Summary
The production of biofuels via microbial biotechnology is a very active field of research. A range of fuel molecule types are currently under consideration: alcohols, ethers, esters, isoprenes, alkenes and alkanes. At the present, the major alcohol biofuel is ethanol. The ethanol fermentation is an old technology. Ongoing efforts aim to increase yield and energy efficiency of ethanol production from biomass. n‐Butanol, another microbial fermentation product, is potentially superior to ethanol as a fuel but suffers from low yield and unwanted side‐products currently. In general, biodiesel fuels consist of fatty acid methyl esters in which the carbon derives from plants, not microbes. A new biodiesel product, called microdiesel, can be generated in engineered bacterial cells that condense ethanol with fatty acids. Perhaps the best fuel type to generate from biomass would be biohydrocarbons. Microbes are known to produce hydrocarbons such as isoprenes, long‐chain alkenes and alkanes. The biochemical mechanisms of microbial hydrocarbon biosynthesis are currently under study. Hydrocarbons and minimally oxygenated molecules may also be produced by hybrid chemical and biological processes. A broad interest in novel fuel molecules is also driving the development of new bioinformatics tools to facilitate biofuels research.
Introduction
Society in the early 21st century appears to be undergoing an unprecedented transition with respect to the fundamental source of its materials and energy. Petroleum, the fuel that has been driving modern society for one century, is showing signs of scarcity (Grant, 2005). Demand for petroleum is increasing, but discoveries of fresh deposits are dwindling. The complex hydrocarbon mixtures found in crude oil deposits were formed under conditions of low heat and pressure over millions of years (Berner, 2003). Conventional petroleum is essentially non‐renewable. Intertwined with this practical impediment, there is an apparent moral dilemma arising from petroleum usage. It has become widely accepted that the combustion of long‐sequestered petroleum carbon is strongly contributing to the observed increase in atmospheric carbon dioxide, with concomitant global warming effects. The convergence of market pressure and concern for the environment is driving a headlong rush to new fuels that are largely bio‐based. But are biofuels really new?
History and needs
For millennia, human societies depended on biological materials for energy and materials. Plant material was combusted for heat, used for building materials and clothing; animal power was harnessed for transportation. A little more than a century ago, society underwent another transition: from horse to automobile, from whale oil to crude oil. The 1890s was similar to today in that many fuel sources were being tested, production was in flux, and the industry was not yet integrated and consolidated. A similar situation exists today with the transitioning to a renewable energy society.
Not all of the technology is new; for example, humans have been purposely making ethanol and fatty acid derivatives for millenia. But current molecular biological tools are opening new doors for old technology, and making radical new technologies seem possible. The biology of today is vastly different from that of decades ago and most of the new technology is microbially based. So it is not surprising that microbial metabolic activities are coming into focus for solutions to our fuel needs for the 21st century.
One of the important questions as we enter this stage of transition is, ‘What constitutes a good motor fuel?’ The answer is partly engine dependent but there are certain rules of thumb that span across different engine types. It is generally considered that a desirable fuel should be: (i) a liquid, (ii) highly combustible but not explosive, (iii) something with a high energy to mass ratio, (iv) stable on long‐term storage, (v) transportable by pipeline and (vi) inexpensive. A range of molecules meet most of those criteria (Fig. 1); it is expected that current microbiological research will lead to additional ones. Of those shown in Fig. 1, hydrogen, methane and propane are gaseous at 20oC, but they are important fuel molecules. Hydrogen suffers from storage problems making it unable to support long‐distance travel currently. Methane, in the form of natural gas, is used principally for heating and electricity generation. Propane is used with construction vehicles, particularly forklifts (AmeriGas, America's Propane Company, 2007), but is not widely used for general transportation. Of the moderate‐sized liquid fuels (C2–C5), about 20 billion pounds of methyl‐t‐butyl ether (MTBE) has been used annually but its use is declining due to water contamination issues (Suffet, 2007). Currently, the alcohol fuels, ethanol and n‐butanol, are being developed as alternative biomass‐derived oxygenated fuels. Of the two alcohols, ethanol is most widely used globally, with Brazil and the USA accounting for approximately 90% of ethanol production (Goldemberg, 2007).
Figure 1.
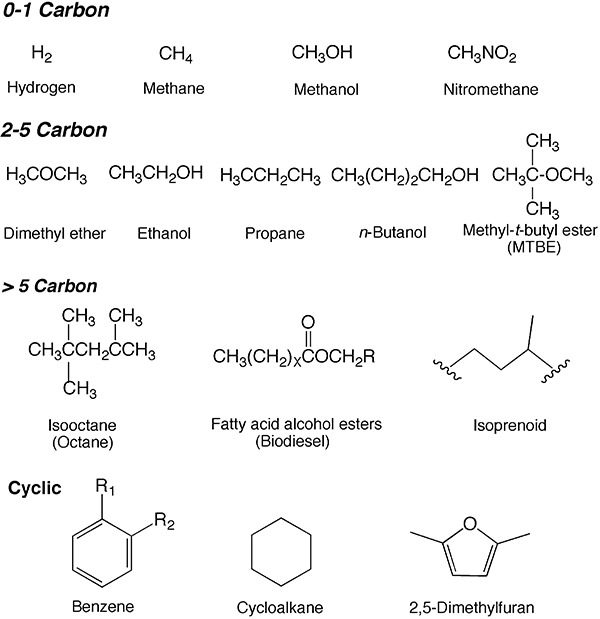
Gallery of fuel molecules in current, or proposed, use in spark ignition, diesel or construction vehicles.
The motor fuel of choice for spark ignition engines has consisted of, among other components, liquid, branched‐chain alkanes exemplified by 2,2,4‐trimethylpentane, or isooctane, which is ranked as 100 on the ‘octane scale’ for measuring suitability of molecules for fuel purposes. For diesel engines, a higher chain length of alkanes is desirable, and hexadecane is the comparable gold standard molecule. The use of longer‐carbon‐length hydrocarbons is due to the compression ignition in diesel engines; a spark plug is used to ignite a fuel–air mixture in spark ignition, or gasoline, engines. Both conventional gasoline and diesel hydrocarbons currently derive from crude oil, although recent microbial research is investigating a bio‐based replacement as discussed in subsequent sections on alkanes and alkenes. Petroleum‐based fuels are complex mixtures, consisting of benzene ring compounds, simple and alkylated cycloalkanes, alkanes and some heterocycles (Fig. 1).
Biodiesel, which serves as a replacement for hydrocarbon fuels in diesel engines, is generally comprised of fatty acid alcohol esters. The fatty acid precursors derive largely from animal and plant fatty acids (typically C8–C22) that are part of fats such as triacylglycerides, an esterified polyalcohol. Triacylglycerides themselves are not good fuels, but switching the alcohol moiety of a glycerol triester to make individual methanol or ethanol monoesters generates molecules that are excellent as a fuel for diesel engines. The chemical process used to switch the alcohol component is known as transesterification (Huber et al., 2006). The upsurge in biodiesel production has resulted in a market glut of glycerol, a by‐product of the transesterification process. However, new and creative uses for glycerol as a fermentation feedstock are emerging as a result of the artificially low price created by increased biodiesel production.
Creative approaches are also emerging to take biomass and use chemical catalysts to dehydrate and reduce sugars for generating molecules that are sufficiently energy‐dense to be used as fuels. An interesting recent example of this, described by Dumesic and co‐workers (Roman‐Leshkov et al., 2007), is a process to generate 2,5‐dimethylfuran, a molecule that is expected to have good combustion properties in vehicular engines. Such developments are indicative of the current perception in the biofuels arena. As we shift from petroleum, it has become unclear what the fuel molecule ‘winners’ will be. Which will have the requisite properties as fuels coupled with low production costs, and will the process of converting biomass be chemical, biological, or some mixture of both (Schmidt and Dauenhauer, 2007)? It is generally accepted that ethanol alone is not going to provide a long‐term solution to meet society's energy needs (Hill et al., 2006). It suffers from a somewhat low energy density, inability to be transported through pipelines and fairly high cost for extraction from fermentation broths. This is opening the door to developing many other molecules as replacements for ethanol and thus, discovering new fuel molecules to be produced via microbial biotechnology.
The desire for very different molecular structural class (hydrocarbons, alcohols, ethers, esters) is driving the idea that the field of synthetic biology will impact biofuels research. Synthetic biology is based on the premise that constructing biological systems from a ‘parts lists’ will lead to a greater understanding and utility of biology. A major tool of synthetic biology is genomics. Presently, the complete genome sequences for over 2000 microorganisms are in the public domain (GOLD, 2007). A significant number of the sequenced microbial strains have some significance in biofuels research due to the prominent role of the United States Department of Energy in choosing and funding the public sequencing of non‐pathogenic microbes. A representative list is shown in Table 1.
Table 1.
Representative microorganisms for which genome sequences have been completed or are in progress that are important in the context of biofuels research.
| Microorganism | Significance for biofuels |
|---|---|
| Anaebaena variabilis | Cyanobacterium producing hydrogen |
| Cladicellulosiruptor saccharolyticus | Degrades various polysaccharides; produces hydrogen |
| Clostridium acetobutylicum | Major organism producing n‐butanol |
| Clostridium phytofermentans | Ferments pectin, cellulose, xylan producing ethanol and hydrogen gas |
| Clostridium thermocellum | Thermophilic ethanol producer |
| Methanosaeta thermophila | Common methanogen; converts acetate to methane |
| Micrococcus luteus | Produces long‐chain alkenes |
| Pichia stipitis | Ethanol producing yeast fermenting xylose |
| Rhodospeudomonas palustris | Phototroph producing hydrogen gas |
| Saccharomyces cerevisiae | Major production organism for ethanol currently |
| Saccharophagus degradans | Degrades many biopolymers |
| Thermoanaerobacter pseudoethanolicus | Thermophilic ethanol producer |
| Vibrio furnissii M1 | Reported to produce high levels of n‐alkanes |
| Zymomonas mobilis | Ethanol fermentation with high ethanol tolerance |
With respect to biofuels, ethanol, butanol and alkyl esters are biologically common. In contrast, many fuel molecules in current usage (Fig. 1) are not composed of biologically common functional groups. However, using synthetic biology, these or similar molecules could conceivably be constructed by exploiting known enzyme catalysis in new ways. This could be used to make new biomolecules, or make known molecules in much larger quantities than their natural abundance. This premise underlies a number of the emerging start‐up biofuels companies that seek to create the next generation of bio‐based fuel. The price of the product they would sell is much lower than a pharmaceutical product but the market is huge, and thus, potentially very lucrative.
Alcohols
Ethanol
Ethanol production is based on an old technology, if one considers the production and consumption of alcoholic beverages by human societies. A 6000‐year‐old Sumerian tablet depicts people drinking alcoholic beverages. Much more recently, investigations into the alcoholic fermentation were instrumental in establishing the early fields of microbiology and biochemistry. In 1839, Leibig ridiculed the cell theory and the proposition that grape juice fermentation was a biological process (Liebig, 1839). About 20 years later, Pasteur firmly associated the presence of yeast cells with ‘good’ fermentations that made ethanol (Pasteur, 1857; 1860). Some 6000 years after initiating the ethanol fermentation, humans had some understanding of the cellular basis underlying the process. In 1897, Buchner prepared a cell‐free extract from Saccharomyces cerevisiae that transformed glucose to ethanol (Buchner, 1897). This was important for establishing a new way of biochemical investigation and it also showed conceptually that metabolism could occur in the absence of a living cell. This paved the way for the measurement of cell‐free enzyme activities and the general principles of enzyme kinetics (Michaelis and Menten, 1913). In fact, the field of enzymology was born out of studies on the ethanol fermentation.
It is somewhat astounding that ethanol produced for automobile engines in the current era is made using the same microorganism (Saccharomyces) and in similar titre (less than 15% of the aqueous fermentation broth) as has been done for centuries. The overall biochemical process used is also the same. Thus, 1 mole of glucose is converted to 2 moles of ethanol and 2 moles of carbon dioxide, with a net lost of one‐third of the carbon atoms in generating the fuel. Bacterial alternatives to Saccharomyces yeast are being studied, principally Zymomonas mobilis, Escherichia coliand Klebsiella oxytoca (Ingram et al., 1987; Dien et al., 2003); however, these other organisms are not widely used commercially. The limitation in the titre of ethanol is due to its toxic effects on microorganisms at concentrations above 5%. Thus, ethanol must be obtained via distillation and drying, which are energy‐intensive and somewhat costly steps. As a result, bioethanol production from corn starch is marginally energy yielding when one considers all the steps in the process (Hill et al., 2006). Ethanol production from sugarcane, as practiced in Brazil and other tropical areas, has a better net energy yield.
Some major features of ethanol fermentation are currently the subject of intensive research. Specifically, the use of hemicellulosic material will require the fermentation of both hexose (glucose, mannose and galactose) and pentose (xylose and arabinose) sugars. Currently, there are no naturally occurring ethanol fermenting strains that handle all of the necessary sugars. This problem could be solved by appropriate metabolic engineering (Dien et al., 2003). A novel approach to this problem would be to conduct a homoacetogenic fermentation, using strains that handle hexoses and pentoses simultaneously (Eggeman and Verser, 2006). In this approach, the resultant acetic acid is esterified with ethanol to make ethyl acetate, which is relatively water insoluble and thus readily recoverable. Hydrogenation of 1 mole of ethyl acetate, using well‐known industrial chemical methods, produces 2 moles of ethanol. An advantage of this approach is the overall stochiometry; 1 mole of hexose sugar yields 3 moles of ethanol. This contrasts with the traditional ethanol fermentation in which two of the hexose carbon atoms are lost as carbon dioxide.
Another critical issue in making the ethanol fermentation cheap and energy efficient is for integration of the different elements in transforming biomass to ethanol. As currently envisioned, the overall process involves four steps: the production of saccharolytic enzymes, the hydrolysis of biomass carbohydrate polymers, the fermentation of hexoses and the fermentation of pentoses (Lynd et al., 2005). It is hoped that genome sequencing will contribute to developing biological agents for a consolidated bioprocessing of cellulosic biomass. Specifically, organisms that are thermophilic, saccharolytic and ethanologenic may be used directly, or after genetic engineering, to develop a one‐pot fermentation process for taking biomass to ethanol (Table 1).
Butanol
Butanol is another microbial fermentation product, studied principally with the anaerobe Clostridium acetobutylicum. Clostridium acetobutylicumwas cultivated on an industrial scale nearly a century ago, although the desired compound in that process was the fermentation co‐product acetone. Acetone was in great demand in 1915 because of its utility in the manufacture of cordite, an explosive used by countries warring against Germany. Germany had been the major manufacturer of acetone prior to World War I but acetone export was cut off with the war. Chaim Weizmann led the British effort to produce acetone using C. acetobutylicum, a process that met the war needs of England (Dixon, 1997). The large‐scale fermentation process was shared with England's allies. By the end of the war, 22 fermentors of 30 000 gallon capacity were operating continuously in Canada (Kluyver, 1957). After World War I, however, the petroleum industry expanded and supplanted the bio‐acetone process.
With the re‐emergence of interest in producing fuels from biomass, there is again a focus on the acetone‐butanol‐ethanol fermentation of C. acetobutylicum. In the present day, n‐butanol is the most desired product. Clostridium acetobutylicum is also of interest because it is saccharolytic, thus it could potentially hydrolyse starch directly, removing a need for pre‐treatment. The fermentation pathway from glucose to ethanol is similar to that occurring in S. cerevisiae, proceeding through pyruvate and acetaldehyde (Fig. 2). Acetone is produced from 2 moles of acetyl‐CoA, via acetoacetyl‐CoA, thus there is a 50% loss of glucose carbon in generating one equivalent of acetone. A better carbon balance is obtained with n‐butanol, which derives from reduction of acetoacetyl‐CoA via butyryl‐CoA, thus capturing four of the carbon atoms of glucose in the end‐product. Recent fibrous bed reactor technology with C. acetobutylicum, developed by Huang and colleagues (2004), makes n‐butanol as the major component in the fermentation broth with a productivity of 4.6 g l−1 h−1 and a yield of 0.42 g g−1. These developments are beginning to make biobutanol an attractive fuel alternative to bioethanol.
Figure 2.
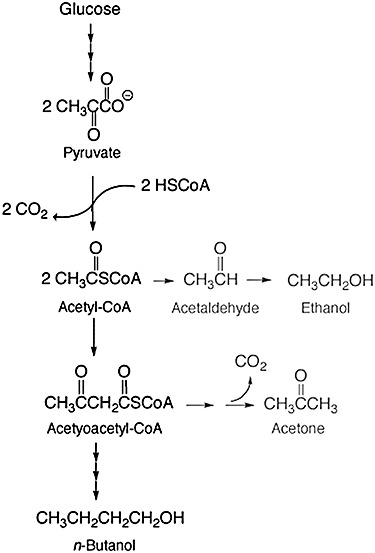
Butanol synthesis by the fermentation pathway of Clostridium acetobutylicum.
As a motor fuel, n‐butanol is superior to ethanol in several properties (Huber et al., 2006). n‐Butanol has a nearly 50% higher energy density than ethanol and is almost comparable to gasoline in this regard. n‐Butanol carries less water than ethanol and could be transported through pipelines, unlike ethanol. Although the performance properties of butanol in car engines is still controversial, it clearly could be used in fuel blends, if not used directly as a motor fuel. British Petroleum and DuPont have recently teamed up in a major effort to further develop biobutanol for use as a motor fuel (Biobutanol, 2007).
Methanol
While methanol is not always considered as a serious contender in the alcohol fuel race, it does have history on its side. Methanol was used in blended motor fuels in the USA in the 1970s. Indy racing cars burn 100% methanol because methanol can run at extremely high compression ratios and thus generate maximal engine power. George Olah, hydrocarbon chemist and Nobel Laureate, has promoted a methanol‐based economy, with methanol use as a fuel and chemical feedstock (Olah and Molnar, 1995). Currently, methanol is produced largely from natural gas, methane principally, by chemical catalytic processes that are mulistep. First, carbon monoxide and hydrogen are produced, and then those are reformed to generate methanol. There has been some examination of the microbial counterpart to this chemistry that generates methanol from methane in one step with 100% yield. The enzyme catalysing this reaction, methane monooxygenase, is a multicomponent enzyme system biosynthesized by methanotrophic bacteria, those that grow on methane as a carbon and energy source (Colby et al., 1979). The monooxygenase component is a µ‐oxo‐bridged di‐iron protein that has been studied in structural and mechanistic detail (Lipscomb, 1994; Sazinsky and Lippard, 2005). One major issue in biotechnological applications is the relative instability of the enzyme to oxidative damage in vitro. In vivo, methanotrophs oxidize methanol completely to carbon dioxide as part of their overall energy metabolism. Recombinant methane monooxygenase systems suffer from low activity in comparison with that observed in native hosts (Wood, 2002).
Alcohol esters of fatty acids (biodiesel)
The current production strategy for biodiesel does not involve microbial biotechnology. As mentioned previously in the section on fuel molecules, the fatty acid carbon atoms in biodiesel come from animal fat waste or from plant oils. Lipids, principally triacylglycerides, are transesterified in a chemical process to produce fatty acid esters that constitute biodiesel (Fig. 3A). However, recent developments suggest that microbial biotechnology could yield a breakthrough in biodiesel production. This is because there are some current limitations in producing biodiesel. Primary among them is the limitation of oil‐producing crops; there is only so much production capacity for rapeseed in Europe and soybean in the USA. Also, the methanol used for transesterification largely derives from natural gas. Thus, the resultant biodiesel is only partly derived from renewable sources. It would be more desirable to make fatty acid esters directly from cheaper and more widely available sugars such as glucose using bacteria as the catalysts for the entire transformation (Fig. 3B). This would dovetail with extensive work on bio‐based cellulose processing to sugars and make for a potential one pot process from biomass to biodiesel. An important step to accomplishing this goal was recently taken.
Figure 3.
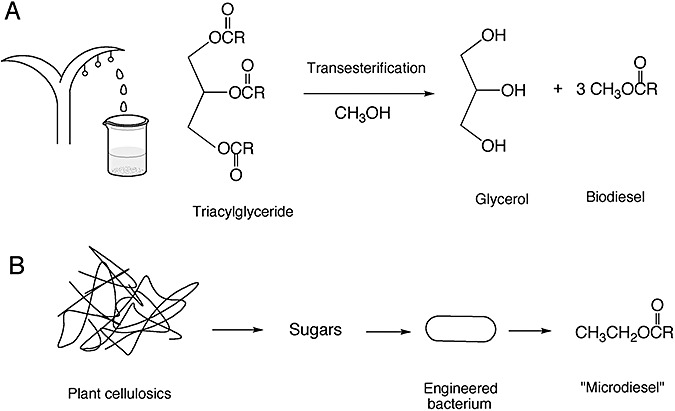
Processes for making biodiesel (A) chemically and (B) biologically.
While microorganisms do not typically make fatty acid methyl or ethyl esters, some microorganisms make copious amounts of storage lipids in the form of triacylglycerides and wax esters (Holdsworth and Ratledge, 1991; Kalscheuer et al., 2007). One bacterium, Acinetobacter baylyi strain ADP1, makes esters via an enzyme that catalyses acyltransfers to make wax esters and triacylglycerols and has very broad specificity with respect to the fatty acids and alcohols it binds (Kalscheuer and Steinbüchel, 2003). The observation that it would use ethanol as the acyl group acceptor, albeit at a lower rate than longer‐chain alcohols, provided the idea that a biodiesel‐producing bacterium could be engineered (Kalscheuer et al., 2006). Thus, the engineered E. coli strain contained the Acinetobacterwax ester synthase gene and ethanol‐production genes from Z. mobilis. The recombinant strain, when fed exogenous fatty acids, produced fatty acid ethyl esters, up to 26% of the bacterial dry mass under ideal conditions (Fig. 3B). This was an excellent demonstration of feasibility. The product was called ‘microdiesel’ as this was a microbially produced fuel, and it was different from the majority of biodiesel by consisting of ethyl esters. In the chemical process, methanol has been used more often as that is cheaper than ethanol. To make microdiesel economically, the fatty acids would need to be biosynthesized de novo from sugars by the E. coli strain. That would require further genetic engineering. Another obstacle to a cheap bioprocess is the intracellular accumulation of the fatty acid esters. Harvesting the desired fuel product would require cell rupture and separation, a potentially expensive process step. It is unclear if a bioprocess can be created in which highly hydrophobic molecules, like fatty acid ethyl esters, can be engineered for biological excretion outside the cell.
Ethers
Low‐molecular‐weight ethers have suitable properties as fuel molecules in terms of stability, water insolubility and combustibility. The major ethers currently in use for fuel applications are MTBE and dimethyl ether. MTBE has been used as a gasoline additive. It serves to reduce carbon dioxide emissions and raise the octane number of fuels. Its use has been declining in the USA because it is reasonably soluble in water and imparts an unpleasant taste and odour when present in drinking water at very low levels (Suffet, 2007). Dimethyl ether is more recently being used as a diesel fuel, for example, powering buses in Sweden (Wu et al., 2006). Both MTBE and dimethyl ether are synthesized by chemical catalytic processes.
While biofuel ethers are not currently in general use, ether biosynthetic reactions, relevant to fuel type molecule, are becoming more fully understood. One example is the biosynthesis of ether lipids by methanogenic bacteria (Koga and Morii, 2007). In this case the alkenyl chains are attached to a glycerol moiety via an ether linkage. The enzymatic condensation of an activated hydrocarbon alcohol to another alcohol might be co‐opted to make molecules that resemble biodiesel, but have superior properties.
Hydrocarbons
Hydrocarbons of the appropriate size are ideal fuels, meeting all the previously indicated fuel criteria: being (i) a liquid, (ii) highly combustible but not explosive, (iii) something with a high energy to mass ratio, (iv) stable on long‐term storage, (v) transportable by pipeline and (vi) inexpensive. The last property, being inexpensive, has been predicated on readily available petroleum resources. As society is increasingly confronted with scarce petroleum reserves, there is widespread interest in developing a bio‐based fuel that consists partly or wholly of hydrocarbons. It is widely believed that petroleum hydrocarbons derive from more highly oxygenated biological molecules deposited underground millions of years ago (Peters et al., 2005). An alternative non‐biogenic origin of petroleum has been proposed by Thomas Gold and others (Gold, 1985). As proposed for the biogenic origins of petroleum, long‐duration conditions of low heat and moderate to high pressure have given rise to less oxygenated molecules, including alkyl, cycloalkyl and aromatic hydrocarbons (Blumer, 1976). Thus, oil companies have often searched for biomarkers, bio‐derived indicator molecules, as indices of oil deposit origin, size and composition (Moldowan and Seifert, 1979). Over the last 50 years, there have been numerous, scattered reports of direct hydrocarbon biosynthesis by living things, animals, plants and microbes (Kolattukudy, 1976; Ladygina et al., 2006). The mechanisms underlying hydrocarbon biosynthesis have not been in the forefront of biochemistry, but these past reports are now being re‐examined with great interest. There is ample opportunity to delve more deeply into the biochemical mechanisms of hydrocarbonbiosynthesis and apply the more recently acquired tools of synthetic biology.
Isoprenoid compounds
Microorganisms make many hydrocarbon molecules; some are found within the large class of molecules known as isoprenoid compounds, of which there are over 50 000 currently known (Walsh, 2007; Withers and Keasling, 2007). The simplest example is isoprene itself, 2‐methylbuta‐1,3‐diene (Kuzma et al., 1995). The physiological function of bacterial unsubstituted isoprene formation is unknown but it is proposed to be enzymatic (Fall and Copley, 2000). Recently, the genes underlying isoprene production by Bacillus were identified (Julsing et al., 2007). The much larger class of isoprenoid compounds derive from the five‐carbon precursors isopentyl diphosphate (IPP) or dimethylallyl diphosphate (DMAPP), the two isomeric building blocks for this class of molecules. Thus, the compounds typically consist of carbon atoms in multiples of five. There can be one condensation reaction to generate a C10 molecules or as many as 21 cycles to generate a C110 molecule (Walsh, 2007). While most known products arise from head‐to‐tail condensation reactions, other variations of IPP and DMAPP coupling have now been identified and this contributes to the enormous diversity of more than 50 000 molecules observed in natural systems.
Isoprenoid compounds are commonly known as terpenes and carotenoids. The term terpene arises from the high abundance of these compounds in turpentine; a carotenoid pigment is the major orange pigment in carrots. Isoprenoid compounds are typically important in nature as chemical signals; they comprise pigments or odorants that might attract or deter another organism (Harborne, 1988). For human applications, isoprenoid compounds comprise many important food additives, fragrances and medicinal compounds. For these reasons, and the emerging potential for biofuel applications, there is burgeoning interest in genetically engineering microbial host strains to make large quantities of isoprenoid compounds. The host organisms used are generally those that have been most well studied with respect to molecular genetics: E. coli, S. cerevisiae and Arabidopsis thaliana.
Simple isoprene compounds have good potential for use as fuels if they can be produced on a large scale and cheaply. The company Amyris is developing a process to generate the anti‐malarial drug artemisinin (Amyris, 2007). Artemisinin is a C15 isoprenoid compound that is effective in inhibiting the malarial parasite Plasmodium falciparum(Cumming et al., 1997). Amyris’ goal is the production of isoprenoid compounds cheaply and abundantly. They recognize that a collateral benefit may be the production of novel biofuel molecules. Before petroleum was in widespread use, the leading fuel in the USA was known as camphene, a mixture of ethyl alcohol and turpentine (Kovarik, 2007). In this context, the use of isoprenoid compounds as a fuel has precedence in an earlier age.
The simplest alkane – methane
It has long been appreciated that microbes generate methane and that natural gas, principally methane, can be used as a fuel (Conrad, 1996). Methane has been used largely in home heating and cooking and for electrical generation at municipal power plants. Commercially, methane is obtained largely from extraction of natural gas fields which are often associated with petroleum deposits. Biosynthetic methane is used in some local applications, meeting energy needs for farms and anaerobic digestor facilities. However, there are significant impediments for generating methane to use as a major biofuel. Methane is generated by strictly anaerobic bacteria, methanogens, that grow relatively slowly on biomass as part of complex anaerobic ecosystems. There are generally numerous end‐products of these anaerobic fermentations that include volatile alkanoic acids in addition to methane. Moreover, the methane in the gas phase is mixed with nitrogen and carbon dioxide making it costly to purify. With present technology, biologically generated methane is probably most applicable to commerce on a small scale.
Longer‐chain alkanes
In general, alkanes power most vehicles today, although the chain length is significantly longer than methane (C1). The ‘gold standard’ fuel for spark combustion engines is isooctane (Fig. 1) and for diesel engines it is hexadecane. These derive almost totally from petroleum at present. Petroleum is believed to derive from biological molecules that have been reformed and become more reduced over millions of years by diagenic processes (Blumer, 1976). Generally, it is not considered that living things make petroleum‐like alkanes. However, there have been reports over many years demonstrating alkane biosynthesis by animals, plants and microbes (Kolattukudy, 1976). In most cases, the amount detected has been very low and the biochemical mechanisms have remained obscure. One of the more well‐documented mechanisms for generating alkanes biologically has been studied in plants and occurs via decarbonylation of fatty acid aldehydes (Schneider‐Belhaddad and Kolattukudy, 2000). In another example, a decarbonylase activity was purified from a microsomal membrane fractions of the alga Botryococcus braunii (Dennis and Kolattukudy, 1992). The purified enzyme was reported to transform octadecanal to carbon monoxide and the corresponding alkane heptadecane. It was suggested that the enzyme responsible contains a cobalt‐containing porphyrin cofactor. Botryococcus is known to produce and accumulate a range of hydrocarbons and hydrophobic ether lipids (Metzger and Largeau, 2005).
Most well‐documented studies of microbial alkane production have been conducted with marine eukaryotic algae. In one survey, Youngblood and Blumer (1973) reported that n‐pentadecane was the major alkane in the brown algae tested, while n‐heptadecane was found to predominate in red algae. Another species of green algae was reported to contain a C17‐cylcopropylalkane. Dunalliella salina has been reported to produce 6‐methyl hexadecane and 4‐methyl octadecane (Tornabene, 1980). Similar internally methyl branched alkanes have been reported in cyanobacteria (Han et al., 1968; Han and Calvin 1969; Fehler and Light, 1970; Gelpi et al., 1970)
Terrestrial microorganisms also produce alkanes other than methane (Jankowski and Zobell, 1948; Davis, 1968; Jones, 1969; Naccarato et al., 1974). The reported alkanes are generally normal chain with a range of C16–C30. However, the following caveat is found in a review by T.G. Tornabene, who studied microbial hydrocarbon biosynthesis for several decades, ‘Small amounts of nonisoprenoid hydrocarbons can be found in extracts from most bacterial cells. However, with appropriate precautions to eliminate extrinsic sources of hydrocarbons from the cultivation, extraction and analytical procedures, it is generally found that hydrocarbon biosynthesis is restricted to a relatively small number of bacteria’ (Tornabene, 1980).
Most recently, a bacterium, Vibrio furnissii M1, was reported to make substantial levels of intermediate to long‐chain alkanes (C16–C28) when grown on sugars or organic acids (Park et al., 2001; 2005). Vibrio furnissii was obtained from activated sludge at a sewage disposal plant located in Japan and observed, in the laboratory, to produce an extensive floating layer on top of liquid cultures. The extracted polar and non‐polar lipids were reported to consist of 48% alkanes (Park et al., 2001). Further research demonstrated the range of alkanes made by V. furnissii M1. A later paper reported that cell‐free membrane fractions catalysed the reduction of hexadecanoic acid to hexadecane (Park, 2005). In addition, a patent was filed in Japan describing alkane biosynthesis by V. furnissii M1 and other V. furnissii strains obtained from the American Type Culture Collection (ATCC) and other sources (Miyamato, 2001).
The reports were widely noted by biofuels researchers for several reasons. Alkanes are a superior fuel and, being water insoluble, would be much cheaper to extract than ethanol. The reports by Park and co‐workers indicated that a significant amount of the alkanes were outside the cells, and thus could potentially be recovered without expensive separation procedures. It was also notable that V. furnissiiM1 produced alkanes when grown on renewable carbon sources such as sugars and polysaccharides, for example, starch, chitin and xylan. Moreover, the titre of alkanes was significant, accounting for as much as 30% of the carbon consumed. It was also very significant that all of the carbon atoms of the alkanes were conserved from their respective fatty acid precursors (Park, 2005), unlike decarbonylation in which a carbon atom is lost.
In light of these findings, Wackett and colleagues (2007) conducted research with V. furnissii M1 with the hope of furthering research on bacterial alkane production. Initially, the strain was confirmed to be V. furnissii based on 16S rRNA sequence and phenotypic characteristics. The organism showed many properties consistent with the reports of Park et al. with respect to growth properties of the bacterium. In subsequent work (Wackett et al., 2007), the complete genome of V. furnissii M1 was sequenced at a coverage of 21‐fold. The genome annotation effort failed to identify genes for enzymes likely to be involved in alkane biosynthesis; however, not all such genes are likely known. Genome annotation also failed to provide evidence for any known alkane oxidation genes. A series of experiments were conducted to screen for alkanes, either associated with the cells or free in the medium. All such experiments were negative. Other V. furnissii strains were tested, including one ATCC strain reported in the Japanese patent to make alkanes (Miyamato, 2001). This also proved negative. Low levels of alkanes were observed in extracts in preliminary experiments but these were shown to derive from glassware, stopcock grease and solvent contamination. In light of these observations, the high‐level production of alkanes by Vibrio species remains to be verified.
Alkenes
Bacteria are also reported to produce alkenyl, waxy hydrocarbons, a class of molecules that might prove interesting in the context of fuel or specialty chemical applications. The most notable reports are with bacteria of the genus Micrococcusand related genera such as Kocuria. Micrococci are commensual residents of human skin from which various strains have been isolated in pure culture. Micrococcusspecies are high % G+C, Gram‐positive cocci. At this stage, the physiological function of alkenes in Micrococcus species is unknown but alkenes have been documented to occur in many different members of the genus (Tornabene, 1980).
There is information on the structure of the alkenes but no detailed information on the mechanism of their biosynthesis. The biosynthetic pathway does not produce a single alkene, but rather a family of alkene products. The alkenes generally range from C21 to C29 (Fig. 4). The double bond is at or near the middle carbon(s) of the hydrocarbon chain. The alkenyl chains are subterminally methylated. In total, these observations provide clues as to plausible biosynthetic mechanisms. The alkenes are proposed to derive from branched chain fatty acids which are common bacterial lipids. The mechanism by which fatty acid chains may condense to make alkenes is more obscure. There have been reports of fatty acid head‐to‐head condensation reactions based on radiolabelling studies (Kolattukudy, 1976). In vitrostudies conducted with crude protein extracts from Micrococcus luteusshowed that label is lost when fatty acid precursors contain 14C in carbon 1, but not carbon 16, consistent with the loss of a carboxy group during the condensation reaction (Albro and Dittmer, 1969). Additional studies are warranted to elucidate further mechanistic details and potentially engineer hydrocarbon production by Micrococcus strains.
Figure 4.
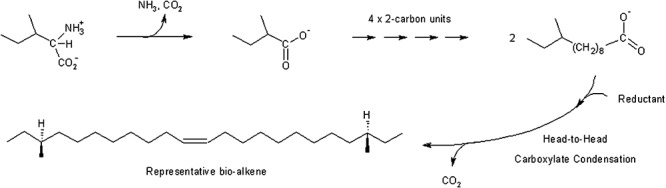
Proposed pathway for alkene biosynthesis by Micrococcus sp.
Other fuels – hydrogen
Hydrogen is both ideal and problematic as a fuel choice. With hydrogen, issues of carbon dioxide or partially combusted atmospheric pollutants, carbon monoxide, for example, are skirted. Hydrogen reacts with oxygen to release considerable heat and generates water as an end‐product. Hydrogen is gaseous even at very low temperatures such that storage density is an issue, especially in any potential vehicular fuel application. Creative efforts to overcome this problem focus on using porous solid materials that can serve as reservoir for hydrogen that can be released as a fuel stream (Rosi et al., 2003). Despite these developments, there are still strong critics of using hydrogen, especially as a motor fuel (Zubrin, 2007).
How might hydrogen be generated in a future society that developed ways to use this energy source? Of course, hydrogen can be generated readily by electrolysis but this uses a high‐grade energy source, electricity, to generate a lower‐grade one, hydrogen, with only a 50% energy conversion efficiency. There are several practical scenarios being considered. For example, electricity could be generated at a remote site from a free resource, like wind, and hydrogen can be generated to store that energy chemically. Hydrogen can be made from biomass, both chemically and biologically. Salge and colleagues (2006) recently described the controlled combustion of vegetable oils and other biomass over a catalyst surface to generate hydrogen, carbon monoxide and small organic molecules, such as alkenes. Alternatively, biological systems are known to generate hydrogen. Biohydrogen derives from fermentation processes, but this is typically in low yield because of thermodynamic considerations (Nath and Das, 2004). Perhaps a more attractive possibility is the potential for ‘bioeletrolysis’ of water driven by photosynthesis. It is known that cyanobacteria and eukaryotic algae generate hydrogen gas. There is consideration of processes that might harvest hydrogen continuously from sunlight‐driven biochemical reactions (Surzycki et al., 2007).
Biohydrogen derives from nitrogenase (Einsle et al., 2002), an accidental hydrogen generator, and hydrogenase, an enzyme class evolved to generate or consume hydrogen (Shima and Thauer, 2007). Nitrogenase evolved to reduce atmospheric nitrogen to ammonia, an impressive six‐electron reduction. The iron‐molybdenum cofactor is a remarkable catalyst but its selectivity is imperfect; every 14 turnovers, approximately, nitrogenase will reduce protons to make hydrogen gas. This catalytic imperfection is being explored by some investigators for its potential to produce hydrogen biologically (Larimer et al., 2004). Naturally, there are also investigations into using nature's designated hydrogen‐generating catalyst, hydrogenase. Hydrogenases typically contain iron‐sulfur clusters, some contain nickel and one is devoid of metal or other cofactors. To reduce protons, the redox centres must be low potential and electrons derive from low‐potential donors, such as ferredoxins. A major issue with hydrogenase utilization and engineering is their sensitivity to molecular oxygen; in some cases this is due to destruction of the iron‐sulfur clusters. Hydrogenase oxygen sensitivity is particularly an issue for photosynthetically powered hydrogen evolution. In this context, hydrogenases are being engineered for oxygen insensitivity (McTavish, 1998; Ghirardi et al., 2005). Another bioengineering challenge is to link photosynthesis directly to hydrogen production rather than carbon fixation, the major biological outcome of photoenergy capture.
Biomass conversion
Regardless of the process or fuel, the source of biomass carbon and hydrogen is an important component of the biofuel equation. Generally, land plants are used to capture solar energy, make carbon molecules and give up the molecules in a transformable state. Ideally, the plants should grow quickly and not require large inputs of chemicals or labour. The most transformable molecules currently are glucose, fructose and starches and thus, commonly used plants are sugarcane, sugar beets and corn. There is currently intensive research to use cellulosic carbon efficiently (Lynd et al., 2005). Cellulose is the most common biopolymer on earth and its efficient utilization would open the door to a much greater variety of biomass resources. One approach is to add cellulase enzymes after mechanically grinding biomass as an initial step of a bioprocess. However, commercial cellulase preparations are currently expensive and only moderately effective. The structure of plant cellulosics is complex, interwoven with lignin polymers and hemicellulose. Most strategies envision using cellulose and hemicellulose sugars following depolymerization, and burning lignin to provide energy for the processing plant. The strong heterogeneous lignin polymer is problematic however, as it blocks access to the usable cellulosic polymers. One emerging approach is to genetically engineer plants to be defective in lignin biosynthesis to make sugars more available and increase fuel yields per unit biomass (Chen and Dixon, 2007).
The use of hemicellulose will require fermentation of xylose and other five‐carbon sugars. A more limited number of microorganisms grow on xylose, compared with glucose, and ferment the sugar to a usable fuel‐like ethanol. Very recently, the genome sequence of a lignocellulose‐converting, xylose‐fermenting yeast, Pichia stipitis, has been obtained (Jeffries et al., 2007). A concept is emerging that organisms must be used in a comprehensive way to minimize unit operations in a bioprocessing plant. There might be benefits in efficiency as well. For example, Clostridium thermocellum is an anaerobic, thermophilic bacterium that breaks down cellulose and ferments the resultant sugars to ethanol (Lu et al., 2006). There is an apparent synergistic effect of having the cellulase enzyme complex, the bacterium and the biomass substrate as a tripartite combination to enhance the rate of cellulose hydrolysis. Clearly, more research is needed to better understand these synergistic effects to best employ them for biofuel production. There are excellent reviews on the major issues in biomass conversion (Himmel et al., 2007).
Chemical processing versus microbial biotechnology
Biologists, chemists and engineers alike all believe that bio‐based carbohydrates will provide the carbon for the fuels of the future. There is much more uncertainty however about the conversion mechanism(s) and the ultimate fuel(s) that derive from those mechanisms. Along with widespread ongoing research pertaining to microbial biochemistry, there is a parallel line of research investigating chemical catalytic conversion of biomass to fuel molecules (Huber et al., 2006; Schmidt and Dauenhauer, 2007). The overall guiding principles are the same; the molecules generated must be stable, energetic and economically produced. The challenge for chemical processing is to remove most of the oxygen atoms in starting materials such as sugars and ultimately produce molecules optimized for use as a fuel. The traditional chemical catalytic approach has been to break biomolecules down into single‐carbon compounds like carbon monoxide and then build up larger molecules from those intermediates. There is a substantial entropic loss of energy in this process leading to inefficiency. Despite this problem, there is an attractiveness to chemical processes because they could, at least in theory, operate quickly and be amenable to large‐scale processing.
A very recent study combined some of the best features of biological and chemical processing to generate a novel biofuel molecule with reasonable efficiency (Roman‐Leshkov et al., 2007). In this process, biomass can be treated enzymatically to release glucose that can be isomerized enzymatically to fructose. Using chemical catalysts, fructose deoxygenation is accomplished in two steps. First, there is a dehydration, eliminating three molecules of water to produce 5‐hydroxymethylfurfural. The second step removes two more oxygen atoms by hydrogenolysis to produce 2,5‐dimethylfuran, the structure of which is shown in Fig. 1. 2,5‐Dimethylfuran (DMF) has a research octane number of 119 and a high energy density of 30 kJ cm−3. Moreover, DMF can be hydrogenated using a ruthenium catalyst to generate 2,5‐diemthyltetrahydrofuran (DTHF), which has a higher energy content and may prove to be more stable to long‐term storage. More research would be needed prior to use of DMF or DTHF as motor fuels, both with respect to engine compatibility and potential toxicological effects on humans and ecosystems.
An alternative approach is the production of synthesis gas (syngas), carbon monoxide and hydrogen, from biomass. Syngas can be converted via Fischer–Tropsch chemistry to form chemical feedstocks and fuels such as linear alkanes (Demirbas, 2007). Syngas can also be converted biologically using a range of organisms (Henstra et al., 2007). There are a growing number of microbes known to oxidize carbon monoxide. Among the most interesting microbes for applications are the carboxydotrophic hydrogenogens that produce carbon dioxide and hydrogen from carbon monoxide. There are also a large class of bacteria containing the acetyl‐CoA synthesis pathway involving carbon monoxide (CO) dehydrogenase. CO dehydrogenase condenses carbon monoxide, a methyl group fragment, and coenzyme A; it also catalyses the reversible oxidation of carbon monoxide to carbon dioxide. Bacteria containing CO dehydrogenase produce acetate, ethanol, butyrate and butanol. Metabolic engineering could be used to develop strains generating high titres of single products.
Overall, this research exemplifies the creativity that will be required to use biomass efficiently, using the best of what biology and chemistry have to offer, and generating new molecules for use in vehicular fuel applications.
Biofuels database, internet resources and knowledge integration
Biofuels research is exploding with new researchers bringing widely divergent expertise to the field. Each researcher will generally publish research findings in the scientific journals of their respective disciplines: chemical engineering, chemistry, biochemistry, genomics, plant biology, microbiology. There is a need to bring this divergent research knowledge together in one place. For example, biomass is often plant material; yield and molecular content is very important for feasibility as a fuel feedstock. Additionally, plant material must be processed to fermentation precursors, requiring chemical, biochemical and engineering knowledge. Manipulation of fermentation organisms will require a knowledge of genomics and molecular biology. Researchers working on biological aspects will need to appreciate the chemical and physical properties necessary for an ideal gasoline replacement. The introduction of new biofuels will also be influenced by economic, environmental and social factors.
To help meet these divergent information needs, the University of Minnesota Biofuels Database (UM‐BFD) is a freely available web database developed to integrate research across fields and provide easy access to web resources pertaining to biofuels (Fig. 5). The database is highly curated. Links are provided to information that is frequently updated by other databases, for example, gene and protein sequence data. For these and other data as appropriate, the UM‐BFD will link to the University of Minnesota Biocatalysis/Biodegradation Database (Bornscheuer, 2001; Leslie, 2005; Ellis et al., 2006). This will also serve to link fuel biosynthesis with biodegradation, allowing users to investigate the impacts of fuel molecules should they spill, as they invariably will, into environmental compartments.
Figure 5.
Homepage of the Biofuels Database (http://www.biofuelsdatabase.org).
A major issue that the UM‐BFD will address is nomenclature pertaining to biofuel molecules. Improper nomenclature impedes the development of clear thinking and new advances (DeTar, 2007). For example, current biodiesel largely consists of methyl esters of fatty acids. This derives from the component alcohol and fatty acids that are most available today but other esters might prove to be superior fuels when tested in engines. For example, the alcohol could be ethanol or butanol. Or the ester bond could be reversed if acetic acid were esterified to a long‐chain alcohol. To our knowledge, many of these new combinations could be generated via metabolic engineering, but have never been tested as motor fuels. Depicting these different molecules, in a chemically correct nomenclature, will further people's understanding of what is possible to generate and test as motor fuels.
Synthetic biology in biofuels research – building hydrocarbons
Nature has evolved biochemical pathways and reactions to generate hydrocarbons that could be excellent fuels. The natural evolutionary process is based on selective pressure. Hydrocarbons are used biologically to impart water resistance to duck feathers, to coat fly wings and to dissolve insect toxins (Eisner et al., 2005). The type and amount of hydrocarbon made by each organism arose via changes in the genetic material under the guiding force of selective pressure. Thus, the hydrocarbon(s) produced aided in the survival of the organism. In contrast, the ideal biosynthetic route to a biofuel might consist of a somewhat different hydrocarbon end‐product and production in much greater amounts. The tools of synthetic biology could come into play, using nature's building blocks, to engineer superior hydrocarbon fuels in high titre.
The first step in constructing a synthetic pathway is for a human to plan the pathway. There are many different combinatorial pathways that one could envision, using different types of reactions and genes. Envisioning the pathways would require an extensive knowledge of metabolic biochemistry, knowledge that for almost everyone is incomplete at best. Moreover, there might be combinations of reactions that could generate hydrocarbons in theory that have never been used in combination in naturally occurring organisms. Such combinations may not be obvious even to an experienced metabolic biochemist. This planning process could be enhanced by the use of computational tools that search and combine metabolic reaction types to generate novel metabolic pathways.
An in silico metabolism prediction tool is now freely available on the world wide web; it is known as the Pathway Prediction System (PPS) (Hou et al., 2003). The PPS was devised to predict the metabolic breakdown, or biodegradation, of chemicals by microorganisms; its major use is for predicting the fate of chemicals in the environment. Each predicted metabolic reaction is derived from a biotransformation rule. Each biotransformation rule represents a particular metabolic reaction type, for example, the oxidation of an alcohol to an aldehyde. There are a finite number of general metabolic reaction types. The current PPS rule set consists of slightly more than 200 rules. Theoretically, a rule set of several hundred rules could depict any known metabolic transformation and thus represent millions of plausible metabolic pathways, some of which exist in nature and some do not. The current PPS serves as a tool to ‘envision’ biodegradation pathways that might occur forany given chemical substance that the user enters into the PPS.
It is plausible to use the same metabolic rule base, chemical structure recognition software and output software to predict biosynthetic pathways leading to molecules of interest, including fuel hydrocarbons. The main difference is that the software would be directed to biosynthesize molecules, rather than to degrade them. Accomplishing this imposes some new computational challenges. However, there has been a similar logic already developed for computer‐aided chemical synthesis, a process named by its developers as ‘retro‐synthesis’ (Corey and Wipke, 1969). In retro‐synthesis, a desired end‐product is broken apart into fragments that might serve as building blocks. The software provides the synthetic chemist with ideas as to what fragments and reagents can be used to build up the larger molecule, and thus make it as efficiently as possible. In a similar manner, the metabolic logic of the PPS and the chemical construction logic of retro‐synthesis could be combined to design molecules for biofuels and other applications.
Acknowledgments
I acknowledge the financial support of the Institute for Renewable Energy and Environment Grants SG‐B6‐2005 and LG‐B13‐2005 and a Discovery grant from the Institute on the Environment at the University of Minnesota. I thank Marc vonKeitz for his critical reading of the manuscript.
References
- Albro P.W., Dittmer J.C. The biochemistry of long‐chain, nonisoprenoid hydrocarbons. IV. Characteristics of synthesis by cell‐free preparation of Sarcina lutea. Biochemistry. 1969;8:3317–3324. doi: 10.1021/bi00836a028. [DOI] [PubMed] [Google Scholar]
- AmeriGas, America's Propane Company. 2007. ) 14 August 2007 [WWW document]. URL http://www.amerigas.com/
- Amyris. 2007. ) Artemisinin Project at Amyris Biotechnologies, 16 August 2007 [WWW document]. URL http://www.amyrisbiotech.com/projects_artemisinin.html.
- Berner R.A. The long‐term carbon cycle, fossil fuels and atmospheric composition. Nature. 2003;426:323–326. doi: 10.1038/nature02131. [DOI] [PubMed] [Google Scholar]
- Biobutanol. 24 July 2007 [WWW document]. URL http://www2.dupont.com/Biofuels/en_US/index.html.
- Blumer M. Polycyclic aromatic hydrocarbons in nature. Sci Am. 1976;234:36–45. [PubMed] [Google Scholar]
- Bornscheuer U. The University of Minnesota Biocatalysis/Biodegradation Database. Angew Chem. 2001;113:3817. [Google Scholar]
- Buchner E. Alkoholische Gährung ohne Hefezellen. Ber Dt Chem Ges. 1897;30:117–124. [Google Scholar]
- Chen F., Dixon R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotech. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Colby J., Dalton H., Whittenbury R. Biological and biochemical aspects of microbial growth on C1 compounds. Annu Rev Microbiol. 1979;33:481–517. doi: 10.1146/annurev.mi.33.100179.002405. [DOI] [PubMed] [Google Scholar]
- Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60:609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey E.J., Wipke W.T. Computer‐assisted design of complex organic synthesis. Science. 1969;166:178–192. doi: 10.1126/science.166.3902.178. [DOI] [PubMed] [Google Scholar]
- Cumming J.N., Ploypradith P., Posner G.H. Antimalarial activity of artemisinin (qinghaosu) and related trioxanes: mechanism(s) of action. Adv Pharmacol. 1997;37:253–297. doi: 10.1016/s1054-3589(08)60952-7. [DOI] [PubMed] [Google Scholar]
- Davis J.F. Paraffinic hydrocarbons in the sulfate‐reducing bacterium Desulfovibrio desulfuricans. Chem Geol. 1968;3:155–160. [Google Scholar]
- Demirbas A. Progress and recent trends in biofuels. Prog Energy Comb Sci. 2007;33:1–18. [Google Scholar]
- Dennis M., Kolattukudy P.E. A cobalt‐porphyrin enzyme converts a fatty aldehyde to a hydrocarbon and CO. Proc Natl Acad Sc USA. 1992;89:5306–5310. doi: 10.1073/pnas.89.12.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTar M.B. Biofuel nomenclature. Chem Eng News. 2007;85:4. [Google Scholar]
- Dien B.S., Cotta M.A., Jeffries T.W. Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol. 2003;63:258–266. doi: 10.1007/s00253-003-1444-y. [DOI] [PubMed] [Google Scholar]
- Dixon B. Spektrum Academic Publishers; 1997. [Google Scholar]
- Eggeman T., Verser D. The importance of utility systems in today's biorefineries and a vision for tomorrow. Appl Biochem Biotechnol. 2006;129:361–381. doi: 10.1385/abab:130:1:361. , and 132. [DOI] [PubMed] [Google Scholar]
- Einsle O., Tezcan F.A., Andrade S.L., Schmid B., Yoshida M., Howard J.B., Rees D.C. Nitrogenase Mo‐Fe‐protein at 1.16 A resolution: a central ligand in the FeMo‐cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- Eisner T., Eisner M., Siegler M. Harvard University Press; 2005. [Google Scholar]
- Ellis L.B.M., Roe D., Wackett L.P. The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. 2006;34:D517–D521. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall R., Copley S.D. Bacterial sources and sinks of isoprene, a reactive atmospheric hydrocarbon. Environ Microbiol. 2000;2:123–130. doi: 10.1046/j.1462-2920.2000.00095.x. [DOI] [PubMed] [Google Scholar]
- Fehler S.W.G., Light R.J. Biosynthesis of hydrocarbons in Anabaena variabilis. Incorporation of [methyl‐14C] and [methyl‐2H2] methionine into 7‐ and 8‐methyl‐heptadecane. Biochemistry. 1970;9:418–422. doi: 10.1021/bi00804a032. [DOI] [PubMed] [Google Scholar]
- Gelpi E., Schneider H., Mann J., Oro J. Hydrocarbons of geochemical significance in microscopic algae. Phytochem. 1970;9:603–612. [Google Scholar]
- Ghirardi M.L., King P.W., Posewitz M.C., Maness P.C., Federov A., Kim K. Approaches to developing biological H(2)‐photoproducing organisms and processes. Biochem Soc Trans. 2005;33:70–72. doi: 10.1042/BST0330070. et al. [DOI] [PubMed] [Google Scholar]
- Gold T. The origin of natural gas and petroleum and prognosis for future supplies. Annu Rev Energy. 1985;10:53–77. [Google Scholar]
- GOLD. 2007. ) Genomes OnLine Database, v. 2, 14 August 2007 [WWW document]. URL http://www.genomesonline.org/
- Goldemberg J. Ethanol for a sustainable energy future. Science. 2007;315:808–810. doi: 10.1126/science.1137013. [DOI] [PubMed] [Google Scholar]
- Grant L. When will the oil run out? Science. 2005;309:52–54. doi: 10.1126/science.309.5731.52. [DOI] [PubMed] [Google Scholar]
- Han J., Calvin M. Hydrocarbon distribution of algae and bacteria, and microbiological activity in sediments. Proc Natl Acad Sci USA. 1969;64:436–443. doi: 10.1073/pnas.64.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., McCarthy E.D., Calvin M., Benn M.H. Hydrocarbon constituents of the blue‐green algae Nostoc muscorum, Anacystis nidulans, Phormidium luridum, and Chloroghea fritschii. J Chem Soc. 1968;C:2785–2791. [Google Scholar]
- Harborne J. 3rd. Academic Proess; 1988. [Google Scholar]
- Henstra A.M., Sipma J., Rinzema A., Stams A.J.M. Microbiology of synthesis gas fermentation for biofuel production. Curr Opin Biotechnol. 2007;18:1–7. doi: 10.1016/j.copbio.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Hill J., Nelson E., Tilman D., Polasky S., Tiffany D. Environmental, economic and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel M.E., Ding S‐Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W., Foust T.D. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Holdsworth J.E., Ratledge C. Triacylglycerol synthesis in the oleaginous yeast Candida curvata D. Lipids. 1991;26:111–118. doi: 10.1007/BF02544004. [DOI] [PubMed] [Google Scholar]
- Hou B.K., Wackett L.P., Ellis L.B.M. Microbial pathway prediction: a functional group approach. J Chem Inf Comput Sci. 2003;43:1051–1057. doi: 10.1021/ci034018f. [DOI] [PubMed] [Google Scholar]
- Huang W.C., Ramey D.E., Yang S.T. Continuous production of butanol by Clostridium acetobutylicum immobilized in a fibrous bed bioreactor. Appl Biochem Biotechnol. 2004;113:887–898. doi: 10.1385/abab:115:1-3:0887. , and 116. [DOI] [PubMed] [Google Scholar]
- Huber G.W., Iborra S., Corma A. Synthesis of transportation fuels from biomass: chemistry, catalysts and engineering. Chem Rev. 2006;106:4044–4098. doi: 10.1021/cr068360d. [DOI] [PubMed] [Google Scholar]
- Ingram L.O., Conway T., Clark D.P., Sewell G.W., Preston J.F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski G.J., Zobell C.E. Hydrocarbon production by sulfate‐reducing bacteria. J Bacteriol. 1948;47:447. [Google Scholar]
- Jeffries T.W., Grigoriev I.V., Grimwood J., Laplaza J.M., Aerts A., Salamov A. Genome sequence of the lignocellulose‐bioconverting and xylose‐fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. et al. [DOI] [PubMed] [Google Scholar]
- Jones J.G. Studies on lipids of soil micro‐organisms with particular reference to hydrocarbons. J Gen Microbiol. 1969;59:145–152. doi: 10.1099/00221287-59-2-145. [DOI] [PubMed] [Google Scholar]
- Julsing M.K., Rijpkema M., Woerdenbag H.J., Quax W.J., Kayser O. Functional analysis of genes involved in biosynthesis of isoprene in Bacillus subtilis. Appl Microbiol Biotechnol. 2007;75:1377–1384. doi: 10.1007/s00253-007-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R., Steinbüchel A. A novel bifunctional wax ester synthase/acyl‐CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem. 2003;278:8075–8082. doi: 10.1074/jbc.M210533200. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Stolting T., Steinbuchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152:2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Stöveken T., Malkus U., Reichelt R., Golyshin P.N., Sabirova J.S. Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol. 2007;189:918–928. doi: 10.1128/JB.01292-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver A.J. Microbiology and industry. In: Kamp A.F., La Riviere J.W.M., Verhoeven W., editors. North‐Holland; 1957. pp. 165–185. [Google Scholar]
- Koga Y., Morii H. Biosynthesis of ether‐type polar lipids and evolutionary considerations. Microbiol Mol Biol Rev. 2007;71:97–120. doi: 10.1128/MMBR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy P.E. Elsevier; 1976. [Google Scholar]
- Kovarik W. 2007. ) Henry Ford, Charles Kettering and the ‘Fuel of the Future’ [WWW document]. URL http://www.radford.edu/~wkovarik/papers/fuel.html.
- Kuzma J., Nemecek‐Marshall M., Pollock W.H., Fall R. Bacteria produce the volatile hydrocarbon isoprene. Curr Microbiol. 1995;30:97–103. doi: 10.1007/BF00294190. [DOI] [PubMed] [Google Scholar]
- Ladygina N., Dedyukhina E.G., Vainshtein M.B. A review on microbial synthesis of hydrocarbons. Process Biochem. 2006;41:1001–1014. [Google Scholar]
- Larimer F.W., Chain P., Hauser L., Lamerdin J., Malfatti S., Do L. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol. 2004;22:55–61. doi: 10.1038/nbt923. et al. [DOI] [PubMed] [Google Scholar]
- Leslie M. DATABASE: eating pollution. Science. 2005;309:1795. [Google Scholar]
- Liebig J. Ueber die Erscheinungen der Gährung, Fäulnis und Verwesung, un ihre. Ursachen Annalen der Physik und Chemie. 1839;48:106–150. [Google Scholar]
- Lipscomb J.D. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- Lu Y., Zhang Y.P., Lynd L.R. Enzyme‐microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci USA. 2006;103:16165–16169. doi: 10.1073/pnas.0605381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd L.R., Van Zyl W.H., McBride J.E., Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- McTavish H. Hydrogen evolution by direct electron transfer from photosystem I to hydrogenases. J Biochem (Tokyo) 1998;123:644–649. doi: 10.1093/oxfordjournals.jbchem.a021986. [DOI] [PubMed] [Google Scholar]
- Metzger P., Largeau C. Botryococcus braunii: a rich source for hydrocarbons and related ether lipids. Appl Microbiol Biotechnol. 2005;66:486–496. doi: 10.1007/s00253-004-1779-z. [DOI] [PubMed] [Google Scholar]
- Michaelis L., Menten M.L. Die Kinetik der invertinwirkung. Biochem Zool. 1913;49:333–369. [Google Scholar]
- Miyamato K. 2001.
- Moldowan J.M., Seifert W.K. Head‐to‐head linked isoprenoid hydrocarbons in petroleum. Science. 1979;204:169–171. doi: 10.1126/science.204.4389.169. [DOI] [PubMed] [Google Scholar]
- Naccarato W.F., Gilbertson J.R., Gelman R.A. Effects of different culture media and oxygen upon lipids of Escherichia coli K‐12. Lipids. 1974;9:322–327. doi: 10.1007/BF02533108. [DOI] [PubMed] [Google Scholar]
- Nath K., Das D. Improvement of fermentative hydrogen production: various approaches. Appl Microbiol Biotechnol. 2004;65:520–529. doi: 10.1007/s00253-004-1644-0. [DOI] [PubMed] [Google Scholar]
- Olah G.A., Molnar A. John Wiley & Sons; 1995. [Google Scholar]
- Park M.O. New pathway for long‐chain n‐alkane synthesis via 1‐alcohol in Vibrio furnissii M1. J Bacteriol. 2005;187:1426–1429. doi: 10.1128/JB.187.4.1426-1429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.O., Tanabe M., Hirata K., Miyamoto K. Isolation and characterization of a bacterium that produces hydrocarbons extracellularly which are equivalent to light oil. Appl Microbiol Biotechnol. 2001;56:448–452. doi: 10.1007/s002530100683. [DOI] [PubMed] [Google Scholar]
- Park M.O., Heguri K., Hirata K., Miyamoto K. Production of alternatives to fuel oil from organic waste by the alkane‐producing bacterium, Vibrio furnissii M1. J Appl Microbiol. 2005;98:324–331. doi: 10.1111/j.1365-2672.2004.02454.x. [DOI] [PubMed] [Google Scholar]
- Pasteur L. Memoire sur la fermentation appelee lactique. Comptes rendus de l’Academie des sciences. 1857;45:913–916. [Google Scholar]
- Pasteur L. Memoire sur la fermentation alcoölique. Annales de Chimie et de Physique. 1860;58:323–426. [Google Scholar]
- Peters K.E., Walters C.C., Moldowan J.M. 2nd. Cambridge University Press; 2005. [Google Scholar]
- Roman‐Leshkov Y., Barrett C.J., Liu Z.Y., Dumesic J.A. Production of dimethylfuran for liquid fuels from biomass‐derived carbohydrate. Nature. 2007;447:982–985. doi: 10.1038/nature05923. [DOI] [PubMed] [Google Scholar]
- Rosi N.L., Eckert J., Eddaoudi M., Vodak D.T., Kim J. Hydrogen storage in microporous metal‐organic frameworks. Science. 2003;300:1127–1129. doi: 10.1126/science.1083440. [DOI] [PubMed] [Google Scholar]
- Salge J.R., Dreyer B.J., Dauenhauer P.J., Schmidt L.D. Renewable hydrogen from nonvolatile fuels by reactive flash volatilization. Science. 2006;314:801–804. doi: 10.1126/science.1131244. [DOI] [PubMed] [Google Scholar]
- Sazinsky M.H., Lippard S.J. Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the alpha‐subunit. J Am Chem Soc. 2005;127:5814–5825. doi: 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- Schmidt L.D., Dauenhauer P.J. Chemical engineering: hybrid routes to biofuels. Nature. 2007;447:914–915. doi: 10.1038/447914a. [DOI] [PubMed] [Google Scholar]
- Schneider‐Belhaddad F., Kolattukudy P. Solubilization, partial purification, and characterization of a fatty aldehyde decarbonylase from a higher plant, Pisum sativum. Arch Biochem Biophys. 2000;377:341–349. doi: 10.1006/abbi.2000.1798. [DOI] [PubMed] [Google Scholar]
- Shima S., Thauer R.K. A third type of hydrogenase catalyzing H2 activation. Chem Rec. 2007;7:37–46. doi: 10.1002/tcr.20111. [DOI] [PubMed] [Google Scholar]
- Suffet I.H. A re‐evaluation of the taste and odour of methyl tertiary butyl ether (MTBE) in drinking water. Water Sci Technol. 2007;55:265–273. doi: 10.2166/wst.2007.188. [DOI] [PubMed] [Google Scholar]
- Surzycki R., Cournac L., Peltier G., Rochaix J.‐D. Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. Proc Natl Acad Sci USA. 2007;104:17548–17553. doi: 10.1073/pnas.0704205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornabene T.G. Formation of hydrocarbons by bacteria and algae. Basic Life Sci. 1980;18:421–438. doi: 10.1007/978-1-4684-3980-9_24. [DOI] [PubMed] [Google Scholar]
- Wackett L.P., Frias J., Seffernick J., Sukovich D., Cameron S. Vibrio furnissii M1: genomic and biochemical studies demonstrating the absence of an alkane‐producing phenotype. Appl Environ Microbiol. 2007;73:7192–7198. doi: 10.1128/AEM.01785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.T. Revealing coupling patterns in isoprenoid alkylation. ACS Chem Biol. 2007;2:296–298. doi: 10.1021/cb700094s. [DOI] [PubMed] [Google Scholar]
- Withers S.T., Keasling J.D. Biosynthesis and engineering of isoprenoid small molecules. Appl Microbiol Biotechnol. 2007;73:980–990. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- Wood T.K. Active expression o soluble methane monooxygenase from Methylosinus trichosporium OB3b in heterologous hosts. Microbiology. 2002;148:3328–3329. doi: 10.1099/00221287-148-11-3328. [DOI] [PubMed] [Google Scholar]
- Wu M., Wu Y., Wang M. Energy and emission benefits of alternative transportation liquid fuels derived from switchgrass: a fuel life cycle assessment. Biotechnol Prog. 2006;22:1012–1024. doi: 10.1021/bp050371p. [DOI] [PubMed] [Google Scholar]
- Youngblood W.W., Blumer M. Alkanes and alkenes in marine benthic algae. Marine Biol. 1973;21:163–172. [Google Scholar]
- Zubrin R. The hydrogen hoax. New Atlantis. 2007;15:9–20. [Google Scholar]


