In the past few years, the number of microbial genome sequencing projects worldwide has rapidly increased, both of single species and microbial consortia (metagenomes). The development of several new high‐throughput sequencing platforms (Hall, 2007; Marsh, 2007), and an enormous reduction in costs, means we can expect to have thousands of complete and incomplete genomes sequences available to us in the coming years. Many of these microbial genomes are of biotechnological interest, and several have spectacular properties in relation to their growth requirements, the metabolites they produce, their potential for environmental clean‐up or survival in extreme environments. One of the ideas behind sequencing and analysis of whole genomes or substantial parts is that it will be used to enable a more targeted construction of mutant strains for improvement of industrial processes. This is in contrast to the more common procedure of production of random mutations and then screening for the desired phenotype.
The sheer number of newly completed genomes, estimated at about 1 per day in 2008, makes it impossible to publish all this information in regular scientific journals. So how do we keep track of which genome sequences are known or are upcoming, and where can we find all this sequence data to do data mining and comparative genomics in search of leads for our own research on biotechnologically interesting microbes?
Genome sequencing and databases
To make genome datasets publicly available, they are initially submitted to the public sequence data repositories GenBank (Benson et al., 2008), EMBL (Cochrane et al., 2008) and DDBJ (Sugawara et al., 2008). Then this genome data is further processed in different ways by curation, annotation, and comparison and ends up in a variety of microbial genome data resources, as reviewed recently (Markowitz, 2007). A very complete and up‐to‐date status of genome sequencing can be found in the Genomes Online Database (GOLD; http://www.genomesonline.org) (Liolios et al., 2008), a World Wide Web resource for comprehensive access to information regarding complete and ongoing genome projects, as well as metagenomes and metadata, around the world. The entry page links to the GOLD tables, each containing a summary of different kinds of sequencing projects: completed genomes, ongoing genomes (archaeal, bacterial or eukaryote) or metagenomes. Links are provided to each organism, genome sequence, institution, funding agency, scientific journal publication, and much much more. By clicking on the button ‘Download’ at the top of a table, access is gained to a wealth of metadata for each microbial genome, such as species/strains/serovars, phenotype, habitat, origin of isolation, pH and temperature regimes, etc.
The GOLD statistics report that most of recent genome sequencing data of bacteria and archaea comes from large high‐throughput sequencing centers such as the Joint Genome Institute (25%) and the J. Craig Venter Institute (23%) in the USA. Many of these genomes are part of major large‐scale microbial sequencing programs funded by government agencies such as National Institutes of Health (NIH), National Science Foundation (NSF), and the Department of Energy (DOE) in the USA.
Unpublished public genomes
At the end of 2007, over 700 completed genomes were listed that can be accessed in public databases, and the large majority of those were of bacterial and archaeal origin. ‘Complete’ means single complete sequences for each chromosome. Up to 2004, nearly all of these complete genomes were also reported in scientific journals, and these are referred to as ‘published public’ genomes (Figure 1). After 2004, the number of newly published public genomes has remained rather steady at 60–70 per year, while the number of ‘unpublished public’ genomes has increased rapidly. Last year, over 200 new genomes were released to public databases, but two‐thirds of those did not appear in scientific publications. These are the genomes that remain ‘invisible’ to the general reader who relies only on PubMed searches or other literature alert services. One way of getting a quick insight into recent ‘unpublished public’ genomes is to read Michael Galperin's two‐monthly brief summaries in the Genomics Update section of Environmental Microbiology (Galperin, 2007a,b).
Figure 1.
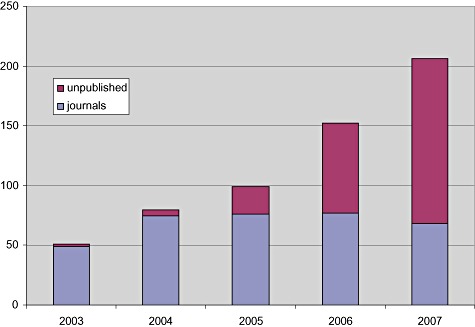
Number of microbial genomes made public annually from 2003–2007 (source GOLD On‐line Database v 2.0; http://www.genomesonline.org).
It is understandable that the more recent depositions may have no publication accompanying them yet, but it is surprising that almost 41% (243) of the completely sequenced microbial genomes catalogued in GOLD remain as yet unpublished. These organisms were sequenced to be used in comparative genomics studies, but either these analyses are still on‐going or they have been accomplished and the findings not reported. As the genomes are all in public databases it would be possible to do the comparison ‘in house’. Some of the sequenced genomes have been carefully investigated, and although not published in the scientific literature they have been used in patent applications submitted by the commissioning scientists and organizations.
Over 1500 additional genomes of bacteria and archaea were listed as ‘ongoing’ or incomplete at the end of 2007 in the GOLD tables, and none of those are reported yet in the scientific literature. Many of these genomes can also be considered as ‘public unpublished’ because access is provided to preliminary sequence data, usually consisting of multiple sequence contigs. So this is the place to go to, to find out what is being sequenced, who is doing this, and what is the status of each sequencing project.
Biotechnological relevance
GOLD also ranks microbial genomes according to biomedical, biotechnological, environmental, agricultural, or evolutionary relevance (with some overlap of categories) (Figure 2). For readers of this journal the category ‘Biotechnological relevance’ is the most interesting to scrutinize in more detail. In the last 6 months of 2007, the GOLD table lists 28 such genomes, of which 24 are still ‘unpublished’ (Table 1). Some interesting examples are Fervidobacterium nodosum from hot springs whose amylolytic enzymes have great potential, Alkaliphilus (Clostridium) oremlandii which reduces arsenate to arsenite, making it potentially useful in bioremediation of contaminated soils and waters, and Petrotoga mobilis from 60°C water near oil wells, which may help in cleaning up oil contaminations. Properties of a few other relevant microbes and their applications are described in more detail below.
Figure 2.
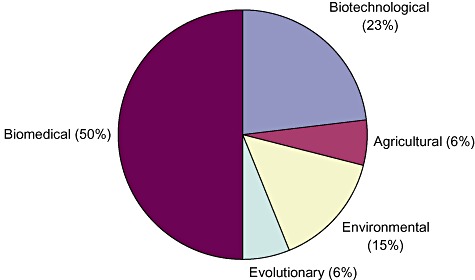
Funding relevance of microbial genome projects (source GOLD On‐line Database v 2.0; http://www.genomesonline.org).
Table 1.
Microbial genomes of biotechnological relevance made public in July–December 2007 (adapted from GOLD On‐line Database v 2.0; http://www.genomesonline.org).
| Domain | Organism | Strain | Phenotype | Habitat | Oxygen requirement | Temperature range | Publication |
|---|---|---|---|---|---|---|---|
| A | Caldivirga maquilingensis | IC‐167 | Sulfate reduction | Aquatic, Hot spring | Aerobe | Hyperthermophile | Unpublished |
| A | Candidatus Methanoregula boonei | 6A8 | Methanogen, Acidophile | Aquatic, Peat bog | Anaerobe | Mesophile | Unpublished |
| A | Ignicoccus hospitalis | Kin4/I | Chemolithoautotrophic, sulfidogenic | Hydrothermal vent | Anaerobe | Hyperthermophile | Unpublished |
| A | Methanococcus maripaludis | C6 | Hydrogenotrophic, Methanogen, Nitrogen fixation | Aquatic, Sediment | Obligate anaerobe | Mesophile | Unpublished |
| A | Nitrosopumilus maritimus | SCM1 | Ammonia oxidizer | Aquatic | Aerobe | Mesophile | Unpublished |
| B | Acaryochloris marina | MBIC11017 | Symbiont | Marine | Aerobe | Mesophile | Unpublished |
| B | Actinobacillus succinogenes | 130Z | Carbon dioxide‐loving | Bovine rumen | Facultative | Mesophile | Unpublished |
| B | Alkaliphilus (Clostridium) oremlandii | OhILAs | Nitrogen fixation, Arsenic metabolizer | Aquatic, Sediment | Anaerobe | Mesophile | Unpublished |
| B | Bacillus amyloliquefaciens | FZB42 | Promotes plant growth | Soil | Aerobe | Mesophile | (Chen et al., 2007) |
| B | Chloroflexus aurantiacus | J‐10‐fl | Carbon dioxide fixation | Aquatic, Hot spring | Anaerobe | Thermophile | Unpublished |
| B | Clostridium kluyveri | DSM 555 | Ethanol and acetate fermentation | Aquatic, Mud | Anaerobe | Mesophile | (Seedorf et al., 2008) |
| B | Clostridium phytofermentans | ISDg | Cellulolytic | Soil | Anaerobe | Mesophile | Unpublished |
| B | Delftia acidovorans | SPH‐1 | Organic acid utilization | Sludge, Soil | Aerobe | Mesophile | Unpublished |
| B | Desulfococcus oleovorans | Hxd3 | Sulfate reducer, Alkane degrader | Aquatic, Oil fields | Anaerobe | Mesophile | Unpublished |
| B | Fervidobacterium nodosum | Rt17‐B1 | Chemoorganotroph | Aquatic, Hot spring | Anaerobe | Thermophile | Unpublished |
| B | Frankia sp. | Mbj2, EAN1pec | Nitrogen fixation | Plant symbiont, Soil | Aerobe | Mesophile | Unpublished |
| B | Herpetosiphon aurantiacus | ATCC 23779 | Motile, filamentous, gilding | Aquatic | Aerobe | Mesophile | Unpublished |
| B | Lactobacillus helveticus | DPC 4571 | Cheese starter | Dairy | Facultative | Mesophile | (Callanan et al., 2008) |
| B | Methylobacterium extorquens | PA1 | Methylanotroph | Plant association | Facultative | Mesophile | Unpublished |
| B | Parvibaculum lavamentivorans | DS‐1 | Surfactant‐degrading | Sludge | Aerobe | Mesophile | Unpublished |
| B | Petrotoga mobilis | SJ95t | Motile, sulfate reducer, halotolerant | Marine, Oil fields | Anaerobe | Thermophile | Unpublished |
| B | Roseiflexus castenholzii | HLO8, DSM 13941 | Photosynthetic, motile | Marine, Hot spring | Facultative | Thermophile | Unpublished |
| B | Salinispora arenicola | CNS205 | Sporulating, halophile | Aquatic, Sediment | Aerobe | Mesophile | Unpublished |
| B | Shewanella baltica | OS195 | Halophile, non‐fermentative | Marine | Facultative | Mesophile | Unpublished |
| B | Shewanella baltica | OS185 | Halophile, non‐fermentative | Marine | Facultative | Psychrotolerant | Unpublished |
| B | Shewanella sediminis | HAW‐EB3T | Non‐sporulating | Aquatic, Sediment | Facultative | Psychrophile | Unpublished |
| B | Sorangium cellulosum | So ce56 | Motile, gliding, cellulolytic | Soil | Aerobe | Mesophile | (Schneiker et al., 2007) |
| B | Thermotoga lettingae | TMOT | Methanol‐degrading | Aquatic | Anaerobe | Thermophile | Unpublished |
A = Archaea; B = Bacteria.
Biofuel production
Cellulose is a complex plant polysaccharide that is not that easy to degrade. Several clostridia achieve this using a mixture of enzymes (endoglucanases and glucanases) which are held together in a large complex on the cell surface known as the cellulosome (Bayer et al., 2004; Doi and Kosugi, 2004). Clostridia are anaerobes mostly isolated from soils where they adhere to decaying plant material. Some also inhabit other niches such as the stomach of ruminants and the human colon. One which has recently had its genome sequenced is Clostridium phytofermentansISDg (ATCC 700394), isolated from a damp silt bed in a forested area (Warnick et al., 2002). This strain is special in that it can anaerobically ferment a vast array of plant sugars, starches and cellulose to produce economically substantial amounts of ethanol and acetate. It produces two to four times more ethanol than acetate and this suggests it contains unusual fermentation pathways. In fact, the genome of Clostridium phylofermentans contains over 100 ABC‐type transport systems and 52 of these appear to be dedicated to transporting carbohydrates into cells. Some of these are monosaccharide transporters but others are involved in the transport of dissacharides (e.g cellobiose), tri‐ and tetrasaccharides (Leuscine and Warnick 2007). The polymer‐hydrolyzing lifestyle of this organism and a distant relative Clostridium thermocellum are currently the object of a comparative genomics effort. The composition of the cellulosome in relation to the substrate that the organism has been adapted to was subject of a proteomics study, which showed that different glucanases were incorporated into the cellulosome (Gold and Martin, 2007). Another genome sequenced but not yet completely assembled is that of Clostridium cellulolyticum H10. Comparative genomics should help to explain the differences in the fermentative capacity of these organisms, which are all very useful as biomass fermenters producing substantial amounts of ethanol but also other compounds such as acetate and lactate. The comparative analysis may also help to explain the differences that occur during biofilm formation with these organisms, as the formation of biofilms may have dramatic effects on subsequent cellulose decomposition. It is possible that in some (Clostridium phytofermentans) it will increase ethanol production and in others (Clostridium cellulolyticum) reduce ethanol formation (Desvaux et al., 2000). The spin‐off company SunEthanol (http://www.sunethanol.com) has been established to exploit the biofuel‐producing potential of Clostridium phytofermentans.
Fine chemicals production
Actinobacillus succinogenes strain 130Z (ATCC 55618) was isolated from the bovine rumen. It is a Gram‐negative, facultatively anaerobic, pleomorphic bacterium, belonging to the family Pasteurellaceae that, in addition to the genus Actinobacillus includes Mannheimia, Haemophilus, and Pasteurella. These bacteria are generally pathogenic or commensal. A. succinogenes is thought to serve a commensal role by producing organic acids that are used as an energy source by the cow. The major end product of its fermentative metabolism is succinate (Guettler et al., 1999), which has many industrial fine chemical uses. It is mostly produced by petrochemical means by butane oxidation at high temperatures with catalysts. Succinic acid can be converted into a number of very important industrially useful chemicals such as butanediol, tetrahydrofuran, γ‐butyrolactone, adipic acid, succinate ester solvents, 2‐pyrrolidone, succinimide, maleic anhydride, and polybutylene succinate. As a specialty chemical, it is a flavour and formulating ingredient in food processing, a pharmaceutical ingredient and has use as a surfactant. The market potential for succinate is substantial and in future it will be used in many white technologies, e.g. for producing bulk chemicals, stronger‐than‐steel plastics, ethylene diamine disuccinate (a biodegradable chelator), and diethyl succinate (a green solvent for replacement of methylene chloride). World‐wide sales of biobased products have increased more than two‐fold in the last 10 years and the projection is for a continued increase (Committee on Biobased Industrial Products, National Research Council 2000).
A. succinogenes is the best known natural succinate producer, and it can utilize a wide range of substrates including glucose, cellobiose, lactose, xylose, arabinose, and fructose. It also has the potential to fix CO2 as every mole of succinate made by A. succinogenes requires a mole of CO2. It should be possible to couple industrial succinate fermentation to industrial ethanol fermentation by capturing the CO2 waste from the ethanol fermentation. The draft genome sequence was put to use in the filing of a patent application (Zeikus et al., 2007a) which claimed the genes from the organism for the production of chemicals from the C4 pathway. The genome sequence has also allowed for modeling of metabolic pathways. This modeling will assist in developing leads in processes which may change metabolic fluxes and control circuits diverting carbon flux away from other endpoints and thereby increasing production of succinate. In another patent application, the genome‐based metabolic model was used to define a minimal growth medium for A.succinogenes (Zeikus et al., 2007b). The genome (Hong et al., 2004) and a genome‐scale metabolic model (Kim et al., 2007) are also available for another succinate producer, Mannheimia succiniciproducens, and it should be interesting to compare their metabolic capacities.
Bioremediation
There are currently three complete sequenced strains of Shewanella baltica (OS195, OS185, OS155), while another (OS233) is in the draft phase. These bacteria were originally isolated from Baltic water and were reclassified from Shewanella putrifaciens to baltica (Ziemke et al., 1998). Many Shewanella have also been isolated from fish kept in cold storage, where they out‐compete other bacterial growth. This family of bacteria is considered as having great value for bioremediation. They have the ability to reduce metals and so could be used to remove contamination from sites with heavy metals. OS195 is highly versatile with respect to its ability to use many electron acceptors and donors. It is fast‐growing, easily cultivated and can survive long periods of starvation and grows quickly once nutrition is supplied. This strain was isolated in deep water in the Baltic Sea from an anoxic basin and formed the most populous clone of Shewanella isolated. The comparative genome analysis of the Shewanella will help in our understanding of biogeochemical potential and the specific ecology of the Baltic Sea, not to mention being potentially very useful as a bioremediation organism.
What to do with all this gold?
All these sequenced genomes and no descriptive publications – it seems a bit like Fort Knox vast vaults of precious metal, but not much being made out of it. The challenge for the comparative genomics field and not just the comparative biotech consortia is to explain what all this sequencing has accomplished, to tell us what it means and what it predicts for the future. There is today much concern that using food stuff for biofuel production is immoral. There is also great concern that in the push to cut dependence upon fossil fuels, that the means of producing the biofuel may be even more damaging on the environment (Cramer Commission Report 2007).
Surely, the comparative analysis of all these biotechnologically relevant micro‐organisms can produce new leads, cleaner methods, less energy demanding processes and sustainable production of biobased products – something which all the world requires.
References
- Bayer E.A., Belaich J.P., Shoham Y., Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch‐Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callanan M., Kaleta P., O’Callaghan J., O'Sullivan O., Jordan K., McAuliffe O. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol. 2008;190:727–735. doi: 10.1128/JB.01295-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I. Comparative analysis of the complete genome sequence of the plant growth‐promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. et al. [DOI] [PubMed] [Google Scholar]
- Cochrane G., Akhtar R., Aldebert P., Althorpe N., Baldwin A., Bates K. Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2008;36:D5–12. doi: 10.1093/nar/gkm1018. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Biobased Industrial Products, National Research Council. 2000.
- Cramer Commission Report. 2007. ) Testing framework for sustainable biomass ( http://www.mvo.nl/biobrandstoffen/download/070427‐Cramer‐FinalReport_EN.pdf.
- Desvaux M., Guedon E., Petitdemange H. Cellulose catabolism by Clostridium cellulolyticum growing in batch culture on defined medium. Appl Environ Microbiol. 2000;66:2461–2470. doi: 10.1128/aem.66.6.2461-2470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R.H., Kosugi A. Cellulosomes: plant‐cell‐wall‐degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- Galperin M.Y. Some bacteria degrade explosives, others prefer boiling methanol. Environ Microbiol. 2007a;9:2905–2910. doi: 10.1111/j.1462-2920.2007.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y. Dark matter in a deep‐sea vent and in human mouth. Environ Microbiol. 2007b;9:2385–2391. doi: 10.1111/j.1462-2920.2007.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold N.D., Martin V.J. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J Bacteriol. 2007;189:6787–6795. doi: 10.1128/JB.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettler M.V., Rumler D., Jain M.K. Actinobacillus succinogenes sp. nov., a novel succinic‐acid‐producing strain from the bovine rumen. Int J Syst Bacteriol. 1999;49(1):207–216. doi: 10.1099/00207713-49-1-207. , and Pt. [DOI] [PubMed] [Google Scholar]
- Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- Hong S.H., Kim J.S., Lee S.Y., In Y.H., Choi S.S., Rih J.K. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol. 2004;22:1275–1281. doi: 10.1038/nbt1010. et al. [DOI] [PubMed] [Google Scholar]
- Kim T.Y., Kim H.U., Park J.M., Song H., Kim J.S., Lee S.Y. Genome‐scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng. 2007;97:657–671. doi: 10.1002/bit.21433. [DOI] [PubMed] [Google Scholar]
- Leuscine S., Warnick T.A. 2007.
- Liolios K., Mavromatis K., Tavernarakis N., Kyrpides N.C. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V.M. Microbial genome data resources. Curr Opin Biotechnol. 2007;18:267–272. doi: 10.1016/j.copbio.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- Schneiker S., Perlova O., Kaiser O., Gerth K., Alici A., Altmeyer M.O. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. et al. [DOI] [PubMed] [Google Scholar]
- Seedorf H., Fricke W.F., Veith B., Bruggemann H., Liesegang H., Strittmatter A. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara H., Ogasawara O., Okubo K., Gojobori T., Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnick T.A., Methe B.A., Leschine S.B. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol. 2002;52:1155–1160. doi: 10.1099/00207713-52-4-1155. [DOI] [PubMed] [Google Scholar]
- Zeikus J.G., McKinlay J.B., Lavieniels M., Vielle C. 2007a. , and ) Genes from Actinobacillus succinogenes 13OZ (ATCC 55618) for production of chemicals from the A. succinogenes C4‐pathway. Patent application WO 2007/019301 A2.
- Zeikus J.G., Vielle C., McKinlay J.B. 2007b. and ) Minimal growth medium for Actinobacillus succinogenes. Patent application WO 2007/035589.
- Ziemke F., Hofle M.G., Lalucat J., Rossello‐Mora R. Reclassification of Shewanella putrefaciens Owen's genomic group II as Shewanella baltica sp. nov. Int J Syst Bacteriol. 1998;48(1):179–186. doi: 10.1099/00207713-48-1-179. [DOI] [PubMed] [Google Scholar]


