Summary
The bioremediation of chloroethene contaminants in groundwater polluted systems is still a serious environmental challenge. Many previous studies have shown that cooperation of several dechlorinators is crucial for complete dechlorination of trichloroethene to ethene. In the present study, we used an explorative functional DNA microarray (DechloArray) to examine the composition of specific functional genes in groundwater samples in which chloroethene bioremediation was enhanced by delivery of hydrogen‐releasing compounds. Our results demonstrate for the first time that complete biodegradation occurs through spatial and temporal variations of a wide diversity of dehalorespiring populations involving both Sulfurospirillum, Dehalobacter, Desulfitobacterium, Geobacter and Dehalococcoides genera. Sulfurospirillum appears to be the most active in the highly contaminated source zone, while Geobacter was only detected in the slightly contaminated downstream zone. The concomitant detection of both bvcA and vcrA genes suggests that at least two different Dehalococcoides species are probably responsible for the dechlorination of dichloroethenes and vinyl chloride to ethene. These species were not detected on sites where cis‐dichloroethene accumulation was observed. These results support the notion that monitoring dechlorinators by the presence of specific functional biomarkers using a powerful tool such as DechloArray will be useful for surveying the efficiency of bioremediation strategies.
Introduction
Chloroethenes are among the most widespread groundwater contaminants that threaten human and environmental health due to their toxicity (Ruder, 2006). Among them, tetrachloroethene (PCE) and trichloroethene (TCE) tend to accumulate at the bottom of aquifers where they form dense non‐aqueous phase liquids (DNAPLs) that serve as reservoirs for long‐term solubilization. Many remediation technologies are available for treating most sites whatever its geochemical properties (Lemming et al., 2010; Pant and Pant, 2010). The ex situ methods (excavation/pumping) are increasingly being replaced by less invasive in situ methods such as biostimulation and bioaugmentation with considerable successes (Major et al., 2002; Sorenson, 2002; Kao et al., 2003; Macbeth et al., 2004; Kennedy et al., 2006).
Enhanced reductive dechlorination (ERD) has been successfully applied to many aquifers to improve chlorethene degradation (He et al., 2002; Scheutz et al., 2008). This biostimulation technology involves the addition of electron donors or highly fermentable nutrients directly to the groundwater and can improve biodegradation if the indigenous microorganisms are capable of complete dechlorination. Indeed, while microorganisms from many genera have been shown to catalyse the dechlorination of PCE and TCE to dichloroethene (DCE) (see reviews by McCarty, 1997; Futagami et al., 2008), only a few strains from Dehalococcoides genus are known to be able to reduce DCE and vinyl chloride (VC) to the non‐toxic end‐product ethene (Maymó‐Gatell et al., 1997; Cupples et al., 2003; He et al., 2003; Duhamel et al., 2004). Thus, the reductive dechlorination can stall at an intermediate stage, which typically results in an accumulation of cis‐DCE and VC daughter products known to be more toxic than PCE and TCE when these strains are not present (McCarty, 1997). In this case, bioaugmentation strategies, i.e. the addition of exogenous bacterial communities, can be used to achieve complete dechlorination at higher rates (Pant and Pant, 2010).
Currently, several diagnostic tools are available to help in answering the question of whether biostimulation, bioaugmentation or both may be necessary at chlorinated ethene DNAPL sites (Maphosa et al., 2010). Most of them rely on the identification of Dehalococcoides species using the gene encoding the 16S rRNA (Hendrickson et al., 2002; Lu et al., 2006; Rahm et al., 2006). However, such approaches still do not access the functional diversity of microorganisms (Taşet al., 2010) and cannot determine whether the detected microbial populations have the specific reductive dehalogenases (RDases) involved in the final step from DCE to ethene that is required for complete dechlorination. Hence, new molecular tools, designed to detect well‐known RDase genes, provide a more complete diagnostic of dechlorination capacities (Müller et al., 2004; Regeard et al., 2004; Taşet al., 2009; van der Zaan et al., 2010).
A better understanding of the biodegrading population distribution as well as microbial interactions at contaminated sites is crucial to achieve effective bioremediation strategies. In this study, the functional ‘DechloArray’, dedicated to the dechlorination processes (Dugat‐Bony et al., 2011a), was used to assess the functional diversity and the spatial distribution of dechlorinating populations from four French chlorinated solvent‐contaminated sites under ERD treatment. For this objective, the microarray was improved by targeting all known genes encoding enzymes involved in three major metabolic biodegradation pathways of chloroethenes: anaerobic reductive dechlorination (Vogel et al., 1987), aerobic oxidation (Mattes et al., 2010) and aerobic cometabolic oxidation (Arp et al., 2001). Among the four sites studied here, one (site B) was regularly monitored during 1 year of biostimulation. The sites F, G and H were also treated by biostimulation, but only sampled at the end of their treatment. Molecular biology and chemical analyses were conducted at each site to monitor the biodegradation process. With the analysis of 30 samples from these sites with their different pollutant loadings and ERD treatments, this study: (i) provided insights into shifts in the spatial and temporal distribution of dechlorinating populations within a contaminated area but also in relation to the specific ERD treatment and (ii) identified key physico‐chemical parameters involved in the changes of bacterial communities. Our results highlight the importance of such surveys to better understand the growth and activity of degrading microorganisms based on their location within a contaminated area, and thus, to better assess the ERD treatment efficiency.
Results
Overall impact of the ERD treatment on site B
Site B was mainly contaminated with TCE resulting from repeated accidental spills from a solvent storage tank between the years of 2003 and 2007. Almost 100 mg l−1 of product was detected at the source in the underlying groundwater (solubility of TCE in water = 1 g l−1), suggesting the formation of DNAPLs. The subsurface lithology was composed of successive layers of sandy and clayey loams. The ERD treatment, consisting of two injections of lactate at 6 month intervals (at days 67 and 256 respectively, over a period of 16–18 days), was applied to target the deeper aquifer (Fig. 1A). An amount of 360 kg of lactate was directly injected into four injection wells (30% of volume in P2, 30% in P3, 30% in P3S and 10% in P6) drilled around the contamination source (Fig. 1B) in order to cover the entire source zone with approximately 3 g l−1 of lactate. The estimated impact radius of the injection varied from 5 to 7.5 m depending on the well. Monitoring was performed between May 2009 and June 2010 through a total of five sampling periods (Fig. 1A): C1 (T = 0), C2 (T = 104 days), C3 (T = 231 days), C4 (T = 291 days) and C5 (T = 378 days) at four monitoring wells, i.e. one upstream from the contamination source (P1), one in the contamination source (P2) and two downstream from the contamination source (P3 and P4) (Fig. 1B).
Figure 1.
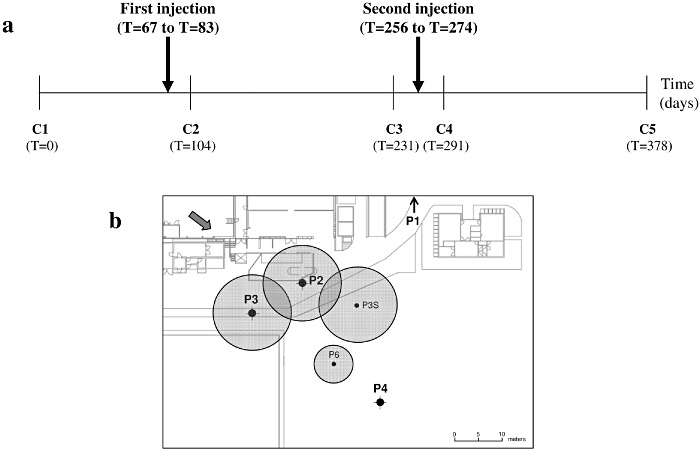
Overview of ERD strategic scheme applied on site B. A. Chronological representation of sampling periods and biostimulation treatment by lactate injections. B. Location of injection and monitoring wells and boundary distribution of lactate immediately after injection (grey circles). P2, P3, P3S and P6: injection wells. P1, P2, P3 and P4: monitoring wells. The upstream monitoring well P1 is located out of the map. The arrow indicated the groundwater flow direction.
In order to assess the evolution of the groundwater geochemistry and the electron donors availability during ERD, analytical measurements were performed on the 19 groundwater samples collected on site B (Table 1). Before biostimulation, the groundwater underlying the pollution source area was aerobic with an average redox potential (ORP) of +541 and +692.2 mV in injection wells P2 and P3 respectively. Lactate addition immediately resulted in average ORP values decreasing to −79.9 and −268.3 mV (first injection) and −95 and −114.6 mV (second injection) in both wells. Dissolved oxygen (DO) consumption also indicated the establishment of reducing conditions. The oxygen depletion was accompanied by a rapid decrease in nitrate, which can be used first as alternative electron acceptors by certain bacterial populations in response to electron donor loading. This was followed in well P3 by a significant decrease in sulfate and the production of methane (CH4) and ethene. After each injection, lactate was fermented rapidly resulting in an increase of total organic carbon (TOC), which then disappeared during the following weeks probably due to the diffusion and consumption of the fermentation products. These results indicated that lactate injections led to the establishment of strongly reductive, oxygen‐free and nutrient‐rich conditions in the contaminated area.
Table 1.
Key parameters analysed for wells P2 and P3 on site B, both used as injection and monitoring wells, before and after lactate injections (360 kg in solution)
| Sampling periods (T in days) | ORP (mV) | DO (mg l−1) | Nitrate (mg l−1) | Sulfate (mg l−1) | Methane (µg l−1) | Ethene (µg l−1) | TOC (mg l−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P2 | P3 | P2 | P3 | P2 | P3 | P2 | P3 | P2 | P3 | P2 | P3 | P2 | P3 | |
| C1 (T = 0) | 541 | 692.2 | 1.44 | 8.08 | 7.6 | 27 | 56 | 56 | 0.03 | 0.02 | <0.01 | <0.01 | 6 | 5 |
| Lactate (T = 67) | ||||||||||||||
| C2 (T = 104) | −79.9 | −268.3 | 0.6 | 0.6 | <0.2 | <0.2 | 7.6 | 13 | <0.01 | <0.01 | 0.03 | <0.01 | 1000 | 910 |
| C3 (T = 231) | −50 | −49 | 0.87 | 1.92 | <0.45 | <0.45 | 9 | 18 | <0.01 | 0.03 | 0.54 | 0.19 | 380 | 7.4 |
| Lactate (T = 256) | ||||||||||||||
| C4 (T = 291) | −95 | −114.6 | 0.77 | 1.1 | <0.75 | <0.75 | 65 | <5 | NA | NA | 150 | 92 | 2000 | 1200 |
| C5 (T = 378) | −91 | −173 | NA | 0.44 | <0.75 | 0.9 | 56 | <5 | 64 | 29 | 1100 | 1500 | 74 | 270 |
NA, data not available.
Chloroethene and ethene concentration trends were also analysed. As expected, only a negligible amount of TCE was detected at the upstream monitoring well P1 (< 7.6 × 10−3–1.9 × 10−2 µmol l−1 throughout the five sampling periods, data not shown). Before biostimulation, the TCE concentration was greatest at the source well P2 (502 µmol l−1) whereas it was only negligible at the more distant well P4 (0.45 µmol l−1). Upon lactate addition, TCE dechlorination to cis‐DCE was evident in both P2 and P3 wells. Throughout the experiment, significant reduction in TCE occurred at the source P2 with the molar concentration decreasing to reach 167 µmol l−1 after 378 days (C5) whereas it disappeared completely in the nearby well P3 after 231 days (by sampling period C3) (Fig. 2). In the more distant well P4, the daughter product cis‐DCE was converted into VC continually during the treatment (Fig. 2). In monitoring wells, P2, P3 and P4, ethene appeared at the end of the treatment with a higher concentration in well P4 after 291 days (sampling period C4); this appearance correlated with the onset of CH4 detection (Fig. 2, Table 1). Thus, anaerobic conditions created by substrate injections were favourable to chlorinated solvent reductive dechlorination and stimulated the biodegradation in the entire contaminated zone. Furthermore, the rapid evolution of the chloroethene composition clearly reflected the spatial configuration of the contaminated site with (i) a source zone close to the well P2 where the main reaction consists of TCE reduction in its daughter products (cis‐DCE and VC), and (ii) a plume zone with wells P3 and P4 where the biodegradation consisted mainly of the conversion of cis‐DCE to VC and ethene.
Figure 2.
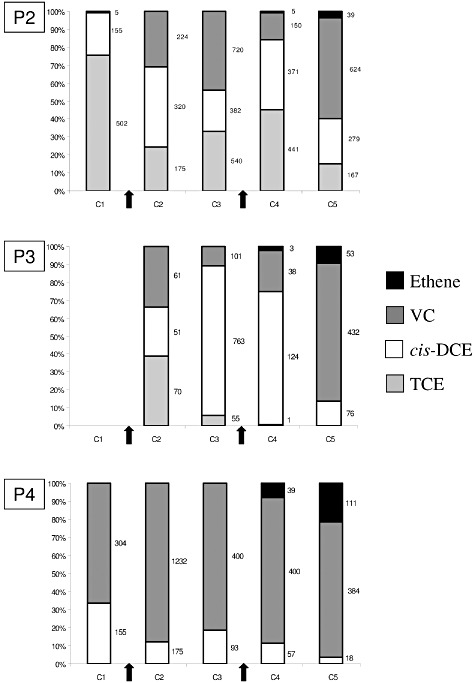
Chlorinated ethene and ethene trends on site B in the three monitoring wells P2, P3 and P4. Data were provided for sampling periods C1–C5, when available. Proportion of each compound is expressed in percentage. Arrows indicated when lactate injections were realized and numbers at right, the concentration of the different chloroethenes in µmol l−1. NA, data not available.
Molecular detection of functional gene diversity involved in chloroethene degradation at site B
To understand whether the biostimulation process affected the composition and structure of the functional genes associated with the dechlorination, the 19 gDNA samples collected throughout the experiment (sampling periods C1 to C5) were hybridized on the DechloArray. The results showed that all genes showing a positive hybridization signal were only involved in the reductive dechlorination pathway except the etnC gene, which encodes an alkene monooxygenase enzyme (Fig. 3A). Indeed, this enzyme, first isolated from Mycobacterium species, but also found in Nocardioides species, is involved in the aerobic oxidation pathway (Mattes et al., 2010). This gene was detected without major signal variation in many samples including those from the uncontaminated well P1 even after the lactate injections (Fig. 3A). Hence, these findings suggested that indigenous species harbouring this gene might not be involved in the dechlorination process.
Figure 3.
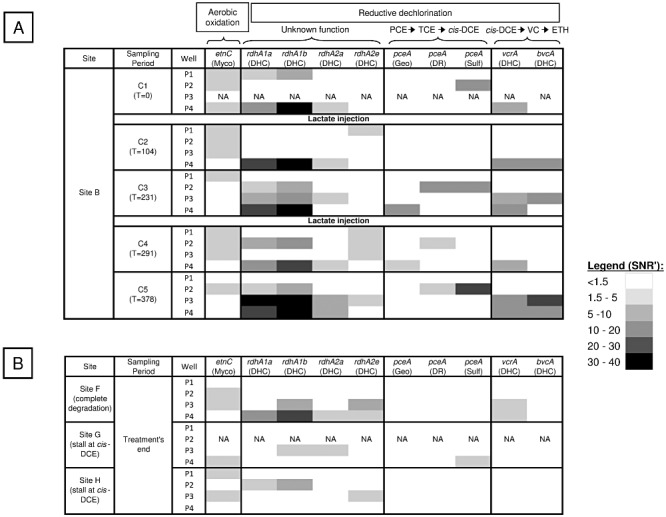
Gene responses from DechloArray data obtained with gDNA samples collected (A) on site B during ERD demonstration and (B) on sites F, G and H at the end of their biostimulation treatment. Gene response levels (mean signal to noise ratio SNR’) are indicated by using a black and white gradient. Metabolic pathways, gene name and affiliations are indicated for each gene. Myco: Mycobacterium; DHC: Dehalococcoides; Geo: Geobacter; DR: Dehalobacter/Desulfitobacterium; Sulf: Sulfurospirillum, VC: vinyl chloride, ETH: ethene.
At sampling period C1, before the biostimulation treatment, gene detection distinguished between two dehalorespiring groups with different biodegradation capabilities in distinct zones (Fig. 3A): (i) species that may belong to Sulfurospirillum (pceA gene was detected) in the source well P2, probably responsible of TCE natural attenuation and (ii) in the plume well P4, species that may belong to Dehalococcoides (vcrA and other putative rdhA genes detected), the only microorganisms known to dechlorinate cis‐DCE and VC, two compounds highly present in this area before the first lactate injection (Fig. 2). These results are consistent with data obtained from 16S rRNA gene (rrs) amplicon pyrosequencing of five samples retrieved from both the P2 and P4 wells (Table 2) that shows presence of Sulfurosprillum mainly in the source zone and Dehalococcoides in the plume. All these genes were detected throughout the bioremediation process with an increase in probe signal intensity for both pceA and vcrA after the lactate injections. This supports the favourable effect of the ERD treatment on these dechlorinators. Furthermore, as shown in Fig. 3A, each lactate addition decreased the number of genes detected during the following sampling periods (C2 and C4) in the injection wells P2 and P3 whereas not in the distant well P4 used only for monitoring. Thus, the injection of a large amount of substrate could result in a strong disturbance of the microbial community for a relatively long period (greater than 3 weeks).
Table 2.
Summary statistics from 16S rRNA gene 454 pyrosequencing data obtained from both P2 source and P4 plume zones on site B at different sampling campaigns
| % of reads per sample | Samples | |||||
|---|---|---|---|---|---|---|
| P2 | P4 | |||||
| Campaign C3 | Campaign C4 | Campaign C5 | Campaign C3 | Campaign C4 | ||
| 2301 reads | 3255 reads | 23 334 reads | 3174 reads | 2577 reads | ||
| Dehalobacter | Dehalobacter unclassified | 0 | 0 | 0 | 0 | 0.116414435 |
| Dehalococcoides | Dehalococcoides sp. BAV1 | 0.217296827 | 0.399385561 | 0.068569469 | 0.882167612 | 1.280558789 |
| Dehalococcoides sp. VS | 0 | 0 | 0 | 0.598613737 | 0.543267365 | |
| TOTAL | 0.217296827 | 0.399385561 | 0.068569469 | 1.480781348 | 1.823826154 | |
| Desulfiobacterium | Desulfitobacterium hafniense | 0.086918731 | 0.030721966 | 0.025713551 | 0 | 0 |
| Geobacter | Geobacter bemidjiensis | 0 | 0 | 0 | 0.157529931 | 0.194024059 |
| Geobacter lovleyi | 0 | 0 | 0 | 0.126023945 | 0.038804812 | |
| Geobacter Pelobacter propionicus | 0 | 0 | 0.004285592 | 0 | 1.590997284 | |
| Geobacter psychrophilus | 0.260756193 | 0.737327189 | 0.077140653 | 0.945179584 | 0.42685293 | |
| Geobacter sp. FRC‐32 | 0 | 0 | 0 | 0.252047889 | 0.077609624 | |
| Geobacter sp. M21 | 0.260756193 | 0.061443932 | 0 | 0.756143667 | 1.047729919 | |
| Geobacter sulfurreducens | 0 | 0 | 0 | 0 | 0.116414435 | |
| Geobacter unclassified | 0.043459365 | 0.215053763 | 0.017142367 | 0.315059861 | 0.42685293 | |
| Geobacter uraniireducens | 0 | 0.122887865 | 0 | 0 | 0 | |
| TOTAL | 0.564971751 | 1.13671275 | 0.098568612 | 2.551984877 | 3.919285991 | |
| Sulfurospirillum | Sulfurospirillum barnesii | 1.043024772 | 1.53609831 | 0.509985429 | 0.031505986 | 0.271633683 |
Depending on the sample, up to six different RDase genes were simultaneously detected (Fig. 3A) with significant variations in the spatial and temporal distribution of three key RDase genes (pceA, vcrA and bvcA). As the DechloArray was designed to discriminate between bacterial groups based on their distinct functional gene sequences, our data highlighted possible associations between different dehalorespiring groups, based on sample location (e.g. Dehalobacter/Desulfitobacterium group and Sulfurospirillum in the source; and Dehalococcoides and Geobacter in the plume) and based on the sampling date (e.g. detection of the Dehalobacter/Desulfitobacterium group and Geobacter only after lactate injections). Pyrosequencing of the 16S rRNA gene (rrs) demonstrated the same trends. The Dehalobacter and Desulfitobacterium pceA genes share > 99% identity, and therefore, they cannot be distinguished with the designed probes. However, PCR amplification of their 16S rRNA gene followed by cloning‐sequencing and identification (data not shown, GenBank accession numbers JQ031637 and JQ031638) and 16S rRNA pyrosequencing (Table 2) suggested that they were both present at site B. In addition, the sporadic detection of genes encoding putative RDases that may belong to Dehalococcoides species (rdhA) in many samples could reflect the widespread occurrence and the importance of this functional group in the dechlorination process (Fig. 3A).
Redundancy analysis (RDA) of gene patterns and geochemical parameters observed at site B
The RDA showed that variation in the detection of genes involved in dechlorination could be explained by different geochemical parameters (P ≥ 0.05). The first principal canonical axis (F1) accounted for 51.25% of the variation; together with the second canonical axis (F2), this value increased to 70.15% (Fig. 4). On the basis of the sample distribution on the F1 and F2 axis, three sectors were identified. In the first sector (Group A), high TCE concentrations were positively correlated with Sulfurospirillum and Dehalobacter/Desulfitobacterium pceA genes that encode enzymes responsible for the reduction of both PCE and TCE to cis‐DCE (Neumann et al., 1998; Maillard et al., 2003) and that were identified in all samples not perturbed by lactate injections collected at the source zone P2 (sampling periods C1, C3 and C5). In contrast, in the second sector (Group B), the three samples unaffected by lactate injections from the plume well P3 (sampling periods C1, C3 and C5), and those from the plume well P4 were associated with high VC, ethene and methane concentrations and showed positive correlation with the detection of VC‐reductase genes vcrA and bvcA. The genes encoding putative RDases (rdhA1a, rdhA1b and rdhA2a) with orthologues in several Dehalococcoides genomes (Table S1) were also associated with this group. Finally, this group showed a strong negative correlation with ORP, sulfate and nitrate concentrations. The sporadic detection of pceA from Geobacter species in well P4 could suggest its importance in the removal of traces of TCE remaining in this plume. The last section (Group C) grouped all samples for which no or few degradation abilities were detected: P1 samples and those from P2 and P3 affected by lactate injections (sampling periods C2 and C4). This last group was also positively correlated with high sulfate and nitrate concentrations. This highlights that environmental parameters, particularly electron acceptors (nitrate, sulphate, TCE, cis‐DCE or VC) and reductive conditions (ORP, methanogenesis), could control the spatial and temporal establishment of key microbial populations involved in the biodegradation process during the bioremediation treatment. In addition, functional gene patterns identified in each sample appeared to be closely related with the degradation reactions occurring in the nearby area, suggesting that these fingerprints could serve as indicator of site conditions and allow optimizing microbial activity for improved bioremediation processes.
Figure 4.
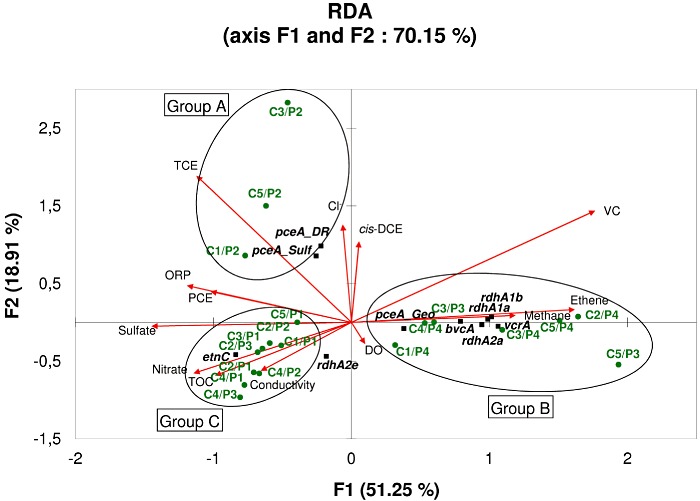
Redundancy analysis (RDA) of gene response levels as one set of variables (species data) and geochemical data as the explaining set of variables (environmental data) for site B. The length of the arrow is correlated with the strength of relation between the response variables. The arrows point in the direction of the maximum change for the associated variable. Black squares: individual genes. Green stars: groundwater samples.
Study of biodegradation capacities at sites F, G and H
Three additional industrial sites with very different pollutant loadings and distinct ERD treatments were also investigated. On the highly contaminated sites, F and G (> 15 mg l−1 TCE in the underlying groundwater at the pollution source), the biodegradation was enhanced by three injections of molasses in the plume at site F and in the source zone at site G between August 2006 and October 2008. In total, seven groundwater samples were collected almost 10 months after the end of the treatment (July 2009) from monitoring wells located as described for site B, four at site F (P1, P2, P3 and P4) and three at site G (P1, P3 and P4) respectively. Although the physico‐chemical assays performed at the end of the bioremediation process confirmed the presence of favourable reductive conditions (Table S2), the biostimulation led to the degradation of the chlorinated compounds at site F, but to an accumulation of cis‐DCE at site G. In contrast, at the last site, H, a former industrial facility where historical practices resulted in multi‐contamination with BTEX (benzene, toluene, ethyl‐benzene and xylenes), many hydrocarbons and chloroethenes (lower concentrations than the other sites), two remediation treatments were applied: a vacuum extraction between 2000 and 2002 (physical treatment), then followed by an injection of an oil/lactate mixture in 2009. These treatments resulted in a build‐up of cis‐DCE, but to a lower extent than at site G (Table S2). Four samples were collected as described previously (wells P1, P2, P3 and P4) almost a year after the treatments (January 2010).
After DechloArray hybridization with all gDNA samples retrieved from these sites, no RDase genes were detected in the upstream well P1 regardless of the site, whereas the etnC gene was widely distributed in many wells. In contrast, the six RDase genes involved in reductive dechlorination already identified from site B, were differently detected in contamination source and downstream wells, depending on the site (Fig. 3B). For site F, both putative rdhA and vcrA genes were detected in the treated area (wells P3 and P4), reflecting complete dechlorination, but not in the non‐treated source zone P2 where reducing conditions were not established (negative ORP and slight DO) (Table S2). For both sites G and H, the apparent stalling of reductive dechlorination at the cis‐DCE intermediate could be explained by the absence of essential genes encoding VC‐reductases (vcrA and bvcA). Surprisingly, no known gene able to catalyse TCE dechlorination into cis‐DCE was detected on all sites, except the pceA gene from Sulfurospirillum, which was present at one sampling point on site G. In summary, our findings indicated that, on the basis of rdhA genes, Dehalococcoides populations could be widespread at the three polluted sites although only site F hosted populations carrying VC‐reductase genes (vcrA and/or bvcA) that enable dechlorination past cis‐DCE.
Discussion
ERD treatment efficacy at site B
In this study, ERD treatment was used to cleanup a DNAPL source zone at an industrial contaminated site (site B), using lactate injections to increase the concentration of organic carbon available in the groundwater and to establish reductive conditions favourable to the growth of anaerobic populations such as certain dechlorinating bacteria. After almost 1 year of treatment, TCE was gradually and massively replaced in the source zone (P2) by cis‐DCE and VC by‐products, whereas it disappeared from the plume (P3 and P4) concurrent with the appearance of ethene (Fig. 2). However, total TCE degradation in the source zone should take a long time given the quantity discharged and would require additional substrate additions to maintain favourable conditions for bacterial growth, as shown by previous studies (Major et al., 2002; Song et al., 2002; Lendvay et al., 2003; Kennedy et al., 2006).
Spatial distribution of dechlorinating populations before ERD treatment
RDase genes are considered major indicators of dechlorination potential and are recommended for monitoring the presence of dechlorinators (Ritalahti et al., 2006; Lee et al., 2008). In the present study, RDase gene diversity was observed on the site B before implementation of ERD treatment (Fig. 3A), indicating the presence and the heterogeneous distribution of indigenous dechlorinating consortia. Indeed, whereas Dehalococcoides strains were identified at several points (on the basis of vcrA and putative rdhA genes) including both uncontaminated and slightly contaminated zones (P1 and P4), Sulfurospirillum (pceA gene) was only detected in the most contaminated source zone (P2). This highlights a broader natural occurrence of Dehalococcoides, as described in previous studies (Futagami et al., 2009; van der Zaan et al., 2010). On contaminated sites, total degradation of TCE to ethene usually results from multiple degraders associations in which the most frequently found are: (i) Desulfitobacterium and Dehalococcoides (Yang et al., 2005; Bunge et al., 2007; Rouzeau‐Szynalski et al., 2011), (ii) Dehalobacter and Dehalococcoides (Daprato et al., 2007), (iii) Geobacter and Dehalococcoides (Duhamel and Edwards, 2007; Amos et al., 2009) or (iv) a mix of Dehalococcoides strains (Duhamel et al., 2002; Holmes et al., 2006). In contrast, the association of Sulfurospirillum and Dehalococcoides is rather rare, described only in an enrichment culture from a bioreactor treating PCE‐contaminated groundwater (Maillard et al., 2011) and in situ on a French chloroethene contaminated site (Dugat‐Bony et al., 2011a).
Temporal succession of bacterial communities during ERD treatment
On site B, soon after the first lactate injection, higher diversity and abundance of RDase genes belonging to new dechlorinators were observed with Dehalobacter, Desulfitobacterium rather located in the highly contaminated source zone and Geobacter only present in the slightly contaminated downstream zone (P4) (Fig. 3A). This is the first report that shows the coexistence of Sulfurospirillum, Dehalobacter, Desulfitobacterium, Geobacter and Dehalococcoides species into a single area during an ERD treatment. All those genera were also detected by 16S rRNA gene (rrs) amplicon pyrosequencing approach (Table 2). In addition, as supported by previous studies (Becker, 2006; Dowideit et al., 2010) and by our results (Fig. 4), their heterogeneous spatial distribution into the contaminated zone could depend on their own specific requirements such as electron acceptors’ nature and concentrations (TCE, cis‐DCE, VC), environmental conditions (redox) or nutrient availability, but also from the inter species competitions.
Furthermore, our findings demonstrated the detection of supplemental RDase genes from Dehalococcoides group, i.e. bvcA and additional putative rdhA genes immediately after lactate injections (Fig. 3A). Thus, the concomitant detection of both bvcA and vcrA suggests the coexistence of distinct Dehalococcoides populations at some locations in the contaminated groundwater, as no strain was described to host both genes in its genome. This finding is consistent with the pyrosequencing data (Table 2). Hence, as shown in previous studies, the coordinated function of many Dehalococcoides species could be essential for a more efficient dechlorination (Behrens et al., 2008; Nishimura et al., 2008).
Impact of substrate injections on biodegradation reactions
Although lactate addition favours the biological dechlorination processes, the substrate injections itself (periods C2 and C4) cause immediately significant disruptions of the groundwater around the injection wells that are visible during several weeks (until periods C3 and C5) with strong disturbances in molecular analyses (Fig. 3A). This lag period could be essential for the restoration of groundwater conditions and the growth of microorganisms, certainly diluted by the massive addition of lactate solution. Thus, the addition of substrate upstream the contaminated zone could be preferable, leading to a lower disturbance of the environment where dechlorination needs to occur and to an acceleration of the decontamination process. This hypothesis was reinforced by the observation of a high degradation capacity in the most distant plume well P4, not used for injections, with both appearance of bvcA and higher signal intensity of vcrA immediately after the first lactate addition (C2) (Fig. 3A). These results reflected the fast diffusion of fermentation products and quick adaptation of degrading populations to environmental changes. For these reasons, it appears crucial to monitor all the reactions that occur in the contaminated zone at several points (spatial coverage) throughout the treatment (temporal coverage).
Potential application of functional gene array (FGA) for bioremediation assistance
Many environmental factors affect both the structure and the function of the microbial community. As shown in several case studies, an inadequate electron donor distribution but also the absence or the low abundance of essential dechlorinators can result in an incomplete dechlorination with a build‐up of intermediate degradation products such as cis‐DCE (Hendrickson et al., 2002). Among the three additional contaminated sites assessed in the present study, two exhibited a stalling of reductive dechlorination (sites G and H). As favourable reductive conditions were demonstrated for at least site G, the non‐detection of genes involved in the late‐stage dechlorination reactions (vcrA or bvcA) using the DechloArray might explain this finding (Fig. 3B). So, in these cases, the bioaugmentation might be the most relevant way for complete dechlorination rather than biostimulation. Indeed, many studies have demonstrated successful decontamination of polluted sites when adding the missing Dehalococcoides species (Ellis et al., 2000; Major et al., 2002).
Microbially mediated reduction of chlorinated solvents is a promising strategy for the remediation of highly contaminated groundwater. A better understanding of microbial community structure and function in relation to environmental conditions is important for designing a successful bioremediation strategy (Lovley, 2003). The use of our DechloArray for the degradation gene detection is a relevant approach to assess the performance of an in situ bioremediation process more rapidly and more accurately than most other tools. Indeed, microcosm testing needs long time to reveal biodegradation capabilities and did not reflect performance of microbial populations in real environmental conditions (Xu, 2006). Furthermore, because FGA enables to simultaneously target many genes (92 for the DechloArray), this technique has the unparallel potential to survey more functions than other molecular tools such as qPCR assays (He et al., 2008) especially when using explorative probe design strategies (Dugat‐Bony et al., 2011b). This tool is also better adapted than metagenomic approach for routine studies requiring rapid analysis of many environmental samples, although the latter gives a deeper view of microbial communities (Roh et al., 2010). Hence, even if this study do not demonstrate the predictive power of the DechloArray, this capacity could be tested in future applications by using it to determine the presence of indigenous dechlorinating populations on contaminated site before the treatment implementation, which is also called diagnostic. However, only the survey of many contaminated sites before and after treatment should answer if this application is conceivable.
Experimental procedures
Groundwater sample collection and analytical methods
No industrial activity and no specific geographical location can be disclosed for the four sites studied due to confidentiality reasons. At each site (B, F, G and H), groundwater samples were collected by SITA Remediation Company (Lyon, France) from the monitoring wells drilled in the deep aquifer i.e. between −20 and −30 m depth for the site B, −7 and −12 m for the site F, −7 and −10 m for the site G and −3 and −6 m for the site H. Wells were sampled by low‐flow purging methods using a peristaltic pump. Three litres of water were collected per well and then stored on ice during transport (from 24 to 48 h). Finally, 30 groundwater samples were collected from all sites (19 from site B, 4 from sites F and H respectively, and 3 from site G) for further analysis.
Chlorinated compounds were analysed by gas chromatography/mass spectrometry (GC/MS Thermo TRACE DSQ, Thermo Fisher Scientific, Villebon sur Yvette, France) through a headspace sampler (150°C for the injection) and a GC column (TRACE TR‐V1, 20 m × 0.18 mm × 1 µm, Thermo Fisher Scientific). The column was held for 6 min at 40°C, followed by a ramp rate of 18°C min−1 until 100°C and 40°C min−1 until 200°C. The carrier gas (helium) had a flow rate of 1 ml min−1.
Gas composition (methane and ethene) was analysed by gas chromatography with a 7890A Gas Chromatograph (Agilent, Santa Clara, California, USA) equipped with a headspace sampler, a flame ionization detector and a bonded polystyrene‐divinylbenzene‐based column (HP‐PLOT Q, 30 m × 0.53 mm × 40 µm, Agilent). The following temperature programme was used: 5 min at 60°C with the injector at 250°C. The carrier gas (nitrogen) had a flow rate of 3 ml min−1.
Total organic carbon and ions (dissolved bromide, chloride, fluoride, nitrate, nitrite, orthophosphate and sulfate) were analysed by ALcontrol Laboratories Company (Hawarden, UK) based on the Nederlands Normalisatie‐instituut (NEN, http://www.nen.nl) norms NEN‐EN 1484, NEN‐EN‐ISO 10304 and NEN‐EN‐ISO 11732.
Temperature, pH, conductivity, DO and the oxidation‐reduction potential (ORP) were measured onsite in groundwater with a portable multiparameter WTW and appropriate probes (VWR International LLC, West Chester, Pennsylvania, USA).
DNA extraction
Genomic DNA (gDNA) was extracted from all groundwater samples based on the experimental protocol described by Dugat‐Bony and colleagues (2011a). Briefly, 5–250 ml of water samples (depending on solids concentration) were filtered under vacuum onto a 47 mm 0.2 µm pore size GTTP filter (Millipore, Billerica, Massachusetts, USA) immediately after the sample reception. Then, filters were stored at −80°C in extraction buffer (see Dugat‐Bony et al., 2011a) until processing or directly subjected to bead beating disruption followed by phenol chloroform extraction. gDNA concentrations were measured using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, North Carolina, USA).
Sample labelling
The BioPrime Total Genomic Labelling System (Invitrogen, Carlsbad, California, USA) was used to label the gDNA with Alexa Fluor 3 or 5 fluorescent dyes. The reactions used 1 µg of gDNA template following the manufacturer's instructions and multiplied the amount of DNA by at least sixfold. The effectiveness of the labelling method was assessed by calculating the amount of labelled nucleotides using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies).
Microarray experimental method
The functional gene array used in this study was an improved version of the fully validated DechloArray previously developed by Dugat‐Bony and colleagues (2011a). The new version contained 760 oligonucleotide probes of 50‐mer length and targeted 92 genes sequences corresponding to 24 different proteins (Table S1). Five distinct regions per gene were targeted and each oligonucleotide probe was synthesized in situ with eight replicates. Microarrays were produced in a 12 plex format (Roche NimbleGen).
Two micrograms of purified labelled products were used for the microarray hybridization process using the standard Nimblegen methods. The targets were hybridized for 72 h at 42°C using a 4‐bay hybridization system (Roche NimbleGen). Array washes were performed as recommended by NimbleGen and slides were scanned at 2 µm resolution using the InnoScan 900 and Mapix software (Innopsys, Carbonne, France). Finally, pixel intensities were extracted using NimbleScan v2.5 software (Roche NimbleGen) and pair reports containing signal intensity data for every spot on the array linked to its corresponding probe identifier were generated.
Microarray data normalization and analysis
Specific scripts developed in this study with the Delphi language were used to establish a four‐step automated method to analyse the pair reports generated by NimbleScan v2.5 software. The first two steps, namely local background evaluation and intra‐array normalization by SNR’ calculation, were as described by Dugat‐Bony and colleagues (2011a). The third step allowed inter‐array normalization of the result distributions using the quantile method originally described by Bolstad and colleagues (2003). This method was implemented in a R function as part of the preprocessCore library of the Bioconductor available at http://www.bioconductor.org/packages/release/bioc/. Finally, the last step of the method consisted of a gene response evaluation. Each probe showing a median SNR’ ≥ 1.5 from the eight replicates was considered a positive response. Furthermore, all genes for which less than four distinct regions of the five available were detected were considered negative. Finally, gene response level was calculated as the mean of the highest probe response in each region. The microarray data discussed in this publication are available at the GEO web site (http://www.ncbi.nlm.nih.gov/geo/) under Accession No.: GSE28609.
454GS FLX Titanium sequencing and read analysis
Pyrosequencing of 16S rRNA gene fragments amplified with the universal bacterial primers 27F and 1492R from five selected gDNA samples (campaigns C2, C3 and C5 for source well P2 as well as campaigns C3 and C4 for the plume well P4 from site B) and sequence analysis were performed by MR DNA (http://www.mrdnalab.com) using a 454 GS FLX Titanium platform and proprietary analysis pipeline. Operational taxonomic units were defined by clustering at 97% similarity and taxonomically classified using BLASTn against a curated GreenGenes database (DeSantis et al., 2006). The sequence data from 16S rRNA gene amplicon pyrosequencing were deposited in the NCBI as a Short Read Archive (SRA) project under Accession No. SRA049465.
RDA
The RDA was performed with XLSTAT software (http://www.xlstat.com/) on data collected on site B for all monitoring wells from all sampling periods except for P3 sample at the period C1 for which no microarray data were available (19 samples). Table Y (species data) contained response levels for the 10 positive genes detected and Table X (environmental data) consisted of values or concentrations obtained for the 13 following parameters: ORP, conductivity, DO, PCE, TCE, cis‐DCE, VC, ethene, methane, TOC, nitrate, sulfate and chloride. Unrestricted Monte Carlo permutation tests were performed with 499 random permutations and a significance level (P) of 0.05.
Acknowledgments
This work was supported by the Grant ID 2598 from the ‘Agence De l'Environnement et de la Maîtrise de l'Energie’ (ADEME, France) and the Grant ANR‐07‐ECOT‐005‐05 for the program PRECODD Evasol from ‘Agence Nationale de la Recherche’ (ANR, France). We are grateful to our undergraduate students Gaëtan Guillaume, Stéphane Freitas and Anne‐Sophie Yvroud (IUT Génie Biologique, Clermont‐Ferrand) for their contribution on this work. We thank the transcriptomic platform of INRA (Crouël, France) for giving access to the microarray hybridization material.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Description of the 92 gene sequences targeted by the improved version of the DechloArray.
Table S2. Physico‐chemical data obtained for the three contaminated sites F, G and H. NA, data not available.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Amos B.K., Suchomel E.J., Pennell K.D., Löffler F.E. Spatial and temporal distributions of Geobacter lovleyi and Dehalococcoides spp. during bioenhanced PCE‐NAPL dissolution. Environ Sci Technol. 2009;43:1977–1985. doi: 10.1021/es8027692. [DOI] [PubMed] [Google Scholar]
- Arp D.J., Yeager C.M., Hyman M.R. Molecular and cellular fundamentals of aerobic cometabolism of trichloroethylene. Biodegradation. 2001;12:81–103. doi: 10.1023/a:1012089908518. [DOI] [PubMed] [Google Scholar]
- Becker J.G. A modeling study and implications of competition between Dehalococcoides ethenogenes and other tetrachloroethene‐respiring bacteria. Environ Sci Technol. 2006;40:4473–4480. doi: 10.1021/es051849o. [DOI] [PubMed] [Google Scholar]
- Behrens S., Azizian M.F., McMurdie P.J., Sabalowsky A., Dolan M.E., Semprini L., Spormann A.M. Monitoring abundance and expression of ‘Dehalococcoides’ species chloroethene‐reductive dehalogenases in a tetrachloroethene‐dechlorinating flow column. Appl Environ Microbiol. 2008;74:5695–5703. doi: 10.1128/AEM.00926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bunge M., Kleikemper J., Miniaci C., Duc L., Muusse M.G., Hause G., Zeyer J. Benzoate‐driven dehalogenation of chlorinated ethenes in microbial cultures from a contaminated aquifer. Appl Microbiol Biotechnol. 2007;76:1447–1456. doi: 10.1007/s00253-007-1097-3. [DOI] [PubMed] [Google Scholar]
- Cupples A.M., Spormann A.M., McCarty P.L. Growth of a Dehalococcoides‐like microorganism on vinyl chloride and cis‐dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69:953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daprato R.C., Löffler F.E., Hughes J.B. Comparative analysis of three tetrachloroethene to ethene halorespiring consortia suggests functional redundancy. Environ Sci Technol. 2007;41:2261–2269. doi: 10.1021/es061544p. [DOI] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowideit K., Scholz‐Muramatsu H., Miethling‐Graff R., Vigelahn L., Freygang M., Dohrmann A.B., Tebbe C.C. Spatial heterogeneity of dechlorinating bacteria and limiting factors for in situ trichloroethene dechlorination revealed by analyses of sediment cores from a polluted field site. FEMS Microbiol Ecol. 2010;71:444–459. doi: 10.1111/j.1574-6941.2009.00820.x. [DOI] [PubMed] [Google Scholar]
- Dugat‐Bony E., Missaoui M., Peyretaillade E., Biderre‐Petit C., Bouzid O., Gouinaud C. HiSpOD: probe design for functional DNA microarrays. Bioinformatics. 2011a;27:641–648. doi: 10.1093/bioinformatics/btq712. et al. [DOI] [PubMed] [Google Scholar]
- Dugat‐Bony E., Peyretaillade E., Parisot N., Biderre‐Petit C., Jaziri F., Hill D. Detecting unknown sequences with DNA microarrays: explorative probe design strategies. Environ Microbiol. 2011b;14:356–371. doi: 10.1111/j.1462-2920.2011.02559.x. et al. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Edwards E.A. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2‐dichloroethane. Environ Sci Technol. 2007;41:2303–2310. doi: 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Wehr S.D., Yu L., Rizvi H., Seepersad D., Dworatzek S. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis‐dichloroethene and vinyl chloride. Water Res. 2002;36:4193–4202. doi: 10.1016/s0043-1354(02)00151-3. et al. [DOI] [PubMed] [Google Scholar]
- Duhamel M., Mo K., Edwards E.A. Characterization of a highly enriched Dehalococcoides‐containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70:5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.E., Lutz E.J., Odom J.M., Buchanan R.J., Bartlett C.L., Lee M.D. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ Sci Technol. 2000;34:2254–2260. et al. [Google Scholar]
- Futagami T., Goto M., Furukawa K. Biochemical and genetic bases of dehalorespiration. Chem Rec. 2008;8:1–12. doi: 10.1002/tcr.20134. [DOI] [PubMed] [Google Scholar]
- Futagami T., Morono Y., Terada T., Kaksonen A.H., Inagaki F. Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl Environ Microbiol. 2009;75:6905–6909. doi: 10.1128/AEM.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Sung Y., Dollhopf M.E., Fathepure B.Z., Tiedje J.M., Löffler F.E. Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene‐contaminated sites. Environ Sci Technol. 2002;36:3945–3952. doi: 10.1021/es025528d. [DOI] [PubMed] [Google Scholar]
- He J., Ritalahti K.M., Aiello M.R., Löffler F.E. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl Environ Microbiol. 2003;69:996–1003. doi: 10.1128/AEM.69.2.996-1003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z.L., Van Nostrand J.D., Wu L.Y., Zhou J.Z. Development and application of functional gene arrays for microbial community analysis. Trans Nonferrous Met Soc China. 2008;18:1319–1327. [Google Scholar]
- Hendrickson E.R., Payne J.A., Young R.M., Starr M.G., Perry M.P., Fahnestock S. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene‐contaminated sites throughout North America and Europe. Appl Environ Microbiol. 2002;68:485–495. doi: 10.1128/AEM.68.2.485-495.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes V.F., He J., Lee P.K., Alvarez‐Cohen L. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol. 2006;72:5877–5883. doi: 10.1128/AEM.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.M., Chen Y.L., Chen S.C., Yeh T.Y., Wu W.S. Enhanced PCE dechlorination by biobarrier systems under different redox conditions. Water Res. 2003;37:4885–4894. doi: 10.1016/j.watres.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Kennedy L.G., Everett J.W., Becvar E., DeFeo D. Field‐scale demonstration of induced biogeochemical reductive dechlorination at Dover Air Force Base, Dover, Delaware. J Contam Hydrol. 2006;88:119–136. doi: 10.1016/j.jconhyd.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Lee P.K.H., Macbeth T.W., Sorenson K.S., Jr, Deeb R.A., Alvarez‐Cohen L. Quantifying genes and transcripts to assess the in situ physiology of ‘Dehalococcoides’ spp. in a trichloroethene‐contaminated groundwater site. Appl Environ Microbiol. 2008;74:2728–2739. doi: 10.1128/AEM.02199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemming G., Hauschild M.Z., Chambon J., Binning P.J., Bulle C.C., Margni M., Bjerg P.L. Environmental impacts of remediation of a trichloroethene‐contaminated site: life cycle assessment of remediation alternatives. Environ Sci Technol. 2010;44:9163–9169. doi: 10.1021/es102007s. [DOI] [PubMed] [Google Scholar]
- Lendvay J.M., Löffler F.E., Dollhopf M., Aiello M.R., Daniels G., Fathepure B.Z. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ Sci Technol. 2003;37:1422–1431. et al. [Google Scholar]
- Lovley D.R. Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- Lu X., Wilson J.T., Kampbell D.H. Relationship between Dehalococcoides DNA in ground water and rates of reductive dechlorination at field scale. Water Res. 2006;40:3131–3140. doi: 10.1016/j.watres.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Macbeth T.W., Cummings D.E., Spring S., Petzke L.M., Sorenson K.S., Jr Molecular characterization of a dechlorinating community resulting from in situ biostimulation in a trichloroethene‐contaminated deep, fractured basalt aquifer and comparison to a derivative laboratory culture. Appl Environ Microbiol. 2004;70:7329–7341. doi: 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty P.L. Breathing with chlorinated solvents. Science. 1997;276:1521–1522. doi: 10.1126/science.276.5318.1521. [DOI] [PubMed] [Google Scholar]
- Maillard J., Schumacher W., Vazquez F., Regeard C., Hagen W.R., Holliger C. Characterization of the corrinoid iron‐sulfur protein tetrachloroethene reductive dehalogenase of Dehalobacter restrictus. Appl Environ Microbiol. 2003;69:4628–4638. doi: 10.1128/AEM.69.8.4628-4638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard J., Charnay M.‐P., Regeard C., Rohrbach‐Brandt E., Rouzeau‐Szynalski K., Rossi P., Holliger C. Reductive dechlorination of tetrachloroethene by a stepwise catalysis of different organohalide respiring bacteria and reductive dehalogenases. Biodegradation. 2011;22:949–960. doi: 10.1007/s10532-011-9454-4. [DOI] [PubMed] [Google Scholar]
- Major D.W., McMaster M.L., Cox E.E., Edwards E.A., Dworatzek S.M., Hendrickson E.R. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol. 2002;36:5106–5116. doi: 10.1021/es0255711. et al. [DOI] [PubMed] [Google Scholar]
- Maphosa F., de Vos W.M., Smidt H. Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide‐respiring bacteria. Trends Biotechnol. 2010;28:308–316. doi: 10.1016/j.tibtech.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Mattes T.E., Alexander A.K., Coleman N.V. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev. 2010;34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- Maymó‐Gatell X., Chien Y.‐T., Gossett J.M., Zinder S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- Müller J.A., Rosner B.M., von Abendroth G., Meshulam‐Simon G., McCarty P.L., Spormann A.M. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A., Wohlfarth G., Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Ebisawa M., Sakihara S., Kobayashi A., Nakama T., Okochi M., Yohda M. Detection and identification of Dehalococcoides species responsible for in situ dechlorination of trichloroethene to ethene enhanced by hydrogen‐releasing compounds. Biotechnol Appl Biochem. 2008;051:1–7. doi: 10.1042/BA20070171. [DOI] [PubMed] [Google Scholar]
- Pant P., Pant S. A review: advances in microbial remediation of trichloroethylene (TCE) J Environ Sci (China) 2010;22:116–126. doi: 10.1016/s1001-0742(09)60082-6. [DOI] [PubMed] [Google Scholar]
- Rahm B., Chauhan S., Holmes V., Macbeth T., Jr, Sorenson K., Alvarez‐Cohen L. Molecular characterization of microbial populations at two sites with differing reductive dechlorination abilities. Biodegradation. 2006;17:523–534. doi: 10.1007/s10532-005-9023-9. [DOI] [PubMed] [Google Scholar]
- Regeard C., Maillard J., Holliger C. Development of degenerate and specific PCR primers for the detection and isolation of known and putative chloroethene reductive dehalogenase genes. J Microbiol Methods. 2004;56:107–118. doi: 10.1016/j.mimet.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Ritalahti K.M., Amos B.K., Sung Y., Wu Q., Koenigsberg S.S., Loffler F.E. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol. 2006;72:2765–2774. doi: 10.1128/AEM.72.4.2765-2774.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S.W., Abell G.C.J., Kim K.‐H., Nam Y.‐D., Bae J.‐W. Comparing microarrays and next‐generation sequencing technologies for microbial ecology research. Trends Biotechnol. 2010;28:291–299. doi: 10.1016/j.tibtech.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Rouzeau‐Szynalski K., Maillard J., Holliger C. Frequent concomitant presence of Desulfitobacterium spp. and ‘Dehalococcoides’ spp. in chloroethene‐dechlorinating microbial communities. Appl Microbiol Biotechnol. 2011;90:361–368. doi: 10.1007/s00253-010-3042-0. [DOI] [PubMed] [Google Scholar]
- Ruder A.M. Potential health effects of occupational chlorinated solvent exposure. Ann NY Acad Sci. 2006;1076:207–227. doi: 10.1196/annals.1371.050. [DOI] [PubMed] [Google Scholar]
- Scheutz C., Durant N.D., Dennis P., Hansen M.H., Jørgensen T., Jakobsen R. Concurrent ethene generation and growth of Dehalococcoides containing vinyl chloride reductive dehalogenase genes during an enhanced reductive dechlorination field demonstration. Environ Sci Technol. 2008;42:9302–9309. doi: 10.1021/es800764t. et al. [DOI] [PubMed] [Google Scholar]
- Song D.L., Conrad M.E., Sorenson K.S., Alvarez‐Cohen L. Stable carbon isotope fractionation during enhanced in situ bioremediation of trichloroethene. Environ Sci Technol. 2002;36:2262–2268. doi: 10.1021/es011162d. [DOI] [PubMed] [Google Scholar]
- Sorenson K.S. Enhanced bioremediation for treatment of chlorinated solvent residual source areas. In: Henry S.M., Warner S.D., editors. American Chemical Society; 2002. pp. 119–131. [Google Scholar]
- Taş N., van Eekert M.H.A., Schraa G., Zhou J., de Vos W.M., Smidt H. Tracking functional guilds: ‘Dehalococcoides’ spp. in European river basins contaminated with hexachlorobenzene. Appl Environ Microbiol. 2009;75:4696–4704. doi: 10.1128/AEM.02829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N., Van Eekert M.H.A., De Vos W.M., Smidt H. The little bacteria that can – diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb Biotechnol. 2010;3:389–402. doi: 10.1111/j.1751-7915.2009.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T.M., Criddle C.S., McCarty P.L. Transformations of halogenated aliphatic compounds. Environ Sci Technol. 1987;21:722–736. doi: 10.1021/es00162a001. [DOI] [PubMed] [Google Scholar]
- Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- Yang Y., Pesaro M., Sigler W., Zeyer J. Identification of microorganisms involved in reductive dehalogenation of chlorinated ethenes in an anaerobic microbial community. Water Res. 2005;39:3954–3966. doi: 10.1016/j.watres.2005.07.010. [DOI] [PubMed] [Google Scholar]
- van der Zaan B., Hannes F., Hoekstra N., Rijnaarts H., de Vos W.M., Smidt H., Gerritse J. Correlation of Dehalococcoides 16S rRNA and chloroethene‐reductive dehalogenase genes with geochemical conditions in chloroethene‐contaminated groundwater. Appl Environ Microbiol. 2010;76:843–850. doi: 10.1128/AEM.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Description of the 92 gene sequences targeted by the improved version of the DechloArray.
Table S2. Physico‐chemical data obtained for the three contaminated sites F, G and H. NA, data not available.


