Summary
The three most common pathogenic species of Vibrio, Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus, are of major concerns due to increased incidence of water‐ and seafood‐related outbreaks and illness worldwide. Current methods are lengthy and require biochemical and molecular confirmation. A novel label‐free forward light‐scattering sensor was developed to detect and identify colonies of these three pathogens in real time in the presence of other vibrios in food or water samples. Vibrio colonies grown on agar plates were illuminated by a 635 nm laser beam and scatter‐image signatures were acquired using a CCD (charge‐coupled device) camera in an automated BARDOT (BActerial Rapid Detection using Optical light‐scattering Technology) system. Although a limited number of Vibrio species was tested, each produced a unique light‐scattering signature that is consistent from colony to colony. Subsequently a pattern recognition system analysing the collected light‐scatter information provided classification in 1−2 min with an accuracy of 99%. The light‐scattering signatures were unaffected by subjecting the bacteria to physiological stressors: osmotic imbalance, acid, heat and recovery from a viable but non‐culturable state. Furthermore, employing a standard sample enrichment in alkaline peptone water for 6 h followed by plating on selective thiosulphate citrate bile salts sucrose agar at 30°C for ∼ 12 h, the light‐scattering sensor successfully detected V. cholerae, V. parahaemolyticus and V. vulnificus present in oyster or water samples in 18 h even in the presence of other vibrios or other bacteria, indicating the suitability of the sensor as a powerful screening tool for pathogens on agar plates.
Introduction
Vibrio species belong to the family Vibrionaceae and are usually found in aquatic environments, including seawater, fish, sea plankton, coral reefs and chitinous sea animals. They can also be found on the body surface and in the gastrointestinal tract of marine animals. The genus Vibrio contains more than 50 species. The taxonomy is ever changing because new additions to the species are identified by molecular taxonomic techniques (Tantillo et al., 2004). The main human pathogenic vibrios are Vibrio cholerae (Singh et al., 2001), Vibrio parahaemolyticus (Nair et al., 2007) and Vibrio vulnificus (Gulig et al., 2005), all three of which cause diseases owing to the consumption of contaminated water or food (especially raw fish and seafood), and to exposure of skin lesions to marine environments (Thompson et al., 2004). Although V. cholerae has been responsible for several major epidemics and continues to be a problem in South‐East Asia and in select countries in South America (Huq et al., 2005), it is seldom seen in the USA. Instead, illnesses from V. parahaemolyticus and V. vulnificus are more prevalent in the USA. In 2005, hurricane Katrina and later by Rita flooded New Orleans and its surrounding areas with seawater resulted in an increase of V. vulnificus infections (Rhoads, 2006; Nigro et al., 2011). Vibrio parahaemolyticus is responsible for as much as 50% of foodborne illness in Japan; the high incidence is attributed to the high consumption of raw fish. Several V. parahaemolyticus‐related outbreaks have been reported to be caused by the consumption of raw oysters (McLaughlin et al., 2005), and raw shellfish and shrimp (Cabanillas‐Beltran et al., 2006). Vibrio vulnificus infections are rare, but extremely dangerous in immunocompromised individuals, causing septicaemia and death (Gulig et al., 2005; Daniels, 2011). As the popularity of raw fish and sushi increases in the USA, the incidence of infection due to Vibrio will likely increase (Nair et al., 2007).
Vibrio vulnificus and V. parahaemolyticus are of the greatest concern to the shellfish industry and to consumers because they are often found at high concentrations in bivalve shellfish, such as clams, oysters and mussels, owing to the shellfish's filter‐feeding process. Water temperature, salinity and nutrient availability influence Vibrio occurrence in the aquatic environment. Survival mechanisms under stressed environments (Roszak and Colwell, 1987) and entrance to the viable but non‐culturable (VBNC) state allow the bacteria to cope with a changing aquatic environment. Cells can be resuscitated from the VBNC state once the adverse conditions are removed (Oliver et al., 1995; Whitesides and Oliver, 1997).
Traditional biochemical techniques as well as molecular tools, including ribotyping, pulsed‐field gel electrophoresis and PCR (Kong et al., 2002; Thompson et al., 2004; Jones et al., 2012), are used to identify Vibrio species. Conventional culture‐based methods include pre‐enrichment, plating onto selective solid media, and then morphological, biochemical, molecular biological and/or serological characterization to identify the bacteria (Ottaviani et al., 2003). Conventional assays for monitoring vibrios are labour intensive, time consuming and relatively costly.
Our earlier work demonstrated the applicability of forward light scattering for identification and differentiation of colonies formed by Listeria spp. (Bayraktar et al., 2006; Bae et al., 2007; 2008; Banada et al., 2007). The described system used scatter patterns formed by the complex interaction of laser light with the structure of colonies to quantify differences in phenotypes of colonies. The idea of recognizing microorganism on the basis of colony phenotypes (morphotypes) has been investigated by many research groups for quite some time; automated image‐analysis systems have been shown to work for classification of colonies belonging to different enterococcal strains (Qamer et al., 2003), Escherichia coli and Streptococci (Chen and Zhang, 2009), as well as direct identification of pure Penicillium species (O'Brien et al., 2008). However, the traditional approach to colony recognition based on implementation of automated image‐analysis systems and pattern‐recognition techniques to automate visual recognition of colonial morphology remains limited to only few specific cases, and it cannot be easily generalized. Our technique based on detection of multi‐angle light‐scatter patterns, rather than simply on colony images, allows for wider applicability, better accuracy and impressive specificity. An important advantage of the forward‐scatter approach to colony phenotyping is that features extracted from scatter patterns are, in principle, readily quantified. The polar nature of the scattering signatures as exemplified in Fig. 1 suggest decomposition using a set of orthogonal polynomials with radial characteristics, the most robust and sensitive of which are the pseudo‐Zernike polynomials. Further, biophysical models using diffraction theory proved that morphological differences modulated the optical amplitude and phase of the incoming laser beam and created unique forward‐scattering signatures that are seen in different species (Bae et al., 2007). Because the forward‐scatter patterns are not ideally rotationally invariant, the pseudo‐Zernike polynomials are augmented with Haralick texture descriptors (Xia et al., 2007). The scatter patterns represented by a number of selected numerical features are subsequently classified using Fisher's linear discriminant, support vector machine with linear kernel (SVM‐L), and support vector machine with radial‐basis function kernel (SVM‐RBF). The classification system follows the implementation described previously (Bayraktar et al., 2006; Banada et al., 2007). Although the BARDOT (BActerial Rapid Detection using Optical light‐scattering Technology) approach requires modestly complex data analysis and numerical processing software, it remains simple in term of sample handling. It is also non‐invasive and does not require any probes or labelling reagents, and the identification results are obtained from bacterial colonies in seconds (3 s per colony).
Figure 1.
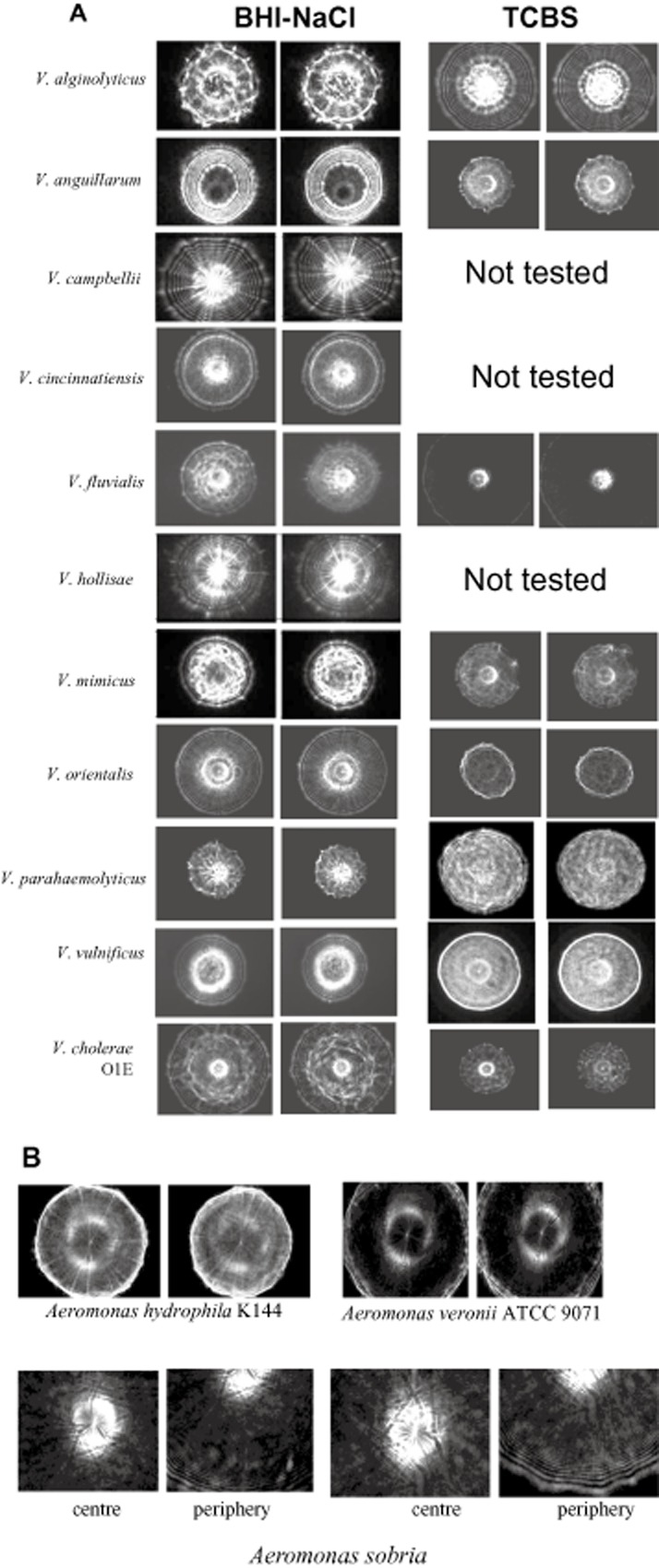
Optical forward light‐scattering images of different (A) Vibrio species grown on BHI‐NaCl and TCBS, and (B) Aeromonas species on BHI‐NaCl agar plates. Scatter images of two representative colonies from over 100 colonies of each species are presented.
In the present study, we used a commercial prototype of BARDOT (Advanced Bioimaging Systems, West Lafayette, IN) to identify Vibrio. An improved pattern‐analysis and image‐processing algorithm was used to extract features from the scatter images to identify and differentiate the various Vibrio species, especially V. cholerae, V. parahaemolyticus and V. vulnificus from other vibrios. We demonstrate the application of BARDOT in detection and identification of Vibrio species from pure cultures and from oyster and spiked water samples.
Results
Identification of Vibrio species with BARDOT
In the BARDOT system, illuminating laser on the centre of the colony on the Petri dish generated scatter image that was identified after matching with the scatter image library. Petri dishes containing test samples were incubated at 30°C for 12–18 h. Colony diameter of 1.3 ± 0.2 mm was used as a fixed parameter for each test culture before analysis with the light scatterometer. Representative forward‐scatter patterns for different Vibrio species on brain‐heart infusion (BHI) containing 1% NaCl (BHI‐NaCl) or thiosulphate citrate bile salts sucrose (TCBS) plates from Table 1 are presented in Fig. 1. The greyscale representation of the scatter pattern shows that they appear specific for a given species and visually differentiable irrespective of the media used. The scatter patterns for a given organism on BHI‐NaCl and TCBS were also distinct. The visual characteristics that can be used to differentiate the collected patterns include concentric rings, apparent spokes, bright central spot, or spiked ring outlining the central scatter region. For the purpose of automated detection and classification the described visual features were quantified using pseudo‐Zernike moments and Haralick texture features as described previously.
Table 1.
Vibrio species used in this study
| Pathogen | Strain designation and sourcea | Origin | Culture confirmation based on 16S rDNA sequence (Accession No.)b |
|---|---|---|---|
| Vibrio alginolyticus | CECT521 | Fish | V. alginolyticus strain VM122 (AF 537959) |
| Vibrio anguillarum | CECT522 | Fish | EU 851049c |
| Vibrio mimicus | CECT4218 | Human | EU 851050c |
| Vibrio orientalis | CECT629 | Seawater | Not tested |
| Vibrio campbellii | CECT523 | Water | V. campbelli strain BL9 (DQ 980029) |
| Vibrio cincinnatiensis | CECT4216 | Human | EU 851051c |
| Vibrio fluvialis | CECT4217 | Human | V. fluvialis isolate C2T1BB1 (DQ 005220) |
| Vibrio hollisae | CECT5069 | Human | V. hollisae strain LMG17719T (AJ 514909) |
| Vibrio parahaemolyticus | CECT511 | Human | V. parahaemolyticus V7 (EU 636231) |
| V. parahaemolyticus, tdh+/trh+ O1:Kuk | FDA DIE 12052499 | Environmental | Not tested |
| V. parahaemolyticus, tdh+/trh+ | FDA SPRC 10290 | Unknown | Not tested |
| V. parahaemolyticus, tdh+/trh‐, O3:K6 | FDA TX 2103 | Human | Not tested |
| V. parahaemolyticus, tdh+/trh‐, O4:K8 | FDA CPA 7081699 | Environmental | Not tested |
| V. parahaemolyticus, tdh‐/trh+, O3:K6 | FDA AQ4037 | Human | Not tested |
| V. parahaemolyticus, tdh‐/trh‐, O4:K55 | FDA BAC‐98–3547 | Human | Not tested |
| V. parahaemolyticus | ATCC 17803 | Unknown | Not tested |
| Vibrio vulnificus | MLT352 | Human stool | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF 546248) | |||
| V. vulnificus | MLT362 | Sediment | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT364 | Saltwater pond | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT367 | Seawater | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT403 | Oysters | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT404 | Oysters | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT1009 | Human blood | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF546248) | |||
| V. vulnificus | MLT LL728 | Human | V. vulnificus clone01_A10 (EF 546246) |
| V. vulnificus clone02_A02 (EF 546248) | |||
| V. vulnificus | MLT1003 | Human blood | V. vulnificus strain 1003BG‐OR (AY 676130) |
| Vibrio cholerae, O1E | CDC | Human | Not tested |
| V. cholerae, O139 | ICDDR‐B CA SHD | Human | Not tested |
| V. cholerae, O1 | FDA 6707 | Human | Not tested |
| V. cholerae, non‐O1 | FDA 140‐58 | Environmental | Not tested |
| Aeromonas hydrophila | USDA K144 | Unknown | A. hydrophila (AB 034761) |
| Aeromonas veronii | ATCC 9071 | Frog | A. veronii HC050630A‐1 (EF 669480) |
| Aeromonas sobria | ATCC 35993 | Activated sludge | A. sobria (AY 827494) |
ATCC, American Type Culture Collection, Manassas, VA; CECT, Coleccion Espanola de Cultivos Tipo, Valencia, Spain provided by Covadonga Arias, Auburn University, Auburn, AL; CDC, Centers for Disease Control and Prevention, Atlanta, GA; FDA, provided by Angelo DePaola, U.S. Food and Drug Administration, Dauphin Island, AL; ICDDR‐B, International Centre for Diarrhoeal Disease Research‐Bangladesh, Dhaka, Bangladesh; USDA, United States Department of Agriculture, Agriculture Research Service, Wyndmoor, PA; MLT, provided by Mark L. Tamplin, while at the University of Florida, Gainesville, FL.
Accession number for 16S rDNA sequences of Vibrio spp. from NCBI database that showed high sequence similarity.
16s rDNA sequences were generated in this study and were submitted to the NCBI database, because no previous sequence existed in the database for these strains.
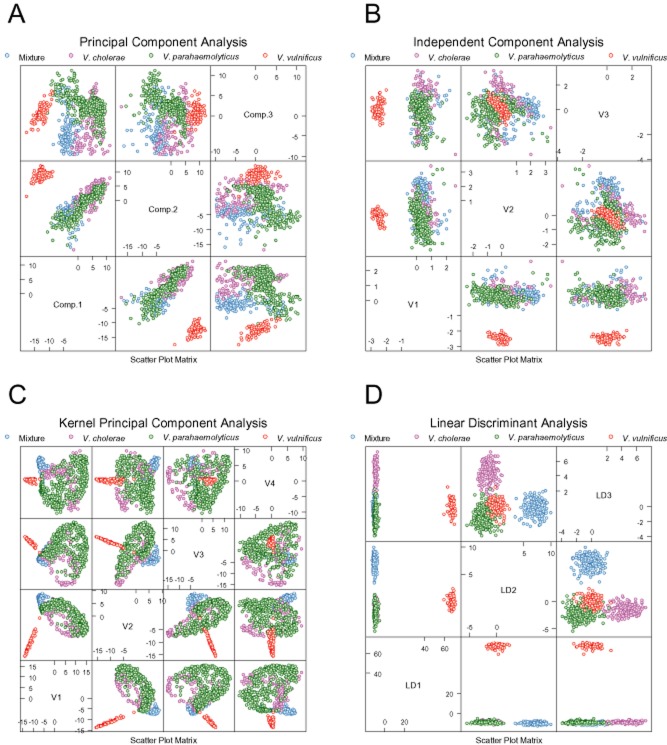
One hundred and ninety features were extracted from every collected forward‐scatter pattern. These features were used directly for exploratory analysis of the acquired results. Fig. 2A–C show principal component analysis (PCA), independent component analysis (ICA), and kernel PCA (KPCA) mapping of the data. Two major clusters of data points are clearly identifiable on the PCA plot. One of these clusters is identical with the V. vulnificus population, whereas the other contains colonies belonging to all other classes. The KPCA plot created using an RBF kernel shows three distinguishable clusters – one identical with the V. vulnificus class, another showing the ‘Mixture’ class consisting of Vibrio alginolyticus, Vibrio anguillarum, Vibrio mimicus and Vibrio orientalis, and the third containing V. cholerae and V. parahaemolyticus. The unsupervised exploratory analysis shows easily discernable differences between patterns formed by V. vulnificus and the remaining scatter patterns. The experimental data were used to construct and cross‐validate single‐instance supervised training systems. The classifier operated in a single‐instance fashion, meaning that it classified a single colony (rather than a plate or a sample) at a time. Three classifiers were evaluated: linear discriminant analysis (Fig. 2D), linear SVM and SVM‐RBF. 10× cross‐validation was used to assess the quality of the classifiers. The following measures of classification success are reported in Tables 2 and 3: sensitivity (true‐positive rate), specificity (1‐ false‐positive rate), accuracy and AUC [area under the ROC (receiver operating curves) curve for a binary classifier, which classifies a given class as ‘positive’ and all the other classes as ‘negative']. The classifiers were trained assuming that every tested colony represents an independent instance. The single‐colony classification results are summarized in Table 2; Fig. 3 shows the ROC plot for the SVM‐RBF classifier.
Figure 2.
Exploratory analysis of Vibrio scatter patterns grown on BHI‐NaCl plates: (A) Principal component plot, (B) Independent component plot, (C) Kernel principal component plot. (D) Linear discriminant plot showing separation of Vibrio classes in linear space. For the purpose of supervised classification four classes were created: Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, and ‘Mixture’ class containing colonies of Vibrio alginolyticus, Vibrio anguillarum, Vibrio mimicus and Vibrio orientalis. The classifiers were optimized to detect and recognize patterns of V. cholerae, V. parahaemolyticus and V. vulnificus.
Table 2.
Classification results for single‐instance classification
| Class | Classifier | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|
| Mixturea | FLD | 0.93 | 0.99 | 0.97 | 1.00 |
| SVM‐L | 0.93 | 0.98 | 0.97 | 0.99 | |
| SVM‐RBF | 0.96 | 0.99 | 0.98 | 1.00 | |
| Vibrio cholerae | FLD | 0.77 | 0.98 | 0.94 | 0.97 |
| SVM‐L | 0.88 | 0.98 | 0.96 | 0.99 | |
| SVM‐RBF | 0.94 | 0.99 | 0.98 | 1.00 | |
| Vibrio parahaemolyticus | FLD | 0.95 | 0.90 | 0.93 | 0.98 |
| SVM‐L | 0.95 | 0.94 | 0.95 | 0.99 | |
| SVM‐RBF | 0.97 | 0.96 | 0.97 | 1.00 | |
| Vibrio vulnificus | FLD | 1.00 | 1.00 | 1.00 | 1.00 |
| SVM‐L | 1.00 | 1.00 | 1.00 | 1.00 | |
| SVM‐RBF | 0.99 | 1.00 | 1.00 | 1.00 |
Mixture represents colonies of Vibrio alginolyticus, Vibrio anguillarum, Vibrio mimicus and Vibrio orientalis.
AUC, area under receiver operating curve; FLD, Fisher's linear discriminant; SVM‐L, support vector machine with linear kernel; SVM‐RBF, support vector machine with radial‐basis function kernel.
Table 3.
Detection success results for automated plate classification
| Species | Sensitivity | Specificity |
|---|---|---|
| Vibrio cholerae | 1.00 | 1.00 |
| Vibrio parahaemolyticus | 0.96 | 1.00 |
| Vibrio vulnificus | 1.00 | 1.00 |
Figure 3.
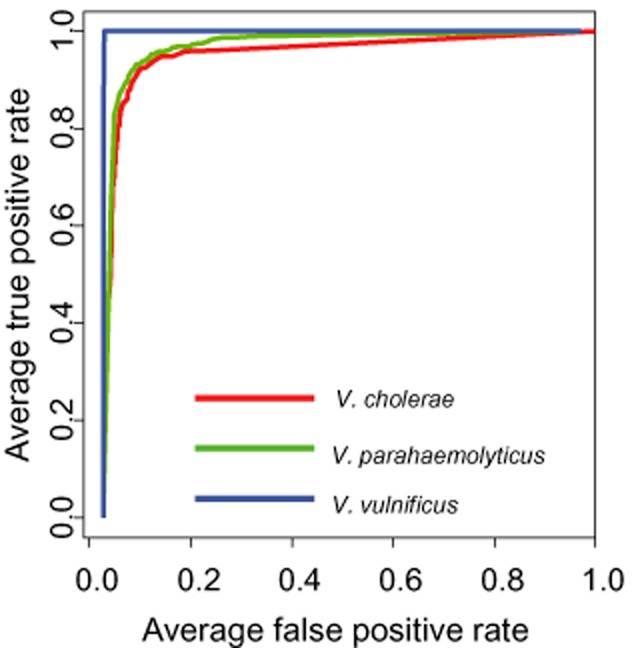
ROC for SVM‐RBF classifiers trained to detect colonies belonging to Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus.
Subsequently, the detection capability of the best classifier (SVM‐RBF) was evaluated with a more realistic approach in which a sample is represented by multiple instances (multiple colonies) present on a plate. However, no true multiple‐instance learning algorithms were used in this project (i.e. the classifiers did not take advantage of useful correlations between classes present on the plates). Instead, a classifier optimized for traditional supervised learning was employed in the multiple‐instance setting. To evaluate the classifier performance, an extended version of a cross‐validation procedure was designed. We generated in silico 100 virtual plates containing random combination of colonies for each of the tested detection tasks. Out of 100, the first group of 50 plates contained one or more colonies representing the class of interest, and another 50 did not. For instance, when V. cholerae detection capability was tested, 50 plates contained one or more V. cholerae colonies, whereas the remaining 50 plates contained colonies other than V. cholerae. The number of colonies on each plate was randomly assigned assuming that it followed Poisson distribution with a mean of 30. The prevalence of every class was normalized. The test was repeated six times for each species of interest. In the described setting, a sample (represented by a single plate) was considered positive if at least one colony in that sample belonged to the class of interest), and negative if all colonies on the plate were something other than the target. Separate accuracy scores were computed for each classification system individually optimized for detection of each of the classes. The results demonstrate very high sensitivity (96–100%) and specificity (100%) of the proposed method. Out of 50 plates containing one or more colonies of V. cholerae and 50 plates containing V. vulnificus, all were tagged as positive for V. cholerae and V. vulnificus respectively in each of the six repeats. In the case of V. parahaemolyticus, 96% of the contaminated plates were detected as positive. In all cases the plates which did not contain contamination were correctly labelled as negative. The results of this classification experiments are summarized in Table 3.
The identity of each Vibrio was initially confirmed by biochemical (API; bioMerieux, Inc.) testing but was further confirmed by sequencing of amplified PCR products of 16S rDNA. A majority of cultures exhibited very high sequence similarity (up to 99%) among 500 nucleotides with those of identified species from the GenBank database and are summarized in Table 1. However, 16S rDNA sequences from V. anguillarum and V. mimicus were not in the database; therefore these were deposited in the National Center of Biotechnology Information (NCBI) database: V. anguillarum (Accession No. EU 851049), and V. mimicus (EU 851050). Among the V. vulnificus strains, most showed very high similarities to two V. vulnificus strains with accession number EF546246 or EF546248 in the database, except V. vulnificus MLT1003, which showed a high similarity to V. vulnificus strain 1003BG‐OR, Accession No. AY 676130.
We also examined whether BARDOT is capable of differentiating capsular polysaccharide (CPS)‐producing V. vulnificus strains from non‐producing strains (Rosche et al., 2006). CPS‐positive strains tend to have a darker central spot in the scatter images whereas non‐CPS‐producing strains have a very bright central spot, owing to variations in the colony opacity (Fig. 4). However, BARDOT images among V. vulnificus strains exhibited the same characteristic patterns with many concentric rings starting at the centre of the colony and extending outward. Such rings are shared among V. vulnificus CPS‐producing or non‐producing strains and are distinct from the other species.
Figure 4.
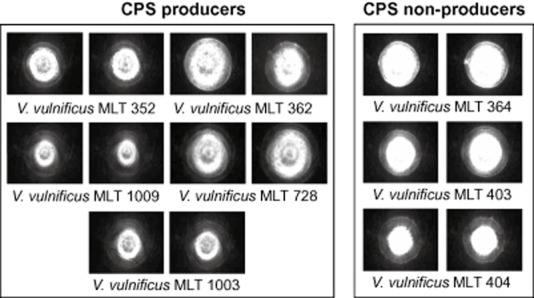
Differences in scatter images of CPS‐producing and CPS non‐producing strains of Vibrio vulnificus shown for two representative colonies of each strain on BHI‐NaCl. Central scatter features are absent for non‐CPS‐producing strains.
Physical and chemical stress and VBNC state do not affect BARDOT‐based detection
Exposure to stress conditions (pH, heat and osmotic stress) and subsequent resuscitation on BHI‐NaCl, heart‐infusion (HI) or TCBS agar did not alter the scatter patterns of any of the tested Vibrio spp. when compared with the untreated controls (Fig. S1). After resuscitation, VBNC V. vulnificus (Fig. S2) showed no discernable difference in the scatter patterns, except the smaller diameter compared to the control (Fig. S2). This demonstrates the ability of the method to recover and identify the pathogens using a light‐scattering sensor with a high degree of specificity even when the bacteria were subjected to possible stress encountered during processing and handling.
BARDOT efficiently detected Vibrio species from oyster and water samples
The ability of BARDOT to detect and identify V. cholerae, V. parahaemolyticus, and V. vulnificus in the presence of other vibrios or other microflora in oyster or water was examined. BARDOT accurately identified V. cholerae colonies on a BHI‐NaCl agar plate in the presence of 10‐fold higher concentrations of V. orientalis and Vibrio fluvialis in a mixed culture set up (Fig. 5).
Figure 5.
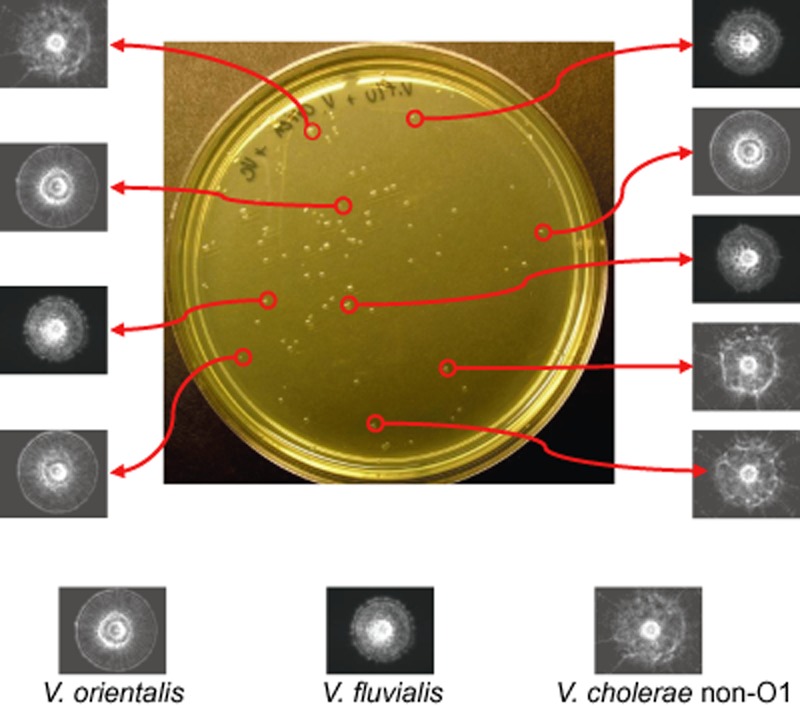
BARDOT‐based detection and identification of Vibrio cholerae non‐O1 colonies on BHI‐NaCl plate grown at 30°C for 18 h in presence of Vibrio orientalis and Vibrio fluvialis colonies.
Vibrio parahaemolyticus and V. vulnificus testing from oysters required approximately 18 h of total growth [6 h of enrichment in alkaline peptone water (APW) followed by 12 h growth on HI or TCBS agar plates] to obtain 1.3 ± 0.2 mm bacterial colonies. BARDOT analysis of the colonies took about 3 s for each colony and the scatter patterns from recovered colonies matched the reference controls (Fig. 6A). BARDOT also generated scatter patterns from multiple colonies that did not match with either V. vulnificus or V. parahaemolyticus. These unmatched colonies were considered background bacteria, isolated more from HI than TCBS (Fig 6B). Select colonies were also tested by colony PCR, which were in agreement with the BARDOT data for both V. vulnificus and V. parahaemolyticus (Fig 6C). All nine uninoculated control oysters were found to be positive for both V. vulnificus and V. parahaemolyticus, and albeit all nine inoculated oysters were also positive (Table 4). Vibrio parahaemolyticus was also successfully detected from spiked sterile tap water (103 CFU ml−1) and negative controls for the tap water (non‐spiked) did not produce any colonies (Fig. S3).
Figure 6.
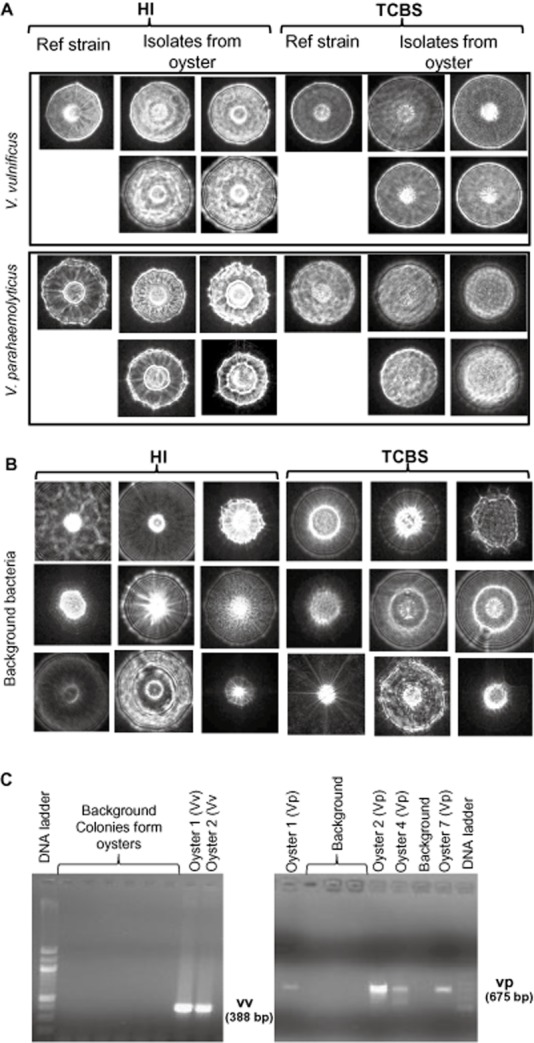
A. Isolation and identification of Vibrio vulnificus and Vibrio parahaemolyticus from oyster on HI and TCBS agar plates using BARDOT.
B. Scatter images of representative background bacterial colonies are shown.
C. PCR confirmation of scatter images of V. parahaemolyticus and V. vulnificus colonies from select oyster samples using vpm (V. parahaemolyticus)‐ and cyt (V. vulnificus)‐specific gene primers. Selected background colonies from B did not produce any amplified products specific to V. parahaemolyticus or V. vulnificus.
Table 4.
Vibrio detection from naturally contaminated or inoculated oysters plated on TCBS platesa
| Oyster # | Colony ID | Vibrio vulnificus | Vibrio parahaemolyticus | ||
|---|---|---|---|---|---|
| BARDOT | PCR | BARDOT | PCR | ||
| Uninoculated | |||||
| 1 | CO‐1‐1 | + | + | + | + |
| 2 | CO‐2‐1 | + | + | + | + |
| 3 | CO‐3‐1 | + | + | + | NT |
| 4 | CO‐1–2 | + | NT | + | + |
| 5 | CO‐2‐2 | + | + | + | + |
| 6 | CO‐3‐2 | + | + | + | + |
| 7 | CO‐4‐2 | + | + | + | + |
| 8 | CO‐5‐2 | + | + | + | NT |
| 9 | CO‐6‐2 | + | NT | + | NT |
| Inoculated | |||||
| 10 | EO‐1‐1 | + | NT | + | + |
| 11 | EO‐2‐1 | + | + | + | + |
| 12 | EO‐3‐1 | + | NT | + | + |
| 13 | EO‐1–2 | + | NT | + | + |
| 14 | EO‐2‐2 | + | + | + | + |
| 15 | EO‐3‐2 | + | NT | + | + |
| 16 | EO‐4‐2 | + | NT | + | + |
| 17 | EO‐5‐2 | + | + | + | + |
| 18 | EO‐6‐2 | + | NT | + | NT |
Oysters were enriched in alkaline peptone water for 6 h at 30°C and then plated on HI and TCBS agar plates and incubated for 12 h. Both plates were examined by BARDOT for Vibrio and data for TCBS plates are presented. Select positive colonies were examined by colony PCR using cyt primers for V. vulnificus and vpm primers for V. parahaemolyticus.
Discussion
In this study, BARDOT was employed to differentiate various cultures of Vibrio at the species level, especially to accurately detect and identify V. parahaemolyticus, V. vulnificus and V. cholerae. The BARDOT system has proved to be a powerful tool allowing for non‐invasive and non‐destructive differentiation of Vibrio spp. The analysis is performed without the use of biochemical or molecular techniques, ensuring that the colonies are available for further analysis. BARDOT does not require any probes or labelling reagents. The sample preparation is simple and the sensor can be employed directly with the colonies that result from a conventional culture‐based assay. Consistency of scatter patterns produced by bacterial colonies is supported by optical light‐scattering theory, which states that the scatter signal observed from the illuminated source depends on the properties (refractive index, size, shape) of the involved scatterer (in this case, a colony). Therefore, samples showing similar biophysical properties should produce similar scatter patterns, and samples with identical properties are expected to produce matching patterns. The scatter patterns can be related to the genotype, cellular arrangement and metabolic by‐products produced during colony growth on agar plates (Banada et al., 2009).
The collected patterns for each species of Vibrio appear to be visually distinct from each other and the patterns also varied depending on the media used (Fig. 1). This is consistent with our previous observation where colony phenotype and the resulting scatter signatures are largely dependent on the substrate/nutrient utilization by a microorganism (Banada et al., 2009). The measurements were consistent and highly reproducible. The raw greyscale representations of scatter magnitude can be used directly as a quick and easy way to distinguish Vibrio species.
We tested whether BARDOT is able to differentiate CPS‐producing V. vulnificus from CPS non‐producing strains. CPS‐positive strains produce opaque colonies, while CPS‐negative bacteria produce translucent colonies (Simpson et al., 1987; Smith and Siebeling, 2003). BARDOT analysis revealed that the scatter images produced by CPS‐positive strains contained a diffused spot at the centre of their pattern, whereas, CPS‐negative strains produced a completely bright centre (Fig. 4). CPS production has been associated with virulence of V. vulnificus (Simpson et al., 1987; Smith and Siebeling, 2003); thus it is possible that BARDOT could be used to differentiate virulent from avirulent V. vulnificus strains. While these preliminary results look intriguing, in‐depth analysis is needed to conclusively demonstrate the suitability of BARDOT for this task.
Pathogens surviving within a food matrix are often subjected to many stress factors such as reduced water activity (Aw), acidity, osmotic stress resulting from exposure to high salt content, and the stress endured during processing or retort procedures. We demonstrated that the stress conditions applied to Vibrio did not influence the respective scatter patterns, as long as the condition of the agar, which the bacterial colony was allowed to recover on, remained consistent (Fig. S1). This remains true for temperature‐, pH‐ and osmotic‐related stress.
Some species of Vibrio, such as V. vulnificus, are known to enter a VBNC state and may persist in that state in some seafood matrices (Oliver et al., 1995). We have shown that cells that have experienced the VBNC state (Fig. S2) generate after resuscitation scatter patterns that are indistinguishable from the pre‐VBNC patterns (Fig. S1). These observations support an important notion that BARDOT‐derived patterns remain unaltered regardless of the bacteria isolation procedure or the conditions endured by bacteria. Thus, BARDOT may serve as a system for identification of vibrios and other bacteria exposed to harsh environmental conditions.
We further explored the applicability of the BARDOT system for detecting bacteria in natural samples. Uninoculated or inoculated oysters and water samples were analysed by BARDOT. From a deliberate mixed culture, V. cholerae colonies can be detected and identified on an agar plate containing two other Vibrio spp. (Fig. 5). Similarly, V. parahaemolyticus and V. vulnificus were successfully detected from naturally contaminated oysters by BARDOT and cultures were confirmed by PCR. Surprisingly all nine uninoculated oysters were found to be positive for both. Even though, incidence rate for these two pathogens in oyster could be very high (22–89%) (Cook et al., 2002; Parvathi et al., 2004; Chen et al., 2010) but in this experiment since all uninoculated oysters were maintained in the same tank for 24 h before testing, pathogens may have spread from the contaminated oyster(s) to others. Nevertheless, all uninoculated or inoculated oysters were found to be positive confirming BARDOT's ability to detect Vibrio from naturally contaminated oyster and spiked tap water in presence of background bacteria within 18 h (Fig. 6, Table 4, Fig. S3). Scatter patterns of colonies on HI and TCBS that did not match with either V. vulnificus or V. parahaemolyticus are referred to as the background bacteria, whose population was much higher in non‐selective HI than selective TCBS plates. Possibility of some of those background bacteria to be vibrios other than V. vulnificus or V. parahaemolyticus cannot be ruled out since the conventional enrichment (APW) and plating (TCBS) methods were used for BARDOT based analysis. Thus, for Vibrio detection by BARDOT, it is recommended that the samples are enriched in standard enrichment broth (APW for at least 6 h) before plating onto TCBS at 30°C for 12 h.
The recognition/classification success of the BARDOT system was analysed in detail using standard statistical measures such as sensitivity, specificity, and AUC in order to compare the proposed technology to other methods. Recently a number of new label‐free biosensors have been reported in the literature (Lazcka et al., 2007; Bhunia, 2008; Velusamy et al., 2010). These tools are label free in the sense of not employing fluorescence labels, but use alternative detection modalities such as surface plasmon resonance, amperometric and potentiometric measurements, or electrochemical impedance spectroscopy. However, these biosensors still utilize traditional biological recognition elements: enzymes, antibodies and nucleic acids. The only well‐researched and broadly utilized techniques capable of true reagentless fingerprinting of bacteria are vibrational spectroscopic methods (Raman and IR) (Naumann et al., 1991; Rösch et al., 2003), and autofluorescence‐based observations (Estes et al., 2003). Unfortunately, evaluation of sensitivity and specificity of spectroscopic (and hence chemometric‐based systems) label‐free biodetectors in order to make a comparison with the BARDOT methodology is very difficult. In many reports on the use of spectroscopic methods only unsupervised learning was employed (such as clustering), and performance of the systems was not evaluated using any objective statistical measure (Sandt et al., 2003; Maquelin et al., 2006; Willemse‐Erix et al., 2009). In other published reports the use of supervised methods was not followed by detailed performance analysis that would allow conclusions regarding the false‐positive and false‐negative rates across a broad range of classes (Goodacre et al., 1998; Yang and Irudayaraj, 2003; Rebuffo‐Scheer et al., 2008). The classification success of the BARDOT system for single‐colony measurement is very high (AUC ∼ 0.98), and the specificity and sensitivity of our system as applied to multiple colony samples is close to 100% for all the tested Vibrio classes. Regardless of the accuracy of other spectral‐based methods, these results place BARDOT on the same level with highly complex and sophisticated molecular techniques.
In summary, we have demonstrated a method for the identification of Vibrio species using a forward light‐scattering technique. This method offers a fast and reliable way to detect Vibrio on BHI‐NaCl, HI and TCBS agar plates. Owing to BARDOT's non‐invasive nature, colonies may be retained for further verification or characterization. Scatter images of E. coli, Salmonella, Staphylococcus and Listeria are being obtained using BARDOT, all of which are showing distinct patterns (Banada et al., 2009). We have applied this technology to food and water samples to verify its robustness, and we foresee BARDOT being used routinely for rapid real‐time detection of bacterial colonies growing on a Petri dish without further sample processing. Screening for specific Vibrio spp. from 100 colonies on an agar plate may be made within minutes; therefore, the speed of Vibrio identification by BARDOT is unparalleled by any known existing sensor or conventional methodology. Further identification of features which characterize light‐scatter patterns may lead to future refinements in the assay and enhanced accuracy for this technique.
Experimental procedures
Media, culture conditions and acquisition of scatter patterns for Vibrio species
The bacterial cultures used (Table 1) were prepared as follows unless otherwise stated. Cultures from frozen stock were grown in tryptic soy broth containing 1% sodium chloride (NaCl) for 18−24 h at 30°C and subcultured in BHI broth (Acumedia, Neogen, MI) containing 1% NaCl (BHI‐NaCl). The cultures were serially diluted and spread‐plated on the surface of BHI‐NaCl agar (BHI‐NaCl with 1.5% agar), HI (Acumedia), or Vibrio selective TCBS (Acumedia) plates to obtain 30–100 colonies per plate. The plates were incubated at 30°C until the diameter of each colony reached 1.3 ± 0.2 mm (typically 12–18 h). Because growth rates are variable among species, colony size was used as a fixed parameter. Scatter patterns of colonies were acquired and images were analysed using the BARDOT (Advanced Bioimaging Systems, West Lafayette, IN). The identity of bacterial cultures was confirmed by Gram‐staining, testing with API 20E (BioMerieux, Hazelwood, MO), and 16S rDNA sequencing.
Bacterial confirmation by 16S rDNA sequencing
Cultures of Vibrio species (Table 1) were confirmed by 16s rDNA sequencing of PCR‐amplified products (Nilsson et al., 2003). 16S rDNA‐specific primer pairs, UFUL and URUL were used to amplify the target gene. The PCR conditions include: an initial denaturation at 94°C for 2 min, followed by 30 cycles consisting of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, and final extension at 72°C for 5 min. PCR products were sequenced at the Purdue Genomics Core Facility (Purdue University, W. Lafayette, IN) and the 16S rDNA nucleotide sequences were compared to the NCBI database by BLAST search (http://blast.ncbi.nlm.nih.gov/).
Effect of physiological stress on forward light‐scattering patterns
Cultures of V. parahaemolyticus CECT511, V. vulnificus MLT362 and V. anguillarum were subjected to the following stresses for 3 h: low pH (4.0), osmotic stress (5% NaCl) or heat stress (42°C) (Hahm and Bhunia, 2006). Overnight cultures were inoculated into BHI‐NaCl broth after adjusting to pH 4.0 using concentrated HCl or into BHI broth containing 5% NaCl and held at 30°C for 3 h. For heat stress, cultures in BHI‐NaCl broth were incubated at 42°C for 3 h. Each culture was washed and re‐suspended in 20 mM phosphate buffered saline (PBS, pH 7.2) and plated onto BHI‐NaCl agar plates for BARDOT analysis.
Detection of bacteria recovered from viable but non‐culturable state
Viable but non‐culturable state in V. vulnificus strains (MLT 362 and MLT 364) were achieved by following procedure described before (Smith and Oliver, 2006). Briefly, cultures were grown in HI broth at room temperature (RT) for 24 h with agitation, subcultured in HI until log phase (OD610 = 0.13–0.2), and inoculated into pre‐chilled (4°C) ½ strength artificial sea water (ASW) (NaCl 24.7 g l−1, KCL 0.67 g l−1, CaCl·H2O 1.36 g l−1, MgCl2·6H2O 4.66 g l−1, MgSO4·7H2O 6.29 g l−1, NaHCO3 0.18 g l−1) (Wolf and Oliver, 1992) and maintained under that condition. Entrance to VBNC state was monitored by analysing 0.1 ml of the sample (serially diluted in ASW) by plating on HI agar plates and by staining cells by LIVE/DEAD stain (BacLight stain kit: Molecular Probes). When no culturable cells were detected in 0.1 ml of test sample, then 5 ml of sample was passed through a 0.22 um nucleopore filter, placed on HI and TCBS agar plates for 2 h then aseptically removed. Plates were incubated at RT for 24 h and monitored for growth. Cultures were deemed VBNC when there was less than 0.1 CFU ml−1 for each strain (Fig. S2). To resuscitate cells, cultures (5 ml) were incubated at RT for 24 h, plated on HI and TCBS, incubated at 30°C for about 12 h and analysed by BARDOT.
Application of BARDOT to detect and identify Vibrio from oyster and water samples
Two dozen oysters (prime grade) from north Atlantic coast were purchased from a local fish monger and transported on ice to the laboratory. Six oysters were discarded because they were either too small or dead. Upon arrival they were placed immediately inside a clear plastic bag and kept in ASW for 30 min. Oysters were rinsed in deionized water, scrubbed and washed in 70% ethanol, and divided into two groups (nine per group): control and experimental, and placed in two separate 5 gallon bucket containing 7 l of sterile ASW, and continually aerated with flowing air. After 2 h, the experimental group received mixed (1:1) culture of V. vulnificus MLT 364 and V. parahaemolyticus CETT5711 to obtain a final concentration of 105 CFU ml−1 and both groups were maintained at RT for 24 h with aeration. Active filtering by oysters were noticed as the ASW became cloudy then cleared multiple times. Tissue and liquor from each oyster were harvested, weighed, placed in stomacher bag built with filter mesh (Nasco Whirlpak®, Fort Atkins, WI) containing APW (pH 8.5) with 1% peptone (Becton Dickinson) to achieve 1:10 dilution, homogenized (for 2 min in a stomacher; Seward, Norfork, UK) and incubated at 30°C for 6 h. Enriched samples were diluted, plated on TCBS and HI, incubated at 30°C for 12 h, and plates were scanned by BARDOT. Colony scatter images were compared with scatter image library consisting of V. vulnificus, and V. parahaemolyticus. Select colonies from representative plates were further verified by colony PCR using primers specific for V. vulnificus and V. parahaemolyticus described below. Entire oyster experiment was conducted under a biosafety cabinet.
Similarly, V. parahaemolyticus CECT511 was also inoculated into 500 ml sterile tap water at about 2 × 103 CFU ml−1 and the samples were passed through a membrane filtration system using 0.45 μm filters (Millipore). The filter was then placed in a centrifuge tube with 10 ml of sterile PBS, vortexed for 2 min, and centrifuged at 5000 g for 10 min. The cell pellet was re‐suspended in 100 μl of PBS and plated onto BHI‐NaCl agar. Plates were incubated at 30°C for 12 h or until colony size reached 1.3 ± 0.2 mm, and colonies were subsequently analysed using BARDOT.
PCR verification of vibrios
Polymerase chain reactions were used to confirm the identity of colonies on BHI‐NaCl plates. Colonies of V. parahaemolyticus were confirmed by using VPM1 and VPM2 primers specific for the metalloprotease gene (Luan et al., 2007). Vibrio vulnificus was confirmed by using Cyt1 and Cyt2 primers targeting the haemolysin/cytolysin gene (Morris et al., 1987). A standard amplification condition was used consisting of initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min and 72°C for 1 min, and final extension at 72°C for 5 min. The amplicons were analysed by agarose gel electrophoresis.
Pattern analysis and classification
Each scatter signature was analysed using an automated pattern processing and classification system (Bayraktar et al., 2006; Banada et al., 2009), which accompanied the BARDOT hardware. A set of radial orthogonal pseudo‐Zernike polynomials, as well as Haralick texture features, was computed for every scatter pattern. The feature reduction and selection procedure was based on three independent wrappers whose output was combined. One hundred and ninety features were analysed using sequential floating forward selection, which is a combination of the sequential forward and the sequential backward methods. The sequential floating forward selection‐based wrapper was used with the k‐nearest neighbour, linear discriminant analysis and recursive‐portioning algorithms. The number of selected features was 23.
Subsequently, the selected best features were used to train the pattern‐recognition system. Three different classifiers were used: linear Fisher's discriminant, SVM‐L and SVM‐RBF. The classifiers were optimized using standard 10× cross‐validation. The quality of detection and classification was evaluated by computing values of sensitivity and specificity, and AUC.
For the purpose of supervised classification four classes were created: V. cholerae, V. parahaemolyticus, V. vulnificus, and ‘Mixture’ class containing colonies of V. alginolyticus, V. anguillarum, V. mimicus and V. orientalis. The classifiers were optimized to detect and recognize patterns belonging to three important classes of known pathogens: V. cholerae, V. parahaemolyticus and V. vulnificus.
Acknowledgments
This research was supported through a cooperative agreement with the Agricultural Research Service of the U.S. Department of Agriculture project number 1935‐42000‐035 and the Center for Food Safety Engineering at Purdue University. The authors thank Covadonga Arias and Mark Tamplin for bacterial cultures, and Amanda Bettasso for technical help. Authors declare no conflict of interest exists.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. A. Scatter images of resuscitated colonies of Vibrio species that were exposed to various stresses [pH (4.0), heat (42°C) and osmotic (5% NaCl)] for 3 h. Cultures were exposed to stress, serially diluted and plated onto BHI containing 1% NaCl, and allowed to grow for 12 h before scatter images were acquired.
B. Scatter images of V. vulnificus MLT 362 and 364 recovered from VBNC state.
Fig. S2. Induction of VBNC state in V. vulnificus MLT 362 and 364 strains suspended in ASW.
A. Viable cell counts were determined by plating on HI agar (HIA) plates and by staining cells with BacLight LIVE/DEAD staining kit.
B. Fluorescence microscopic analysis of live, dead and live/dead V. vulnificus cells taken at 48 h indicating < 1% cells are still viable at that condition and are entering into VBNC state.
Fig. S3. Detection of V. parahaemolyticus on BHI‐NaCl agar plate from spiked water sample using BARDOT.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bae E., Banada P.P., Huff K., Bhunia A.K., Robinson J.P., Hirleman E.D. Bio‐physical modeling of forward scattering from bacterial colonies using scalar diffraction theory. Appl Opt. 2007;46:3639–3648. doi: 10.1364/ao.46.003639. , and . [DOI] [PubMed] [Google Scholar]
- Bae E., Banada P.P., Huff K., Bhunia A.K., Robinson J.P., Hirleman E.D. Analysis of time‐resolved scattering from macroscale bacterial colonies. J Biomed Opt. 2008;13:014010. doi: 10.1117/1.2830655. , and . [DOI] [PubMed] [Google Scholar]
- Banada P.P., Guo S.L., Bayraktar B., Bae E., Rajwa B., Robinson J.P. Optical forward‐scattering for detection of Listeria monocytogenes and other Listeria species. Biosens Bioelectron. 2007;22:1664–1671. doi: 10.1016/j.bios.2006.07.028. et al. [DOI] [PubMed] [Google Scholar]
- Banada P.P., Huff K., Bae E., Rajwa B., Aroonnual A., Bayraktar B. Label‐free detection of multiple bacterial pathogens using light‐scattering sensor. Biosens Bioelectron. 2009;24:1685–1692. doi: 10.1016/j.bios.2008.08.053. et al. [DOI] [PubMed] [Google Scholar]
- Bayraktar B., Banada P.P., Hirleman E.D., Bhunia A.K., Robinson J.P., Rajwa B. Feature extraction from light‐scatter patterns of Listeria colonies for identification and classification. J Biomed Opt. 2006;11:34006. doi: 10.1117/1.2203987. , and . [DOI] [PubMed] [Google Scholar]
- Bhunia A.K. Biosensors and bio‐based methods for the separation and detection of foodborne pathogens. In: Taylor S., editor. London, UK: Elsevier; 2008. pp. 1–44. [DOI] [PubMed] [Google Scholar]
- Cabanillas‐Beltran H., Llausas‐Magana E., Romero R., Espinoza A., Garcia‐Gasca A., Nishibuchi M. Outbreak of gastroenteritis caused by the pandemic Vibrio parahaemolyticus O3:K6 in Mexico. FEMS Microbiol Lett. 2006;265:76–80. doi: 10.1111/j.1574-6968.2006.00475.x. et al. [DOI] [PubMed] [Google Scholar]
- Chen W.‐B., Zhang C. An automated bacterial colony counting and classification system. Inf Syst Front. 2009;11:349–368. , and . [Google Scholar]
- Chen Y., Liu X.M., Yan J.W., Li X.G., Mei L.L., Ma Q.F., Ma Y. Foodborne pathogens in retail oysters in South China. Biomed Environ Sci. 2010;23:32–36. doi: 10.1016/s0895-3988(10)60028-1. , and . [DOI] [PubMed] [Google Scholar]
- Cook D.W., Leary P., Hunsucker J.C., Sloan E.M., Bowers J.C., Blodgett R.J., Depaola A. Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: a national survey from June 1998 to July 1999. J Food Protect. 2002;65:79–87. doi: 10.4315/0362-028x-65.1.79. , and . [DOI] [PubMed] [Google Scholar]
- Daniels N.A. Vibrio vulnificus oysters: pearls and perils. Clin Infect Dis. 2011;52:788–792. doi: 10.1093/cid/ciq251. [DOI] [PubMed] [Google Scholar]
- Estes C., Duncan A., Wade B., Lloyd C., Ellis W., Powers L. Reagentless detection of microorganisms by intrinsic fluorescence. Biosens Bioelectron. 2003;18:511–519. doi: 10.1016/s0956-5663(03)00008-3. , and . [DOI] [PubMed] [Google Scholar]
- Goodacre R., Timmins E.M., Burton R., Kaderbhai N., Woodward A.M., Kell D.B., Rooney P.J. Rapid identification of urinary tract infection bacteria using hyperspectral whole‐organism fingerprinting and artificial neural networks. Microbiology. 1998;144:1157–1170. doi: 10.1099/00221287-144-5-1157. , and . [DOI] [PubMed] [Google Scholar]
- Gulig P.A., Bourdage K.L., Starks A.M. Molecular pathogenesis of Vibrio vulnificus. J Microbiol. 2005;43:118–131. , and . [PubMed] [Google Scholar]
- Hahm B.K., Bhunia A.K. Effect of environmental stresses on antibody‐based detection of Escherichia coli O157:H7, Salmonella enterica serotype Enteritidis and Listeria monocytogenes. J Appl Microbiol. 2006;100:1017–1027. doi: 10.1111/j.1365-2672.2006.02814.x. , and . [DOI] [PubMed] [Google Scholar]
- Huq A., Sack R.B., Nizam A., Longini I.M., Nair G.B., Ali A. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol. 2005;71:4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.L., Hara‐Kudo Y., Krantz J.A., Benner Jr R.A., Smith A.B., Dambaugh T.R. Comparison of molecular detection methods for Vibrio parahaemolyticus and Vibrio vulnificus. Food Microbiol. 2012;30:105–111. doi: 10.1016/j.fm.2011.12.011. et al. [DOI] [PubMed] [Google Scholar]
- Kong R.Y.C., Lee S.K.Y., Law T.W.F., Law S.H.W., Wu R.S.S. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 2002;36:2802–2812. doi: 10.1016/s0043-1354(01)00503-6. , and . [DOI] [PubMed] [Google Scholar]
- Lazcka O., Campo F.J.D., Munoz F.X. Pathogen detection: a perspective of traditional methods and biosensors. Biosensor Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. , and . [DOI] [PubMed] [Google Scholar]
- Luan X.Y., Chen J.X., Zhang X.H., Jia J.T., Sun F.R., Li Y. Comparison of different primers for rapid detection of Vibrio parahaemolyticus using the polymerase chain reaction. Lett Appl Microbiol. 2007;44:242–247. doi: 10.1111/j.1472-765X.2006.02074.x. , and . [DOI] [PubMed] [Google Scholar]
- McLaughlin J.B., DePaola A., Bopp C.A., Martinek K.A., Napolilli N.P., Allison C.G. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med. 2005;353:1463–1470. doi: 10.1056/NEJMoa051594. et al. [DOI] [PubMed] [Google Scholar]
- Maquelin K., van der Dijkshoorn L., Reijden T.J.K., Puppels G.J. Rapid epidemiological analysis of Acinetobacter strains by Raman spectroscopy. J Microbiol Methods. 2006;64:126–131. doi: 10.1016/j.mimet.2005.04.028. , and . [DOI] [PubMed] [Google Scholar]
- Morris J.G., Wright A.C., Roberts D.M., Wood P.K., Simpson L.M., Oliver J.D. Identification of envrionmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin hemolysin gene. Appl Environ Microbiol. 1987;53:193–195. doi: 10.1128/aem.53.1.193-195.1987. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G.B., Ramamurthy T., Bhattacharya S.K., Dutta B., Takeda Y., Sack D.A. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann D., Helm D., Labischinski H. Microbiological characterizations by FT‐IR spectroscopy. Nature. 1991;351:81–82. doi: 10.1038/351081a0. , and . [DOI] [PubMed] [Google Scholar]
- Nigro O.D., Hou A., Vithanage G., Fujioka R.S., Steward G.F. Temporal and spatial variability in culturable pathogenic Vibrio spp. in lake Pontchartrain, Louisiana, following Hurricanes Katrina and Rita. Appl Enriron Microbiol. 2011;77:5384–5393. doi: 10.1128/AEM.02509-10. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson W.B., Paranjype R.N., DePaola A., Strom M.S. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J Clin Microbiol. 2003;41:442–446. doi: 10.1128/JCM.41.1.442-446.2003. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien M., Egan D., O'Kiely P., Forristal P.D., Doohan F.M., Fuller H.T. Morphological and molecular characterisation of Penicillium roqueforti and P. paneum isolated from baled grass silage. Miicrobiol Res. 2008;112:921–932. doi: 10.1016/j.mycres.2008.01.023. , and . [DOI] [PubMed] [Google Scholar]
- Oliver J.D., Hite F., McDougald D., Andon N.L., Simpson L.M. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl Environ Microbiol. 1995;61:2624–2630. doi: 10.1128/aem.61.7.2624-2630.1995. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D., Masini L., Bacchiocchi S. A biochemical protocol for the isolation and identification of current species of Vibrio in seafood. J Appl Microbiol. 2003;95:1277–1284. doi: 10.1046/j.1365-2672.2003.02105.x. , and . [DOI] [PubMed] [Google Scholar]
- Parvathi A., Kumar H.S., Karunasagar I., Karunasagar I. Detection and enumeration of Vibrio vulnificus in Oysters from two estuaries along the southwest coast of India, using molecular methods. Appl Environ Microbiol. 2004;70:6909–6913. doi: 10.1128/AEM.70.11.6909-6913.2004. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamer S., Sandoe J.A.T., Kerr K.G. Use of colony morphology to distinguish different enterococcal strains and species in mixed culture from clinical specimens. J Clin Microbiol. 2003;41:2644–2646. doi: 10.1128/JCM.41.6.2644-2646.2003. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebuffo‐Scheer C., Dietrich J., Wenning M., Scherer S. Identification of five Listeria species based on infrared spectra (FTIR) using macrosamples is superior to a microsample approach. Anal Bioanal Chem. 2008;390:1629–1635. doi: 10.1007/s00216-008-1834-1. , and . [DOI] [PubMed] [Google Scholar]
- Rhoads J. Post‐hurricane Katrina challenge: Vibrio vulnificus. J Am Acad Nurse Pract. 2006;18:318–324. doi: 10.1111/j.1745-7599.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Rösch P., Schmitt M., Kiefer W., Popp J. The identification of microorganisms by micro‐Raman spectroscopy. J Mol Struct. 2003;661–662:363–369. , and . [Google Scholar]
- Rosche T.M., Smith B., Oliver J.D. Evidence for an intermediate colony morphology of Vibrio vulnificus. Appl Environ Microbiol. 2006;72:4356–4359. doi: 10.1128/AEM.02937-05. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D.B., Colwell R.R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandt C., Sockalingum G.D., Aubert D., Lepan H., Lepouse C., Jaussaud M. Use of Fourier‐transform infrared spectroscopy for typing of Candida albicans strains isolated in intensive care units. J Clin Microbiol. 2003;41:954–959. doi: 10.1128/JCM.41.3.954-959.2003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L.M., White V.K., Zane S.F., Oliver J.D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.V., Matte M.H., Matte G.R., Jiang S., Sabeena F., Shukla B.N. Molecular analysis of Vibrio cholerae O1, O139, non‐O1, and non‐O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol. 2001;67:910–921. doi: 10.1128/AEM.67.2.910-921.2001. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.B., Siebeling R.J. Identification of genetic loci required for capsular expression in Vibrio vulnificus. Infect Immun. 2003;71:1091–1097. doi: 10.1128/IAI.71.3.1091-1097.2003. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B., Oliver J.D. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol. 2006;72:1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo G.M., Fontanarosa M., Di Pinto A., Musti M. Updated perspectives on emerging vibrios associated with human infections. Lett Appl Microbiol. 2004;39:117–126. doi: 10.1111/j.1472-765X.2004.01568.x. , and . [DOI] [PubMed] [Google Scholar]
- Thompson F.L., Iida T., Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velusamy V., Arshak K., Korostynska O., Oliwa K., Adley C. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv. 2010;28:232–254. doi: 10.1016/j.biotechadv.2009.12.004. , and . [DOI] [PubMed] [Google Scholar]
- Whitesides M.D., Oliver J.D. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl Environ Microbiol. 1997;63:1002–1005. doi: 10.1128/aem.63.3.1002-1005.1997. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse‐Erix D.F.M., Scholtes‐Timmerman M.J., van Jachtenberg J.‐W., Leeuwen W.B., Horst‐Kreft D., Bakker Schut T.C. Optical fingerprinting in bacterial epidemiology: raman spectroscopy as a real‐time typing method. J Clin Microbiol. 2009;47:652–659. doi: 10.1128/JCM.01900-08. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P.W., Oliver J.D. Temperature effects on the viable but non‐culturable state of Vibrio vulnificus. FEMS Microbiol Ecol. 1992;101:33–39. , and . [Google Scholar]
- Xia T., Zhu H., Shu H., Haigron P., Luo L. Image description with generalized pseudo‐Zernike moments. J Opt Soc Am A. 2007;24:50–59. doi: 10.1364/josaa.24.000050. , and . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Irudayaraj J. Rapid detection of foodborne microorganisms on food surface using Fourier transform Raman spectroscopy. J Mol Struct. 2003;646:35–43. , and . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A. Scatter images of resuscitated colonies of Vibrio species that were exposed to various stresses [pH (4.0), heat (42°C) and osmotic (5% NaCl)] for 3 h. Cultures were exposed to stress, serially diluted and plated onto BHI containing 1% NaCl, and allowed to grow for 12 h before scatter images were acquired.
B. Scatter images of V. vulnificus MLT 362 and 364 recovered from VBNC state.
Fig. S2. Induction of VBNC state in V. vulnificus MLT 362 and 364 strains suspended in ASW.
A. Viable cell counts were determined by plating on HI agar (HIA) plates and by staining cells with BacLight LIVE/DEAD staining kit.
B. Fluorescence microscopic analysis of live, dead and live/dead V. vulnificus cells taken at 48 h indicating < 1% cells are still viable at that condition and are entering into VBNC state.
Fig. S3. Detection of V. parahaemolyticus on BHI‐NaCl agar plate from spiked water sample using BARDOT.