Summary
Activated sludge used for wastewater treatment globally is composed of a high‐density microbial community of great biotechnological significance. In this study the presence and purpose of quorum sensing via N‐acylated‐l‐homoserine lactones (AHLs) in activated sludge was explored. The presence of N‐heptanoyl‐l‐homoserine lactone in organic extracts of sludge was demonstrated along with activation of a LuxR‐based AHL monitor strain deployed in sludge, indicating AHL‐mediated gene expression is active in sludge flocculates but not in the bulk aqueous phase. Bacterial isolates from activated sludge were screened for AHL production and expression of phenotypes commonly but not exclusively regulated by AHL‐mediated gene transcription. N‐acylated‐l‐homoserine lactone and exoenzyme production were frequently observed among the isolates. N‐acylated‐l‐homoserine lactone addition to sludge upregulated chitinase activity and an AHL‐ and chitinase‐producing isolate closely related to Aeromonas hydrophila was shown to respond to AHL addition with upregulation of chitinase activity. N‐acylated‐l‐homoserine lactones produced by this strain were identified and genes ahyI/R and chiA, encoding AHL production and response and chitinase activity respectively, were sequenced. These experiments provide insight into the relationship between AHL‐mediated gene expression and exoenzyme activity in activated sludge and may ultimately create opportunities to improve sludge performance.
Introduction
Wastewater treatment processes commonly rely on the biological activity of microorganisms in activated sludge to consume organic materials and convert them into biomass. The bulk of this biomass, in the form of flocs, can then be separated through settling. Activated sludge is of interest because it is a biotechnology fundamental to human health and represents a model high population density ecosystem for studying microbial interactions (Daims et al., 2006).
Flocs in activated sludge are composed of a dense microbial consortium in a matrix of extracellular polymeric substances produced primarily from lysed cell components (protein, humic substances, carbohydrates, nucleic acids and lipids) and adsorbed wastewater particles (Frolund et al., 1996). As such, parallels are drawn between activated sludge flocs and biofilms, with activated sludge flocs regarded as biofilms without surface association. Bacteria typically abundant in activated sludge flocs include the Alpha‐, Beta‐ and Gammaproteobacteria, as well as the Bacteroidetes and the Actinobacteria (Wagner and Loy, 2002). The high cell density of flocculates is likely to result in a multitude of interactions between cells, but very little is known about these interactions and the impacts they have on activated sludge structure and function.
N‐acyl‐l‐homoserine lactone (AHL)‐mediated gene expression is a cell density‐dependent gene expression mechanism. It involves the production of small membrane diffusible metabolites, AHLs, by an AHL synthase (LuxI homologue), interacting with their cognate receptor protein (LuxR homologue) when a threshold AHL concentration accumulates in the local environment thereby orchestrating gene expression (Manefield and Whiteley, 2007). Evidence has been generated suggesting that thin layers of cells can retard AHL diffusion (Mason et al., 2005) and it is clear that AHL‐mediated gene expression is active in biofilms harbouring AHL‐producing bacteria (McLean et al., 1997).
N‐acyl‐l‐homoserine lactone‐mediated gene expression is encoded by three out of five classes of Proteobacteria (alpha‐, beta‐ and gamma‐), with approximately 7% of genera within these classes containing known AHL‐producing representatives (Manefield and Turner, 2002). While restricted to the Proteobacteria within the bacterial phylogenetic tree, AHL producers tend to be abundant in the environment with activated sludge being no exception (Manefield and Whiteley, 2007).
N‐acyl‐l‐homoserine lactone‐mediated gene expression regulates the secretion of extracellular degradative enzymes (Givskov et al., 1997) in pathogenic bacteria to liberate resources from plants (McGowan et al., 1995) and animals (Whiteley et al., 1999). N‐acyl‐l‐homoserine lactones also control the synthesis of antibiotics (carbapenem, pyocyanin) to reduce competition for resources (McGowan et al., 1995). Siderophore production, biosurfactant production and the conjugal transfer of plasmids are other known AHL‐regulated activities (Fuqua and Winans, 1996; Lindum et al., 1998; Stintzi et al., 1998). It is conceivable that any of these phenotypes could affect microbial community structure and function in activated sludge.
Relatively little is known about the role of AHL‐mediated gene expression in activated sludge. A number of AHL‐producing bacteria have been isolated from activated sludge (Morgan‐Sagastume et al., 2005) and changes have been observed in community structure and phenol degradation rates when N‐3‐oxo‐hexanoyl‐l‐homoserine lactone concentrations were artificially elevated in sludge (Valle et al., 2004). More recently, studies have shown AHLs can be detected in biological material deposited on membrane bioreactors (Yeon et al., 2009).
In this study a combination of bioassays and analytical chemistry and molecular biology techniques have been used to generate compelling evidence suggesting AHL‐mediated gene expression is active in activated sludge and plays a role in regulating activity of extracellular degradative enzymes. Evidence is also presented suggesting Aeromonas hydrophila regulates chitinase activity through AHL‐mediated gene expression.
Results
Identification and AHL profiling of activated sludge isolates
This study was initiated through the production of a bacterial culture collection of activated sludge isolates. Activated sludge was plated on R2A agar and 80 colonies were randomly selected and characterized based on 16S rRNA gene sequencing and AHL production profiles. Of the 80 isolates selected, 52 of them were distinct with respect to phylogeny or phenotype. The collection includes 17 Aeromonas, 6 Acinetobacter, 5 Citrobacter, 4 Klebsiella, 2 Neisseria, 2 Malikia and 2 Pseudomonas lineages.
To confirm previous observations of AHL‐producing activity in sludge isolates, solvent extracted cultures of the collection were tested for AHL‐like activity using five bioassays based on four AHL response regulator enzymes (CviR, LuxR, TraR and LasR). The results are presented in Table 1. Of the 52 distinct isolates only 12 (23%) did not produce AHLs or AHL‐like activity detectable in the bioassays used (metabolites other than AHLs are known to generate false positives). Of the 40 isolates that displayed AHL‐like activity, 28 (70%) were positive in more than one assay. Fourteen distinct AHL production profiles were observed with the most common profile (excluding non‐AHL producers) activating LasR and inhibiting CviR. This was observed in six isolates, including the two Pseudomonas isolates, and likely indicates long‐chain AHL production. The second most common profile, observed in five isolates, activated CviR, TraR and LuxR, which is indicative of shorter‐chain AHLs. All five isolates with this profile were aeromonads. It is important to recognize that these values and proportions relate to the culturing approach used. The data do not necessarily reflect predominance of phylotypes or AHLs produced in situ.
Table 1.
Activated sludge isolates and their AHL activity and AHL‐associated phenotypes in various bioassays
| Isolate identifier | Closest relative | CviR | LuxR | TraR | LasR | CviR‐ | Lipase | Cellulase | Elastase | Chitinase | Antimicrobial | Surfactant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GC1 | Aeromonas sp. 17m clone G09 | + | + | + | − | − | + | − | − | + | − | − |
| GC2 | Aeromonas sp. ‘CDC 862‐83’ | − | − | − | − | − | + | − | − | + | + | + |
| GC3 | Aeromonas punctata VITSCA01 | + | + | + | − | − | + | + | − | + | − | − |
| GC4 | Aeromonas punctata VITSCA01 | − | − | − | + | − | + | − | + | − | − | − |
| GC5 | Aeromonas sp. TH088 | + | + | − | − | − | + | − | − | − | − | − |
| GC6 | Aeromonas sp. TH088 | − | − | − | − | + | − | − | − | − | − | + |
| GC7 | Aeromonas sp. TH088 | − | − | + | − | + | + | − | − | + | − | + |
| GC8 | Aeromonas sp. TH095 | − | − | − | + | + | − | − | − | − | − | − |
| GC9 | Aeromonas sp. TH095 | − | − | − | − | − | + | − | − | + | − | − |
| GC10 | Aeromonas hydrophila | + | + | + | − | − | + | − | − | − | − | + |
| GC11 | Aeromonadaceae sp. NJ‐40 | − | − | + | − | − | + | − | + | + | + | − |
| GC12 | Aeromonadaceae sp. NJ‐40 | − | − | + | − | − | + | − | − | − | + | − |
| GC13 | Aeromonadaceae sp. NJ‐40 | − | − | − | − | + | + | − | − | + | + | − |
| GC14 | Aeromonadaceae sp. NJ‐40 | − | − | − | + | − | − | + | + | + | − | + |
| GC15 | Aeromonas allosaccharophila PIC2 | + | + | + | − | − | + | − | − | + | − | − |
| GC16 | Aeromonas media | + | + | + | − | − | + | − | − | + | − | − |
| GC17 | Aeromonas media PW28 | − | − | + | − | − | + | − | − | + | + | − |
| GC18 | Citrobacter sp. AzoR‐4 | − | + | + | − | + | − | − | − | − | − | − |
| GC19 | Citrobacter sp. AzoR‐4 | − | − | − | + | + | − | − | − | − | − | − |
| GC20 | Citrobacter sp. AzoR‐4 | − | + | + | + | + | − | − | − | − | − | − |
| GC21 | Citrobacter sp. AzoR‐4 | − | + | + | − | + | − | − | − | − | − | + |
| GC22 | Citrobacter sp. AzoR‐4 | − | + | − | + | + | − | − | − | − | − | + |
| GC23 | Acinetobacter johnsonii CONC8 | − | + | − | − | − | − | − | − | − | − | − |
| GC24 | Acinetobacter johnsonii CONC8 | − | + | − | + | − | − | − | − | − | − | − |
| GC25 | Acinetobacter johnsonii CONC8 | − | − | − | − | − | − | − | − | − | + | − |
| GC26 | Acinetobacter johnsonii CONC8 | − | + | − | + | − | − | − | − | − | − | + |
| GC27 | Acinetobacter sp. Hi7 clone G07 | − | − | − | − | − | − | − | − | − | − | − |
| GC28 | Acinetobacter johnsonii FR2_89con | − | − | − | − | − | + | − | + | − | − | − |
| GC29 | Klebsiella sp. | − | − | − | − | − | − | + | + | − | + | − |
| GC30 | Klebsiella sp. 141 clone D03 | − | + | + | + | + | − | − | + | − | + | − |
| GC31 | Klebsiella sp. clone TM2_6 | − | + | − | + | + | − | + | + | − | − | + |
| GC32 | Klebsiella oxytoca clone C06 | − | + | + | + | + | − | − | − | − | − | + |
| GC33 | Neisseria sp. GRW59 | − | − | − | + | − | − | + | + | − | − | − |
| GC34 | Neisseria sp. GRW59 | − | − | − | + | − | − | + | − | − | − | − |
| GC35 | Pseudomonas sp. XQ3e | − | − | − | + | + | − | − | + | − | + | − |
| GC36 | Pseudomonas sp. R‐35723 | − | − | − | + | + | − | − | + | − | − | + |
| GC37 | Shigella sp. 4096 | − | + | − | + | + | − | − | + | − | − | − |
| GC38 | Vitreoscilla stercoraria | − | − | − | − | − | − | − | − | − | − | − |
| GC39 | Gordonia Australia A554 | − | − | − | − | − | − | − | + | − | − | − |
| GC40 | Klebsiella sp. 141 | − | − | − | − | + | + | − | − | − | − | − |
| GC41 | Enterococcus faecalis D023 | − | − | − | − | − | − | − | + | − | + | − |
| GC42 | Microbacterium paraoxydans M2 | − | + | − | + | + | − | − | − | − | − | − |
| GC43 | Chitinimonas taiwanensis fA3 | − | + | − | − | − | − | + | − | − | − | − |
| GC44 | Uncultured ß‐Proteobacterium | − | − | − | − | − | + | − | + | − | − | − |
| GC45 | Malikia spinosa | − | − | − | + | + | − | − | − | − | − | − |
| GC46 | Malikia spinosa | − | + | − | + | + | − | − | − | − | − | − |
| GC47 | Pantoea agglomerans 3I2 | − | + | + | + | − | − | − | + | − | − | + |
| GC48 | Acidovorax sp. PPs‐5 | − | − | − | − | − | − | − | − | − | − | + |
| GC49 | Raoultella terrigena m30 | − | + | + | + | + | − | + | − | − | − | + |
| GC50 | Microbacterium sp. KSL5401‐069 | − | + | − | + | − | − | − | − | − | − | − |
| GC51 | Wautersiella falsenii genomovar 1 | − | − | − | − | − | − | − | − | − | − | − |
| GC52 | Paenibacillus sp. P33 | − | − | − | + | + | − | + | − | − | − | − |
| Total positive | 6 | 23 | 16 | 23 | 21 | 17 | 9 | 15 | 11 | 10 | 14 | |
| Percentage positive | 12 | 44 | 31 | 44 | 40 | 33 | 17 | 29 | 21 | 19 | 27 |
N‐acyl‐l‐homoserine lactone production in activated sludge
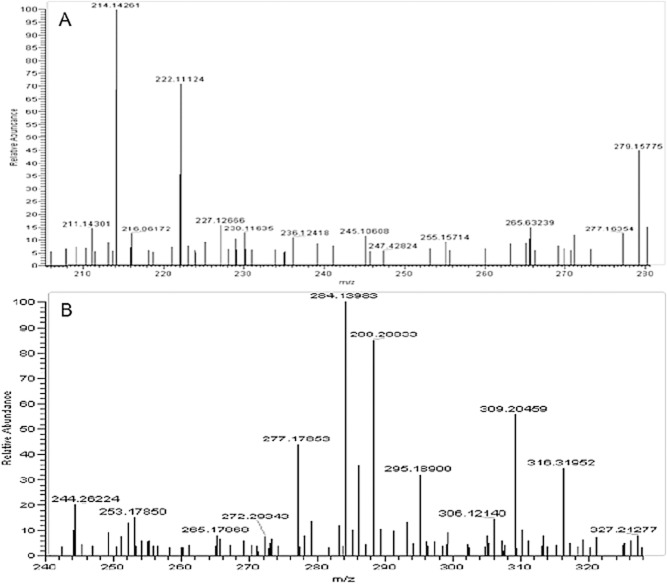
The isolation of AHL‐producing bacteria from activated sludge raises the question of whether AHLs are produced in activated sludge at biologically relevant concentrations. Ethyl acetate extracts of activated sludge were analysed by LC‐ESI‐MS revealing the presence of a molecule with peaks at 214.14261 (M+H)+ and 236.12418 (M+Na)+ and a retention time corresponding to synthetic N‐heptanoyl‐l‐homoserine lactone applied as a standard (Fig. 1A). LC‐ESI‐MS analysis also identified a compound with a retention time slightly longer than synthetic N‐dodecanoyl‐l‐homoserine lactone (36 versus 32 min) and peaks with m/z ratios of 284.13983 (M+H)+ and 306.12140 (M+Na)+, similar but not identical to the N‐dodecanoyl‐l‐homoserine lactone standard (Fig. 1B). The extract was also subjected to thin‐layer chromatography and assessed for AHL‐like compounds using CviR and TraR‐based overlays. While the CviR overlay assay did not detect any compounds with AHL‐like activity, the TraR‐based assay revealed a compound with stimulatory activity migrating between N‐decanoyl‐l‐homoserine lactone and N‐dodecanoyl‐l‐homoserine lactone (Fig. 2). It is tentatively suggested that this unknown compound observed by both MS and TLC is N‐3‐oxo‐undecanoyl‐l‐homoserine lactone, although a standard was not available to verify this. While these analyses confirm the presence of AHLs in activated sludge amid a complex background of solvent extractable organic matter, they do not address questions of biological relevance. Hence, an alternative strategy was devised to assess AHL production in activated sludge based on a fluorescent AHL biosensor strain.
Figure 1.
Electrospray ionization mass spectra showing the presence of (A) N‐heptanoyl‐l‐homoserine lactone as evidenced by peaks at 214.14261 (M+H)+ and 236.12418 (M+Na)+ and a retention time matching the synthetic standard and (B) an unidentified AHL as evidenced by peaks at 284.13983 and 306.12140 possibly corresponding to N‐3‐oxo‐undecanoyl‐l‐homoserine lactone.
Figure 2.
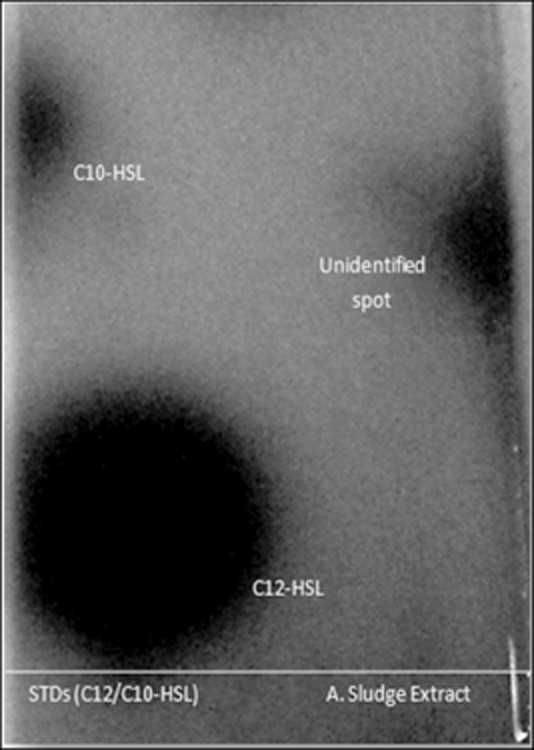
Thin‐layer chromatography with Agrobacterium tumefaciensA136 overlay showing response to synthetic N‐dodecanoyl‐l‐homoserine lactone (C12‐HSL), N‐decanoyl‐l‐homoserine lactone (C10‐HSL) and an unidentified compound with AHL‐like activity (possibly N‐3‐oxo‐undecanoyl‐l‐homoserine lactone).
An Aeromonas sp. isolated from activated sludge was selected as a carrier for a LuxR‐dependent Gfp‐based AHL sensor plasmid (pBB–LuxR). This strain was selected because it: (i) does not produce AHLs as evidenced by inactivity in LuxR, CviR, TraR or LasR‐based AHL bioassays and a negative PCR for the ahyRI operon, (ii) belongs to a genus known to be abundant in sludge (Kampfer et al., 1996) and (iii) formed cellular aggregates or flocs in culture suggesting it has the capacity to adhere to or associate with sludge flocs. The plasmid pBB–LuxR was transferred into the Aeromonas sp. host by conjugation, checked for stability and dose response, and deployed into activated sludge samples.
Activated sludge samples were inoculated with 3 × 106 cells per millilitre Aeromonas sp. (pBB–LuxR) and observed by epifluorescence microscopy over time. Approximately 4 h after inoculation of the monitor strain into activated sludge, green fluorescent cells were clearly detected associated with flocs but not in the bulk aqueous phase of the sludge (Fig. 3). Variations in the intensity of the fluorescence in different flocs and in different parts of the same floc were observed, independent of floc size, suggesting AHL production and/or accumulation was spatially heterogeneous among the flocs. The addition of N‐3‐oxo‐hexanoyl‐l‐homoserine lactone to the samples revealed the monitor cells were abundant in both the bulk aqueous phase and the aggregated biomass of the sludge (Fig. 3). No AHL‐like activity was detected when the monitor strain was inoculated into autoclaved activated sludge samples or sludge at pH 9 or both (Fig. 3). These experiments suggest that AHLs or functionally equivalent molecules are present in activated sludge flocs but not in the bulk aqueous phase, at concentrations high enough to activate the LuxR transcriptional regulator encoded by the Aeromonas sp. (pBB–LuxR) monitor strain. While this finding raises the intriguing question of the function of AHL‐mediated gene expression in activated sludge flocs, it provides no reliable information regarding the identity of the activating molecules, their concentrations, the identities of the producers or the identities of regulated phenotypes.
Figure 3.
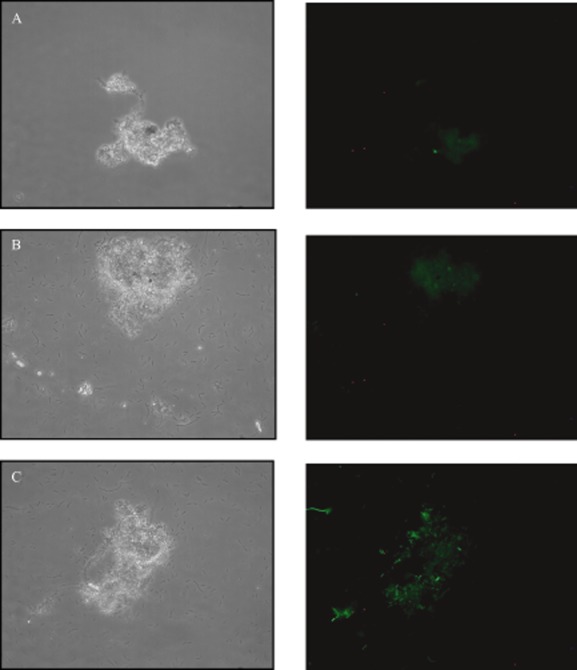
Phase contrast (left) and epifluorescence (right) microscopy images of the same field of view of activated sludge flocs. A shows activated sludge without augmentation with the AHL monitor strain Aeromonas sp. (pBB–LuxR). B shows autoclaved sludge with prior incubation at pH 9 augmented with the monitor strain. C shows live activated sludge augmented with the monitor strain. The monitor strain generated green fluorescence in association with flocs but not in the planktonic phase. Images are representative.
N‐3‐oxo‐hexanoyl‐l‐homoserine lactone addition to activated sludge increases chitinase activity
To approach the challenge of discovering a phenotype linked to AHL‐mediated gene expression in activated sludge, samples were treated hourly with 10 μM N‐3‐oxo‐hexanoyl‐l‐homoserine lactone and supernatants subsequently tested for the products of six known AHL‐regulated phenotypes. Extracellular lipase and cellulase activity were detected through observation of zones of influence on agar plates. Extracellular surfactant activity was observed by the collapse of the meniscus of a drop of culture supernatant on a Petri dish. Extracellular antimicrobial production was observed as a zone of clearing of a lawn of Escherichia coli ESS on agar plates. Cellulase and chitinase activity were detected using colourimetric assays.
Elevated N‐3‐oxo‐hexanoyl‐l‐homoserine concentrations generated a rapid 10‐fold increase in extracellular chitinase activity between 60 and 90 min (Fig. 4). After 2 h the chitinase activity began to decrease and resembled the untreated control after 4 h. The cause of the drop in chitinase production is currently unknown, although may relate to upregulation of acylase or lactonase activities. None of the remaining phenotypes tested showed any response to exogenous N‐3‐oxo‐hexanoyl‐l‐homoserine lactone addition.
Figure 4.
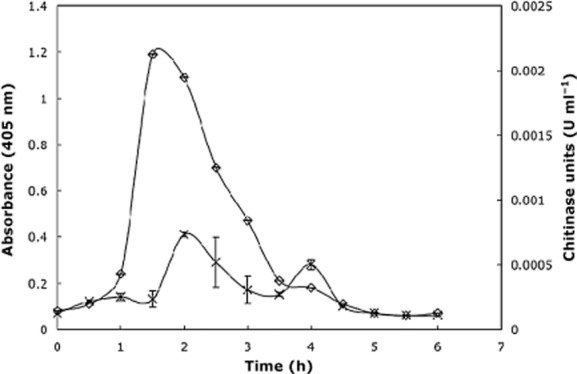
Extracellular chitinase activity in activated sludge in response to hourly addition of N‐3‐oxo‐hexanoyl‐l‐homoserine lactone. A strong increase in chitinase activity is evident in response to AHL addition. Treatments were performed in triplicate and error bars represent standard deviation.
Chitinase production is AHL‐responsive in sludge isolate Aeromonas sp. strain GC1
We additionally tested our isolate collection for the six known AHL‐regulated phenotypes tested above. The results from all six assays for the 52 different strains isolated are presented in Table 1. Known AHL‐regulated phenotypes were common. The most frequent phenotypes detected were lipase production (33% of isolates tested positive), elastase production (29% of isolates tested positive) and surfactant production (27% of isolates tested positive). Chitinase activity was detected in 21% of isolates. Fourteen out of the 17 aeromonad strains isolated produced lipase while 11 of the 17 produced chitinase. Of the five aeromonads displaying the same AHL production profile (activating CviR, TraR and LuxR), all of them produced lipase and four of them produced chitinase.
Six Aeromonas strains isolated from sludge were additionally screened for shifts in chitinase activity in response to exogenous addition of 10 μM N‐butanoyl‐l‐homoserine lactone, N‐3‐oxo‐hexanoyl‐l‐homoserine lactone or N‐hexanoyl‐l‐homoserine lactone in the presence of chitin after 6 h incubation. While AHL addition to four isolates had no impact on chitinase activity, two isolates, Aeromonas sp. strain GC1 and Aeromonas sp. strain GC40, showed elevated chitinase activity in response to N‐butanoyl‐l‐homoserine lactone. Aeromonas sp. strain GC1 was further examined for its response to N‐butanoyl‐l‐homoserine lactone and N‐3‐oxo‐hexanoyl‐l‐homoserine lactone addition over time. Figure 5 shows the stimulation of chitinase activity in response to N‐butanoyl‐l‐homoserine lactone and N‐3‐oxo‐hexanoyl‐l‐homoserine lactone, suggesting that AHL concentration has a regulatory impact on chitinase activity in Aeromonas lineages isolated from activated sludge.
Figure 5.
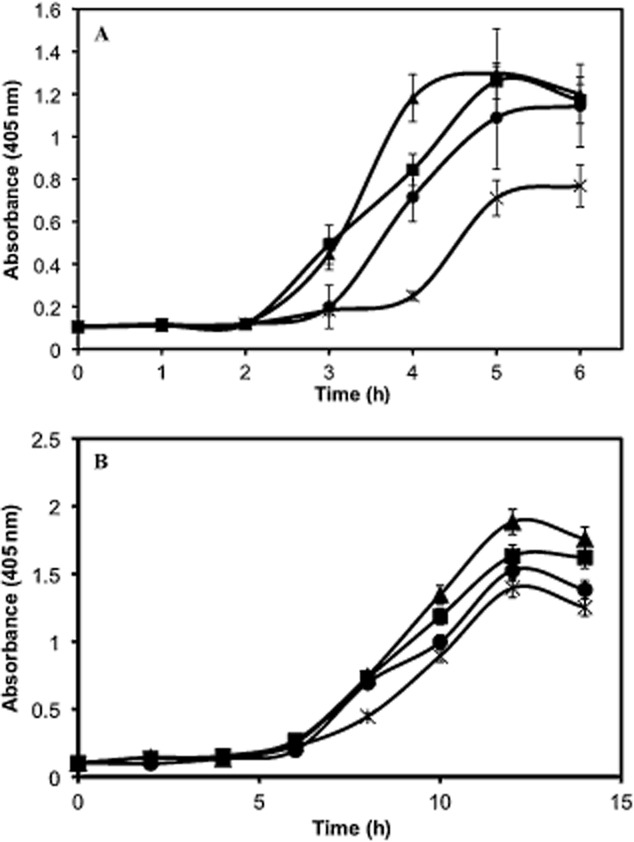
Extracellular chitinase activity in activated sludge isolate Aeromonas sp. strain GC1 in response to exogenous AHL addition.
A. Stimulation of extracellular chitinase activity (cell‐free) in response to 0, 10, 100 and 1000 nM N‐butanoyl‐l‐homoserine lactone addition (crosses, circles, squares and triangles respectively).
B. Stimulation of cell‐associated chitinase activity (cells included) in response to 0, 0.1, 1 or 10 μM N‐oxo‐hexanoyl‐l‐homoserine lactone (crosses, circles, squares and triangles respectively). Treatments were performed in triplicate. Error bars represent standard deviation.
Quorum sensing and chitinase genes in Aeromonas sp. strain GC1
PCR amplification of Aeromonas sp. strain GC1 DNA using three primer sets previously shown to amplify components of the divergently transcribed ahyR/I operon of A. hydrophila ATCC7966 gave products of expected size. Sequencing of these amplicons confirmed that Aeromonas sp. strain GC1 encodes both ahyR and ahyI gene homologues with 99% identity with A. hydrophila (GenBank Accession Numbers JN987188, JN987189 and JN987190). Congruent with the detection of and response to AHLs, this analysis suggests that Aeromonas sp. strain GC1 carries an AHL synthase and an AHL response regulator.
Aeromonas species have previously been shown to produce chitin‐degrading enzymes (Chen et al., 1991). PCR was applied with primer sets targeting the chi1 gene of Aeromonas caviae and the chiA gene of A. hydrophila encoding chitinase enzyme homologues. Both primer sets generated products of expected size, and the sequences of these products over common regions were identical to each other and the chiA gene of A. hydrophila and 97% identical to the chi1 gene of A. caviae (GenBank Accession Numbers JN987186 and JN987187). Interestingly, PCR amplification of a gene encoding β‐N‐acetylglucosaminidase from A. hydrophila, which is required to degrade the chitin monomer, did not generate a product.
Aeromonas sp. strain GC1 produces unsubstituted C4, C6, C8 and C12 AHLs
Cross‐streaking and reverse‐phase thin‐layer chromatography overlay assays indicated that the activated sludge isolate Aeromonas sp. strain GC1 produces four molecules with AHL‐like activity, three of which could be putatively identified based on R f values as N‐butanoyl‐l‐homoserine lactone, N‐hexanoyl‐l‐homoserine lactone and N‐dodecanoyl‐l‐homoserine lactone. NSI‐MS analysis of the culture extract identified hydrogen and sodium adducts of N‐butanoyl‐l‐homoserine lactone with m/z ratios of 172.096 and 194.078 (Fig. 6A). LC‐ESI‐FTMS analysis identified N‐hexanoyl‐l‐homoserine lactone at a retention time of 17.85 min with m/z ratios of [M+H] 200.127 and [M+Na] 222.109 (Fig. 6B) and N‐dodecanoyl‐l‐homoserine lactone at a retention time of 31.41 min with m/z ratios of [M+H] 284.221 and [M+Na] 306.203 (Fig. 6C). A fourth AHL was identified as N‐octanoyl‐l‐homoserine lactone with a retention time of 24.62 min and m/z ratios of [M+H] 228.158 and [M+Na] 250.143 (Fig. 6D). All retention times matched relevant synthetic AHL standards applied.
Figure 6.
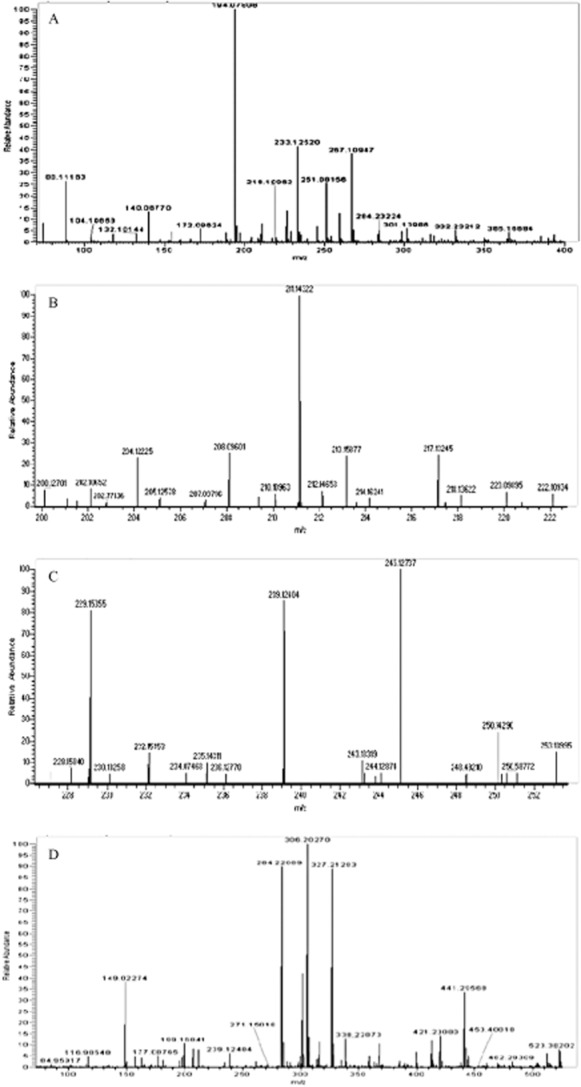
Mass spectra of ethyl acetate culture supernatant extracts of Aeromonas sp. strain GC1 showing (A) hydrogen and sodium adducts of N‐butanoyl‐l‐homoserine lactone with m/z ratios of 172.096 and 194.078 respectively, (B) hydrogen and sodium adducts of N‐hexanoyl‐l‐homoserine lactone with m/z ratios of 200.127 and 222.109 respectively, (C) hydrogen and sodium adducts of N‐octanoyl‐l‐homoserine lactone with m/z ratios of 228.158 and 250.143 and (D) hydrogen and sodium adducts of N‐dodecanoyl‐l‐homoserine lactone with m/z ratios of 284.221 and 306.203 respectively.
Discussion
Activated sludge represents a model high‐density microbiological community of enormous biotechnological significance. While much is known about process engineering and microbial metabolism in sludge, surprisingly little is known about the role of interactions between bacterial cells in dictating community composition and function. This study sought to address this knowledge void.
Evidence is presented to suggest that the well‐studied quorum sensing signalling molecules, AHLs, are present in activated sludge at biologically relevant concentrations. Numerous AHL‐producing strains were isolated from the sludge under investigation. LCMS analysis of sludge extracts detected a molecule with an LC retention time and m/z ratios corresponding to N‐heptanoyl‐l‐homoserine lactone, previously shown to be produced by Erwinia and Edwardsiella species (Morohoshi et al., 2004; Pomini et al., 2005). A TLC overlay assay revealed the presence of an unidentified compound activating TraR‐dependent gene expression. Mass spectral data suggested the presence of a compound that may be N‐3‐oxo‐undecanoyl‐l‐homoserine lactone, which has not previously been observed. Activation of a LuxR/gfp‐based AHL biosensor deployed into activated sludge indicated the presence of short‐chain AHLs. Taken together, these results suggest both short‐ and long‐chain AHLs accumulate in activated sludge flocs at concentrations high enough to activate AHL‐mediated gene transcription.
Many questions remain regarding AHL profiles in activated sludge and it is important to recognize that the identities and concentrations of AHLs present in activated sludge flocs are likely to vary over time based on sludge age and changes in the abundance and activity of AHL‐producing and AHL‐degrading bacteria. Indeed, like snowflakes, no two flocs are identical and it is expected that stochastic variations in AHL profiles exist within and between flocs and between different batches of sludge. What is clear from the data presented here is that AHLs are present in the matrix of activated sludge flocs as opposed to the bulk aqueous phase of sludge at concentrations high enough to activate expression of AHL‐regulated phenotypes. This is consistent with observations of high AHL concentrations in high cell density biofilm biomass (Charlton et al., 2000).
Some of the AHL‐producing strains isolated and identified in this study belonged to genera with known AHL producers such as Citrobacter, Klebsiella (Wang et al., 2006), Acinetobacter (Sarkar and Chakraborty, 2008), Aeromonas and Pseudomonas (Morgan‐Sagastume et al., 2005), and are usually pathogenic isolates with AHL‐mediated phenotypes associated with exoprotease production or biofilm formation. However, no isolates from the genera Chitinimonas, Malikia and Raoultella have previously been reported to produce AHLs. Furthermore, members of the genera from Microbacterium and Paenibacillus, known to possess AHL‐degrading activity (Medina‐Martinez et al., 2007; Morohoshi et al., 2009), and Shigella, known to respond to but not produce AHLs (Subramoni and Venturi, 2009), were also among the isolates showing AHL‐like activity. Interestingly, there was a lot of variation in AHL production between closely related strains, supporting the contention that dynamic evolutionary processes govern the maintenance of quorum sensing machinery (Manefield and Turner, 2002). Regardless, activated sludge clearly harbours bacteria with a rich diversity of quorum sensing systems ripe for investigation into the future.
Activities not exclusively but often observed to be AHL‐dependent, such as production of the extracellular degradative enzymes lipase, chitinase, elastase and cellulase (Givskov et al., 1997; Barnard et al., 2007), were found to be common among the bacterial strains isolated from activated sludge in this study. Activated sludge is well known as a source of such enzyme activities although little is known about the regulation of their expression (Barbara et al., 1992; Frolund et al., 1995; Gessesse et al., 2003; Xia et al., 2007; Yu et al., 2007).
Because chitinase activity was responsive to N‐3‐oxo‐hexanoyl‐l‐homoserine lactone addition to sludge and because chitinase activity was common among Aeromonas species isolated in abundance from sludge, additional experimentation focused on the role of AHL‐mediated gene expression in regulation of chitinase activity in Aeromonas species. While Aeromonas species are generally overrepresented in sludge culture collections owing to the rich media used, cultivation‐independent approaches have shown they do indeed represent one of the most abundant clades constituting roughly 2% of the biomass in this environment (Kampfer et al., 1996).
Two AHL and chitinase‐positive Aeromonas species were identified in which chitinase activity could be upregulated by increasing AHL concentration. Both N‐butanoyl‐l‐homoserine lactone and N‐3‐oxo‐hexanoyl‐l‐homoserine lactone generated dose‐dependent increases in chitinase activity in Aeromonas sp. strain GC1. This strain has a 16S rRNA sequence 99% identical to A. hydrophila ATCC 7966T, which carries the luxI/R homologues ahyI/R and the chitinase‐encoding gene chiA on its genome (Seshadri et al., 2006). The presence of these genes in the strain isolated here was confirmed by PCR and sequencing and the AHLs produced were identified (predominantly N‐butanoyl‐l‐homoserine lactone when grown in LB, consistent with previous studies on Aeromonas species) (Swift et al., 1997). N‐acyl‐l‐homoserine lactone‐dependent chitinase activity has previously been demonstrated in Pseudomonas, Chromobacterium and Serratia species (Chernin et al., 1998; Folders et al., 2001; Christensen et al., 2003). These data represent the first evidence suggesting that Aeromonas species regulate chitinase activity through AHL‐mediated quorum sensing.
Chitin and chitosan (deacetylated chitin) are commonly used as flocculants for efficient coagulation of suspended particles in wastewater treatment systems (Chi and Cheng, 2006). Fungal cell walls, crustacean and insect debris are also sources of chitin in wastewater (Merzendorfer, 2006). Based on these results it is speculated that quorum sensing may be involved in the depolymerization and hence liberation of carbon and energy from chitin in activated sludge.
The experiments conducted in this study provide insight into the role of AHL‐mediated gene expression in activated sludge. It is speculated that AHL‐dependent production of extracellular chitinase underpins the success of some aeromonad lineages in activated sludge. This study also raises the question of whether activated sludge flocs are in fact surface‐associated biofilms anchored to particulate matter such as chitin. This fundamental knowledge may ultimately be applied in the development of stable flocculate scaffolds through provision of particulate organic matter and manipulation of AHL‐mediated gene expression. Development of an experimental model for floc formation based on the colonization of chitin by AHL‐ and chitinase‐producing Aeromonas species is currently underway.
Experimental procedures
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli harbouring the plasmid pJBA357 was a gift from M. Givskov (University of Copenhagen). The plasmid encodes the Vibrio fischeri lux machinery with an unstable variant of the green fluorescence protein reporter gene fused to the AHL‐inducible PluxI promoter. Sludge organisms were grown in R2A medium at 30°C unless otherwise specified. Strains used in AHL bioassays were grown in ABT minimal medium [AB minimal medium (Clark and Maaloe, 1967) with thiamine at 2.5 mg l−1], supplemented with 0.5% (w/v) glucose and 0.2% (w/v) casamino acids. Activated sludge samples were incubated in artificial wastewater (AWW) medium containing (per litre of deionized water) 0.2 g of NH4Cl, 0.15 g of CaCl2.2H2O, 0.33 g of KCl, 0.3 g of NaCl, 3.15 g of MgCl2.6H2O, 1.26 g of K2HPO4, 0.42 g of KH2PO4, 0.25 g of yeast extract, 1 ml of trace elements and 1 ml of vitamin solution, supplemented with 0.5% (w/v) glucose. Vitamin and trace element solutions were prepared as previously described (So and Young, 1999). All cultures were incubated at 30°C with shaking at 160 r.p.m. The following concentrations of antibiotics were used as appropriate: kanamycin 50 μg ml−1, tetracycline 20 μg ml−1, ampicillin 100 μg ml−1, streptomycin 25 μg ml−1 and gentamycin 20 μg ml−1. N‐acyl‐l‐homoserine lactones were purchased from the laboratory of Prof. Paul Williams, Nottingham University, UK and stored as 1 mM stocks in appropriate solvents at −20°C.
Isolation of bacterial strains from activated sludge
Activated sludge samples were obtained from St Mary's Wastewater Treatment Plant in Sydney, Australia. Activated sludge was serially diluted in R2A medium, and 100 μl of aliquots were plated onto R2A agar (Difco). Plates were incubated for 24 h at 30°C.
To identify activated sludge isolates, 16S rRNA gene fragments were amplified using primers 8f (5′‐AGAGTTTGATCCTGGCTCAG‐3′) and 926r (5′‐CCGTCAATTCCTTTRAGTTT‐3′) (Liu et al., 1997) and sequenced with an ABI Prism BigDye kit and an ABI model 310 genetic analyser (Perkin‐Elmer Applied Biosystems). Oligonucleotides were synthesized by Sigma‐Aldrich. The resulting sequences were deposited in GenBank (GU136494–GU136545).
Assays for detection of AHL molecules
Activated sludge isolates were screened for AHL production by cross‐streaking against AHL biosensors, Chromobacterium violaceum CV026 and Agrobacterium tumefaciens A136 on ABT medium with 0.7% (w/v) agar as previously described (McClean et al., 1997). For the C. violaceum CV026 inhibition assay, ABT agar was supplemented with 500 nM N‐3‐oxo‐hexanoyl‐l‐homoserine lactone. The presence of long‐chained AHLs was detected by inhibition of the induced C. violaceum CV026.
Culture supernatants of activated sludge isolates were tested for AHL activity using E. coli (pJBA357) and E. coli (pMHLas). Sludge isolates, E. coli (pJBA357) and E. coli (pMHLas) were grown in ABT medium with the appropriate antibiotics, for 16 h at 30°C. Escherichia coli (pJBA357) and E. coli (pMHLas) were diluted (1:5) in fresh ABT medium, incubated with shaking for 30 min at 30°C, and then each strain was dispensed in 100 μl of aliquots into wells of a flat‐bottomed microtiter plate (Sarstedt Australia). Cell‐free supernatants of sludge isolates were prepared by filtration through a 0.22 μm Millex filter unit (Millipore). A 100 μl portion of the cell‐free culture supernatants was incubated with E. coli (pJBA357) and E. coli (pMHLas) with shaking for 3 h. A similar protocol was used to characterize the dose response of the AHL monitor strain constructed in this study to N‐3‐oxo‐hexanoyl‐l‐homoserine lactone. After pre‐incubation and dilution, 100 μl of aliquots of the monitor strain were treated with 0–100 nM N‐3‐oxo‐hexanoyl‐l‐homoserine lactone. The impact of the density of non‐fluorescing cells on the apparent fluorescence output as d etected by the fluorometer was assessed in the same format with E. coli (pSM10) cells added to microtiter wells. Fluorescence measurements were taken hourly for 3 h using a microtiter plate fluorometer (Wallac Victor2) (excitation, 485 nm; emission, 535 nm). Fluorescence values were corrected for autofluorescence.
Thin‐layer chromatography assays in combination with biosensor overlays were used to further characterize AHL production (Shaw et al., 1997). Culture extracts and AHL standards were spotted on reversed‐phase C18 TLC glass plates coated with silica (10 × 10 cm2, RP‐18, Merck Darmstadt, Germany). Plates were developed with a methanol and MilliQ water mixture (60:40 v/v) and allowed to dry at room temperature for 15 min before being overlaid with overnight cultures of CV026 or A136 mixed with LB or ABT medium containing 1.2% of agar (wt/vol) at 42°C. X‐gal and appropriate antibiotics were included in A. tumefaciens A136 assays. Plates were incubated for 24 h at 30°C. N‐acyl‐l‐homoserine lactones were identified by characteristic R f values.
Construction of the activated sludge AHL monitor strain
An activated sludge isolate with a 16S rRNA gene sequence with 99% similarity to A. hydrophila over 700 bp (Accession No. GU136514) was tested for the absence of endogenous AHL production and the absence of ahyI/R sequence and ultimately chosen as a host for a luxR‐based gfp(ASV) AHL reporter plasmid. For cloning and reporter strain construction, routine protocols for plasmid DNA purifications and DNA fragment isolation were followed (Sambrook et al., 1989) using the QIAprep spin miniprep kit (Qiagen) and QIAquick gel extraction kit and restriction enzymes from New England Biolabs. A 2.8 kb BamHI–EcoRV fragment from pJBA132, containing the luxR–PluxI–gfp(ASV)–T0–T1 cassette, was ligated into a stable broad host range vector pBBR1MCS2 via compatible restriction sites to create pBB–LuxR. While ultimately not exploited in this study, pBBR1MCS2 has the ability to replicate in Acinetobacter species, while p15A (pJBA)‐derived plasmids do not (Figueiredo et al., 2009). The ligated construct was then chemically transformed into competent E. coli BW20767 cells subsequently used as a donor for conjugal transfer of the construct into the sludge isolate via the filter mating technique (DeLorenzo et al., 1990). Kanr sludge isolates were selected on R2A agar containing Kan and Amp (isolate is inherently Ampr). Kanr isolates were further screened for a response to N‐3‐oxo‐hexanoyl‐l‐homoserine lactone by fluorimetry and tested for the presence of the construct by restriction digestion. The minimum concentration of N‐3‐oxo‐hexanoyl‐l‐homoserine lactone yielding fluorescence observable by epifluorescence microscopy was 10 nM.
N‐acyl‐l‐homoserine lactone detection in sludge samples
Overnight cultures of the AHL monitor strain were diluted (1:5) in fresh AWW medium, and 100 μl of aliquots (6 × 106 cells per millilitre) were added to microtiter plate wells. Sludge samples were washed by concentrating the sludge followed by resuspension in an equal volume of AWW media and 100 μl added to wells containing the AHL monitor strain. N‐3‐oxo‐hexanoyl‐l‐homoserine lactone (20 nM) was added to positive controls. Autoclaved sludge samples or sludge samples incubated overnight at pH 9 in AWW medium and washed in AWW medium at pH 7 were used as negative controls. Green fluorescence was observed using an axioplan epifluorescence microscope (Leica model DMR). The microscope was equipped was a 100 W mercury lamp and filter set No. 10 (Carl Zeiss) to visualize GFP. A slow‐scan charge‐coupled device (CCD) camera CH250 (Photometrics) equipped with KAF 1400 chip (pixel size 608 × 608 μm) was used for capturing images.
Phenotypic characterization of sludge isolates and sludge
Cellulase activity was tested by adding 100 μl of culture supernatant to small wells cut from 0.8% agar plates containing 0.1% (w/v) soluble sodium carboxymethyl cellulose (CMC‐Na) substrate and 0.004% (w/v) Congo red. After 48 h incubation at 37°C, the agar medium was flooded twice with an aqueous solution of 1 M NaCl for 15 min to destain, and zones of hydrolysis were visualized by changes in dye colour (from red to orange) around the well, indicating enzyme activity. Lipolytic activity was determined by the opacity zones formed around the wells of 0.8% agar containing 2% Tween80® and 0.01% (w/v) Victoria Blue B dye, with 100 μl of culture supernatant, after 48 h incubation at 30°C. Surfactant production was assessed through observation of the collapse of a 10 μl aliquot of bacterial culture supernatant placed on the lid of a Petri dish, when compared with Serratia liquefaciens MG1 (positive control) (Lindum et al., 1998) and culture media (negative control). Antimicrobial activity was assayed on R2A agar plates employing the beta‐lactam supersensitive E. coli ESS strain. Chitinase and elastase activities were assayed by measuring spectrophotometric changes in solutions of 4‐Nitrophenyl N‐acetyl‐beta‐d‐glucosaminide (405 nm) (Sigma Cat. No. CS0980) and Elastin‐Congo red substrate (590 nm) (Sigma Cat. No. E0502) respectively. Sludge was incubated shaking at room temperature and supernatant samples were taken from sludge treated with 10 μM N‐3‐oxo‐hexanoyl‐l‐homoserine lactone or sludge alone [with equivalent volume (10 μl) of DMSO used to dissolve N‐3‐oxo‐hexanoyl‐l‐homoserine lactone in the untreated sludge control] every hour and tested for the phenotypes of interest. All assays were conducted in triplicate.
Molecular detection of quorum sensing and chitinase genes
Genomic DNA was isolated from pure cultures grown in LB at 30°C with aeration. DNeasy Blood & Tissue Kit (QIAGEN) was used for DNA extraction. Extracted DNA was quantified using a Nanodrop1000 spectrophotometer (Thermo Fischer Scientific). Primers used for amplification of chitinase and quorum sensing genes are listed in Table S2 along with thermocycling protocols. PCRs were set up as follows: 20 μl of reactions contained 10 μl of Promega Master Mix (Promega, Madison), 0.8 μl of each primer at 10 μM, 1–5 ng of DNA template and molecular biology grade water. PCR products were purified using the DNA Clean & Concentrator Kit (Zymo Research) and sequenced according to protocols described above.
NSI‐MS and LC‐ESI‐MS analysis
Strains were cultured in LB10 at 30°C overnight and culture samples (20 ml) were centrifuged to remove the bulk of the biomass and filtered through 0.20 μm Millipore filters. Cultures and activated sludge supernatants were extracted with equal volume of acidified ethyl acetate (0.5% formic acid) three times and solvent free extracts were dissolved in 100 μl of methanol. Synthetic AHL standards were prepared in methanol. N‐acyl‐l‐homoserine lactones were analysed with a LTQ Orbitrap XL hybrid Fourier Transform Mass Spectrometer (Thermo Fisher Scientific) equipped with static nanospray ion source for NSI‐MS and electrospray ionization source connected to an Accela liquid chromatography (LC) system (Thermo Fischer Scientific) for ESI‐LC‐MS. Volumes of 3 μl of standards and extracts were loaded into the nano‐electrospray needle and the voltage adjusted to 1.0 V. Signals were recorded for a period of 60 s.
For LC‐ESI‐MS analysis, 25 μl of standards and extracts were chromatographed with a Phenomenex reversed‐phase C18 (150 × 2 mm × 5 μm) column. Mobile phases A and B were water containing 0.1% formic acid and HPLC grade methanol respectively. Elution was performed at a flow rate of 200 μl min−1. The following gradient was used for the elution: 0–30 min from 5% B to 95% B; 6 min maintained at 95% B; 4 min from 95% percent B to 5% B; 5 min to 0% B. The column was re‐equilibrated 10 min with 70% B. Eluted samples were ionized by positive ion mode in a full‐scan type. Sample results were compared with standards analysed with the same LC‐ESI‐MS parameters.
Acknowledgments
This work was partially supported by grants from the Centre for Marine Bio‐Innovation, and the Environmental Biotechnology Cooperative Research Centre.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Bacterial strains and plasmids.
Table S2. Primers and thermocycling protocols for molecular detection of quorum sensing and chitinase genes.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Barbara A.B., William M.B., Robert J.L. Characterization of enzyme activity in activated sludge using rapid analyses for specific hydrolases. Water Environ Res. 1992;64:792–797. [Google Scholar]
- Barnard A.M.L., Bowden S.D., Burr T., Coulthurst S.J., Monson R.E., Salmond G.P.C. Quorum sensing, virulence and secondary metabolite production in plant soft‐rotting bacteria. Philos Trans R Soc Lond B Biol Sci. 2007;362:1165–1183. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton T.S., de Nys R., Netting A., Kumar N., Hentzer M., Michael Givskov M., Kjelleberg S. A novel and sensitive method for the quantification of N‐3‐oxoacyl homoserine lactones using gas chromatography mass spectrometry: application to a model bacterial biofilm. Environ Microbiol. 2000;2:530–541. doi: 10.1046/j.1462-2920.2000.00136.x. [DOI] [PubMed] [Google Scholar]
- Chen J.P., Nagayama F., Chang M.C. Cloning and expression of a chitinase gene from Aeromonas hydrophila in Escherichia coli. Appl Environ Microbiol. 1991;57:2426–2428. doi: 10.1128/aem.57.8.2426-2428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernin L.S., Winson M.K., Thompson J.M., Haran S., Bycroft B.W., Chet I. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J Bacteriol. 1998;180:4435–4441. doi: 10.1128/jb.180.17.4435-4441.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F.H., Cheng W.P. Use of chitosan as coagulant to treat wastewater from milk processing plant. J Polym Environ. 2006;14:411–417. [Google Scholar]
- Christensen A.B., Riedel K., Eberl L., Flodgaard L.R., Molin S., Gram L., Givskov M. Quorum‐sensing‐directed protein expression in Serratia proteamaculans B5a. Microbiology. 2003;149:471–483. doi: 10.1099/mic.0.25575-0. [DOI] [PubMed] [Google Scholar]
- Clark J.D., Maaloe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- Daims H., Taylor M., Wagner M. Wastewater treatment: a model system for microbial ecology. Trends Biotechnol. 2006;24:483–489. doi: 10.1016/j.tibtech.2006.09.002. [DOI] [PubMed] [Google Scholar]
- DeLorenzo V., Herrero M., Jakubzik U., Timmis K. Mini‐Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram‐negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo S., Poirel L., Croize J., Recule C., Nordmann P. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1‐mediated overexpression of the natural blaOXA‐66 oxacillinase gene. Antimicrob Agents Chemother. 2009;53:2657–2659. doi: 10.1128/AAC.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folders J., Algra J., Roelofs M.S., van Loon L.C., Tommassen J., Bitter W. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J Bacteriol. 2001;183:7044–7052. doi: 10.1128/JB.183.24.7044-7052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolund B., Griebe T., Nielsen P.H. Enzymatic activity in the activated‐sludge floc matrix. Appl Microbiol Biotechnol. 1995;43:755–761. doi: 10.1007/BF00164784. [DOI] [PubMed] [Google Scholar]
- Frolund B., Palmgren R., Keiding K., Nielsen P.H. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 1996;30:1749–1758. [Google Scholar]
- Fuqua C., Winans S.C. Conserved cis‐acting promoter elements are required for density‐dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessesse A., Dueholm T., Petersen S.B., Nielsen P.H. Lipase and protease extraction from activated sludge. Water Res. 2003;37:3652–3657. doi: 10.1016/S0043-1354(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Givskov M., Eberl L., Molin S. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol Lett. 1997;148:115–122. [Google Scholar]
- Kampfer P., Erhart R., Beimfohr C., Bohringer J., Wagner M., Amann R. Characterization of bacterial communities from activated sludge: culture‐dependent numerical identification versus in situ identification using group‐ and genus‐specific rRNA‐targeted oligonucleotide probes. Microb Ecol. 1996;32:101–121. doi: 10.1007/BF00185883. [DOI] [PubMed] [Google Scholar]
- Lindum P.W., Anthoni U., Christophersen C., Eberl L., Molin S., Givskov M. N‐Acyl‐L‐homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Marsh T.L., Cheng H., Forney L.J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean K.H., Winson M.K., Fish L., Taylor A., Chhabra S.R., Camara M. Quorum sensing and Chromobacterium violaceum: exploitation of violaceum production and inhibition for the detection of N‐acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. et al. [DOI] [PubMed] [Google Scholar]
- McGowan S.J., Sebaihia M., Jones S., Yu B., Bainton N.J., Chan P.F. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology. 1995;141:541–550. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- McLean R.J.C., Whiteley M., Stickler D.J., Fuqua C. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Lett. 1997;154:259–263. doi: 10.1111/j.1574-6968.1997.tb12653.x. [DOI] [PubMed] [Google Scholar]
- Manefield M., Turner S.L. Quorum sensing in context: out of molecular biology and into microbial ecology. Microbiology. 2002;148:3762–3764. doi: 10.1099/00221287-148-12-3762. [DOI] [PubMed] [Google Scholar]
- Manefield M., Whiteley A.S. Acylated homoserine lactones in the environment: chameleons of bioactivity. Philos Trans R Soc Lond B Biol Sci. 2007;362:1235–1240. doi: 10.1098/rstb.2007.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason V.P., Markx G.H., Thompson I.P., Andrews J.S., Manefield M. Colonial architecture in mixed species assemblages affects AHL mediated gene expression. FEMS Microbiol Lett. 2005;244:121–127. doi: 10.1016/j.femsle.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Medina‐Martinez M.S., Uyttendaele M., Rajkovic A., Nadal P., Debevere J. Degradation of N‐acyl‐L‐homoserine lactones by Bacillus cereus in culture media and pork extract. Appl Environ Microbiol. 2007;73:2329–2332. doi: 10.1128/AEM.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzendorfer H. Insect chitin synthases: a review. J Comp Physiol [B] 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- Morgan‐Sagastume F., Boon N., Dobbelaere S., Defoirdt T., Verstraete W. Production of acylated homoserine lactones by Aeromonas and Pseudomonas strains isolated from municipal activated sludge. Can J Microbiol. 2005;51:924–933. doi: 10.1139/w05-077. [DOI] [PubMed] [Google Scholar]
- Morohoshi T., Inaba T., Kato N., Kanai K., Ikeda T. Identification of quorum‐sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J Biosci Bioeng. 2004;98:274–281. doi: 10.1016/S1389-1723(04)00281-6. [DOI] [PubMed] [Google Scholar]
- Morohoshi T., Someya N., Ikeda T. Novel N‐acylhomoserine lactone‐degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum‐quenching properties. Biosci Biotechnol Biochem. 2009;73:2124–2127. doi: 10.1271/bbb.90283. [DOI] [PubMed] [Google Scholar]
- Pomini A.M., Manfio G.P., Araújo W.L., Marsaioli A.J. Acyl‐homoserine lactones from Erwinia psidii R. IBSBF 435T, a guava phytopathogen (Psidium guajava L.) J Agric Food Chem. 2005;53:6262–6265. doi: 10.1021/jf050586e. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. 2nd edn. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarkar S., Chakraborty R. Quorum sensing in metal tolerance of Acinetobacter junii BB1A is associated with biofilm production. FEMS Microbiol Lett. 2008;282:160–165. doi: 10.1111/j.1574-6968.2008.01080.x. [DOI] [PubMed] [Google Scholar]
- Seshadri R., Joseph S.W., Chopra A.K., Sha J., Shaw J., Graf J. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P.D., Ping G., Daly S.L., Cha C., Cronan J.E., Rinehart K.L., Farrand S.K. Detecting and characterizing N‐acyl‐homoserine lactone signal molecules by thin‐layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.M., Young L.Y. Isolation and characterization of a sulfate‐reducing bacterium that anaerobically degrades alkanes. Appl Environ Microbiol. 1999;65:2969–2976. doi: 10.1128/aem.65.7.2969-2976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Evans K., Meyer J.M., Poole K. Quorum‐sensing and siderophore biosynthesis in Pseudomonas aeruginosalasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- Subramoni S., Venturi V. LuxR‐family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155:1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- Swift S., Karlyshev A.V., Fish L., Durant E.L., Winson M.K., Chhabra S.R. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N‐acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle A., Bailey M.J., Whiteley A.S., Manefield M. N‐acyl‐L‐homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ Microbiol. 2004;6:424–433. doi: 10.1111/j.1462-2920.2004.00581.x. [DOI] [PubMed] [Google Scholar]
- Wagner M., Loy A. Bacterial community composition and function in sewage treatment systems. Curr Opin Microbiol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- Wang H., Cai T., Weng M., Zhou J., Cao H., Zhong Z., Zhu J. Conditional production of acyl‐homoserine lactone‐type quorum‐sensing signals in clinical isolates of enterobacteria. J Med Microbiol. 2006;55:1751–1753. doi: 10.1099/jmm.0.46756-0. [DOI] [PubMed] [Google Scholar]
- Whiteley M., Lee K.M., Greenberg E.P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Kong Y., Nielsen P.H. In situ detection of protein‐hydrolysing microorganisms in activated sludge. FEMS Microbiol Ecol. 2007;60:156–165. doi: 10.1111/j.1574-6941.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- Yeon K.M., Cheong W.S., Oh H.S., Lee W.N., Hwang B.K., Lee C.H. Quorum sensing: a new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ Sci Technol. 2009;43:380–385. doi: 10.1021/es8019275. et al. [DOI] [PubMed] [Google Scholar]
- Yu G.H., He P.J., Shao L.M., Lee D.J. Enzyme activities in activated sludge flocs. Appl Microbiol Biotechnol. 2007;77:605–612. doi: 10.1007/s00253-007-1204-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Bacterial strains and plasmids.
Table S2. Primers and thermocycling protocols for molecular detection of quorum sensing and chitinase genes.