Summary
l‐DOPA (3,4‐dihydroxyphenyl‐l‐alanine) is an extensively used drug for the treatment of Parkinson's disease. In the present study, optimization of nutritional parameters influencing l‐DOPA production was attempted using the response surface methodology (RSM) from Brevundimonas sp. SGJ. A Plackett–Burman design was used for screening of critical components, while further optimization was carried out using the Box–Behnken design. The optimized levels of factors predicted by the model were pH 5.02, 1.549 g l−1 tryptone, 4.207 g l−1 l‐tyrosine and 0.0369 g l−1 CuSO4, which resulted in highest l‐DOPA yield of 3.359 g l−1. The optimization of medium using RSM resulted in a 8.355‐fold increase in the yield of l‐DOPA. The anova showed a significant R2 value (0.9667), model F‐value (29.068) and probability (0.001), with insignificant lack of fit. The highest tyrosinase activity observed was 2471 U mg−1 at the 18th hour of the incubation period with dry cell weight of 0.711 g l−1. l‐DOPA production was confirmed by HPTLC, HPLC and GC‐MS analysis. Thus, Brevundimonas sp. SGJ has the potential to be a new source for the production of l‐DOPA.
Introduction
l‐DOPA is considered as the most potent drug available in the market for the treatment of Parkinson's disease (Kofman, 1971; Rani et al., 2007). l‐DOPA is marketed as tablets under various brand names, such as Sinemet®, Atamet®, Parcopa® and Stalevo® (Ali and Haq, 2006). The world market for l‐DOPA amounts to about 250 tons year−1, and the total market volume is about $101 billion year−1 (Koyanagi et al., 2005). Tyrosinase catalyses the conversion of l‐tyrosine to l‐DOPA. Tyrosinases (E.C.1.14.18.1) are copper‐containing enzymes widely distributed in plants, animals and microorganisms. They are involved in two steps of melanin synthesis, that is, from l‐tyrosine to l‐DOPA and then to l‐DOPAchrome. Tyrosinase catalyses two successive reactions: cresolase and catecholase (Claus and Decker, 2006).
Although the chemical synthesis of l‐DOPA was reported, it was produced by various biological systems. l‐DOPA is conventionally extracted from the seeds of Mucuna pruriens and Vicia faba (Chattopadhyay et al., 1994). l‐DOPA production been reported from plant sources like Portulaca grandiflora and banana (Bapat et al., 2000; Rani et al., 2007). The bacterial and fungal sources have been reported earlier for the production of l‐DOPA, including Erwinia herbicola (Koyanagi et al., 2005), Aspergillus oryzae (Ali and Haq, 2006), Yarrowia lipolytica (Ali et al., 2007), Acremonium rutilum (Krishnaveni et al., 2009) and Bacillus sp. JPJ (Surwase and Jadhav, 2011). In addition, l‐DOPA has also been produced by immobilized tyrosinase (Gabriela et al., 2000). The chemical process in l‐DOPA production involves harsh conditions; hence, the eco‐friendly bioconversion of l‐tyrosine to l‐DOPA is highly important. Aginomoto started the commercial production of l‐DOPA using Erwinia herbicola in a fed‐batch process, which has many advantages over the classical chemical process (Krishnaveni et al., 2009).
The high production cost and high commercial value of l‐DOPA have motivated many researchers to search for alternative sources for its synthesis. The optimal design of the culture medium is a very important aspect in the development of fermentation processes. The conventional method for medium optimization involves changing one parameter at a time while keeping all others constant. This method may be very expensive and time‐consuming. In addition, it fails to determine the combined effect of different factors (Lee et al., 2003; Zhang et al., 2007). Statistical experimental designs have been used to address these problems, such as the response surface methodology (RSM). In the present investigation, a Plackett–Burman design was used for screening of process parameters, while optimization of the critical factors for l‐DOPA production was carried out using RSM with a Box–Behnken design.
Results and discussion
Plackett–Burman design for screening of critical factors
Microorganisms have been exploited as an alternative source of l‐DOPA. Given the potential uses of l‐DOPA and the high demand for it, there exists a need to develop low‐cost industrial media formulations. Statistical analysis using a Plackett–Burman design indicated that pH (X1), tryptone (X3), l‐tyrosine (X7) and CuSO4 (X8) were significantly affected the l‐DOPA production, with P‐values less than the significance level of 0.1. The remaining components, including temperature (X2), yeast extract (X4), beef extract (X5), glucose (X6), MgSO4 (X9), K2HPO4 (X10) and NaCl (X11), were found to be insignificant, with P‐values above 0.05. The experimental runs and their respective l‐DOPA yield are presented in Table S1. Statistical analysis of the responses was performed, as shown in Table 1. The model F‐value of 28.48 implies that the model is significant; there was only 0.01% chance that a model F‐value this large could occur due to noise. The values of P < 0.05 indicate that the model terms are significant. ‘Adeq Precision’ measures the signal‐to‐noise ratio, with a ratio greater than 4 regarded as desirable (Anderson and Whitcomb, 2005). The ‘Adeq Precision’ ratio of 4.389 obtained in this study indicates an adequate signal. Thus, this model can be used to navigate the design space. Regression analysis was performed on the results, and a first‐order polynomial equation was derived (Eq. 1), representing l‐DOPA production as a function of the independent variables:
| (1) |
Table 1.
Statistical analysis of the model by Plackett–Burman design for l‐DOPA production.
| Source | Sum of squares | d.f. | Mean square | F‐value | P‐value P > F |
|---|---|---|---|---|---|
| Model | 0.072669 | 4 | 0.018167 | 28.4801 | 0.0002* |
| X1 – pH | 0.007651 | 1 | 0.007651 | 11.99379 | 0.0105* |
| X2 – tryptone | 0.00913 | 1 | 0.00913 | 14.31288 | 0.0069* |
| X7 – l‐tyrosine | 0.049024 | 1 | 0.049024 | 76.85316 | < 0.0001* |
| X8 – CuSO4 | 0.006864 | 1 | 0.006864 | 10.76056 | 0.0135* |
| Residual | 0.081013 | 1 | 0.081013 | ||
| Corrected total | 0.004465 | 7 |
Significant P‐values at P ≤ 0.05.
Statistical analysis showed that it is not possible to evaluate the relationship between significant independent variables and the response by a first‐order equation. Thus, the first‐order model is not appropriate to predict the response. Indeed, further investigation could be conducted through a second‐order model.
Medium optimization by response surface methodology
Medium optimization using the Box–Behnken design was carried out with the components found to be significant from the Plackett–Burman design, including pH (X1), tryptone (X3), l‐tyrosine (X7) and CuSO4 (X8). Table S2 presents the design matrix and the results of the 29 experiments carried out using the Box–Behnken design. The results obtained were submitted to anova using the Design expert software (version 8.0, Stat‐Ease, USA), and the regression model was given as:
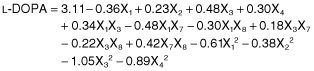 |
(2) |
where X1 is pH, X3 is tryptone, X7 is l‐tyrosine and X8 is CuSO4. The anova of the quadratic regression model (Table 2) demonstrated that Eq. 2 is a highly significant model (P = 0.001). The model F‐value of 29.068 implies that the model is significant. There was only a 0.01% chance that a model F‐value this large could occur due to noise. The goodness of fit of the model was checked using the determination coefficient (R2). In this case, the value of the determination coefficient was R2 = 0.9667. The value of the adjusted determination coefficient (Adj R2 = 0.9335) was in reasonable agreement with the predicted R2 (0.8217). The lack‐of‐fit value for regression Eq. 2 was not significant (0.9280), indicating that the model equation was adequate for predicting the l‐DOPA production under any combination of values of the variables. ‘Adeq Precision’ measures the signal‐to‐noise ratio, with a ratio greater than 4 considered as desirable (Anderson and Whitcomb, 2005). The ‘Adeq Precision’ ratio of 15.968 obtained in this study indicates an adequate signal. Thus, this model can be used to navigate the design space (Table 2).
Table 2.
Analysis of variance (anova) for the fitted quadratic polynomial model of l‐DOPA production.
| Source | Sum of squares | d.f. | Mean square | F‐value | P‐value P > F |
|---|---|---|---|---|---|
| Model | 19.84159 | 14 | 1.417257 | 29.06809 | < 0.0001* |
| X1 – pH | 1.521544 | 1 | 1.521544 | 31.20704 | < 0.0001* |
| X2 – tryptone | 0.608851 | 1 | 0.608851 | 12.4876 | 0.0033* |
| X7 – l‐tyrosine | 2.79271 | 1 | 2.79271 | 57.27879 | < 0.0001* |
| X8 – CuSO4 | 1.112034 | 1 | 1.112034 | 22.80794 | 0.0003* |
| X1X3 | 0.457652 | 1 | 0.457652 | 9.386498 | 0.0084* |
| X1X7 | 0.937024 | 1 | 0.937024 | 19.21847 | 0.0006* |
| X1X8 | 0.349281 | 1 | 0.349281 | 7.163792 | 0.0181* |
| X3X7 | 0.134689 | 1 | 0.134689 | 2.762486 | 0.1187 |
| X3X8 | 0.189225 | 1 | 0.189225 | 3.881026 | 0.0689 |
| X7X8 | 0.693056 | 1 | 0.693056 | 14.21466 | 0.0021* |
| X12 | 2.448766 | 1 | 2.448766 | 50.22446 | < 0.0001* |
| X32 | 0.952366 | 1 | 0.952366 | 19.53313 | 0.0006* |
| X72 | 7.215174 | 1 | 7.215174 | 147.984 | < 0.0001* |
| X82 | 5.102503 | 1 | 5.102503 | 104.6529 | < 0.0001* |
| Residual | 0.68259 | 14 | 0.048756 | ||
| Lack of fit | 0.617689 | 10 | 0.061769 | 3.80695 | 0.1047 |
| Pure error | 0.064901 | 4 | 0.016225 | 29.06809 | < 0.0001* |
| Corrected total | 20.52418 | 28 | 1.417257 | 31.20704 | < 0.0001* |
Significant P‐values at P ≤ 0.05.
Response surface curves
Graphical representation provides a method to visualize the relationship between the response and experimental levels of each variable and the type of interactions between test variables to deduce the optimum conditions. These techniques have been widely adopted for optimizing the processes of enzymes and peptides, solvents, polysaccharides, etc. (Wang and Lu, 2005). Three‐dimensional (3D) graphs were generated for the pairwise combination of the four factors while keeping the other two at their optimum levels for l‐DOPA production. The graphs are given here to highlight the roles played by various factors in the final yield of l‐DOPA.
The interaction of two variables, pH and tryptone, is shown in Fig. 1A, the 3D response surface plot indicates that interaction of these components moderately affected the production of l‐DOPA. The higher and lower levels of these components did not affect the l‐DOPA yield drastically, but mid‐levels provide a maximum yield. The acidic and alkaline pH results in lower l‐DOPA yields might be because of inhibition of tyrosinase activity and cell viability. Also at alkaline pH, less l‐DOPA yield resulted due to the conversion of l‐DOPA into further metabolites such as dopaquinone and melanin (Ali and Haq, 2006). The pH 3.5 and 5.4 has been reported for bioconversion of l‐DOPA by using biomass of Aspergillus oryzae and Yarrowia lipolytica respectively (Ali and Haq, 2006; Ali et al., 2007). The shape of the 3D response surface curve of the interaction between pH and l‐tyrosine depicted in Fig. 1B shows that l‐DOPA production was drastically affected by a slight change in the levels of these two factors. The higher and lower concentrations of both factors resulted in lesser l‐DOPA yield.
Figure 1.
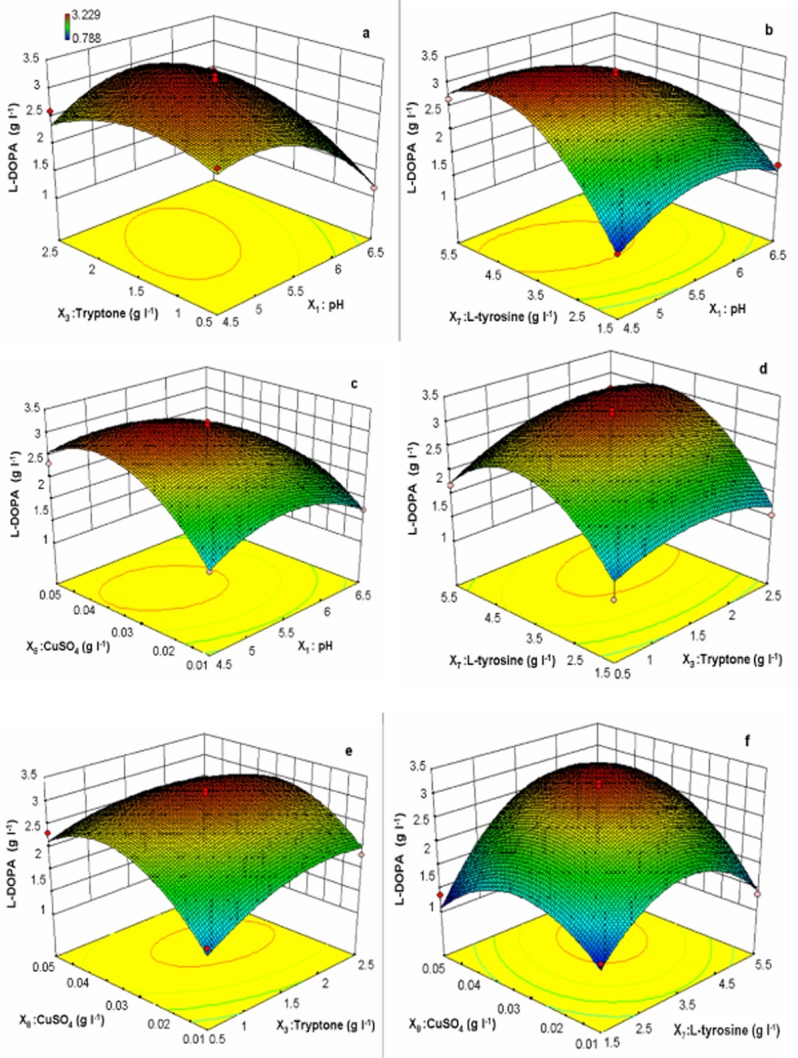
Three‐dimensional response surface curve showing the effect of interactions of (A) pH and tryptone, (B) pH and l‐tyrosine, (C) pH and CuSO4, (D) tryptone and l‐tyrosine, (E) tryptone and CuSO4, (F) l‐tyrosine and CuSO4 on l‐DOPA production.
The response surface curve is shown in Fig. 1C, illustrating that the interaction between pH and CuSO4 moderately affected the yield of l‐DOPA. This is because the tyrosinase involved in the conversion of l‐tyrosine to l‐DOPA is a copper‐containing enzyme (Claus and Decker, 2006). The use of CuSO4 in the media for l‐DOPA production by Acremonium rutilum has been reported earlier (Krishnaveni et al., 2009). Figure 1D shows the quadratic effect of tryptone and l‐tyrosine on l‐DOPA productivity, which indicates that the l‐DOPA yield was gradually enhanced by increasing the concentration of both components to their optimum level, after which the yield decreased steadily. The statistical analysis (Table 2) shows that insignificant interaction occurred with these components.
The effect of the interaction between tryptone and CuSO4 on the production of l‐DOPA is shown in Fig. 1E, which indicates that the l‐DOPA yield was not highly altered by changes in the concentration of both media components. The shape of the response surface curve and statistical analysis (Table 2) indicate that insignificant interaction occurred between these ingredients. The response surface curve and contour of l‐tyrosine and CuSO4 shown in Fig. 1F indicate a positive effect on l‐DOPA production. The lower concentration of CuSO4 resulted in a lower yield of l‐DOPA, while the higher concentration of l‐tyrosine inhibited the l‐DOPA production due to its decreased solubility (Ali et al., 2007; Surwase and Jadhav, 2011).
Validation of the experimental model
Validation was carried out under conditions predicted by the model. The optimized levels predicted by the model were pH 5.02, 1.549 g l−1 tryptone, 4.207 g l−1 l‐tyrosine and 0.0369 g l−1 CuSO4. The predicted yield of l‐DOPA with these concentrations was 3.361 g l−1, while the actual yield obtained was 3.359 g l−1. A close correlation between the experimental and predicted values was observed, which validates this model.
l‐DOPA yield, biomass trend and tyrosinase activity
The l‐DOPA production before and after optimization is illustrated in Fig. 2A, which indicates that in the medium before optimization (nutrient broth with 1 g l−1 l‐tyrosine), l‐DOPA production started after the 6th hour with a yield of 0.068 g l−1, gradually increased to 0.402 g l−1 at the 18th hour, and then decreased to 0.194 g l−1 at the 24th hour. In contrast, in the medium optimized by RSM, l‐DOPA production started at the 6th hour with a yield of 0.427 g l−1, gradually increased to 3.359 g l−1 at the 18th hour, and finally decreased to 1.582 g l−1. The decrease in the l‐DOPA yield after the 18th hour was due to the conversion of l‐DOPA to further metabolites, such as dopaquinone and melanin (Ali et al., 2007; Surwase and Jadhav, 2011). Thus, the medium optimization by RSM resulted in a 8.355‐fold increase in the l‐DOPA yield over the yield before optimization. The literature survey revealed that single‐ and multiple‐stage cell suspension cultures of Mucuna pruriens have been reported to yield 0.028 g l−1 l‐DOPA within 15 and 30 days respectively (Chattopadhyay et al., 1994). Portulaca grandiflora has been reported to produce 0.488 g l−1 of l‐DOPA at the 16th hour (Rani et al., 2007); Acremonium rutilum produced 0.89 g l−1 l‐DOPA, whereas Egyptian black yeast yielded 0.064 g l−1 (Krishnaveni et al., 2009; Mahmoud and Bendary, 2011). The biomass of Aspergillus oryzae and Yarrowia lipolytica has been reported to produce 1.686 and 2.960 g l−1 l‐DOPA respectively (Ali and Haq, 2006; Ali et al., 2007). Thus, Brevundimonas sp. SGJ in the present study produced the highest yield of l‐DOPA (3.359 g l−1).
Figure 2.
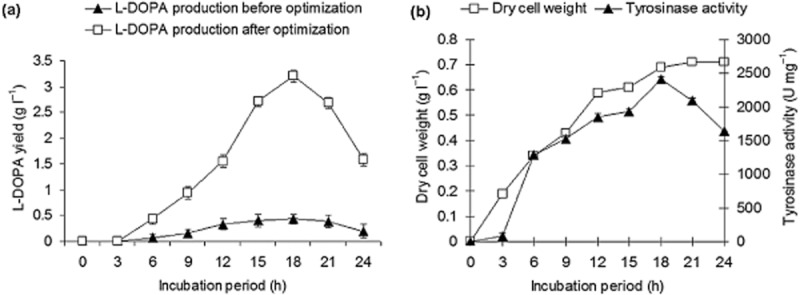
(A) l‐DOPA yield using conditions before optimization and using the medium optimized by RSM.
(B) Biomass trend and tyrosinase activity during l‐DOPA production using conditions optimized by RSM.
The biomass trend and tyrosinase activity during l‐DOPA production with optimized medium are depicted in Fig. 2B, which shows that the dry cell weight increased gradually up to the 15th hour (0. 614 g l−1) and then remained nearly constant, with a final weight of 0.711 g l−1. Meanwhile, tyrosinase activity increased slowly up to 2471 U mg−1 at the 18th hour and then decreased suddenly to 1629 U mg−1 at the 24th hour. This observed decrease in tyrosinase activity may be due to the conversion of l‐tyrosine to l‐DOPA and of l‐DOPA to dopaquinone; substrates for tyrosinase were not available further, during further pathway (Ali et al., 2007; Surwase and Jadhav, 2011).
l‐DOPA analysis
The HPTLC peak profile (Fig. S1A) and the HPTLC plate (Fig. S1B) of the cell‐free broth showed a distinct peak and band at the Rf 0.24, which was identical to standard l‐DOPA (0.23). These results primarily confirmed the l‐DOPA production in the medium. The HPLC elution profile of standard l‐DOPA showed a peak at the retention time 2.723 min (Fig. S2A), while the HPLC elution profile of the broth after incubation showed a prominent peak at the retention time 2.725 min (Fig. S2B). This analysis confirmed the production of l‐DOPA. GC‐MS analysis of the extracted cell‐free broth showed a distinct mass peak at m/z 197, which corresponded to the molecular weight of l‐DOPA, confirming the production of l‐DOPA by Brevundimonas sp. SGJ (Fig. S2C).
The Brevundimonas sp. SGJ reported here produced maximum l‐DOPA and has several advantages over the plant, fungal and bacterial sources used earlier, such as a short incubation period, efficient production and requirement of simple medium components. The l‐DOPA produced previously by bacterial sources like Erwinia herbicola used pyrocatechol as substrate, which is a toxic phenolic compound, and required polyacrylamide gel, which is an expensive chemical (Koyanagi et al., 2005; Surwase and Jadhav, 2011). Thus, the present study contributes to the optimization of the nutritional requirements that will be most useful for large‐scale production of l‐DOPA using Brevundimonas sp. SGJ.
Experimental procedures
Chemicals and microorganisms
l‐tyrosine and l‐DOPA were purchased from Sigma‐Aldrich (St. Louis, MO, USA), and all other chemicals were purchased from Himedia (India). The bacterial strain producing l‐DOPA was isolated from soil samples collected from Shivaji University, Kolhapur region, using a serial dilution technique. The nutrient broth (Himedia, India) used for the isolation with 1 g l−1 l‐tyrosine. The bacterium was identified as Brevundimonas sp. SGJ (NCBI GenBank ID: HM998899) by 16S rDNA analysis.
Inoculum preparation and l‐DOPA production
The nutrient broth (Himedia, India) used for the cultivation of the isolated bacterium consisted of 5 g l−1 peptone, 1.5 g l−1 beef extract, 1.5 g l−1 yeast extract and 0.5 g l−1 NaCl, supplemented with 1 g l−1 l‐tyrosine and with a pH of 5.5. The 6 h grown, 2 ml of cell suspension was inoculated in 100 ml of the same medium for l‐DOPA production in 250 ml Erlenmeyer flasks. The flasks were kept in an incubator shaker at 30°C and 120 r.p.m.; l‐DOPA was assayed after 18 h.
Experimental design
Plackett–Burman design
A Plackett–Burman design was used to select the most critical physical and nutritional parameters for l‐DOPA production by Brevundimonas sp. SGJ. The factors affecting the yield of l‐DOPA were selected by screening various carbon sources, nitrogen sources, mineral salts and physical factors, such as pH and temperature. In addition, some of these variables were selected from the primary literature review (Krishnaveni et al., 2009; Mahmoud and Bendary, 2011).
A total of 11 process parameters, including pH (X1), temperature (X2), tryptone (X3), yeast extract (X4), beef extract (X5), glucose (X6), l‐tyrosine (X7), CuSO4(X8), MgSO4 (X9), K2HPO4 (X10) and NaCl (X11), were added at two levels: low (−1) and high (+1). The low and high levels of these factors were taken as pH (4.5 and 6.5), temperature (20°C and 40°C). While levels of media components were (g l−1) tryptone (0.5 and 2.5), yeast extract (0.5 and 2.5), beef extract (0.5 and 2.5), glucose (0.5 and 2.5), l‐tyrosine (1.5 and 5.5), CuSO4 (0.01 and 0.05), MgSO4 (0.001 and 0.005), K2HPO4 (0.5 and 2.5) and NaCl (0.1 and 0.5). This design characterizes a model that identifies the significant variables when no interaction among the factors is expected (Plackett and Burman, 1946; Anderson and Whitcomb, 2005; Wang and Lu, 2005). Therefore, a first‐order multiple regression can model the data properly (Eq. 3):
| (3) |
where Y is the predicted response (l‐DOPA production), β0 is the intercept, and βi is the linear coefficient. The design matrix created using the Design expert software is presented in Table S1. Three replicates at the centre point were also performed to find the curvature that may exist in the model and the pure experimental error, which shows lack of fit. The statistical significance of the first‐order model was identified using Fisher's test for analysis of variance (anova). Moreover, the multiple correlation coefficients (R2) were used to express the fit of this first model.
Box–Behnken design
Once the critical factors were identified via screening, a Box–Behnken design for independent variables was used for further optimization. The variables each at levels with three replicates at the centre points (Box and Behnken, 1960; Anderson and Whitcomb, 2005), was used to fit a polynomial model by a response equation (Eq. 4):
| (4) |
A second‐order model is designed such that the variance of Y is constant for all points equidistant from the centre of the design. The Design expert software was used in the experimental design and data analysis. A multiple regression analysis of the data was carried out to define the response in terms of the independent variables. Response surface graphs were obtained to understand the effect of the variables, individually and in combination, and to determine their optimum levels for maximum l‐DOPA production. All trials were performed in triplicate, and the average l‐DOPA yield was used as response Y.
Biomass trend, tyrosinase activity and l‐DOPA production
After validation of the experiment using the optimum process parameters generated by the Design expert software, the l‐DOPA production was observed in the medium before optimization (nutrient broth with 1 g l−1 l‐tyrosine) and after optimization. The biomass trend, tyrosinase activity and l‐DOPA production were observed at 3 h time intervals for up to 24 h. The biomass trend was obtained by measuring the dry cell weight. The tyrosinase activity was determined by the previously described method (Kandaswami and Vaidyanathan, 1973).
l‐DOPA assay
l‐DOPA produced in the broth was determined according to Arnow's method (Arnow, 1937). The reaction mixture was centrifuged at 5000 r.p.m. for 15 min., and 1 ml supernatant was added with 1 ml of 0.5 N HCl, 1 ml of nitrite molybdate reagent, and 1 ml of 1 N NaOH; the final volume was adjusted to 5 ml with distilled water. The absorbance was measured at 530 nm using a double‐beam UV‐visible spectrophotometer (Shimadzu, Japan).
l‐DOPA analysis
High‐performance thin‐layer chromatography (HPTLC) analysis of the cell‐free broth was performed using a HPTLC system (CAMAG, Switzerland). The conditions used for HPTLC were similar to those in the previously described method (Surwase and Jadhav, 2011). High‐performance liquid chromatography (HPLC) analysis of the cell‐free broth was carried out (Waters model no. 2690) on a C18 column (4.6 mm × 250 mm, Symmetry) using methanol as mobile phase, with a flow rate of 1 g l−1 for 10 min and a UV detector at 280 nm. The standard l‐DOPA and cell‐free broth were prepared in HPLC‐grade water and injected into the HPLC column (Krishnaveni et al., 2009; Surwase and Jadhav, 2011). Gas chromatography‐mass spectroscopy (GC‐MS) analysis of the broth obtained after incubation was centrifuged at 5000 r.p.m. for 15 min, and supernatant was collected. The supernatant was extracted twice with equal volumes of chloroform in a separating funnel. The chloroform fraction was recovered in a new flask and evaporated. The residues were dissolved in methanol and used for further analysis. GC‐MS analysis was carried out with a QP2010 gas chromatography coupled with a mass spectrometer (Shimadzu). The analysis was performed with conditions standardized earlier (Rani et al., 2007; Surwase and Jadhav, 2011).
Conclusion
From the results of this study, it is evident that the use of the RSM helped not only to find the most significant factors in l‐DOPA production but also to locate the optimum levels of these factors with minimum resources and time. Thus, the optimized medium formulation obtained in this study using RSM was proven more effective in improving l‐DOPA production than the classical ‘one factor at a time’ method. In addition, Brevundimonas sp. SGJ presents a promising new source for l‐DOPA production with advantages over traditional sources.
Acknowledgments
Shripad N. Surwase thanks the Lady Tata Memorial Trust, Mumbai, Maharashtra, India, for awarding him with a Junior Research Scholarship for his doctoral research. Sushama A. Patil and Shekhar B. Jadhav thank the Department of Science and Technology, Government of India, for providing them fellowships under the DST‐PURSE program.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. HPTLC analysis. (A) 3D graph generated after scanning of HPTLC plate, (C) standard l‐DOPA, (D) cell‐free broth; (B) HPTLC plate, (E) standard l‐DOPA, (F) cell‐free broth.
Fig. S2. HPLC elution profile. (A) Standard l‐DOPA and (B) cell‐free broth after incubation; (C) GC‐MS analysis of cell‐free broth after incubation.
Table S1. Plackett–Burman design matrix performed for the l‐DOPA production.
Table S2. Box–Behnken design matrix for coded variables along with actual and predicted responses for l‐DOPA production.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ali S., Haq I. Innovative effect of illite on improved microbiological conversion of L‐tyrosine to 3, 4 dihydroxy phenyl L‐alanine (L‐DOPA) by Aspergillus oryzae ME2 under acidic reaction conditions. Curr Microbiol. 2006;53:351–357. doi: 10.1007/s00284-005-0220-x. [DOI] [PubMed] [Google Scholar]
- Ali S., Jeffry S.L., Haq I. High performance microbiological transformation of L‐tyrosine to L‐DOPA by Yarrowia lipolytica NRRL‐143. BMC Biotechnol. 2007;7:50–57. doi: 10.1186/1472-6750-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M., Whitcomb P. New York, USA: Productivity press; 2005. [Google Scholar]
- Arnow L.E. Colorimetric determination of the components of L‐3, 4 dihydroxyphenylalanine‐tyrosine mixture. J Biol Chem. 1937;118:531–537. [Google Scholar]
- Bapat V., Suprasanna P., Ganapathi T., Rao P. In vitro production of L‐DOPA in tissue cultures of banana. Pharm Biol. 2000;38:271–273. doi: 10.1076/1388-0209(200009)3841-AFT271. [DOI] [PubMed] [Google Scholar]
- Box G.E.P., Behnken D.W. Some new three level designs for the study of quantitative variables. Technometrics. 1960;2:455–475. [Google Scholar]
- Chattopadhyay S., Datta S.K., Mahato S.B. Production of L‐DOPA from cell suspension culture of Mucuna pruriens fpruriens. Plant Cell Rep. 1994;13:519–522. doi: 10.1007/BF00232948. [DOI] [PubMed] [Google Scholar]
- Claus H., Decker H. Bacterial tyrosinases. Syst Appl Microbiol. 2006;29:3–14. doi: 10.1016/j.syapm.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Gabriela M., Carvalho T., Denise M. L‐DOPA production by immobilized tyrosinase. Appl Biochem Biotechnol. 2000;84:791–800. doi: 10.1385/abab:84-86:1-9:791. [DOI] [PubMed] [Google Scholar]
- Kandaswami C., Vaidyanathan C.S. Enzymatic Assay of tyrosinase Catechol Oxidase Activity(EC 1.14.18.1) J BiolChem. 1973;49:4035–4038. [Google Scholar]
- Kofman O. Treatment of Parkinson's disease with L‐DOPA: a current appraisal. Can Med Assoc J. 1971;104:483–487. [PMC free article] [PubMed] [Google Scholar]
- Koyanagi T., Katayama T., Suzuki H., Nakazawab H., Yokozeki K., Kumagai H. Effective production of 3,4‐dihydroxyphenyl‐l‐alanine (L‐DOPA) with Erwinia herbicola cells carrying a mutant transcriptional regulator TyrR. J Biotechnol. 2005;115:303–306. doi: 10.1016/j.jbiotec.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Krishnaveni R., Rathod V., Thakur M.S., Neelgund Y.F. Transformation of L‐tyrosine to L‐DOPA by a novel fungus, Acremonium rutilum, under submerged fermentation. Curr Microbiol. 2009;58:122–128. doi: 10.1007/s00284-008-9287-5. [DOI] [PubMed] [Google Scholar]
- Lee H., Song M.H., Wang S. Optimizing bioconversion of deproteinated cheese whey to mycelia of Ganoderma lucidum. Process Biochem. 2003;38:1685–1693. [Google Scholar]
- Mahmoud D.A.R., Bendary M.A. Production of 3, 4‐dihydroxy phenyl‐L‐ alanine (L‐DOPA) by Egyptian halophilic black yeast. World J Microbiol Biotechnol. 2011;27:39–46. , and . [Google Scholar]
- Plackett R.L., Burman J.P. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. [Google Scholar]
- Rani N., Joy B., Abraham T.E. Cell suspension cultures of Portulaca grandiflora as potent catalysts for biotransformation of L‐tyrosine into L‐DOPA, an anti‐Parkinson's drug. Pharm Biol. 2007;45:48–53. [Google Scholar]
- Surwase S.N., Jadhav J.P. Bioconversion of L‐tyrosine to L‐DOPA by a novel bacterium Bacillus sp. JPJ. Amino Acids. 2011;41:495–506. doi: 10.1007/s00726-010-0768-z. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lu Z. Optimization of processing parameters for the mycelial growth and extracellular polysaccharide production by Boletus spp. ACCC 50328. Process Biochem. 2005;40:1043–1051. [Google Scholar]
- Zhang T., Wen S., Tan T. Optimization of the medium for glutathione production in Saccharomyces cerevisiae. Process Biochem. 2007;42:454–458. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. HPTLC analysis. (A) 3D graph generated after scanning of HPTLC plate, (C) standard l‐DOPA, (D) cell‐free broth; (B) HPTLC plate, (E) standard l‐DOPA, (F) cell‐free broth.
Fig. S2. HPLC elution profile. (A) Standard l‐DOPA and (B) cell‐free broth after incubation; (C) GC‐MS analysis of cell‐free broth after incubation.
Table S1. Plackett–Burman design matrix performed for the l‐DOPA production.
Table S2. Box–Behnken design matrix for coded variables along with actual and predicted responses for l‐DOPA production.


