results
CfaB is the main cyclopropane synthase in P. putida DOT‐T1E
Total chromosomal DNA of P. putida DOT‐T1E was hybridized against cfaA and cfaB gene probes generated by PCR amplification using oligonucleotides based on the cfaA and cfaB gene sequence of P. putida KT2440. We found that the cfaA gene hybridized to a single 20 kb fragment PpuMI or 3.5 kb XcmI fragment, whereas the cfaB gene hybridized to an EcoRI fragment of about 14 kb or a BamHI fragment of about 12 kb in the DNA of the solvent‐tolerant DOT‐TIE strain. We subsequently amplified and sequenced the complete cfaA and cfaB genes from the chromosome of P. putida DOT‐T1E. Translation of the nucleotide sequences into amino acid sequences revealed 98.7% identity between CfaB proteins of P. putida KT2440 and DOT‐T1E, and 99% identity between CfaA proteins in the two organisms, although the translated cfaA in DOT‐T1E is 26 amino acid longer than that deduced from the KT2440 genome sequence. An in silico analysis of the chromosomal region around the cfaA and cfaB genes in P. putida KT2440 suggested that the cfaB gene is transcribed as a monocistronic unit, as it is divergent from the 3′ downstream gene and convergent with the gene located in 5′ (Fig. 1B). In contrast, the originally annotated cfaA gene is in a cluster of seven open reading frames that are transcribed in the same direction (Fig. 1C). This gene organization is identical in both the solvent‐sensitive P. putida KT2440 and solvent‐tolerant P. putida DOT‐T1E strains. We have shown with RT‐PCR that cluster in P. putida DOT‐T1E is transcribed as a single operon (data not shown). The functions of the gene products of this operon are unknown.
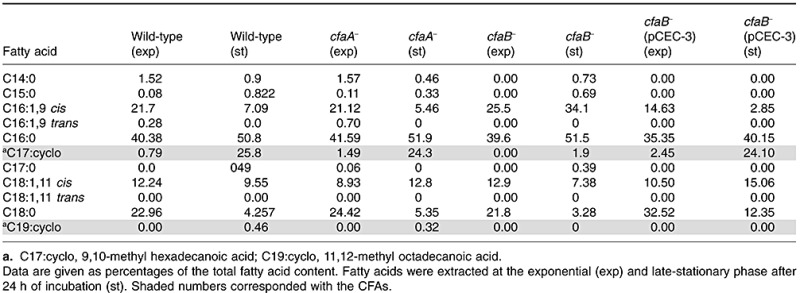
To assign a function to each of the genes in P. putida DOT‐T1E, knockout mutants of the two putative cfa genes were constructed as described in Experimental procedures, and fatty acid profiles were established in cells in the early‐exponential and stationary phases. In the wild‐type strain cyclopropane levels were higher in the stationary phase of growth than in the exponential phase (Table 1). As expected, relative levels of the precursor unsaturated fatty acids (C16:1.9 cis for C17:cyclo and C18:1.11 cis for C19:cyclo) diminished as cyclopropane levels increased. The cfaA mutant contained almost the same amount of CFA as the wild‐type strain, whereas CFA levels in the cfaB mutant were undetectable (Table 1). A double cfaA cfaB mutant had the same fatty acid profile as the cfaB‐ strain (data not shown). To verify that cfaB was responsible for CFA synthesis we complemented the cfaB mutant with plasmid pCEC‐3, which carries the cfaB gene. The results showed that in the complemented strain the pattern of fatty acids in the stationary phase was similar to that in the parental strain. These data support that the cfaB gene product is the one responsible for CFA synthesis in P. putida DOT‐T1E.
Table 1.
Fatty acid composition of P. putida DOT‐T1E and its cfaA and cfaB mutant derivatives.
The cfaB mutant is more sensitive to organic solvent shocks than the wild‐type strain
Cyclopropane fatty acids have been implicated in the resistance to different stresses in E. coli, S. enterica serovar Thyphimurim and P. putida KT2440 (Chang and Cronan, 1999; Kim et al., 2005; Muñoz‐Rojas et al., 2006). To study the role of CfaB in stress resistance in P. putida DOT‐T1E, cultures in the stationary phase (OD ≈ 2.5–3.0) of the wild‐type and cfaB mutant were exposed to different stress conditions and survival was compared. We found that survival of the cfaB mutant strain was nearly 1.5 orders of magnitude lower than in the wild type when exposed to 0.3% (v/v) toluene (Fig. 2). Complementation of the cfaB mutation by pCEC‐3 restored survival to levels similar to those in the wild‐type strain (Fig. 2). Increased organic solvent sensitivity of the cfaB mutant was confirmed when solvent shock was triggered with 0.3% (v/v) xylene, ethylbenzene or propylbenzene instead of toluene. Survival rate of the cfaB mutant was correlated with the theoretical toxicity of the hydrocarbons (that is related with the log Pow value) and was lower than that of the wild‐type strain (Table 2).
Figure 2.
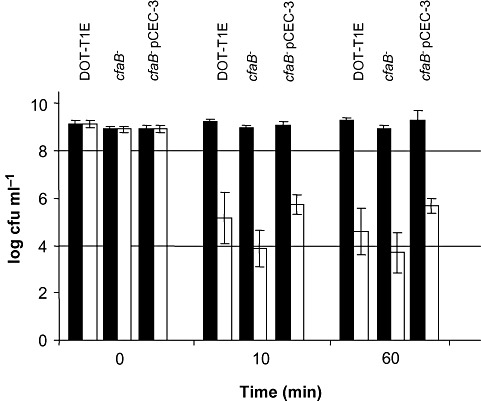
Survival of P. putida DOT‐T1E, its cfaB‐ derivative strain and the cfaB‐ pCEC‐3 complemented strain at 0, 10 and 60 min after the addition of 0.3% (v/v) toluene. Black bars, survival of control cultures; white bars, survival of cultures with toluene.
Table 2.
Survival of the P. putida DOT‐T1E and cfaB‐ cultures after the addition of 0.3% (v/v) m‐xylene, ethylbenzene or propylbenzene.
| log Pow | DOT‐T1E | cfaB‐ | ttgGHI‐ | ttgABC‐ | |
|---|---|---|---|---|---|
| None | 9.3 ± 0 | 9.3 ± 0.6 | 9.4 ± 0.1 | 9.2 ± 0.1 | |
| m‐xylene | 3.2 | 9.4 ± 0.7 | 7.0 ± 0.4 | 7.5 ± 0.3 | 6.6 ± 0.2 |
| Ethylbenzene | 3.4 | 8.8 ± 0.1 | 8.0 ± 0.2 | 6.3 ± 0.1 | 5.9 ± 0.3 |
| Propylbenzene | 3.6 | 9.3 ± 0.2 | 8.8 ± 0.1 | 8.9 ± 0.1 | 8.5 ± 0.1 |
Solvents were added when cultures reached the stationary phase, and cells were plated 10 min after the solvent was added. Survival is expressed as log cfu ml−1. Logarithm of the partition coefficient of an organic solvent in a mixture of octanol/water (log Pow) was used as a toxicity indicator. Organic solvents with log Pow between 1 and 4.5 are very toxic for microorganisms. Within this range, organic solvents with lower log Pow values are more toxic than those with higher values.
Aromatic organic acids such as p‐hydroxybenzoate or protocatechuate are tolerated up to a concentration of 25 mM without affecting survival rate in the wild type (Ramos‐González et al., 2001). This was also the case for the cfaB mutant strain with other aromatic organic acids such as benzoate or salicylate. In E. coli CFAs are involved in acid stress resistance in the stationary phase. To test whether this was the case in P. putida DOT‐T1E, HCl (to 20 mM) was added to cultures of the parental and cfaB mutant strains grown in Luria–Bertani (LB) until the stationary phase. We found that viability of both strains decreased by two orders of magnitude after 24 h. Then we run an experiment where cells grown on minimal medium supplemented with glucose until stationary phase were harvested and HCl was added to obtain pHs of 6, 5.5, 5, 4.5, 4 and 3.5. There was no difference between the wild type and mutant (100% survival at pH 6, 5.5, 5 and 4, and no survival at pH 3.5).
Mutant cfaB cells showed no differences in survival rates compared with the wild type when cultures at the late‐exponential growth phase were exposed to temperature shifts from 30°C to 16°C or from 30°C to 42°C. The mutant was also not more sensitive to a number of antibiotics and heavy metals than the wild type.
The series of results presented above suggested that the cfaB gene product is specifically involved in organic solvent stress, but not in a general stress response.
Transcriptional analysis of the cfa genes
The higher levels of CFA during the stationary phase suggested that cfaB expression might be governed by the RpoS sigma factor. To test this hypothesis we first determined the transcriptional start point of the cfaB gene, which was found 53 nucleotides upstream from the ATG start codon (Fig. 3). Around the −10 region we found a sequence that resembled that proposed to be recognized by RNA polymerase/σ38 (5′‐CTACTCT‐3′). To further study the expression of the cfaB gene we fused the cfaB promoter region to 'lacZ to yield plasmid pMPcfaB. Figure 4 shows that expression of the cfaB gene was clearly dependent on the growth phase, with a sharp increase in expression when the culture entered the late‐exponential phase and persistently high levels throughout the stationary phase. The response of the cfaB promoter was totally different to that of the promoter of the efflux pump TtgGHI (Fig. 4) in which no increase in expression was observed during stationary phase. This increase in expression, together with the finding of a −10 region with high similarity to a consensus sequence for σ38, indicated that expression of the cyclopropane synthase gene was tightly controlled by RpoS. To verify the role of RpoS in the expression of cfaB we introduced plasmid pMPcfaB into a P. putida rpoS ‐deficient strain. Expression of the cfaB promoter was completely shut down in this genetic background (not shown), confirming that RpoS is the sigma factor responsible for transcription of the cfaB gene of P. putida DOT‐T1E. At the same time as expression of β‐galactosidase was measured, we determined CFA levels in the cells, and found that in the wild‐type strain CFAs became detectable as cfaB expression increased, whereas no C17:CFA was detected in the rpoS mutant background at any time.
Figure 3.
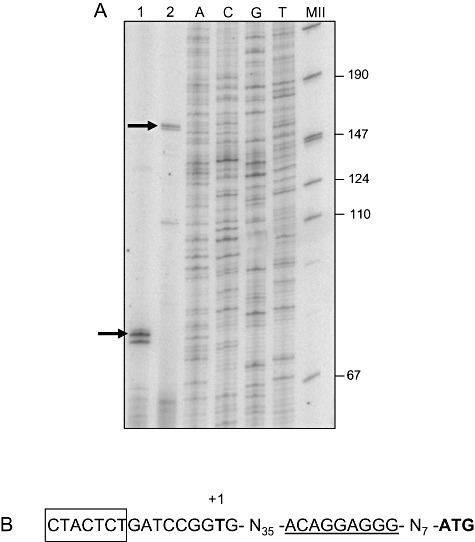
Mapping of the transcription start point of the cfaB gene. A. Primer extension analysis. Lane 1, primer extension using primer p100; lane 2, primer extension using primer p180. B. The + 1 position in the promoter region is in boldface; the putative σ38 recognition sequence is boxed.
Figure 4.
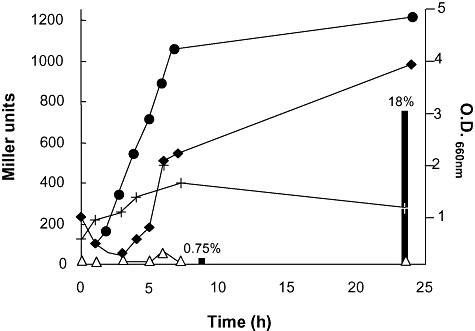
Expression of the cfaB promoter (measured as β‐galactosidase activity) along the P. putida DOT‐T1E growth curve. Circles indicate culture growth. Black diamonds, white triangles and crosses indicate β‐galactosidase activity of pMcfaB, pMP220 without promoter and pMP220 with PttgG::lacZ (promoter of ttgGHI effux pump; Rojas et al., 2003) respectively. Black bars show the percentage of CFAs in the membranes.
cfaB expression under different stress conditions
To determine whether the expression of cfaB was influenced by the above stress conditions, we used a PcfaB:′lacZ fusion. Table 3 shows that in the stationary phase cfaB expression occurred at around 500 Miller units, a level of expression that was not significantly influenced by the presence of toluene or propylbenzene. The addition of HCl to 20 mM did not affect the level of β‐galactosidase either (not shown). However, the addition of 15 mM benzoate led to a 2.5‐fold increase in β‐galactosidase levels (Table 3). No differences in expression were observed when the plasmid was introduced in the rpoS mutant or when the assay was done with the plasmid without promoter; that is, activity levels were always below 40 Miller units (not shown). These results suggested that organic acids rather than inorganic acids were able to induce cfaB expression. To further test this hypothesis we analysed the expression of the promoter when different organic acids were added to the culture medium. None of the linear organic acids (mono‐ and di‐carboxylic) increased expression from the cfaB promoter; however, several aromatic acids such us phenylacetate, salicylate and 2‐ or 3‐toluate led to a modest increase in expression (Table 3). cfaB expression did not change after shifting cells from 30°C to 42°C or to 16°C.
Table 3.
Transcription from the cfaB promoter determined as β‐galactosidase activity using a fusion of the cfaB promoter to ′lacZ.
| Compound | Miller units |
|---|---|
| None | 520 ± 15 |
| Acetate | 500 ± 10 |
| Citrate | 350 ± 5 |
| Succinate | 500 ± 20 |
| Lactate | 260 ± 5 |
| Protocatechuate | 460 ± 30 |
| Benzoate | 1170 ± 20 |
| Phenylacetate | 1290 ± 30 |
| Salicylate | 1050 ± 40 |
| 2‐Toluate | 1230 ± 45 |
| 3‐Toluate | 1210 ± 35 |
| 4‐Toluate | 750 ± 5 |
| 3‐Hydroxybenzoate | 880 ± 35 |
Bacterial cultures were grown until they reached a turbidity at an 660 nm of 2.5–3.0. β‐Galactosidase assays were determined as described by Miller (1972) 3 h after acid addition. Data are average values of three independent assays. All compounds were added to reach a final concentration of 15 mM except for protocatechuate, which was used at a final concentration of 5 mM.
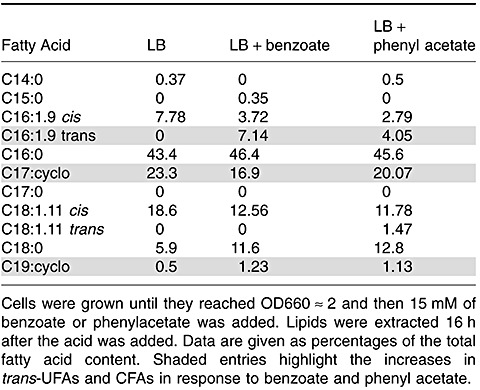
Based on the expression results, we decided to test whether cells growing in the presence of benzoate or phenylacetate would exhibit higher levels of CFA than cells growing only on LB. The results revealed that the modest increase in cfaB expression in the presence of organic acids did not translate into higher levels of CFA, probably because under these conditions the cis–trans isomerase was also competing for the same substrate (Table 4).
Table 4.
Fatty acid composition of P. putida DOT‐T1E membranes grown in the presence of benzoate or phenylacetate.
Membrane fluidity
As described above, lipid membrane composition is an important determinant in tolerance towards different stresses, especially in response to those that affect membrane fluidity. The role of CFA in membrane fluidity has not been studied in detail. To determine membrane fluidity in the wild‐type and cfaB mutant strains, we analysed fluorescence polarization values when the cultures were incubated with the dye 1,6‐diphenyl‐1,3,5‐hexatriene. The P values for all cultures were 0.25 ± 0.04, indicating that there were no significant differences in membrane fluidity between the wild‐type and mutant strains.
Discussion
Although CFAs are present in bacterial cytoplasmic membranes at high levels during the stationary phase of growth (when they can represent up to 25% of the total fatty acid content), their physiological role has not been clearly established. The CFAs are nonessential components of the membranes, at least under laboratory conditions, as mutants unable to synthesize CFAs are viable. Although in P. putida DOT‐T1E two putative open reading frames may encode a cyclopropane synthase, our results suggest that the cfaB gene product is the main enzyme involved in cyclopropane synthesis. Although we subjected the cfaA mutant to the same stresses as the cfaB mutant we, did not obtain any distinctive phenotype for the cfaA mutation. Our results also show that the lack of CFA in the membranes of the P. putida DOT‐T1E cfaB mutant increased toluene sensitivity as determined by the diminished viability (by 1.5 orders of magnitude) of mutant cultures with respect to the wild type. The decrease in solvent tolerance has also been documented when other enzymes responsible for the synthesis of membrane components (such as cis–trans isomerase or cardiolipin synthase) were knocked out (Junker and Ramos, 1999; Bernal et al., 2007a). The fact that the cfaB mutant cultures did not totally loose viability after solvent shock indicates that CFAs are involved in, but not essential for, solvent tolerance. Thus far the viability of mutant cultures after exposure to toluene (log Pow 2.4) has been reported to be lost completely only when some of the efflux pumps directly involved in solvent extrusion have been knocked out (Rojas et al., 2001). Other organic solvents with log Pow values higher than that of toluene are less toxic for bacterial cells and viability of cultures exposed to a sudden shock with these solvents remained high even in the efflux pump mutants (Table 2). Our results confirmed the toxic effect of these solvents, with a higher decrease in solvent tolerance in the cfaB mutant when m‐xylene was added to the culture, subtle effects on cfaB survival with ethylbenzene and almost no differences with propylbenzene.
The involvement of CFA in solvent tolerance has been described in the anaerobic bacteria Clostridium acetobutylicum ATCC 824 (Zhao et al., 2003); overexpression of cyclopropane synthase in this microorganism increased butanol tolerance; however, it decreased butanol production. In C. acetobutylicum butanol was able to increase the CFA content in membranes (Vollherbst‐Schneck et al., 1984), whereas in P. putida DOT‐T1E no increase in CFA content was detected after toluene was added, and cfa gene expression was not induced in the presence of the solvent.
We found no other differences between the wild type and the cfaB mutant with regard to resistance to other stresses. Brown and colleagues (1997) reported a correlation between E. coli strains with a high degree of intrinsic acid tolerance and higher CFA content in the membranes of these strains. In this connection it was also found that cfa null mutants of E. coli and S. enterica were more sensitive to acid stress than the corresponding wild‐type strain (Chang and Cronan, 1999; Kim et al., 2005). We found no pH at which survival of the P. putida DOT‐T1E cfaB mutant was compromised in comparison with the wild‐type strain, suggesting that in P. putida CFA may not be related with survival after acid stress. In E. coli and S. enterica expression of the cfa gene is driven from two promoters (one dependent on σ70 and another on σ38); however, in P. putida DOT‐T1E cfaB gene expression is mediated by RNA polymerase only with σ38. Expression of the cfa gene in E. coli increased in response to short‐chain carboxylic acids such as acetate, which is present in its natural gut niche (Arnold et al., 2001); in contrast with E. coli, short‐chain organic acids did not induce cfaB gene of P. putida DOT‐T1E, although aromatic carboxylic acids did induce P. putida DOT‐T1E cfaB gene expression. Whether the response to organic and inorganic acids reflects different regulatory mechanisms of cyclopropane synthase in pseudomonadaceaes and enteric bacteria remain to be elucidated. The role of CFA in survival of P. putida under environmental conditions is an area that deserves further research, in particular in the rhizosphere of plants, a niche in which P. putida proliferates and in which a large number of aromatic carboxylic acids derived from lignin breakdown are present.
Interestingly, although the cfaB gene in pCEC‐3 was expressed from a plasmid promoter, the CFA levels observed during the exponential phase were only slightly higher than when cfaB was expressed from its own promoter. This indicates that some other regulatory mechanisms operate to control the levels of CFAs in the membranes. In E. coli CFA synthase is unstable because of RpoH‐dependent proteolysis (Chang et al., 2000), and in P. putida rpoH expression is induced in the presence of alkylbenzoates (Domínguez‐Cuevas et al., 2006), so increased cyclopropane synthase degradation might explain the discrepancy between cfaB expression and CFA content in the membranes. Proteolysis of cyclopropane synthase has been suggested to be an important mechanism to avoid consumption of S‐adenosyl‐methionine in the exponential phase during which this compound is required for the synthesis of other molecules. The complexity of the regulation of CFA might be related not only to membrane physical properties, but it might also respond to a general metabolic regulation.
Experimental procedures
Bacterial strains and growth conditions
Pseudomonas putida DOT‐T1E and its mutant derivatives were routinely grown on LB or M9 minimal medium supplemented with the carbon sources indicated in the text (Abril et al., 1989). Cultures were incubated at 30°C and shaken on an orbital platform operating at 200 strokes per minute. The stationary phase was normally reached after 5 h of growth (OD660 ≈ 2.5–3).
Construction of plasmid and mutant strains
The P. putida DOT‐T1E cfaB gene was amplified from chromosomal DNA using oligonucleotides Kpncfa2b (5′‐TAGGTACCGTAGGCGTCGTGGCCAATTA‐3′) and EcoRIcfa (5′‐TGAATTCGGCCGCTCCTGCACAACC‐3′), and digested with EcoRI and KpnI. This DNA was ligated to pBBR1MCS‐5 (Kovach et al., 1995), which had been previously cut with the same enzymes. The resulting plasmid, named pCEC‐3, was transformed in the P. putida DOT‐T1E and P. putida DOT‐T1E cfaB‐ strains. The resulting gentamicin‐resistant colonies were checked for the presence of the plasmid, and one clone of each transformation was selected for further analysis.
To construct a cfaB mutant strain, a small fragment (706 bp) of the cfaB gene was amplified using oligonucleotides CFA1 (5′‐ATGCTTGCTCAACTTCCACC‐3′) and CFA2‐6 (5′‐GTATGGGTGCGACTGTTGGA‐3′). This fragment was cloned into the pMBL‐T vector (Dominion MBL, Spain) to obtain pANA247, which was cut with BamHI, and the 2.2 kb BamHI fragment (containing the Ω::km cassette) of pHP45Ω‐Km (Prentki and Krisch, 1984) was ligated in the linearized plasmid. The resulting plasmid (pANA247K) was introduced into P. putida DOT‐T1E by electroporation (Enderle and Farwell, 1998), and cultures were plated onto LB medium plus kanamycin to select for the single homologous recombination event that disrupts cfaB by incorporating the plasmid into the chromosome. A cfaA mutant strain was constructed as in Muñoz‐Rojas et al. (2006), but using P. putida DOT‐T1E as a recipient strain. To construct the double mutant (P. putida DOT‐T1E cfaA‐cfaB‐) we excised the Ω::Km cassette of plasmid pANA247 with BamHI and replaced it with the GmR cassette of pMS255 (Ko and Park, 2000) previously recovered with BamHI. The new plasmid (pCEC‐1) was electroporated into P. putida DOT‐T1E cfaA‐ strain and gentamicin‐resistant clones were selected. All mutants were checked by Southern blot (not shown).
Survival after stress treatment
Cultures of the wild‐type and mutant strains were grown in LB medium until the stationary phase (OD660 ≈ 2.5). Then cultures were divided into two halves, one of which was treated with the stress agent [temperature shift from 30°C to 42°C or 16°C, addition of 15 mM benzoate, 0.7 M NaCl, 20 mM HCl or 0.3% (v/v) organic solvent] while the other one was kept as a control. Serial dilutions of the cultures were plated onto LB after 30 min, 1 h and 4 h to determined survival rates. In the assays with organic solvents, cells were plated after 10, 30 and 60 min; data represent the media of at least three independent experiments. Acid resistance was also tested as described by Chang and Cronan (1999). Briefly, cells of cultures growing in M9 minimal medium supplemented with glucose in the stationary phase were harvested and resuspended in M9 minimal medium. Hydrochloric acid was added until pH of the cultures reached 6, 5.5, 5, 4.5, 4 or 3.5 and survival was measured after incubation for 1 and 3 h at 30°C.
Fatty acid analysis
Phospholipids were extracted as described by Bligh and Dyer (1959) and trans‐esterified as described before (Bannon et al., 1982). After gas chromatographic separation, the fatty acids were identified by mass spectrometry. Data presented in tables are representative of three experiments.
β‐Galactosidase assays
The cfaB promoter was amplified using oligonucleotides PstIcfaB2 (5′‐AACTGCAGAGCGGCAGATGCAA‐3′) and EcoR1cfaB2 (5′‐GGAATTCGCTGGCAGCTTCGT‐3′). The fragment was then cut with PstI and EcoRI and cloned into pMP220 (Spaink et al., 1987) previously cut with the same enzymes to construct plasmid pMPcfaB. This plasmid was electroporated into P. putida DOT‐T1E and P. putida DOT‐T1E rpoS. Cultures were grown overnight on LB medium plus tetracycline, and on the following morning they were diluted to an OD660 of 0.1. When the cultures reached the stationary phase (OD ≈ 3.0) they were exposed to different stressors, for example, 37°C or 16°C, 20 mM HCl, 15 mM benzoate, 15 mM phenylacetate, 25 mM protecatechuate, 25 mM p‐hydroxybenzoate, 5 mM propylbenzene, 15 mM succinate, 15 mM acetate and 15 mM citrate, and 0.075% (v/v) toluene. β‐Galactosidase activity was assayed 1, 3 and 20 h after stress treatment.
Fluorescence anisotropy measurements
Membrane fluidity was determined by measuring fluorescence polarization of the dye 1,6‐diphenyl‐1,3,5‐hexatriene probe inserted into the lipid bilayer of the cytoplasmic membranes. Experiments were done as described by Bernal and colleagues (2007a) except that cultures were harvested during the early stationary phase.
Primer extension analysis
Cells of P. putida DOT‐T1E grown overnight in LB medium were diluted 1:100 in the same medium. After 12 h of incubation, 15 ml samples were harvested by centrifugation and RNA was extracted as described before (Aranda‐Olmedo et al., 2005). Primer extension analysis of 100 µg of total RNA samples was carried out as described previously (Marqués et al., 1993) using 32P‐end‐labelled oligonucleotides. For this step we used oligonucleotides p180 (5′‐GAATGGTGACCTGAGGACTA‐3′) and p100 (5′‐CAATGCAGGTGGAAGTTGAG‐3′), which were complementary to the coding strands within the cfaB gene. cDNAs were run in urea sequencing gels, and gels were exposed to a phosphor‐imaging screen (Fuji Photo Film) for 5–12 h. Phosphor‐imaging screens were scanned with a phosphor‐imaging instrument (Molecular imager FX; Bio‐Rad).
Gene accession number
The sequence of the P. putida DOT‐T1E cfaB gene has been deposited under number DQ665843. The sequence of the P. putida DOT‐T1E cfaA and surrounding genes has been deposited under number DQ665844.
Acknowledgments
This study was supported grants from the CICYT (BIO2006‐05668) and the Junta de Andalucía (CVI‐344), and ERA‐NET PSYSMO (GEN2006‐27750‐C5‐5E/SYS). C.V.P. is the recipient of a predoctoral fellowship from the Cátedras Volantes program of the Santander‐Central‐Hispano Bank and CSIC. Pseudomonas putida DOT‐T1E rpoS‐ was a gift from Dr M. I. Ramos‐Gonzalez. We thank K. Shashok for improving the use of English in the manuscript.
References
- Abril M.A., Michán C., Timmis K.N., Ramos J.L. Regulator and enzyme specificities of the TOL plasmid‐encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda‐Olmedo I., Ramos J.L., Marqués S. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl Environ Microbiol. 2005;71:4191–4198. doi: 10.1128/AEM.71.8.4191-4198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C.N., McElhanon J., Lee A., Leonhart R., Siegele D.A. Global analysis of Escherichia coli gene expression during the acetate‐induced acid tolerance response. J Bacteriol. 2001;183:2178–2186. doi: 10.1128/JB.183.7.2178-2186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon C.D., Breen G.J., Craske J.D., Hai N.T., Harper N.L., O'Rourke K.L. Analysis of fatty acid methyl esters with high accuracy and readability. III. Literature review of an investigation into the development of a rapid procedure for methoxide catalysed methanolysis of fats and oils. J Chromatogr. 1982;247:71–89. [Google Scholar]
- Bernal P., Muñoz‐Rojas J., Hurtado A., Ramos J.L., Segura A. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ Microbiol. 2007a;9:1135–1145. doi: 10.1111/j.1462-2920.2006.01236.x. [DOI] [PubMed] [Google Scholar]
- Bernal P., Segura A., Ramos J.L. Compensatory role of the cis‐trans isomerase and cardiolipin synthase in the membrane fluidity of Pseudomonas putida DOT‐T1E. Environ Microbiol. 2007b;9:1658–1664. doi: 10.1111/j.1462-2920.2007.01283.x. [DOI] [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Ross T., McMeekin T.A., Nichols P.D. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int J Food Microbiol. 1997;37:163–173. doi: 10.1016/s0168-1605(97)00068-8. [DOI] [PubMed] [Google Scholar]
- Chang Y.‐Y., Cronan J.E. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol. 1999;33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- Chang Y.‐Y., Eichel J., Cronan J.E., Jr Metabolic instability of Escherichia coli cyclopropane fatty acid synthase is due to RpoH‐dependent proteolysis. J Bacteriol. 2000;182:4288–4294. doi: 10.1128/jb.182.15.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez‐Cuevas P., González‐Pastor J.E., Marqués S., Ramos J.L., De Lorenzo V. Transcriptional tradeoff between metabolic and stress‐response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem. 2006;281:11981–11991. doi: 10.1074/jbc.M509848200. [DOI] [PubMed] [Google Scholar]
- Enderle P.J., Farwell M.A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques. 1998;25:954–958. doi: 10.2144/98256bm05. [DOI] [PubMed] [Google Scholar]
- Grogan D.W., Cronan J.E., Jr Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev. 1997;61:429–441. doi: 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper H.J., Diefenbach R., Keweloh H. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol‐degrading Pseudomonas putida P8 from substrate toxicity. Appl Environ Microbiol. 1992;58:1847–1852. doi: 10.1128/aem.58.6.1847-1852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas M.J., Duque E., Molina L., Rosselló‐Mora R., Mosqueda G., Godoy P. Tolerance to sudden organic solvent shocks by soil bacteria and characterization of Pseudomonas putida strains isolated from toluene polluted sites. Env Sci Technol. 2000;34:3395–3400. et al. [Google Scholar]
- Härtig C., Loffhagen N., Harms H. Formation of trans fatty acids is not involved in growth‐linked membrane adaptation of Pseudomonas putida. Appl Environ Microbiol. 2005;71:1915–1922. doi: 10.1128/AEM.71.4.1915-1922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken S., De Bont J.A.M. Active efflux of toluene in a solvent‐resistant bacterium. J Bacteriol. 1996;178:6056–6058. doi: 10.1128/jb.178.20.6056-6058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker F., Ramos J.L. Involvement of the cis/trans isomerase Cti in solvent resistance of Pseudomonas putida DOT‐T1E. J Bacteriol. 1999;181:5693–5700. doi: 10.1128/jb.181.18.5693-5700.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieboom J., Dennis J.J., De Bont J.A.M., Zylstra G.J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- Kim B.H., Kim S., Kim H.G., Lee J., Lee I.S., Park Y.K. The formation of cyclopropane fatty acids in Salmonella enterica serovar. Typhimurium Microbiol. 2005;151:209–218. doi: 10.1099/mic.0.27265-0. [DOI] [PubMed] [Google Scholar]
- Kim K., Lee S., Lee K., Lim D. Isolation and characterization of toluene‐sensitive mutants from the toluene‐resistant bacterium Pseudomonas putida GM73. J Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Park C. Two novel flagellar components and H‐NS are involved in the motor function of Escherichia coli. J Molecular Biology. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., II, Peterson K.M. Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Loffhagen N., Härtig C., Babel W. Pseudomonas putida NCTC 10936 balances membrane fluidity in response to physical and chemical stress by changing the saturation degree and the trans/cis ratio of fatty acids. Biosci Biotechnol Biochem. 2001;68:317–323. doi: 10.1271/bbb.68.317. [DOI] [PubMed] [Google Scholar]
- Marqués S., Ramos J.L., Timmis K.N. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ region. Biochim Biophys Acta. 1993;1216:227–237. doi: 10.1016/0167-4781(93)90149-8. [DOI] [PubMed] [Google Scholar]
- Miller J.H. Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mosqueda G., Ramos‐González M.I., Ramos J.L. Toluene metabolism by the solvent‐tolerant Pseudomonas putida DOT‐T1 strain and its role in solvent impermeabilization. Gene. 1999;232:69–76. doi: 10.1016/s0378-1119(99)00113-4. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Rojas J., Bernal P., Duque E., Godoy P., Segura A., Ramos J.L. Involvement of C17:cyclopropane fatty acid in the response of Pseudomonas putida KT2440 to freeze‐drying. Appl Environ Microbiol. 2006;72:472–477. doi: 10.1128/AEM.72.1.472-477.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K.E., Weinel C., Paulsen I.T., Dodson R.J., Hilbert H., Martins dos Santos V.A. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. et al.. Erratum in: Environ Microbiol. 2003, 5: 630. [DOI] [PubMed] [Google Scholar]
- Pinkart H.C., Wolfram J.W., Rogers R., White D.C. Cell envelope changes in solvent‐tolerant and solvent‐sensitive Pseudomonas putida strains following exposure to o‐xylene. Appl Environ Microbiol. 1996;62:1129–1132. doi: 10.1128/aem.62.3.1129-1132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H.M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ramos‐González M.I., Godoy P., Alaminos M., Ben‐Basset A., Ramos J.L. Physiological characterization of Pseudomonas putida DOT‐TIE tolerance to p‐hydroxybenzoate. Appl Environ Microbiol. 2001;67:4388–4341. doi: 10.1128/AEM.67.9.4338-4341.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Huertas M.J., Haïdour A. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J Bacteriol. 1995;177:3911–3916. doi: 10.1128/jb.177.14.3911-3916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Rodríguez‐Herva J.J., Godoy P., Haïdour A., Reyes F., Fernández‐Barrero A. Mechanisms for solvent tolerance in bacteria. J Biol Chem. 1997;272:3887–3890. doi: 10.1074/jbc.272.7.3887. [DOI] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Godoy P., Segura A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT‐T1E. J Bacteriol. 1998;180:3323–3329. doi: 10.1128/jb.180.13.3323-3329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Duque E., Gallegos M.T., Godoy P., Ramos‐Gonzalez M.I., Rojas A. Mechanisms of solvent tolerance in gram‐negative bacteria. Ann Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. et al. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Hervá J.J., García V., Hurtado A., Segura A., Ramos J.L. The ttgGHI solvent efflux pump operon of Pseudomonas putida DOT‐T1E is located on a large self‐transmissible plasmid. Environ Microbiol. 2007;9:1550–1561. doi: 10.1111/j.1462-2920.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- Rojas A., Duque E., Mosqueda G., Golden G., Hurtado A., Ramos J.L., Segura A. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT‐T1E. J Bacteriol. 2001;183:3967–3973. doi: 10.1128/JB.183.13.3967-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., Segura A., Guazzarone M.E., Terán W., Hurtado A., Gallegos M.T., Ramos J.L. In vivo and in vitro evidence that TtgV is the specific regulador of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J Bacteriol. 2003;185:4755–4763. doi: 10.1128/JB.185.16.4755-4763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura A., Duque E., Mosqueda G., Ramos J.L., Junker F. Multiple responses of Gram‐negative bacteria to organic solvents. Environ Microbiol. 1999;1:191–198. doi: 10.1046/j.1462-2920.1999.00033.x. [DOI] [PubMed] [Google Scholar]
- Sikkema J., De Bont J.A.M., Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M. Homeoviscous adaptation – a homeostatic process that regulates the viscosity of the membrane lipids in Escherichia coli. Proc Natl Acad Sci USA. 1974;71:522–525. doi: 10.1073/pnas.71.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H.P., Okker R.J.H., Wijffelman C.A., Pees E., Lugtenberg B.J.J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- Vollherbst‐Schneck K., Sands J.A., Montenecourt B.S. Effect of butanol on lipid composition and fluidity of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1984;47:193–194. doi: 10.1128/aem.47.1.193-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F.J., De Bont J.A.M. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim Biophys Acta. 1996;1286:225–245. doi: 10.1016/s0304-4157(96)00010-x. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hindorff L.A., Chuang A., Monroe‐Augustus M., Lyristis M., Harrison M.L. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 2003;69:2831–2841. doi: 10.1128/AEM.69.5.2831-2841.2003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


