Summary
The tetralin biodegradation pathway in Rhodococcus sp. strain TFB, a Gram‐positive bacterium resistant to genetic manipulation, was characterized using a proteomic approach. Relative protein expression in cell free extracts from tetralin‐ and glucose‐grown cells was compared using the 2D‐DIGE technique. Identification of proteins specifically expressed in tetralin‐grown cells was used to characterize a complete set of genes involved in tetralin degradation by reverse genetics. We propose a tetralin degradation pathway analogous to that described for Sphingomonas macrogolitabida strain TFA. TFB thn genes are organized into three operons; two contain all of the structural genes and are transcribed in the same direction, while the third operon, thnST, is transcribed in the opposite direction and encodes a two‐component regulatory system, whose transcription is higher in tetralin‐grown cells. In addition to tetralin induction, TFB thn structural genes are subject to glucose repression. Primer extension assays and translational thnA1::gfp and thnS::gfp fusions were used to characterize putative promoter regions. A mutational analysis of the thnA1 promoter region allowed us to define nucleotides within the cis regulatory elements that are important for the control of thn gene expression.
Introduction
Members of the Gram‐positive genus Rhodococcus are known to have important roles in biodegradation because of their broad metabolic diversity and ubiquity in contaminated environments (Bell et al., 1998). However, rhodococcal catabolic pathways have been less well characterized at the molecular level than those of other Gram‐negative bacteria involved in biodegradation, such as Pseudomonas. This is mainly due to the limited availability of genetic tools for Rhodococcus and the fact that some strains are resistant to genetic manipulation. The redundancy of metabolic pathways and genes in this genus, confirmed by analysis of the complete Rhodococcus sp. RHA1 genome sequence (van der Geize and Dijkhuizen, 2004; McLeod et al., 2006), complicates the elucidation of whole biodegradation pathways. As an alternative to genetic analysis, proteomics has been shown to be a powerful tool for high‐throughput analysis of rhodococcal metabolism (Navarro‐Llorens et al., 2005; Patrauchan et al., 2005; Tomás‐Gallardo et al., 2006).
Analysis of Rhodococcus genome sequences has identified a number of transcriptional regulators, the most abundant belonging to the LysR‐type and IclR‐like families. Several two‐component regulatory systems have also been found (see http://www.rhodococcus.ca). Some transcriptional regulators involved in regulating the degradation of aromatic compounds by rhodococcal strains have been characterized. In Rhodococcus erythropolis CCM2595, the IclR‐like transcriptional regulator CatR represses the expression of the catechol degradation operon (Veselýet al., 2007). In Rhodococcus sp. strains M5 (Labbéet al., 1997) and RHA1 (Takeda et al., 2004), a two‐component regulatory system (BphST) has been characterized as the transcriptional regulatory system for biphenyl degradation. The response regulator BphT is a member of the LuxR family of transcriptional regulators. Two‐component regulatory genes are also known to be involved in o‐xylene degradation by Rhodococcus sp. strain DK17 (Kim et al., 2005). However, very little is known about the molecular mechanism by which these regulatory systems control the activity of catabolic pathways in response to environmental signals.
Tetralin (1,2,3,4‐dihydronaphthalene) is a toxic organic solvent, which is composed of an aromatic and an alicyclic ring that share two carbon atoms. A number of bacterial strains able to grow aerobically on tetralin as the sole carbon and energy source have been described previously (Sikkema and de Bont, 1991). The tetralin catabolic pathway of Sphingomonas macrogolitabida strain TFA has been the best characterized at molecular and biochemical levels (Hernáez et al., 1999). In this pathway, the aromatic tetralin ring is initially modified by a ring‐hydroxylating dioxygenase and a dehydrogenase (Moreno‐Ruiz et al., 2003). The resulting catechol derivative is then further metabolized through reactions sequentially catalysed by an extradiol dioxygenase, a hydrolase, a hydratase and an aldolase (Andújar et al., 2000; Hernáez et al., 2000; Hernáez et al., 2002). This set of enzymes is able to cleave both the aromatic and the alicyclic rings of tetralin to render pyruvate and pimelic semialdehyde (Hernáez et al., 2002). Structural and regulatory TFA thn genes are clustered into two closely linked operons, which are transcribed in opposite directions (Hernáez et al., 1999; Moreno‐Ruiz et al., 2003). The regulatory system responsible for the induction of TFA thn genes in response to tetralin includes a LysR‐type transcriptional activator, ThnR and a ferredoxin reductase‐like protein, ThnY (Martínez‐Pérez et al., 2004). In addition, TFA thn genes are subjected to catabolite repression in the presence of preferential carbon sources (Martínez‐Pérez et al., 2004). An integrated response to several inducers through communication between the catabolic pathway and the regulatory system has also been recently reported in strain TFA. This additional level of regulation prevents efficient TFA thn gene induction by molecules that are not substrates of the catabolic pathway, thus preventing gratuitous induction (Martínez‐Pérez et al., 2007).
Previously, we have reported that growth on phthalate, tetralin or naphthalene induces the expression of specific proteins in Rhodococcus sp. strain TFB (Tomás‐Gallardo et al., 2006). In this paper, we extend our initial proteomic approach in order to characterize the tetralin catabolic pathway in Rhodococcus sp. strain TFB at the molecular level.
Results
Proteome analysis of tetralin‐ versus glucose‐grown TFB cells and identification of differentially expressed proteins
Qualitative and quantitative analysis of tetralin‐induced proteins was carried out using the Ettan DIGE fluorescent 2D gel electrophoresis system (GE Healthcare). Equal amounts of soluble protein extract (50 µg) from Rhodococcus sp. strain TFB grown on either glucose or tetralin were labelled with Cy5 or Cy3 dyes respectively. Proteins specifically expressed in tetralin‐ (green spots) or glucose‐grown cells (red spots) were detected after scanning (Fig. 1A). A total of 151 (or 13.5%) out of 1115 distinct spots showed quantitative differences in their expression between glucose‐ and tetralin‐grown cells. Of the 151 spots, 47 could be individually selected from the 2D‐DIGE gel for analysis by MALDI‐MS(MS) (maldi‐assisted laser desorption/ionization tandem mass spectrometry) or ESI‐IT MS/MS (electrospray ionization‐ion trap tandem mass spectrometry), which identified 16 different proteins (Table 1). Of these proteins, we identified eight with probable roles in the conversion of tetralin to a linear compound: the α and β subunits of a dioxygenase most similar to an ethylbenzene dioxygenase (ThnA1 and ThnA2; spots 7 and 12), a ferredoxin reductase (ThnA4, spot 5), a cis‐biphenyl‐2,3‐dihydrodiol‐2,3‐dehydrogenase (ThnB, spots 2 and 11), a catechol 2,3‐dioxygenase (ThnC; spots 8 and 9) and a 4‐(2‐oxocyclohexyl)‐2‐hydroxy‐buta‐2,4‐dienoic acid hydrolase (ThnD, spot 1). We also identified a protein (ThnU, spot 3) that may be involved in sterol transfer and is encoded by a gene previously linked to others involved in biodegradation (Kulakov et al., 2005; Maruyama et al., 2005). Two additional proteins, a beta‐ketoadipyl CoA thiolase (ThnI, spot 13) and an acyl‐CoA dehydrogenase (ThnV, spot 4), were also identified, which could be involved in further processing of the linear tetralin derivative via β‐oxidation.
Figure 1.
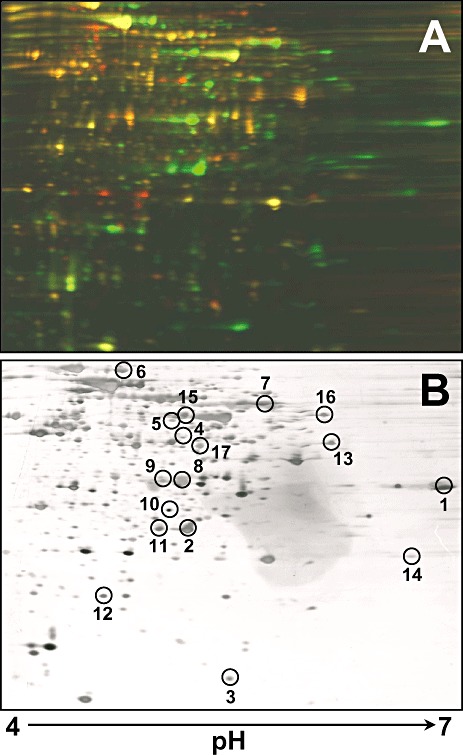
2D‐DIGE gel analysis of glucose‐ (red spots) versus tetralin‐ (green spots) grown cell proteomes (A) and silver staining of the same gel (B). Fifty micrograms of proteins from each condition was labelled with Cy3 or Cy5. As internal standard, 25 µg of each sample was mixed and labelled with Cy2. Numbered spots were digested in‐gel with trypsin and analysed by MALDI‐MS(/MS) or ESI‐IT for protein identification.
Table 1.
Identification of tetralin induced Rhodococcus sp. strain TFB proteins.
| (A) Proteins identified by MALDI‐MS(MS) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | MASCOT score | Accession code | Predicted protein | Organism | Mwa | pI | Peptide sequenceb | thn gene |
| 1 | 53 | gi|10567587 | 4‐(2‐Oxocyclohexyl)‐2‐hydroxy‐buta‐2.4‐dienoic acid hydrolase | Sphingomonas macrogolitabidus TFA | 32 | 6.11 | CGHWAQLER | thnD |
| 2, 11 | 110 | gi|29647412 | Cis‐biphenyl‐2.3‐dihydrodiol‐2.3‐dehydrogenase | Rhodococcus sp. RHA1 | 28 | 4.95 | LDTFVGNAAIWDFSTK | thnB |
| TATGAIINCDGGMGVR | ||||||||
| 3 | 112 | gi|110825065 | Possible sterol transfer protein | Rhodococcus sp. RHA1 | 14 | 5.17 | TPDFVLATK | thnU |
| LDVWRQFADGKLRA | ||||||||
| 5 | 66 | gi|2072113 | Ferredoxin reductase | Escherichia coli K12 | 43 | 5.6 | HLPYERPPLSK | thnA4 |
| 7 | 99 | gi|111026201 | Ethylbenzene dioxygenase alpha subunit | Rhodococcus sp. RHA1 | 51 | 5.28 | VFANSCPHR | thnA1 |
| VCFADAGNR | ||||||||
| MMPVAQVASYK | ||||||||
| 12 | 181 | gi|110825055 | Ethylbenzene dioxygenase beta subunit | Rhodococcus sp. RHA1 | 21 | 5.06 | MAYYNDDLDMIFTR | thnA2 |
| DEDRPLVGSREDTWR | ||||||||
| VYSNFFAFR | ||||||||
| 14 | 137 | gi|42475486 | Hypothetical protein | Rhodococcus rhodochrous K37 | 18 | 5.47 | DMIVVPAGVPR | |
| YVGSGATGNHENDNR | ||||||||
| HEDWDTLGFQAK | ||||||||
| 15 | 103 | gi|52783628 | Isocitrate lyase | Rhodococcus equi 103P+ | 47 | 4.8 | NGIEPCIAR | |
| VLIPTQQHIR | ||||||||
| EGMTAFVDLQER | ||||||||
| LAADVADVPTVVIAR | ||||||||
| TDAEAATLLTSDVDER | ||||||||
| AMIAAGVAGSHWEDQLASEK | ||||||||
| EVGAGYFDSIATTVDPNTSTAALK | ||||||||
| 16 | 122 | gi|706845 | N.N'‐dimethyl‐4‐nitrosoaniline oxidoreductase | Rhodococcus erythropolis NI86/21 | 46 | 5.4 | EAVFEPR | |
| EFHPFPR | ||||||||
| TLLMTTGLR | ||||||||
| VWEYNLPSR | ||||||||
| DVGIPDNFGQVR | ||||||||
| NINEFEGFAK | ||||||||
| NLTTVQAADAAVEAAIR | ||||||||
| YAQLAGALGVDTR | ||||||||
| 17 | 54 | gi|10176610 | Transposase | Bacillus halodurans C‐125 | 50 | 8.9 | YFAPTCVR | |
| (B) Proteins identified by ESI IT MS/MS | ||||||||
| Spot | Accession code | Predicted proteins | Organism | Mwa | pI | Tentative sequence | thn gene | |
| 4 | gi|119718512 | Acyl‐CoA dehydrogenase | Nocardioides sp. JS614 | 41 | 5.06 | VMTLYEGTSQIQK | thnV | |
| 6 | gi|118470801 | ATPases of the AAA+ class | Mycobacterium smegmatis | 65 | 4.85 | SVLDTGAPGLR | ||
| MC2 155 | AIDTESNTGQYL | |||||||
| IKIERPDAESAQDIFSK | ||||||||
| DFNSGAMIQNIVDR | ||||||||
| 8, 9 | gi|63148158 | Catechol 2,3‐dioxygenase | Rhodococcus sp. YU6 | 34 | 4.99 | LLGLEGAVEYK | thnC | |
| DIFGHDNEVEGYGLDPIPLK | ||||||||
| GAVGTPVFMHCNNR | ||||||||
| 10 | gi|16332030 | Undecaprenyl pyrophosphate synthetase | Synechocystis sp.PCC 6803 | 28 | 6.6 | QEIVHVCQAIAR | ||
| 13 | gi|91787128 | Beta‐ketoadipyl CoA thiolase | Polaromonas sp. JS666 | 40 | 5.38 | APFVFPK | thnI | |
Mw and pI were calculated from the predicted protein sequences.
Peptides subjected to MS/MS analysis are underlined.
Mw and pI were calculated from the predicted protein sequences.
The other tetralin‐induced proteins that we have identified (Table 1) do not appear to be directly involved in tetralin catabolism. Spot 6 is similar to a group of proteins with chaperone‐like functions that assist in the assembly or disassembly of protein complexes (Neuwald et al., 1999). Spot 10 was identified as a protein similar to an enzyme involved in cell wall synthesis. We found that spot 14 corresponded to a hypothetical protein encoded by a gene located in a biphenyl degradation operon of Rhodococcus rhodochrous (Taguchi et al., 2004). Spot 15 (Table 1A) was identified as an isocitrate lyase of Rhodococcus equi (Kelly et al., 2002), an enzyme that catalyses the reversible cleavage of isocitrate to glyoxylate and succinate (glyoxylate cycle). Spot 16 was found to match ThcE of R. erythropolis (Nagy et al., 1995), an enzyme induced by atrazine and thiocarbamate herbizides. Finally, spot 17 was found to correspond to a protein similar to a Bacillus halodurans transposase.
Cloning and identification of thn genes of Rhodococcus sp. strain TFB
Based on the peptide sequences obtained from the proteomic analysis, degenerate primers (HIDFw and HIDRv; Table S1) were designed to amplify a region of the putative thnD gene. A 477 bp PCR product, obtained using total DNA from Rhodococcus sp. strain TFB as template, was cloned and sequenced. Its nucleotide sequence shows 70% identity to the corresponding region of the Rhodococcus sp. RHA1 bphD gene (GenBank CP000433) that encodes a 2‐hydroxy‐6‐oxo‐6‐phenylhexa‐2,4‐dienoate hydrolase. We used the putative thnD PCR product to screen a TFB library, identifying three positive cosmids (Fig. 2A). Cosmid pMPO613 was selected for further subcloning and sequencing. A 23 Kb fragment from this cosmid contained 18 ORFs oriented in the same direction. The deduced amino acid sequences of these ORFs were screened against the peptide sequences obtained from the proteome analysis to determine whether these ORFs encoded any of the proteins induced in tetralin‐grown cells. Remarkably, peptides from spots 1, 2, 3, 4, 5, 6, 7, 8, 11, 12 and 13 were perfect matches to sequences of several of these ORFs (Table 1). As this suggests that the 23 Kb cosmid fragment contains one or more operons specifically induced by growth on tetralin medium, we designated each of these sequences as thn genes. Database comparison revealed high similarity between the TFB thn genes and the bph genes of Rhodococcus sp. RHA1 (Table S2). Based on information from previously characterized Thn proteins (GenBank AF157565), we propose a putative tetralin degradation pathway for strain TFB (Fig. 2C), analogous to the pathway that seems to be operating in S. macrogolitabida strain TFA (Fig. 2B). Nevertheless, differences between the genetic organization of the thn genes in both bacteria are evident. In strain TFB, the structural genes are transcribed in the same direction, although seem to be arranged into two operons: one made up of thnA1A2A3UB and another comprising of a perfect duplication of thnA1A2 (thnA1′A2′) plus the thnCGQILWVDEF genes (Fig. 2A). In addition, the thnU gene, which encodes a putative sterol transfer protein, is missing in strain TFA. We also found that a set of five TFB strain genes (thnQILWV) encodes proteins similar to those involved in β‐oxidation pathways. DNA sequences resembling the remnants of transposons were found between both operons.
Figure 2.
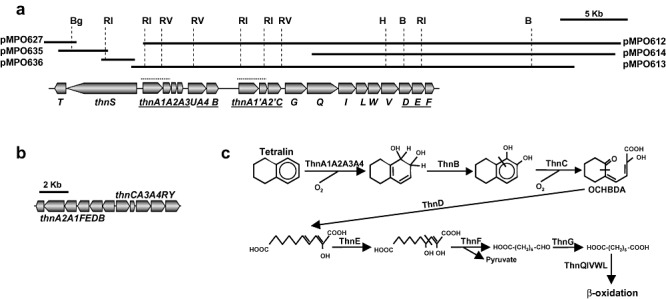
Genetic organization of thn genes. A. Cosmids and plasmids used for TFB thn genes sequencing and genetic organization. Genes functionally homologous to TFA thn genes are underlined. A perfect duplication containing thnA1A2 genes is denoted by a dotted line. B, BamHI; Bg, BglII; H, HindIII; Ri, EcoRI; Rv, EcoRV. B. Genetic organization of S. macrogolitabida strain TFA thn genes. C. Proposed tetralin degradation pathway in TFB.
Analysis of the sequence upstream of thnA1, at the end of the cosmid insert, revealed a partial sequence similar to the two component regulatory system of Rhodococcus sp. RHA1 bphST (Fig. 2A). Inverse PCR was used to obtain a 5.83 kb fragment containing the complete sequence of thnST, as this fragment was not present in the TFB library. Spots corresponding to the protein products of these genes were not found during our proteomic analysis.
Given that the bph genes in Rhodococcus sp. RHA1 are located on the linear plasmids pRHL1 and pRHL2 (Masai et al., 1997; McLeod et al., 2006), we decided to isolate large plasmids from TFB bacteria. Pulse field electrophoresis analysis revealed two large linear plasmids, denoted pTFB1 and pTFB2, with estimated sizes of 1100 and 280 kb respectively (Fig. 3A). Using a thnD DNA fragment as the probe for southern blotting, we found that the thn genes are located on the pTFB1 plasmid (Fig. 3B).
Figure 3.
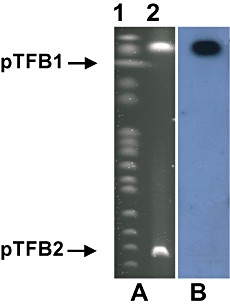
Plasmidic location of thn genes in TFB. A. Agarose pulse‐field gel stained with ethidium bromide. Lane 1, molecular mass marker (New England). Lane 2, TFB linear plasmids pTFB1 and pTFB2. B. Southern blot hybridization using thnD as probe.
Characterization of the ThnD hydrolase strain from TFB
One of the most important enzymes in tetralin degradation is ThnD, a hydrolase able to open the tetralin alicyclic ring, rendering a linear compound, 2‐hydroxydeca‐2,4‐dienedioic acid. In order to characterize its activity, the thnD gene of strain TFB was cloned into Escherichia coli under PT7 control (plasmid pMPO611, Table S1). Crude E. coli extracts showed hydrolytic activity when 2‐hydroxy‐4‐(2‐oxocyclohexyl)‐2,4‐butadienoic acid (OCHBDA) the aromatic ring fission product in the tetralin degradation pathway, see Fig. 2C) or 2‐hydroxy‐6‐oxo‐6‐phenylhexa‐2,4‐dienoic acid (6‐phenyl‐HODA, the aromatic ring fission product in the biphenyl degradation pathway) were used as substrates. However, ThnD hydrolase activity using OCHBDA as a substrate was 40% of that obtained using 6‐phenyl‐HODA (data not shown). To estimate the optimal reaction temperature, enzyme activity assays were performed at different temperatures using 6‐phenyl‐HODA as substrate. The rate of substrate consumption increased with temperature until reaching an optimal temperature of 70°C.
In order to demonstrate the specific role of TFB‐ThnD as a hydrolase in the tetralin biodegradation pathway, we introduced a plasmid (pMPO620) expressing TFB‐thnD under a Ptac promoter into a Thn‐ TFA mutant that bears a KIXX insertion in thnD (K6 mutant; Hernáez et al., 1999). We found that mutant K6 bacteria containing TFB‐thnD were able to grow on a tetralin substrate (data not shown), thus confirming the functional role of TFB‐ThnD in tetralin metabolism.
Transcriptional analysis of thn genes
Total RNA isolated from TFB cells grown to mid‐log phase with glucose or tetralin was used to perform semiquantitative RT‐PCR with different combinations of oligonucleotide pairs to amplify intergenic or intragenic thn regions. The PCR products of the sizes expected for the structural genes were detected only in cells grown in the presence of tetralin (Fig. 4), indicating that expression of the identified thn genes is induced by this compound. From these results we could define three different operons (Fig. 4): one codes for the regulatory genes thnST, a second encompasses the genes from thnA1 to thnB and a third one extends from thnA1′ to thnF.
Figure 4.
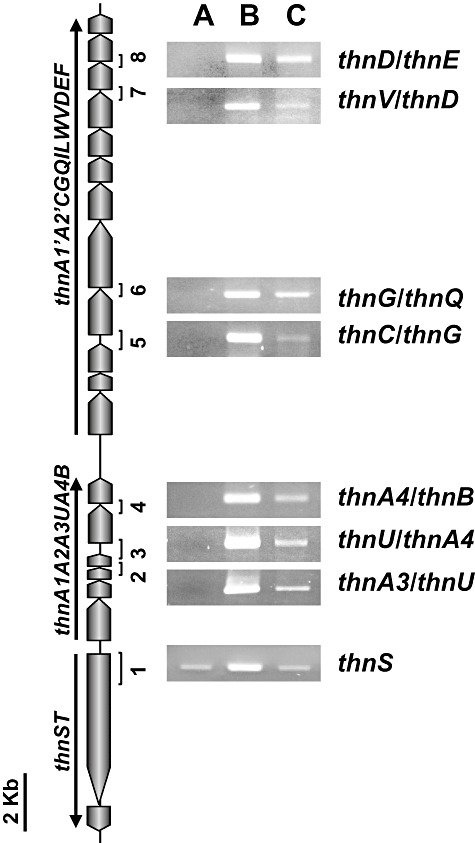
Expression of thn genes in strain TFB analysed by RT‐PCR. RNA was isolated from cells grown on glucose (A), tetralin (B) or glucose plus tetralin (C) as carbon sources. The expected fragments of each PCR reaction are numbered. The three defined operons are denoted by arrows.
In order to determine whether or not the thn genes are subject to catabolite repression, TFB cells were grown to mid‐log phase on tetralin, then incubated in a 2% glucose solution in the presence of tetralin for 2 h. Total RNA was isolated and converted to cDNA for semiquantitative RT‐PCR analysis. Glucose treatment resulted in reduced PCR amplification of all the structural genes tested (Fig. 4), indicating a partial glucose repression of these genes.
The thnST regulatory genes showed higher expression in tetralin‐grown cells compared with glucose or glucose plus tetralin‐grown cells (Fig. 4). However, in order to detect the thnST transcript, the number of PCR cycles had to be increased to 30, indicating a very low expression of these genes even in the presence of tetralin.
Determination of the 5′ ends of thn transcripts and analysis of the promoter regions
Primer extension assays were carried out to characterize the 5′ end of the thnA1 and thnS transcripts. Total RNA extracts from glucose‐ or tetralin‐grown cells were subjected to reverse transcription with primers P‐Ext‐thnSA1 and P‐Ext‐thnS (Table S1). As shown in Fig. 5A, an intense band for thnA1 was obtained only with RNA extracts from tetralin‐grown cells. Comparison with parallel sequencing reactions using the same primer, indicates that the thnA1 mRNA transcript starts at an A (a T in the DNA, Fig. 5A) and has a 132 bp non‐translated leader. As expected, the primer extension assay detected the same start point for the two main thn gene transcripts because of the perfect duplication of the thnA1A2 genes and upstream sequences. For thnS, the transcription start point was detected 35 nucleotides upstream of the first ATG codon (Fig. 5A). Basal transcription of thnS from the same promoter was also detected in glucose‐grown cells (Fig. 5A).
Figure 5.
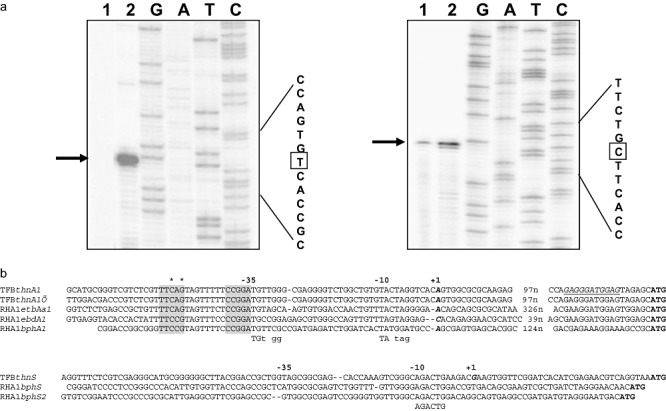
Characterization of thnA1 and thnS promoter regions. A. Primer extension assays for thnA1 (left) and thnS (right) transcriptional start point mapping. Total RNA from glucose‐ (lanes 1) or from tetralin‐ (lanes 2) grown cells were used as templates for primer extension reactions. The transcriptional start point is marked with a square. B. Alignment of the thnA1 and thnS promoter sequences (coding strand) with other RHA1 promoter sequences. In bold and italic, transcriptional start points experimentally determined. Underlined, putative thnA1 ribosome binding site. Proposed −10 and −35 consensus sequences are indicated under the alignment. Putative thn boxes, important for tetralin induction, are shadowed and asterisks denote the nucleotides mutated in plasmids pMPO645 (C‐51A) and pMPO646 (G‐49T).
In silico analysis of the promoter sequences revealed the presence of two nucleotide regions conserved in other Rhodococcus genes (Takeda et al., 2004). These regions, 6 and 33 nucleotides upstream of the thnA1 transcription start point, could be acting as −10 and −35 promoter sequences (Fig. 5B). No equivalent sequences were found within the thnS promoter, but a region at −10 seems to be conserved with the putative RHA1 bphS gene promoters. Additionally, an 18 bp sequence, which matches sequences present in biphenyl‐inducible promoters of Rhodococcus sp. RHA1 (Takeda et al., 2004), was identified 44 bp upstream of thnA1 and thnA1′ (Fig. 5B). This region has been proposed to be specifically involved in the transcriptional regulation of aromatic degradation enzymes in Rhodococcus (Takeda et al., 2004).
Expression of gfp translational fusions in strain TFB and mutational analysis of the thnA1 promoter
In order to study the transcriptional control of thn genes, we developed a translational GFP fusion system for TFB cells. Translational thnA1::gfp (pMPO633) and thnS::gfp (pMPO639) fusions were expressed in TFB cells grown on glucose, glucose plus tetralin or tetralin. Subsequent fluorescence measurements corroborated the expression pattern previously determined by RT‐PCR (Table 2).
Table 2.
Expression of gfp translational fusions in Rhodococcus sp. strain TFB.
| Arbitrary fluorescence units (×104) | |||
|---|---|---|---|
| Glucose | Glucose plus tetralin | Tetralin | |
| pMPO634 (promoterless gfp) | n.d. | 0.3 ± 0.2 | n.d. |
| pMPO633 (WT thnA1::gfp) | 1.4 ± 0.4 | 79 ± 6.4 | 399 ± 24 |
| pMPO645 (C‐51A thnA1::gfp) | 1.3 ± 0.1 | 18 ± 1.2 | 127 ± 16 |
| pMPO646 (G‐49T thnA1::gfp) | 0.98 ± 0.08 | 0.93 ± 0.06 | 1.07 ± 0.08 |
| pMPO639 (WT thnS::gfp) | 1.49 ± 0.35 | 1.63 ± 0.62 | 2.95 ± 0.39 |
| pMPO647 (thnST plus thnA1::gfp) | 5.4 ± 0.3 | 137 ± 1.4 | 449 ± 43 |
n.d., non detected.
Recently, Antunes and colleagues (2008) described the critical nucleotides for LuxR binding sites in Vibrio fisheri. As ThnT is similar to members of the LuxR family of regulators, we postulated that ThnT might act in an analogous way to regulate thnA1 expression. We tested this hypothesis by performing a mutational analysis of the thnA1 promoter region. To this end, we assayed two different point mutations, C(‐51)A and G(‐49)T, using the gfp expression system (see Fig. 5B). The C(‐51)A mutation reduced expression to 30% of that of the wild‐type promoter, but maintained the same regulatory pattern: low tetralin induction in the presence of glucose and higher induction in its absence. In contrast, the G(‐49)T mutation resulted in a constitutive basal expression in all the conditions tested. These results indicate that the upstream region affected by these mutations is likely to be recognized by the transcriptional activator of the thn genes and play a key role in controlling the response of TFB cells to aromatic compounds.
Overexpression of thnST genes
As it is not currently possible to generate mutants in strain TFB we tested the effect of overexpressing thnST in TFB cells on thnA1 expression to test the role of ThnST as transcriptional regulators of the thn genes. To achieve this, we introduced plasmid pMPO647, harbouring the thnST genes expressed from their own promoter, together with the translational thnA1::gfp fusion into TFB cells. Fluorescence was monitored in glucose‐, tetralin‐ and glucose plus tetralin‐grown TFB cells. In all cases, fluorescence was higher in TFB cells carrying extra copies of thnST than in cells that contained thnA1::gfp alone (Table 2). Basal expression of thnA1::gfp in glucose‐grown TFB cells was increased by the presence of thnST in the plasmid, and increased almost twofold in the presence of glucose plus tetralin. Taken together, our results suggest that thnST genes encode positive regulators involved in the induction of TFB thn genes in response to tetralin.
Discussion
Rhodococcus sp. strain TFB is able to grow on different aromatic compounds as its sole carbon and energy source (Tomás‐Gallardo et al., 2006). Previous studies with this strain have shown that growth on such compounds is associated with an induction of specific proteins. Growth on tetralin involves the induction of a significant number of proteins different from those induced by growth on other aromatic carbon sources (e.g. phthalate; Tomás‐Gallardo et al., 2006). Of the 18 thn gene products, MALDI‐MS(/MS) analysis or de novo peptide sequencing of tetralin‐induced spots identified nine of them (ThnA1, ThnA2, ThnU, ThnA4, ThnB, ThnC, ThnI, ThnV and ThnD). The proteomic analysis of tetralin‐grown cells and sequencing of the genes encoding the induced proteins allow us to propose a tetralin degradation pathway operating in strain TFB, which is identical to that described for S. macrogolitabida strain TFA. The aromatic ring of tetralin is cleaved by an extradiol dioxygenase (ThnC) after its dioxygenation and dehydrogenation (catalysed by ThnA1A2A3A4 and ThnB respectively). The alicyclic ring is subsequently cleaved by the serine hydrolase ThnD, thus rendering a linear molecule that is then broken down into central metabolites (Fig. 2C).
One of the most important enzymes in the tetralin degradation pathway is ThnD. This protein hydrolyses a C–C bond of the alicyclic ring of tetralin to produce a dicarboxylate product (Hernáez et al., 2000). Like S. macrogolitabida strain TFA‐ThnD protein, TFB‐ThnD is a thermostable protein with activity at high temperatures, an interesting feature in organic synthesis that might reflect a common origin. However, of the two hydrolases found in Rhodococcus sp. RHA1, ThnD from strain TFA is more closely related to EtbD while TFB‐ThnD is most similar to BphD1. In addition, unlike the hydrolase from strain TFA (Hernáez et al., 2000), TFB‐ThnD is more active towards 6‐phenyl‐HODA (an intermediate in the biphenyl pathway) than towards OCHBDA (the tetralin ring fission product), which supports the notion that this enzyme has been recruited from a biphenyl biodegradation pathway. However, other TFB‐Thn enzymes, like the ring hydroxylating dioxygenase system (ThnA1A2A3A4) or extradiol dioxygenase (ThnC), are more similar to the ethylbenzene dioxygenase (EBDO) and EtbC, from strain RHA1. The combination of enzymes from the ethylbenzene pathway in the TFB tetralin degradation pathway could be explained by the fact that EBDO transforms naphthalene (a molecule with two aromatic rings sharing two carbons) more efficiently than biphenyl (Iwasaki et al., 2007) and that biphenyl dioxygenases transform mono‐aromatic compounds more efficiently than EBDO (Patrauchan et al., 2008).
In addition to proteins directly involved in tetralin metabolism, other proteins were also induced following growth on tetralin. These could reflect a change in the metabolic status of TFB cells during growth on tetralin. Tetralin‐induced isocitrate lyase, a key enzyme of the glyoxylate cycle that cleaves isocitrate into succinate and glyoxylate, indicates that TFB cells adapt their central metabolism depending on the contaminant. In this case, the putative tetralin degradation products (pyruvate and acetyl‐CoA) would be used as carbon sources via the glyoxylate cycle, whose end‐product is succinate. Induction of central metabolism enzymes in response to organic contaminants has been previously described. Growth of strain TFB on phthalate induces a succinate dehydrogenase, a component of the citric acid cycle (Tomás‐Gallardo et al., 2006). Phthalate metabolism renders acetyl‐CoA and succinate, which suggests that the induction of succinate dehydrogenase may be required to deal with the succinate excess, although in this case a glyoxylate bypass, to use phthalate as a carbon source, would not be required. Similar results have been obtained in Rhodococcus sp. strain RHA1 (Navarro‐Llorens et al., 2005; Patrauchan et al., 2005) in which growth on benzoate, phthalate or phenylacetate upregulates citric acid cycle enzymes, while growth on pyruvate upregulates the glyoxylate cycle (Patrauchan et al., 2005). Tetralin is toxic to bacteria at concentrations above 15 µM (Schreiber and Winkler, 1983) and, due to its lipophilic character, may interact with biological membranes causing changes in their structure and function, impairing growth and cell activity (Sikkema et al., 1992; 1994). The induction of proteins involved in cell wall synthesis (spot 10) or with chaperone‐like functions (spot 6) might be a response to tetralin toxicity.
The thn genes from strain TFB are most similar to bph and etb genes from other Rhodococcus strains (see Table S2). Despite their homology, the genetic organization is different. While in Rhodococcus sp. strain RHA1 several copies of bph and etb genes are located on the linear plasmids pRHL1 and pRHL2 (McLeod et al., 2006), TFB thn genes are clustered together on the plasmid pTFB1 (a linear plasmid similar to pRHL1 in size) and no additional copies have been found elsewhere (data not shown). The DNA sequences most closely related to TFB thn genes are the etb genes present on pRHL2 and pDK2 from Rhodococcus sp. strain DK17. In plasmid pRHL2, a duplication of the etbAa1Ab1 genes (homologous to thnA1A2) is also present. In this case, there is only a single nucleotide difference between etbAa1Aba and etbAa2Ab2. This is another example of the genetic rearrangements that can take place between different replicons of members of the Rhodococcus genus.
Our in silico analysis of the promoter regions of TFB‐thn genes and RHA1‐etb‐ebd‐bph‐genes allowed us to define putative −10 and −35 consensus sequences that do not follow the E. coli consensus. However, the −10 sequence of thnS does not match the consensus found in thnA1. This fact could be related to transcriptional differences between regulatory (with a basal expression in glucose) and structural genes (only expressed in the presence of the inducer).
A stretch of conserved nucleotides, centered at −44, which has been described in other etb and bph gene promoters as possibly being involved in a specific transcriptional regulatory mechanism (Takeda et al., 2004), is also present at thnA1 and thnA1′ promoters. This sequence shows a palindromic structure that resembles the DNA binding sites of LuxR‐type transcriptional regulators (White and Winans, 2007; Antunes et al., 2008). As ThnT is similar to this kind of transcriptional regulators, we have functionally analysed the putative thn box within the thnA1 promoter. Our finding that single‐base substitution at nucleotide 3 or 5 of this region reduce or eliminates induction, respectively, indicates the importance of the thn box in ensuring correct thnA1 transcription. Given the conservation of the −44 thnA1 box sequence with promoters of other Rhodococcus genes involved in aromatic compound degradation, which appear to be controlled by homologous two‐component LuxR‐type regulatory systems, we postulate a common regulatory mechanism with conserved binding specificity that operates to modulate gene expression in response to the presence of aromatic compounds. However, in S. macrogolitabida strain TFA, the response to tetralin involves a LysR‐type transcriptional activator, ThnR and a co‐activator, ThnY (Martínez‐Pérez et al., 2004). This difference in the regulatory systems of the same biodegradation pathway is another example of how the evolution of metabolic pathways and their regulatory systems are independent, the latter conferring selective advantage by improving the ability of some bacteria to adapt their catabolic capability and general metabolism to the available resources (Cases and de Lorenzo, 2001; Tropel et al., 2004).
Experimental procedures
Chemicals, bacterial strains, culture conditions, plasmids and primers
Dihydroxytetralin was chemically synthesized (Andújar et al., 2000). Tetralin and 2,3‐dihydroxybiphenyl were purchased from Sigma. The OCHBDA and 6‐phenyl‐HODA were biologically synthesized (Andújar et al., 2000). Bentone MA was a gift of Zeus Química (Barcelona, Spain).
Rhodococcus sp. strain TFB (Tomás‐Gallardo et al., 2006), S. macrogolitabida strain TFA and the thnD mutant derivative K6 lacking the hydrolase activity (Hernáez et al., 1999; Hernáez et al., 2000) were grown on MM medium (Dorn et al., 1974) at 30°C with tetralin concentration in vapour phase, 11.11 mM glucose (for strain TFB) or 40 mM β‐hydroxybutyrate (for strain TFA) as carbon and energy sources. Luria–Bertani broth (LB; Sambrook et al., 1989) or MML (Andújar et al., 2000) were used as rich media for TFB and TFA respectively. Escherichia coli strains were grown in LB medium containing the appropriate antibiotics depending on the plasmid carried (see Table S1). Escherichia coli DH5α (Hanahan, 1983) was used for DNA manipulation. Escherichia coli NCM631/pIZ227 (Govantes et al., 1996) was used to overproduce the tetralin hydrolase ThnD.
Bacterial strains, plasmids and primers used in this work are listed in Table S1.
Conjugation of plasmids to S. macrogolitabida strain TFA was carried out by triparental mating as described previously (de Lorenzo and Timmis, 1994).
ThnD overexpression, crude extract preparation and hydrolase activity assay
For TFB tetralin hydrolase overexpression, the thnD gene was amplified with NdeI‐hid and 608Hid‐Ter primers (Table S1) and cloned in pT7‐7 (Tabor and Richardson, 1985) with NdeI and PstI to make pMPO611. Escherichia coli NCM631/pIZ227 was transformed with pMPO611 and cell free extracts were obtained as described in Hernáez and colleagues (2000). Protein concentration was determined using the Bradford method (Bradford, 1976).
Hydrolase activity towards different ring fission products was assayed as described in Hernáez and colleagues (2000). One unit of enzyme activity was defined as the amount of enzyme that converts 1 µmol of substrate per min.
Protein sample preparation, fluorophore labelling and 2D electrophoresis analysis
TFB cell free extracts, fluorophore labelling, 2D electrophoresis and protein identification were carried out as described by Tomás‐Gallardo and colleagues (2006). For protein quantification, fluorescent gel imagines were obtained by scanning using a Typhoon TM 9400 Scanner and analysed with the DeCyder Differential Analysis software 5.01 version (GE Healthcare) with default parameters. Theoretical molecular masses and isoelectric points of the proteins of interest were calculated using expasy tools.
Screening for genes encoding hydrolases
A total TFB DNA library (Tomás‐Gallardo et al., 2006) was screened by colony hybridization, using a probe targeting serine hydrolase genes. The probe was obtained by PCR using degenerate primers HIDFw and HIDRv (Table S1), whose design was based on the peptide sequence obtained from the proteomic analysis. PCR was carried out using 50 ng of total DNA (isolated as described in Tomás‐Gallardo et al., 2006) as template and the puRe‐Taq Ready‐To‐Go PCR Beads (GE Healthcare) under the following amplification conditions: 95°C for 10 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 50 s. The 477 bp fragment was cloned into EcoRV‐cleaved pBluescript II SK+ (Stratagene) to make pMPO602. This fragment was used to probe a TFB library by Southern analysis (Sambrook et al., 1989). A 23 kb fragment from cosmid pMPO613 was sequenced commercially on both strands.
Plasmids pMPO627, pMPO635 and pMPO636 were obtained by inverse PCR as follows: total DNA from strain TFB was cut with EcoRI, BglII or BamHI and religated. Each religation product was used as template for PCR reactions with primers PCRInvEcoRI619 and PCRInvthnSFw, PCRInvBglII626 and PCRInvBglII627, and PCRInvBamHI624 and PCRInvBamHI635 respectively. The amplification products were cloned in pBluescript SK+.
The obtained DNA sequence was compared with those in the databases using blastn 2.2.6 (Altschul et al., 1997) and submitted to GenBank under Accession Number FJ183471.
Plasmid isolation, pulsed field gel electrophoresis, linearity determination and Southern blot hybridization
For detection of extrachromosomal DNA, plasmid isolation and pulse field gel electrophoresis assays were developed as described by König and colleagues (2004). Yeast Chromosome PFG marker (New England Biolabs) was used as reference. After electrophoresis, DNA was transferred to Hybond‐N+ nylon membranes (GE Healthcare) and subjected to Southern hybridization with the pMPO602 thnD probe.
Total RNA sample preparation, RT‐PCR and primer extension analysis
Total RNA of Rhodococcus sp. strain TFB was isolated following the method described by Tomás‐Gallardo and colleagues (2006). PCR amplification of 16S rDNA and some thn genes with RNA samples as template was conducted to test for DNA contamination.
DNA‐free RNA samples (2 µg) were converted to cDNA using the TaqMan Reverse Transcription kit (Applied Biosystems) following the manufacturer's instructions. Equal amounts of cDNA were used as templates in PCR reactions with primers listed in Table S1. To detect intergenic and intragenic mRNA, combinations of primers were: 608‐5SG with 8836G for thnV‐thnD, 8837G with 9273L for thnD‐thnE, 2‐608ESf570 with β‐ox1rev for thnG‐thnQ; RT‐Exdio with RT‐Aldh for thnC‐thnG; RT‐FerRed with RT‐DihyDehy for thnA4‐thnB, RTfer‐fw with RTorf7‐rev for thnA3‐orf7, RTorf7‐fw with 3‐624rev for thnU‐thnA4, ThnS‐RTPCR‐Fw with ThnS‐RTPCR‐Rev for thnS. PCR reactions were performed with 30 ng of cDNA for structural genes and 5 ng of cDNA for thnS using the puReTaq Ready‐To‐Go PCR beads (GE Healthcare) and consisted of 20 cycles of 30 s at 94°C, 30 s at 59°C and 45 s at 72°C (30 cycles for thnS RT‐PCR). As loading control, primers f27 and r519 (Hugenholtz et al., 1998) were used with 20 pmol of cDNA.
Primer extension reactions were performed as described previously (Govantes et al., 2000), with 40 µg of RNA as a template, 32P‐end‐labelled oligonucleotides P‐Ext‐thnSA1 and P‐Ext‐thnS, and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). Sequencing reactions with the same primers and pMPO647 as template were performed with the Thermo Sequenase cycle sequencing kit (USB, Cleveland, OH), according to the manufacturer's instructions. Samples were run on polyacrylamide sequencing cells. Dried gels were exposed to radiosensitive screens, which were subsequently scanned with a Typhoon 9410 scanner (GE Healthcare).
Construction of a gfp‐based promoter probe vector and gene fusions
A promoter probe vector was constructed using the Rhodococcus‐E. coli shuttle vector, pNC9503 (H. Saeki, Japan Energy Corporation, Japan) and a variant of the GFP (Cormack et al., 1996) as the reporter gene. The gfp gene was cloned as a PCR product with ScaI‐XbaI ends (pMPO634) and the tested promoters were cloned as SalI‐ScaI PCR fragments. The used primers and the obtained plasmids are described in Table S1 and electroporation of the plasmids into TFB cells was carried out as described by Tomás‐Gallardo and colleagues (2006). thnA1 promoter region mutagenesis was carried out by overlapping PCR using plasmid pMPO633 as template. Primers 633C3AFw and GFPXbaIPCRsolap and primers 633C3Arev and thnSA1SalIPCRsolap were used to amplify 210 and 850 bp fragments respectively. These fragments were used as templates with primers thnSA1SalIPCRsolap and GFPXbaIPCRsolap to obtain a 1060 bp fragment that includes the point mutation C‐51A and that was cloned in pMPO634 to obtain pMPO645. The same strategy was applied to obtain pMPO646 (which included G‐49T point mutation) with the overlapping primers 633G5TFw and 633G5Trev.
Fluorescence measurements were carried out with a Typhoon 9410 scanner (GE Healthcare). Cells were grown in mineral medium with glucose, tetralin or glucose plus tetralin as carbon sources to an OD600 of 1.5–2. The excitation and emission wavelength used were 488 and 532 nm respectively. The intensity of fluorescence is expressed as its relative value per unit of optical density at 600 nm (arbitrary units). Basal fluorescence of the plate was subtracted in each experiment. Data were the result of triplicate measurements of independent cultures of strain TFB carrying each plasmid.
Acknowledgments
This work was supported by Spanish Comisión Interministerial de Ciencia y Tecnología Grants BIO2005‐03094 and BIO2008‐01805, by the Andalusian Autonomic Government Grant Proyecto de Excelencia CVI‐131, and by a fellowship from the Spanish Ministerio de Educación y Ciencia to L.T.‐G. We thank J.A. López for his helpful assistance with the proteomic analysis and S.R. Kaschabek and J. Gröning for their technical help with linear plasmid isolation.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Bacterial strains, plasmids and primers.
Database comparison of TFB-thn genes.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andújar E., Hernáez M.J., Kaschabek S.R., Reineke W., Santero E. Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis and purification and characterization of the gene product. J Bacteriol. 2000;182:789–795. doi: 10.1128/jb.182.3.789-795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L.C., Ferreira R.B., Lostroh C.P., Greenberg E.P. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol. 2008;190:4392–4397. doi: 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K.S., Philip J.C., Aw D.W.J., Christoff N. The genus. Rhodococcus. J Appl Microbiol. 1998;85:195–210. doi: 10.1046/j.1365-2672.1998.00525.x. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cases I., De Lorenzo V. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 2001;20:1–11. doi: 10.1093/emboj/20.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia R.H., Falkow S. FACS‐optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H.J. Isolation and characterization of a 3‐chlorobenzoate degrading. Pseudomonas. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Van Der Geize R., Dijkhuizen L. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol. 2004;7:255–261. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Govantes F., Molina‐López J.A., Santero E. Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J Bacteriol. 1996;178:6817–6823. doi: 10.1128/jb.178.23.6817-6823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govantes F., Albrecht J.A., Gunsalus R.P. Oxygen regulation of the Escherichia coli cytochrome d oxidase cydAB operon: roles of multiple promoters and the Fnr‐1 and Fnr‐2 binding sites. Mol Microbiol. 2000;6:1456–1469. doi: 10.1046/j.1365-2958.2000.02100.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hernáez M.J., Reineke W., Santero E. Genetic analysis of biodegradation of tetralin by a Sphingomonas strain. Appl Environ Microbiol. 1999;65:1806–1810. doi: 10.1128/aem.65.4.1806-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernáez M.J., Andújar E., Ríos J.L., Kaschabek S.R., Reineke W., Santero E. Identification of a serine hydrolase which cleaves the alicyclic ring of tetralin. J Bacteriol. 2000;182:5448–5453. doi: 10.1128/jb.182.19.5448-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernáez M.J., Floriano B., Ríos J.J., Santero E. Identification of a hydratase and a class II aldolase involved in biodegradation of the organic solvent tetralin. Appl Environ Microbiol. 2002;68:4841–4846. doi: 10.1128/AEM.68.10.4841-4846.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P., Pitulle C., Hershberger K.L., Pace N.R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T., Takeda H., Miyauchi K., Yamada T., Masai E., Fukuda M. Characterization of two biphenyl dioxygenases for biphenyl/PCB degradation in a PCB degrader, Rhodococcus sp. strain RHA1. Biosci Biotechnol Biochem. 2007;71:993–1002. doi: 10.1271/bbb.60663. [DOI] [PubMed] [Google Scholar]
- Kelly B.G., Wall D.M., Boland C.A., Meijer W.G. Isocitrate lyase of the facultative intracellular pathogen Rhodococcus equi. Microbiology. 2002;148:793–798. doi: 10.1099/00221287-148-3-793. [DOI] [PubMed] [Google Scholar]
- Kim D., Chae J.C., Zylstra G.J., Sohn H.Y., Kwon G.S., Kim E. Identification of two‐component regulatory genes involved in o‐xylene degradation by Rhodococcus sp. strain DK17. J Microbiol. 2005;43:49–53. [PubMed] [Google Scholar]
- Kulakov L.A., Chen S., Allen C.C., Larkin M.J. Web‐type evolution of rhodococcus gene clusters associated with utilization of naphthalene. Appl Environ Microbiol. 2005;71:1754–1764. doi: 10.1128/AEM.71.4.1754-1764.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König C., Eulberg D., Gröning J., Lakner S., Seibert V., Kaschabek S.R., Schlömann M. A linear megaplasmid, p1CP, carrying the genes for chlorocatechol catabolism of Rhodococcus opacus 1CP. Microbiology. 2004;150:3075–3087. doi: 10.1099/mic.0.27217-0. [DOI] [PubMed] [Google Scholar]
- Labbé D., Garnon J., Lau P.C. Characterization of the genes encoding a receptor‐like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl‐degrading bacterium, Rhodococcus sp. strain M5. J Bacteriol. 1997;179:2772–2776. doi: 10.1128/jb.179.8.2772-2776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo V., Timmis K.N. Analysis and construction of stable phenotypes in gram‐negative bacteria with Tn5‐ and Tn10‐derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- McLeod M.P., Warren R.L., Hsiao W.W.L., Araki N., Myhre M., Fernandes C. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Pérez O., Moreno‐Ruiz E., Floriano B., Santero E. Regulation of tetralin biodegradation and identification of genes essential for expression of thn operons. J Bacteriol. 2004;186:6101–6109. doi: 10.1128/JB.186.18.6101-6109.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Pérez O., López‐Sánchez A., Reyes‐Ramírez F., Floriano B., Santero E. Integrated response to inducers by communication between a catabolic pathway and its regulatory system. J Bacteriol. 2007;189:3768–3775. doi: 10.1128/JB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Ishikura M., Taki H., Shindo K., Kasai H., Haga M. Isolation and characterization of o‐xylene oxygenase genes from Rhodococcus opacus TKN14. Appl Environ Microbiol. 2005;71:7705–7715. doi: 10.1128/AEM.71.12.7705-7715.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai E., Sugiyama K., Iwashita N., Shimizu S., Hauschild J.E., Hatta T. The bphDEF meta‐cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. et al. [DOI] [PubMed] [Google Scholar]
- Moreno‐Ruiz E., Hernáez M.J., Martínez‐Pérez O., Santero E. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J Bacteriol. 2003;185:2026–2030. doi: 10.1128/JB.185.6.2026-2030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I., Verheijen S., De Schrijver A., Van Damme J., Proost P., Schoofs G. Characterization of the Rhodococcus sp. NI86/21 gene encoding alcohol: N,N'‐dimethyl‐4‐nitrosoaniline oxidoreductase inducible by atrazine and thiocarbamate herbicides. Arch Microbiol. 1995;163:439–446. doi: 10.1007/BF00272133. et al. [DOI] [PubMed] [Google Scholar]
- Navarro‐Llorens J.M., Patrauchan M.A., Stewart J.R., Davies J.E., Eltis L.D., Mohn W.W. Phenylacetate catabolism in Rhodococcus sp. strain RHA1: a central pathway for degradation of aromatic compounds. J Bacteriol. 2005;187:4497–4504. doi: 10.1128/JB.187.13.4497-4504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: a class of chaperone‐like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Patrauchan M.A., Florizone C., Dosanjh M., William W., Mohn W.W., Davies J., Eltis L.D. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 2005;187:4050–4063. doi: 10.1128/JB.187.12.4050-4063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrauchan M.A., Florizone C., Eapen S., Gómez‐Gil L., Sethuraman B., Fukuda M. Roles of ring‐hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J Bacteriol. 2008;190:37–47. doi: 10.1128/JB.01122-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Winkler U.K. 2nd. Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schreiber A.F., Winkler U.K. Transformation of tetralin by whole cells of Pseudomonas stutzeri AS39. J Appl Microbiol Biotechnol. 1983;18:6–10. [Google Scholar]
- Sikkema J., De Bont J.A.M. Isolation and initial characterization of bacteria growing on tetralin. Biodegradation. 1991;2:15–23. [Google Scholar]
- Sikkema J., Poolman B., Konings W.N., De Bont J.A.M. Effects of the membrane action of tetralin on the functional and structural properties of artificial and bacterial membranes. J Bacteriol. 1992;174:2986–2992. doi: 10.1128/jb.174.9.2986-2992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema J., De Bont J.A.M., Poolman B. Interactions of cyclic hydrocarbons with biological membranes. J Biol Chem. 1994;269:8022–8028. [PubMed] [Google Scholar]
- Tabor S., Richardson C.C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K., Motoyama M., Kudo T. Multiplicity of 2,3‐dihydroxybiphenyl dioxygenase genes in the Gram‐positive polychlorinated biphenyl degrading bacterium Rhodococcus rhodochrous K37. Biosci Biotechnol Biochem. 2004;68:787–795. doi: 10.1271/bbb.68.787. [DOI] [PubMed] [Google Scholar]
- Takeda H., Yamada A., Miyauchi K., Masai E., Fukuda M. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J Bacteriol. 2004;186:2134–2146. doi: 10.1128/JB.186.7.2134-2146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás‐Gallardo L., Canosa I., Santero E., Camafeita E., Calvo E., López J.A., Floriano B. Proteomic and transcriptional characterization of aromatic degradation pathways in Rhodoccocus sp. strain TFB. Proteomics. 2006;6:119–132. doi: 10.1002/pmic.200500422. [DOI] [PubMed] [Google Scholar]
- Tropel D., Bähler A., Globig K., Van Der Meer J.R. Design of new promoters and of a dual‐bioreporter based on cross‐activation by the two regulatory proteins XylR and HbpR. Environ Microbiol. 2004;6:1186–1196. doi: 10.1111/j.1462-2920.2004.00645.x. [DOI] [PubMed] [Google Scholar]
- Veselý M., Knoppová M., Nesvera J., Pátek M. Analysis of catRABC operon for catechol degradation from phenol‐degrading Rhodococcus erythropolis. Appl Microbiol Biotechnol. 2007;76:159–168. doi: 10.1007/s00253-007-0997-6. [DOI] [PubMed] [Google Scholar]
- White C.E., Winans S.C. The quorum‐sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol Microbiol. 2007;64:245–256. doi: 10.1111/j.1365-2958.2007.05647.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial strains, plasmids and primers.
Database comparison of TFB-thn genes.


