Summary
Nitrilase enzymes (nitrilases) catalyse the hydrolysis of nitrile compounds to the corresponding carboxylic acid and ammonia, and have a wide range of industrial and biotechnological applications, including the synthesis of industrially important carboxylic acids and bioremediation of cyanide and toxic nitriles. Nitrilases are produced by a wide range of organisms, including plants, bacteria and fungi, but despite their biotechnological importance, the role of these enzymes in living organisms is relatively underexplored. Current research suggests that nitrilases play important roles in a range of biological processes. In the context of plant–microbe interactions they may have roles in hormone synthesis, nutrient assimilation and detoxification of exogenous and endogenous nitriles. Nitrilases are produced by both plant pathogenic and plant growth‐promoting microorganisms, and their activities may have a significant impact on the outcome of plant–microbe interactions. In this paper we review current knowledge of the role of nitriles and nitrilases in plants and plant‐associated microorganisms, and discuss how greater understanding of the natural functions of nitrilases could be applied to benefit both industry and agriculture.
Introduction
Nitrilase enzymes (nitrilases) catalyse the hydrolysis of nitrile (R‐CN) compounds to the corresponding carboxylic acid and ammonia. These enzymes have been identified and characterized in plants, bacteria and fungi, and homologues have been found in the genomes of animals and yeast (Pace and Brenner, 2001; O'Reilly and Turner, 2003). Since the first identification of nitrilase activity in plants in 1958 (Thimann and Mahadevan, 1958) and in bacteria in 1964 (Hook and Robinson, 1964), over 30 nitrilases have been characterized. However, most bacterial nitrilases have been identified with the aim of elucidating novel mechanisms for chemical synthesis or degradation, rather than deciphering their function in nature (DeSantis et al., 2002; 2003), and the biological role of many of these enzymes remains unknown. Fortunately, as more enzymes have been identified and their substrates and expression patterns determined, clues as to their biological role have been revealed. Nitrile compounds are abundant in the plant environment and current evidence suggests that microbial nitrilases form part of an array of mechanisms that facilitate microbial colonization of plants, with possible roles in plant hormone synthesis, nitrogen utilization, the catabolism of cyanogenic glycosides and glucosinolates and the detoxification of nitriles and cyanide (O'Reilly and Turner, 2003; Kiziak et al., 2005; Howden et al., 2009). As a consequence it seems likely that a greater understanding of nitrilases and their role in plant–microbe interactions could have substantial benefits for a range of biotechnological applications, including plant growth promotion, bioremediation and disease control. In this paper the activity of nitrilases will be reviewed and their potential role in plant–microbe interactions will be discussed.
The nitrilase superfamily
The nitrilase superfamily, also referred to as the CN‐hydrolases, is comprised of enzymes that catalyse the hydrolysis of non‐peptide carbon–nitrogen bonds. Members of the superfamily are divided into 13 branches according to sequence identity and catalytic activity. These branches include the aliphatic amidase, N‐terminal amidase, biotinidase, carbamylase and nitrilase branches, among others (Pace and Brenner, 2001). Nitrilases are perhaps the best characterized of all members of the superfamily with numerous examples identified across kingdoms (O'Reilly and Turner, 2003). These enzymes hydrolyse the CN group of a nitrile compound resulting in the synthesis of the corresponding carboxylic acid and the release of ammonia. The reaction catalysed by nitrilases is shown in Fig. 1. The nitrilase branch also contains the closely related cyanide hydratase and cyanide dihydratase enzymes. Cyanide hydratase enzymes preferentially hydrolyse cyanide to formamide while cyanide dihydratase enzymes specifically hydrolyse cyanide to formic acid and ammonia (O'Reilly and Turner, 2003; Singh et al., 2006).
Figure 1.
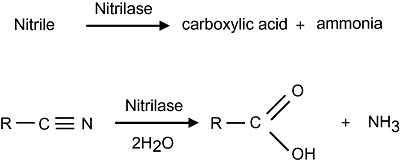
The nitrilase reaction. Nitrilases catalyse the hydrolysis of nitriles to the corresponding carboxylic acid plus ammonia.
All members of the nitrilase superfamily have a catalytic triad of amino acids – glutamic acid, lysine and cysteine. The nitrilase branch can be distinguished from other members of the superfamily by a conserved cysteine‐tryptophan‐glutamic acid motif positioned at the cysteine residue of the catalytic triad. This cysteine residue is thought to form the active site for enzyme activity, and may be the point to which substrate groups attach prior to hydrolysis (Nakai et al., 2000; Pace et al., 2000; Novo et al., 2002). Mutating this cysteine residue causes complete loss of nitrilase activity, as has been observed in Alcaligenes faecalis JM3 and Arabidopsis thaliana (Kobayashi et al., 1993; Vorwerk et al., 2001). In addition, nitrilases have a sulfhydryl group that is essential for catalytic activity and thus nitrilases are classified as thiol enzymes (O'Reilly and Turner, 2003; Podar et al., 2005). Enzyme activity may be inhibited by the presence of thiol binding compounds such as silver nitrate (AgNO3) and copper sulfate (CuSO4), and enhanced in the presence of thiol‐reducing agents such as dithiothreitol (Layh et al., 1998).
Nitrilases are frequently classified into one of three categories according to substrate specificity: aliphatic nitrilases, which act primarily on aliphatic nitriles such as acrylonitrile, glutaronitrile and β‐cyano‐l‐alanine; aromatic and heterocyclic nitrilases, which act primarily on aromatic or heterocyclic nitriles such as benzonitrile and cyanopyridine, and arylacetonitrilases which act primarily on arylacetonitriles such as indole‐3‐acetonitrile (IAN), phenylacetonitrile and phenylpropionitrile (Brenner, 2002; O'Reilly and Turner, 2003). Examples of each class of nitrile compound are shown in Fig. 2. Some nitrilases are extremely substrate specific, such as the nitrilase of Klebsiella pneumoniae sp. ozaenae and NIT4 of A. thaliana (McBride et al., 1986; Piotrowski et al., 2001), which catalyse the hydrolysis of bromoxynil and β‐cyano‐l‐alanine respectively. Other enzymes have a broad substrate range, such as the nitrilase of Bacillus pallidus Dac521, which hydrolyses aromatic, aliphatic and heterocyclic nitriles (Almatawah and Cowan, 1999; Almatawah et al., 1999). Nitrilases with the same substrate specificity often show amino acid sequence similarity and may fall within the same clade in phylogenetic analyses (Robertson et al., 2004; Podar et al., 2005; Howden et al., 2009). Figure 3 shows the phylogenetic relationship of a representative set of characterized nitrilases from different organisms. Within this tree are distinct groupings that in many cases correlate with the active substrate for enzyme activity.
Figure 2.
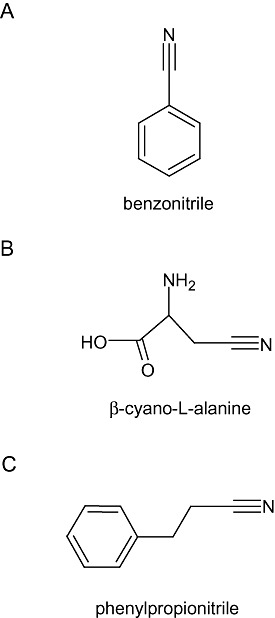
Examples of nitrile compounds. Nitrile compounds can be classified into one of three groups according to their structure: aromatic or heterocyclic (A), aliphatic (B) and arylacetonitrile (C). Chemical structures were drawn using ACD/ChemSketch (http://www.acdlabs.com/download/chemsk.html).
Figure 3.
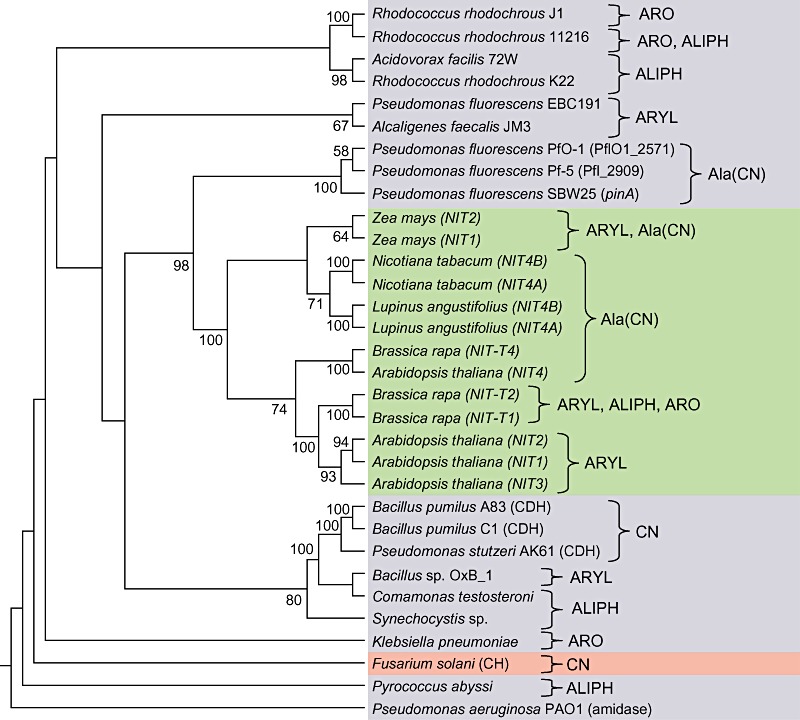
Strict consensus tree of characterized nitrilases. The tree was generated by parsimony analysis and is supported by a bootstrap analysis with 500 replicates. The tree is rooted using the Pseudomonas aeruginosa aliphatic amidase sequence. Shading corresponds to plant genes (green), bacterial genes (blue) and fungal genes (pink). Included in the tree are cyanide hydratase (CH) and cyanide dihydratase (CDH) genes, which are closely related to nitrilases and are found within the nitrilase branch of the superfamily (O'Reilly and Turner, 2003). Gene names, where known, are shown in parentheses. The tree is annotated with the most active substrate or substrate class for each enzyme, where known (ARO, aromatic; ALIPH, aliphatic; ARYL, arylacetonitriles; Ala(CN), β‐cyano‐l‐alanine; CN, cyanide). The tree was generated using the method described by Howden and colleagues (2009).
Nitrile compounds in nature
Cyanolipids, cyanogenic glycosides and glucosinolates
Nitrile compounds are abundant in the natural environment and are synthesized by plants and microbes as intermediates in chemical biosynthesis and degradation (Legras et al., 1990). The widespread occurrence of nitrile compounds may explain the prevalence of nitrilases in prokaryotes and eukaryotes. Indeed, nitrilase activity may be a universal property of all land plants (Piotrowski, 2008). Within plants, nitriles are particularly common in defence pathways and in many cases are linked with the metabolism of cyanide. Members of the Sapindaceae and Boraginaceae plant families produce cyanolipids from the esterification of α‐hydroxylated nitriles and fatty acids. Cyanolipids act as a nitrogen store in the seeds of these plants. In addition they may be used a defensive compound against herbivory, as their hydrolysis results in the production of cyanohydrin which subsequently decomposes to liberate hydrogen cyanide (HCN; Legras et al., 1990; Selmar et al., 1990).
Nitrile compounds are also produced during the metabolism of cyanogenic glycosides and glucosinolates. Both are defence molecules which provide protection to plants against herbivory and pathogen attack (Vetter, 2000; Fahey et al., 2001; Halkier and Gershenzon, 2006). While cyanogenic glycosides are widely distributed in plants, glucosinolates are almost exclusively found in the Capparales order, which includes the Brassicaceae. Nitrile compounds are also found as intermediates during cyanogenic glycoside biosynthesis (Vetter, 2000; Wittstock and Halkier, 2002; Halkier and Gershenzon, 2006). Within the plant, cyanogenic glycosides and glucosinolates are stored in compartments spatially isolated from the enzymes that degrade them. Tissue damage causes enzyme and substrate to mix, resulting in their degradation and the release of a toxic product. Cyanogenic glycosides are degraded by glycosidases to cyanohydrin which is decomposed to HCN and an aldehyde (Dewick, 1984; Vetter, 2000). Glucosinolates are degraded by the activity of myrosinase enzymes to form glucose and an unstable aglycone molecule. Aglycone molecules undergo rearrangement to form either nitriles, isothiocyanates, epithionitriles, thiocyanates or oxazolidine‐2‐thione (Halkier and Gershenzon, 2006).
In addition to their role in plant defence, glucosinolates may also be intermediates in plant hormone synthesis. Ludwig‐Muller and Cohen (2002) hypothesize that in nasturtium (Trapoleum majus) benzylglucosinolate and indole‐3‐methylglucosinolate are degraded by myrosinase to produce phenylacetonitrile and IAN respectively. Phenylacetonitrile and IAN can also be produced directly from aldoxime precursors by aldoxime dehydratase enzymes (Nafisi et al., 2007). These two compounds may subsequently be hydrolysed by nitrilase activity to the auxins phenylacetic acid and indole‐3‐acetic acid (IAA; Ludwig‐Muller and Cohen, 2002). The hydrolysis of IAN to IAA by nitrilase activity is an extremely well‐characterized reaction in plants and bacteria (Kobayashi et al., 1993; Bartel and Fink, 1994; Normanly et al., 1997; Vorwerk et al., 2001). However, the IAN pathway is one of several that have been identified for IAA biosynthesis in plants (Normanly and Bartel, 1999) and the contribution of nitrilase activity to overall IAA production may be less important than first predicted (Piotrowski, 2008). Nafisi and colleagues (2007) suggest that an alternative, or additional, function for IAN may be as an intermediate in synthesis of the phytoalexin camalexin, but a biosynthetic route from IAN to camalexin has not yet been identified.
Cyanide and the synthesis of β‐cyano‐l ‐alanine
Nitriles are also present in plants as intermediates in cyanide metabolism. Cyanide is synthesized by plants during defence responses, as mentioned above, but also as a co‐product of ethylene biosynthesis. Ethylene is synthesized from 1‐aminocyclopropane‐1‐carboxylic acid with the release of CO2 and HCN (Peiser et al., 1984). Free cyanide and cysteine are metabolized to the nitrile β‐cyano‐l‐alanine by the enzyme β‐cyano‐l‐alanine synthase (Floss et al., 1965; Blumenthal et al., 1968). β‐Cyano‐l‐alanine is a potent neurotoxin and may accumulate in the tissues of some plants, such as vetch (Vicia), to be used as an anti‐herbivory agent (Vannesland et al., 1981; Ressler et al., 1997). However, in most plants β‐cyano‐l‐alanine is quickly detoxified by nitrilase activity to aspartic acid, asparagine and ammonia. The reactions linking ethylene biosynthesis with cyanide and β‐cyano‐l‐alanine production are shown in Fig. 4.
Figure 4.
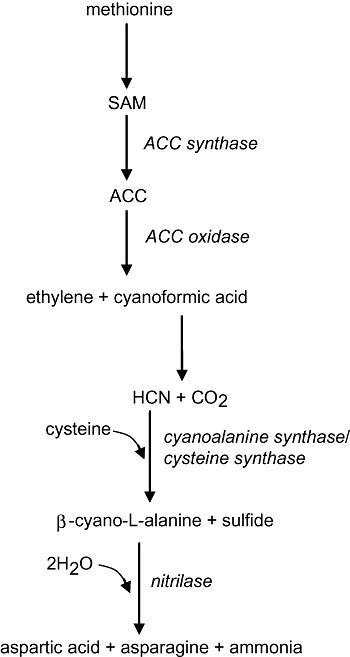
Ethylene, cyanide and β‐cyano‐l‐alanine synthesis in plants. When plants synthesize ethylene they also produce cyanide as a co‐product. Cyanide is converted to β‐cyano‐l‐alanine which is subsequently detoxified by a NIT4‐type nitrilase to aspartic acid and ammonia. Asparagine is also produced in this reaction because NIT4 displays β‐cyano‐l‐alanine‐hydratase activity (Piotrowski et al., 2001). Enzymes are shown in italics. SAM, S‐adenosyl‐l‐methionine; ACC, 1‐aminocyclopropane‐1‐carboxylic acid. This figure was adapted from Davies (1995).
In plants, the hydrolysis of β‐cyano‐l‐alanine is catalysed by the nitrilase NIT4, and NIT4 homologues have been found throughout the plant kingdom (Piotrowski et al., 2001; Jenrich et al., 2007; Piotrowski, 2008). NIT4‐type nitrilases from plants have been shown to have nitrilase and nitrile hydratase activity, which explains the synthesis of asparagine and aspartic acid from β‐cyano‐l‐alanine (Piotrowski et al., 2001). β‐Cyano‐l‐alanine synthases have also been found in a number of bacteria and in insects (Dunnill and Fowden, 1965; Macadam and Knowles, 1984; Meyers and Ahmad, 1991), and a NIT4‐type nitrilase has recently been characterized in bacteria (Howden et al., 2009). β‐Cyano‐l‐alanine synthase and NIT4 are likely to have a dual role in cyanide detoxification and also in the recycling of nitrogen from cyanide into amino acids (Hatzfeld et al., 2000). β‐Cyano‐l‐alanine synthase activity has been found to correlate with levels of ethylene in plant tissues (Goudey et al., 1989), and plants experiencing drought stress have been shown to have enhanced production of ethylene which in turn causes an increase in HCN synthesis and β‐cyano‐l‐alanine synthase activity (Liang, 2003).
In bacteria, nitriles may be formed during the detoxification of endogenous and exogenous cyanide. Bacteria produce cyanide in a process termed cyanogenesis, in which HCN and CO2 are synthesized from glycine by the enzyme HCN synthase. Hydrogen cyanide synthase is encoded by three biosynthetic genes, hcnA, hcnB and hcnC (Laville et al., 1998; Ramette et al., 2003). Hydrogen cyanide synthase has been partially purified and characterized from strains of Pseudomonas aeruginosa and Pseudomonas fluorescens, and this enzyme appears to be present in a number of bacteria (Laville et al., 1998; Ramette et al., 2003). The degree of cyanide production may depend on nutrient supply, the phase of bacterial growth and the level of aeration in the environment. While the function of bacterial cyanogenesis remains unclear, this property may promote competitiveness as a consequence of antagonistic activity towards competitors and predators, or may facilitate mobilization and uptake of metals (Ramette et al., 2003; Faramarzi et al., 2004).
Bacteria that are exposed to, or actively producing cyanide may protect themselves from cyanide toxicity using cyanide‐degrading enzymes, which may have cyanide hydratase, cyanide dihydratase or rhodanese activity. Alternatively, bacteria may incorporate cyanide into nitrile compounds, such as β‐cyano‐l‐alanine, which can then be used by the bacterium as a carbon and nitrogen source through the activity of a nitrilase (Vannesland et al., 1981; Yoshikawa et al., 2000; O'Reilly and Turner, 2003; Baxter and Cummings 2006). Bacteria, like plants, may produce β‐cyano‐l‐alanine through the activity of cysteine synthase enzymes. β‐Cyano‐l‐alanine synthase is related to the enzyme cysteine synthase, and both enzymes have been shown to have overlapping activities (Hatzfeld et al., 2000). Putative cysteine synthase genes have been identified in the β‐cyano‐l‐alanine‐degrading bacterium P. fluorescens SBW25, and it seems likely that this bacterium is able to synthesize β‐cyano‐l‐alanine from cysteine and cyanide as well as hydrolysing β‐cyano‐l‐alanine by NIT4‐type nitrilase activity (Howden et al., 2009).
The regulation of nitrilase activity
Nitrilases have been found to be regulated at the transcriptional and post‐translational level, and gene expression and enzyme activation may depend on environmental conditions and substrate availability. The regulation of nitrilase activity in plants appears to be closely linked to tissue‐specific conditions and certain developmental stages, although at present relatively little is known about the transcription factors and regulatory mechanisms that regulate nitrilase expression in plants. The expression patterns of NIT1, 2 and 3 of A. thaliana are distinct. NIT1 is expressed in all green tissue and is strongly expressed in apical buds, root tips, tissue of developing adventitious roots and the nodal region of adventitious root formation (Hillebrand et al., 1998; Vorwerk et al., 2001). NIT2 on the other hand is strongly expressed in the mature embryo and cotyledons of very young seedlings (Vorwerk et al., 2001), and is induced upon pathogen attack, which is consistent with the idea that one function for these nitrilases is in synthesis or detoxification of defensive metabolites (Bartel and Fink, 1994). NIT3 is expressed in the cotyledons and hypocotyls of germinating seedlings (Vorwerk et al., 2001). Interestingly, NIT4 activity is higher in senescent leaves of A. thaliana compared with non‐senescent leaves (Piotrowski et al., 2001). The upregulation of NIT4 activity during senescence may be linked with increased ethylene biosynthesis and cyanide production. These defined expression patterns suggest that each nitrilase has a precise role within a particular tissue.
In bacteria, some progress has been made towards identifying regulatory proteins involved in transcriptional regulation of nitrilase activity. The nitrilase of Rhodococcus rhodochrous J1, nitA, has a regulatory gene nitR, in close proximity, which is required for nitrile‐dependent activation of nitA (Komeda et al., 1996). While the precise mechanism of nitR activity remains to be elucidated, it has been shown that deleting portions of nitR results in complete loss of nitA activity in R. rhodochrous J1. nitR shows homology to xylS, a positive regulator of xylene metabolism in Pseudomonas putida, and to araC, a positive regulator of arabinose metabolism in Escherichia coli (Komeda et al., 1996), which suggests that it may function in a similar manner to these well‐characterized regulators, and activate gene expression in response to a direct interaction with a nitrile substrate effector (Schleif, 2003; Dominguez‐Cuevas et al., 2008). Bacillus sp. OxB‐1 has also been shown to have a nitR‐type regulatory gene adjacent to its nitrilase (Kato et al., 2000). The P. fluorescens strains SBW25, PfO‐1 and Pf‐5 all have a LysR‐type transcriptional regulator adjacent to the β‐cyano‐alanine nitrilase gene and it is possible that this gene is a positive regulator of nitrilase activity in these bacteria (Howden et al., 2009).
The β‐cyano‐l‐alanine nitrilase of P. fluorescens SBW25 has been shown to be transcriptionally induced by a plant‐derived signal produced by both A. thaliana seedlings and sugar beet seedlings (Gal et al., 2003; Howden et al., 2009). This nitrilase is also induced by the substrate for enzyme activity, β‐cyano‐l‐alanine, and by its precursors cyanide and cysteine (Howden et al., 2009), which suggests that one or more of these chemicals are present at inducing levels in root exudates. Nitrilase expression was found to be induced by nanomolar amounts of β‐cyano‐l‐alanine, compared with micromolar amounts of cyanide and cysteine, which suggests that cyanide or cysteine‐dependent induction may result from the conversion of these chemicals into β‐cyano‐l‐alanine, as discussed above.
Some nitrile‐degrading bacteria have been shown to exhibit both substrate‐specific and environmental regulation of nitrilase activity. For example, R. rhodochrous cells induced with propionitrile hydrolyse a different range of nitriles than cells induced with benzonitrile, and the nitrilase generated upon induction with propionitrile is different from that generated by benzonitrile. The N‐terminal sequence of these enzymes differs at the third residue and the optimal temperature and pH for catalytic activity is different for each enzyme (Hoyle et al., 1998). The nitrilase of P. fluorescens DSM7155 requires arylacetonitriles for enzyme induction, but the presence of ammonium ions represses nitrilase induction, suggesting a degree of catabolite repression, and supporting the hypothesis that the primary role of this nitrilase is in nitrogen assimilation (Layh et al., 1998).
Each subunit of a nitrilase consists of a single polypeptide approximately 40 kDa in size (O'Reilly and Turner, 2003). In most cases the active form of the enzyme is an aggregate of subunits. However, some nitrilases are active as monomers, such as the nitrilase of R. rhodochrous PA34 (Bhalla et al., 1992), while others are active as dimers, such as the nitrilase of K. pneumoniae sp. ozaenae (Stalker et al., 1988a). The nitrilase of R. rhodochrous NCIMB 11216 is active in a multimeric form, comprising of 12 subunits associated to give a 560 kDa protein. Subunit association and subsequent enzyme activation only occur when bacteria are incubated with the substrate. Thus, the enzyme can be classified as an inducible enzyme (Hoyle et al., 1998). Similar observations have been made with a number of characterized nitrilases.
Some nitrilases exist as heterologous co‐polymers where two different subunits make up an active multimeric enzyme. The nitrilase of P. fluorescens DSM7155 consists of 40 kDa and 38 kDa subunits (Layh et al., 1998). In addition, nitrilase proteins may associate with proteins with no nitrile‐hydrolysing activity. The nitrilase of B. pallidus Dac521 associates with a GroEL‐like protein while the nitrilase of P. fluorescens DSM7155 associates with the protein CPN60. Both proteins are chaperonins and may enhance protein folding or enzyme stability (Layh et al., 1998; Almatawah and Cowan, 1999; Almatawah et al., 1999).
Nitrilases in nature and their role in plant–microbe interactions
The biochemical and biological properties of nitrilases and their substrates indicate that these enzymes are likely to have functions in defence, detoxification, nitrogen utilization and plant hormone synthesis. Plant nitrilases are perhaps the best characterized of all nitrilases in relation to their biological functions, particularly NIT1, 2, 3 and 4 of A. thaliana. Plant nitrilases form two distinct groups according to substrate specificity: those with high hydrolytic activity towards arylacetonitriles and those with high hydrolytic activity towards β‐cyano‐l‐alanine. NIT1, 2 and 3 of A. thaliana are arylacetonitrilases and are likely to have roles in the hydrolysis of nitriles produced during the synthesis or degradation of cyanogenic glycosides and glucosinolates. Phenylpropionitrile and other naturally occurring products of glucosinolate metabolism are preferred substrates for NIT1, 2 and 3 (Vorwerk et al., 2001). In addition, all three enzymes have been shown to hydrolyse IAN to IAA, thus linking them to the biosynthesis of auxin (Bartel and Fink, 1994). NIT4 of A. thaliana falls into the second group of plant nitrilases, the NIT4‐type enzymes. NIT4 enzymes are widespread in the plant kingdom and as mentioned earlier, are likely to be important in the cyanide detoxification pathway (Piotrowski et al., 2001; Piotrowski, 2008).
Many microbial nitrilase, cyanide hydratase and cyanide dihydratase enzymes have been identified in organisms isolated from cyanide or nitrile‐contaminated land and water, and have been shown to enhance the cyanide and nitrile tolerance of the organisms that produce them, which supports the idea that these enzymes have functions in cyanide and nitrile detoxification. For example, the cyanide hydratase of Fusarium solani can hydrolyse free or metal‐complexed cyanide, enabling this organism to tolerate concentrations of cyanide that would be toxic to other microorganisms. This hydrolysis may also supplement the organism's nitrogen supply and thus enhance growth (Barclay et al., 1998; 2002). Microbes may also use nitrilase activity for the detoxification and assimilation of nitriles and cyanide present in the plant environment. For example, the cyanide hydratase of the sorghum pathogen Gloeocercospora sorghi may allow this fungal pathogen to colonize its cyanogenic host plant (Wang and VanEtten, 1992; Wang et al., 1992). Sorghum plants store the cyanogenic glycoside dhurrin in vacuoles of leaf epidermal cells. Upon tissue damage dhurrin degradation commences, causing the liberation of cyanide which is thought to act as a defensive compound (Legras et al., 1990). Gloeocercospora sorghi can break down this cyanide by cyanide hydratase activity, and the production of cyanide hydratase by G. sorghi has been shown to correlate with the concentration of cyanogenic compounds in sorghum tissue (Wang and VanEtten, 1992; Wang et al., 1992). The β‐cyano‐l‐alanine nitrilase produced by the plant growth‐promoting rhizobacterium P. fluorescens SBW25 has also been shown to enable this bacterium to tolerate toxic concentrations of this nitrile (Howden et al., 2009). However, it is still unclear whether P. fluorescens SBW25 and other plant‐associated organisms encounter toxic concentrations of microbial or plant‐derived β‐cyano‐l‐alanine in natural environments. β‐Cyano‐l‐alanine nitrilase activity has also been detected in cyanogenic Pseudomonas, such as P. fluorescens Pf‐5 (Howden et al., 2009), and it seems likely that a primary function of this nitrilase is as a mechanism for detoxifying endogenous and exogenous cyanide, rather than β‐cyano‐l‐alanine.
An alternative or additional function for β‐cyano‐l‐alanine nitrilase and for other nitrilases produced by plant‐associated microorganisms may be to allow microorganisms to use plant nitriles as a carbon and nitrogen source (Howden et al., 2009). The nitrilase of P. fluorescens EBC191 has been shown to hydrolyse a number of arylacetonitriles, including mandelonitrile, which is produced from cyanogenic glycosides as a defence against herbivores (Legras et al., 1990; Kiziak et al., 2005). The corresponding nitrilase gene is located in close proximity to genes involved in the mandelate pathway, suggesting that P. fluorescens EBC191 hydrolyses mandelonitrile to obtain nutrients during colonization of the plant environment (Kiziak et al., 2005). A similar nitrilase has been found in the bacterium Alcaligenes faecalis JM3 (Kobayashi et al., 1993).
Bacteria may also hydrolyse arylacetonitriles, such as IAN, in order to synthesize auxins. The ability of microorganisms to produce auxins has been widely documented and may be associated with pathogenicity, symbiosis or plant growth promotion (reviewed by Spaepen et al., 2007). Indole‐3‐acetic acid has been shown to inhibit plant defence mechanisms and to alter plant growth and development. For example, IAA biosynthesis induced by Agrobacterium tumefaciens plays a central role in the formation of plant tumours known as galls (Klee et al., 1984; Kobayashi et al., 1995). Indole‐3‐acetic acid production by both non‐pathogenic and pathogenic microorganisms may also stimulate cell wall elongation, lateral root formation and nutrient release from plant cells, all of which could facilitate invasion and colonization of plant tissues (Spaepen et al., 2007). Genome sequence analysis of the plant pathogenic bacteria Pseudomonas syringae pv. syringae B728a and P. syringae pv. tomato DC3000 has shown that they both contain nitrilase genes that show sequence similarity to those of P. fluorescens EBC191 and Alcaligenes faecalis JM3, which have been shown to hydrolyse arylacetonitriles such as IAN (Kobayashi et al., 1993; Feil et al., 2005; Kiziak et al., 2005). Furthermore, both nitrilase genes are adjacent to a gene that encodes a putative aldoxime dehydratase, which could catalyse the conversion of IAOx into IAN, although the putative acetaldoxime dehydratase of P. syringae pv. tomato DC3000 appears to be a pseudogene, as is an adjacent regulatory gene. While these enzymes are yet to be characterized, it seems likely that they could act to produce auxins from IAOx, IAN and related compounds, and that their activity could contribute towards the pathogenicity of these bacteria.
Industrial applications of nitrilases
In the last section of this review the industrial and biotechnological applications of nitrilases will be discussed, along with the challenges associated with their use. Nitrilases may be used in the synthesis of industrially important carboxylic acids which are otherwise produced by chemical methods requiring extreme conditions of temperature and pH (Kobayashi and Shimizu, 1994; Osswald et al., 2002). For example, the nitrilase of R. rhodochrous J1 can hydrolyse 3‐cyanopyridone to nicotinic acid, a vitamin used in animal feed and medicine (Mathew et al., 1988).
Nitrilases may also be used in the bioremediation of land and water contaminated with toxic nitrile compounds. These compounds enter the environment from a variety of sources. In industry, acetonitrile is used as a solvent while acrylonitrile is used in the synthesis of plastics. Nitrile compounds such as bromoxynil are used as herbicides, and cyanide is used in the manufacture of plastics and the extraction of precious metals (O'Reilly and Turner, 2003; Singh et al., 2006), as well as being released to the environment as a toxic by‐product of mining, metal finishing and organic chemical industries (Baxter and Cummings, 2006). The cyanide dihydratase of Pseudomonas stutzeri (isolated from the effluent of a metal plating plant) has been investigated as a mechanism for the removal of cyanide due to its cyanide‐degrading properties and the ability of this bacterium to tolerate high concentrations of KCN (Sewell et al., 2003). The nitrilase of K. pneumoniae sp. ozaenae is highly specific for the herbicide bromoxynil (McBride et al., 1986). This enzyme has been expressed in plants, and confers herbicide resistance to transgenic lines (Stalker et al., 1988b).
However, despite great potential, few nitrilases have been used for chemical synthesis and bioremediation. One reason for this may be the laborious nature of identifying and characterizing nitrilases, which often relies on a hit‐or‐miss strategy where a range of nitrile compounds are tested as substrates for a potential nitrilase. Recently, scientists have developed high‐throughput strategies for identifying bacterial nitrilases. DNA samples from the environment are transformed into a bacterial expression vector and are screened for nitrilase activity. Using this strategy DeSantis and colleagues (2002) have identified over 200 nitrilase sequences which have been overexpressed and tested for hydrolytic activity, while Robertson and colleagues (2004) have found 137 novel nitrilases, all of which contain the glutamic acid, lysine, cysteine catalytic triad. The availability of genome sequence data has also helped in the discovery of new nitrilases. For example, Heinemann and colleagues (2003a,b) have used genome sequence data to identify a functional nitrilase from the cyanobacterium, Synechocystis sp.
Another reason for lack of progress in developing commercial applications of nitrilases is that these enzymes are often unstable and difficult to purify in an enzymatically active form. The addition of reducing agents such as dithiothreitol and 2‐mercaptoethanol can prevent oxidation of enzyme thiol residues that are important in the hydrolysis reaction (Banerjee et al., 2006). Ammonium sulfate and glycerol have also been found to stabilize nitrilases, possibly by preventing dissociation of enzyme subunits (Kiziak et al., 2005). Such strategies may be useful in improving enzyme activity and stability. Alternatively, scientists may use nitrilases isolated from organisms found growing naturally in extreme conditions, as these enzymes are likely to be more stable under harsh conditions. For example, the nitrilase from the thermophilic bacterium B. pallidus strain Dac521 has a broad substrate range and is extremely stable at high temperatures (Almatawah et al., 1999), while the nitrilase of the hyperthermophilic archeon Pyrococcus abyssi shows catalytic activity at high temperatures and across a wide pH range (Mueller et al., 2006).
Concluding remarks
The aim of this paper was to provide an overview of current knowledge of nitrilase activity, and specifically to review current understanding of the role of nitrilase activity in plant–microbe interactions. Figure 5 summarizes the main hypothesized roles for nitrilase activity in plant‐associated bacteria: detoxification, nutrient assimilation and modulation of plant development and physiology. The diversity and catalytic activity of the nitrilase family make them a useful tool in the catalysis of industrially and environmentally important reactions, but researchers continue to face significant challenges in identifying enzymes with suitable specificities and enzymatic properties for industrial‐scale processes. Research has shown that bioinformatic and phylogenetic analyses can be used to generate broad predictions of enzyme activity, as illustrated in Fig. 3, so further characterization of the sequences and nitrilase activities associated with specific microbial communities, combined with recent developments in high‐throughput sequencing and profiling, could allow researchers to focus isolation efforts on communities that are likely to contain nitrilase activities of interest. Many microbial nitrilases appear to have evolved to degrade plant‐derived nitriles, so it may be possible to identify plant species that are rich in nitriles that show chemical similarities to target compounds, and to investigate the nitrilase activities present in the microorganisms associated with these plants.
Figure 5.
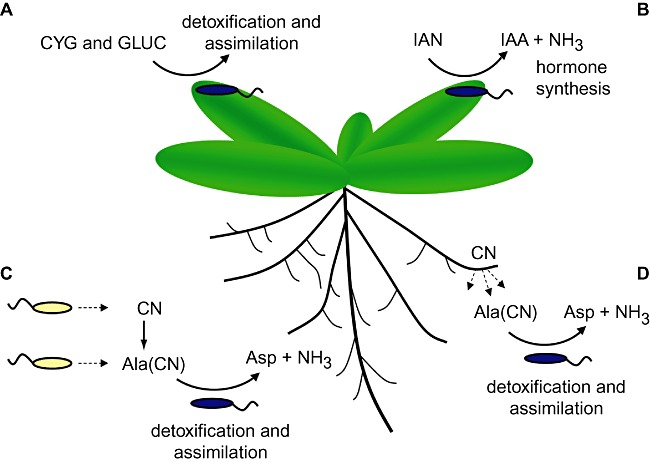
The hydrolysis of nitriles in the plant environment by plant‐associated bacteria. The figure shows four hypothesized roles for bacterial nitrilase enzymes during plant colonization: (A) the hydrolysis of plant nitriles generated during cyanogenic glycoside (CYG) and glucosinolate (GLUC) metabolism, for the purpose of detoxification or assimilation; (B) the hydrolysis of arylacetonitriles such as indole‐3‐acetonitrile (IAN), for the purpose of indole‐3‐acetic acid (IAA) biosynthesis; (C) the detoxification of cyanide (CN) and β‐cyano‐l‐alanine (Ala(CN)) synthesized endogenously or by other microbes, which results in the formation of aspartic acid (Asp) and ammonia (NH3) that can be assimilated as carbon and nitrogen sources; (D) the detoxification and assimilation of CN and Ala(CN) synthesized by plants.
Another important area of nitrilase research involves remediation of cyanide and nitrile contaminated soil. Research into nitrilase regulation has shown that nitrilase activity is subject to environmental regulation and to the presence of appropriate inducing compounds. It may be possible to use knowledge of nitrilase activity and regulatory mechanisms to engineer organisms with increased levels of activity; and to exploit the fact that some nitrilases show evidence of being plant‐induced by implementing combined plant and microbe remediation strategies. Howden and colleagues (2009) showed that overexpression of a β‐cyano‐l‐alanine‐degrading bacterial nitrilase in plant tissues resulted in increased tolerance to the corresponding nitrile, and stimulated root elongation in the absence of the nitrile, possibly as a consequence of increased cyanide detoxification in plant tissues. Introduction of cyanide and nitrile‐degrading plants and bacteria into cyanide and nitrile contaminated sites could provide an effective mechanism to accelerate removal of these chemicals from soil and water.
Finally, it is worth noting that although numerous studies have shown that nitrilase‐related activities, particularly IAA and ethylene synthesis, have a significant effect on plant pathogenesis and plant growth promotion, in many cases the molecular mechanisms underpinning these effects remain unclear (Spaepen et al., 2007). Greater understanding of the role of nitrilases in plant physiology and plant–microbe interactions could be used to develop and deploy plant growth‐promoting organisms with greater effectiveness and to develop new strategies for preventing pathogenesis. Advances in genome sequencing are revealing new nitrilases in a plethora of plant‐associated microorganisms. Discovering what these enzymes do, and why they do it remains an ongoing challenge.
Acknowledgments
G.P. is a Royal Society University Research Fellow. This work was supported by a Biotechnology and Biological Sciences Research Council graduate studentship awarded to A.H.
References
- Almatawah Q.A., Cowan D.A. Thermostable nitrilase catalysed production of nicotinic acid from 3‐cyanopyridine. Enzyme Microb Technol. 1999;25:718–724. [Google Scholar]
- Almatawah Q.A., Cramp R., Cowan D.A. Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles. 1999;3:283–291. doi: 10.1007/s007920050129. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Kaul P., Banerjee U.C. Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol. 2006;72:77–87. doi: 10.1007/s00253-005-0255-8. [DOI] [PubMed] [Google Scholar]
- Barclay M., Tett V.A., Knowles C.J. Metabolism and enzymology of cyanide/metallocyanide biodegradation by Fusarium solani under neutral and acidic conditions. Enzyme Microb Technol. 1998;23:321–330. [Google Scholar]
- Barclay M., Day J.C., Thompson I.P., Knowles C.J., Bailey M.J. Substrate‐regulated cyanide hydratase (chy) gene expression in Fusarium solani: the potential of a transcription‐based assay for monitoring the biotransformation of cyanide complexes. Environ Microbiol. 2002;4:183–189. doi: 10.1046/j.1462-2920.2002.00284.x. [DOI] [PubMed] [Google Scholar]
- Bartel B., Fink G.R. Differential regulation of an auxin‐producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J., Cummings S.P. The current and future applications of microorganism in the bioremediation of cyanide contamination. Antonie Van Leeuwenhoek. 2006;90:1–17. doi: 10.1007/s10482-006-9057-y. [DOI] [PubMed] [Google Scholar]
- Bhalla T.C., Miura A., Wakamoto A., Ohba Y., Furuhashi K. Asymmetric hydrolysis of alpha‐aminonitriles to optically‐active amino‐acids by a nitrilase of Rhodococcus rhodochrous Pa‐34. Appl Microbiol Biotechnol. 1992;37:184–190. [Google Scholar]
- Blumenthal S.G., Hendrickson H.R., Abrol Y.P., Conn E.E. Cyanide metabolism in higher plants. 3. The biosynthesis of beta‐cyanolanine. J Biol Chem. 1968;243:5302–5307. [PubMed] [Google Scholar]
- Brenner C. Catalysis in the nitrilase superfamily. Curr Opin Struct Biol. 2002;12:775–782. doi: 10.1016/s0959-440x(02)00387-1. [DOI] [PubMed] [Google Scholar]
- Davies P.J. Kluwer Academic Publishers; 1995. [Google Scholar]
- DeSantis G., Zhu Z., Greenberg W.A., Wong K., Chaplin J., Hanson S.R. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J Am Chem Soc. 2002;124:9024–9025. doi: 10.1021/ja0259842. et al. [DOI] [PubMed] [Google Scholar]
- DeSantis G., Wong K., Farwell B., Chatman K., Zhu Z., Tomlinson G. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM) J Am Chem Soc. 2003;125:11476–11477. doi: 10.1021/ja035742h. et al. [DOI] [PubMed] [Google Scholar]
- Dewick P.M. The biosynthesis of cyanogenic glycosides and glucosinolates. Nat Prod Rep. 1984;1:545–549. [Google Scholar]
- Dominguez‐Cuevas P., Marin P., Busby S., Ramos J.L., Marques S. Roles of effectors in XylS‐dependent transcription activation: intramolecular domain derepression and DNA binding. J Bacteriol. 2008;190:3118–3128. doi: 10.1128/JB.01784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnill P.M., Fowden L. Enzymatic formation of beta‐cyanoalanine from cyanide by Escherichia coli extracts. Nature. 1965;208:1206–1207. doi: 10.1038/2081206a0. [DOI] [PubMed] [Google Scholar]
- Fahey J.W., Zalcmann A.T., Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Photochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Faramarzi M.A., Stagars M., Pensini E., Krebs W., Brandl H. Metal solubilization from metal‐containing solid materials by cyanogenic Chromobacterium violaceum. J Biotechnol. 2004;113:321–326. doi: 10.1016/j.jbiotec.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Feil H., Feil W.S., Chain P., Larimer F., DiBartolo G., Copeland A. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss H.G., Hadwiger L., Conn E.E. Enzymatic formation of beta‐cyanoalanine from cyanide. Nature. 1965;208:1207–1208. doi: 10.1038/2081207a0. [DOI] [PubMed] [Google Scholar]
- Gal M., Preston G.M., Massey R.C., Spiers A.J., Rainey P.B. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol Ecol. 2003;12:3109–3121. doi: 10.1046/j.1365-294x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Goudey J.S., Tittle F.L., Spencer M.S. A role for ethylene in the metabolism of cyanide by higher plants. Plant Physiol. 1989;89:1306–1310. doi: 10.1104/pp.89.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y., Maruyama A., Schmidt A., Noji M., Ishizawa K., Saito K. Beta‐cyanoalanine synthase is a mitochondrial cysteine synthase‐like protein in spinach and Arabidopsis. Plant Physiol. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Engels D., Burger S., Kiziak C., Mattes R., Stolz A. Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl Environ Microbiol. 2003a;69:4359–4366. doi: 10.1128/AEM.69.8.4359-4366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U., Kiziak C., Zibek S., Layh N., Schmidt M., Griengl H., Stolz A. Conversion of aliphatic 2‐acetoxynitriles by nitrile‐hydrolysing bacteria. Appl Microbiol Biotechnol. 2003b;63:274–281. doi: 10.1007/s00253-003-1382-8. [DOI] [PubMed] [Google Scholar]
- Hillebrand H., Bartling D., Weiler E.W. Structural analysis of the nit2/nit1/nit3 gene cluster encoding nitrilases, enzymes catalyzing the terminal activation step in indole‐acetic acid biosynthesis in Arabidopsis thaliana. Plant Mol Biol. 1998;36:89–99. doi: 10.1023/a:1005998918418. [DOI] [PubMed] [Google Scholar]
- Hook R.H., Robinson W.G. Ricinine nitrilase. II. Purification and properties. J Biol Chem. 1964;239:4263–4267. [PubMed] [Google Scholar]
- Howden A.J., Harrison C.J., Preston G.M. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 2009;57:243–253. doi: 10.1111/j.1365-313X.2008.03682.x. [DOI] [PubMed] [Google Scholar]
- Hoyle A.J., Bunch A.W., Knowles C.J. The nitrilases of Rhodococcus rhodochrous NCIMB 11216. Enzyme Microb Technol. 1998;23:475–482. [Google Scholar]
- Jenrich R., Trompetter I., Bak S., Olsen C.E., Moller B.L., Piotrowski M. Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc Natl Acad Sci USA. 2007;104:18848–18853. doi: 10.1073/pnas.0709315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Nakamura K., Sakiyama H., Mayhew S.G., Asano Y. Novel heme‐containing lyase, phenylacetaldoxime dehydratase from Bacillus sp, strain OxB‐1: purification, characterization, and molecular cloning of the gene. Biochemistry. 2000;39:800–809. doi: 10.1021/bi991598u. [DOI] [PubMed] [Google Scholar]
- Kiziak C., Conradt D., Stolz A., Mattes R., Klein J. Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology. 2005;151:3639–3648. doi: 10.1099/mic.0.28246-0. [DOI] [PubMed] [Google Scholar]
- Klee H., Montoya A., Horodyski F., Lichtenstein C., Garfinkel D., Fuller S. Nucleotide sequence of the tms genes of the pTiA6NC Octopine Ti Plasmid – two gene products involved in plant tumorigenesis. Proc Natl Acad Sci USA. 1984;81:1728–1732. doi: 10.1073/pnas.81.6.1728. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Shimizu S. Versatile nitrilases – nitrile‐hydrolyzing enzymes. FEMS. Microbiol Lett. 1994;120:217–223. [Google Scholar]
- Kobayashi M., Izui H., Nagasawa T., Yamada H. Nitrilase in biosynthesis of the plant hormone indole‐3‐acetic‐acid from indole‐3‐acetonitrile – cloning of the Alcaligenes gene and site‐directed mutagenesis of cysteine residues. Proc Natl Acad Sci USA. 1993;90:247–251. doi: 10.1073/pnas.90.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Suzuki T., Fujita T., Masuda M., Shimizu S. Occurrence of enzymes involved in biosynthesis of indole‐3‐acetic‐acid from indole‐3‐acetonitrile in plant‐associated bacteria, Agrobacterium and Rhizobium. Proc Natl Acad Sci USA. 1995;92:714–718. doi: 10.1073/pnas.92.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda H., Hori Y., Kobayashi M., Shimizu S. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Natl Acad Sci USA. 1996;93:10572–10577. doi: 10.1073/pnas.93.20.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laville J., Blumer C., Von Schroetter C., Gaia V., Defago G., Keel C., Haas D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh N., Parrat J., Willetts A. Characterisation and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. Journal of Molecular Catalysis B-Enzymatic. 1998;5:467–474. [Google Scholar]
- Legras J.L., Chuzel G., Arnaud A., Galzy P. Natural nitriles and their metabolism. World J Microbiol Biotechnol. 1990;6:83–108. doi: 10.1007/BF01200927. [DOI] [PubMed] [Google Scholar]
- Liang W.S. Drought stress increases both cyanogenesis and beta‐cyanoalanine synthase activity in tobacco. Plant Sci. 2003;165:1109–1115. [Google Scholar]
- Ludwig‐Muller J., Cohen J.D. Identification and quantification of three active auxins in different tissues of Tropaeolum majus. Physiol Plant. 2002;115:320–329. doi: 10.1034/j.1399-3054.2002.1150220.x. [DOI] [PubMed] [Google Scholar]
- Macadam A.M., Knowles C.J. Purification and properties of beta‐cyano‐L‐alanine synthase from the cyanide‐producing bacterium, Chromobacterium violaceum. Biochim Biophys Acta. 1984;786:123–132. [Google Scholar]
- McBride K.E., Kenny J.W., Stalker D.M. Metabolism of the herbicide Bromoxynil by Klebsiella pneumoniae subsp. Ozaenae. Appl Environ Microbiol. 1986;52:325–330. doi: 10.1128/aem.52.2.325-330.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew C.D., Nagasawa T., Kobayashi M., Yamada H. Nitrilase‐catalyzed production of nicotinic‐acid from 3‐cyanopyridine in Rhodococcus rhodochrous J1. Appl Environ Microbiol. 1988;54:1030–1032. doi: 10.1128/aem.54.4.1030-1032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers D.M., Ahmad S. Link between L‐3‐cyanoalanine synthase activity and differential cyanide sensitivity of insects. Biochim Biophys Acta. 1991;1075:195–197. doi: 10.1016/0304-4165(91)90252-c. [DOI] [PubMed] [Google Scholar]
- Mueller P., Egorova K., Vorgias C.E., Boutou E., Trauthwein H., Verseck S., Antranikian G. Cloning, overexpression, and characterization of a thermoactive nitrilase from the hyperthermophilic archaeon Pyrococcus abyssi. Protein Expr Purif. 2006;47:672–681. doi: 10.1016/j.pep.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole‐3‐acetaldoxime in camalexin synthesis. Plant Cell. 2007;19:2039–2052. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T., Hasegawa T., Yamashita E., Yamamoto M., Kumasaka T., Ueki T. Crystal structure of N‐carbamyl‐D‐amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure. 2000;8:729–737. doi: 10.1016/s0969-2126(00)00160-x. et al. [DOI] [PubMed] [Google Scholar]
- Normanly J., Bartel B. Redundancy as a way of life – IAA metabolism. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G.R., Bartel B. Arabidopsis mutants resistant to the auxin effects of indole‐3‐acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo C., Farnaud S., Tata R., Clemente A., Brown P.R. Support for a three‐dimensional structure predicting a Cys‐Glu‐Lys catalytic triad for Pseudomonas aeruginosa amidase comes from site‐directed mutagenesis and mutations altering substrate specificity. Biochemical J. 2002;365:731–738. doi: 10.1042/BJ20011714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly C., Turner P.D. The nitrilase family of CN hydrolysing enzymes – a comparative study. J Appl Microbiol. 2003;95:1161–1174. doi: 10.1046/j.1365-2672.2003.02123.x. [DOI] [PubMed] [Google Scholar]
- Osswald S., Wajant H., Effenberger F. Characterization and synthetic applications of recombinant AtNIT1 from Arabidopsis thaliana. Eur J Biochem. 2002;269:680–687. doi: 10.1046/j.0014-2956.2001.02702.x. [DOI] [PubMed] [Google Scholar]
- Pace H.C., Brenner C. The nitrilase superfamily: classification, structure and function. Genome Biol. 2001;2:REVIEWS0001. doi: 10.1186/gb-2001-2-1-reviews0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace H.C., Hodawadekar S.C., Draganescu A., Huang J., Bieganowski P., Pekarsky Y. Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers. Curr Biol. 2000;10:907–917. doi: 10.1016/s0960-9822(00)00621-7. et al. [DOI] [PubMed] [Google Scholar]
- Peiser G.D., Wang T.T., Hoffman N.E., Yang S.F., Liu H.W., Walsh C.T. Formation of cyanide from carbon‐1 of 1‐aminocyclopropane‐1‐carboxylic acid during its conversion to ethylene. Proc Natl Acad Sci USA. 1984;81:3059–3063. doi: 10.1073/pnas.81.10.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M. Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry. 2008;69:2655–2667. doi: 10.1016/j.phytochem.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Piotrowski M., Schonfelder S., Weiler E.W. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode beta‐cyano‐L‐alanine hydratase/nitrilase. J Biol Chem. 2001;276:2616–2621. doi: 10.1074/jbc.M007890200. [DOI] [PubMed] [Google Scholar]
- Podar M., Eads J.R., Richardson T.H. Evolution of a microbial nitrilase gene family: a comparative and environmental genomics study. BMC Evol Biol. 2005;5:ARTN 42. doi: 10.1186/1471-2148-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A., Frapolli M., Defago G., Moenne‐Loccoz Y. Phylogeny of HCN synthase‐encoding hcnBC genes in biocontrol fluorescent Pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol Plant Microbe Interact. 2003;16:525–535. doi: 10.1094/MPMI.2003.16.6.525. [DOI] [PubMed] [Google Scholar]
- Ressler C., Tatake J.G., Kaizer E., Putnam D.H. Neurotoxins in a vetch food: stability to cooking and removal of gamma‐glutamyl‐beta‐cyanoalanine and beta‐cyanoalanine and acute toxicity from common vetch (Vicia sativa) legumes. J Agri Food Chem. 1997;45:189–194. [Google Scholar]
- Robertson D.E., Chaplin J.A., DeSantis G., Podar M., Madden M., Chi E. Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol. 2004;70:2429–2436. doi: 10.1128/AEM.70.4.2429-2436.2004. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. AraC protein: a love‐hate relationship. Bioessays. 2003;25:274–282. doi: 10.1002/bies.10237. [DOI] [PubMed] [Google Scholar]
- Selmar D., Grocholewski S., Seigler D.S. Cyanogenic lipids – utilization during seedling development of Ungnadia speciosa. Plant Physiol. 1990;93:631–636. doi: 10.1104/pp.93.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell B.T., Berman M.N., Meyers P.R., Jandhyala D., Benedik M.J. The cyanide degrading nitrilase from Pseudomonas stutzeri AK61 is a two‐fold symmetric, 14‐subunit spiral. Structure. 2003;11:1413–1422. doi: 10.1016/j.str.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Singh R., Sharma R., Tewari N., Rawat D.S. Nitrilase and its application as a ‘green’ catalyst. Chem Biodivers. 2006;3:1279–1287. doi: 10.1002/cbdv.200690131. [DOI] [PubMed] [Google Scholar]
- Spaepen S., Vanderleyden J., Remans R. Indole‐3‐acetic acid in microbial and microorganism‐plant signalling. FEMS Microbiol Rev. 2007;31:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Stalker D.M., Malyj L.D., McBride K.E. Purification and properties of a nitrilase specific for the herbicide Bromoxynil and corresponding nucleotide‐sequence analysis of the bxn gene. J Biol Chem. 1988a;263:6310–6314. [PubMed] [Google Scholar]
- Stalker D.M., McBride K.E., Malyj L.D. Herbicide resistance in transgenic plants expressing a bacterial detoxification gene. Science. 1988b;242:419–423. doi: 10.1126/science.242.4877.419. [DOI] [PubMed] [Google Scholar]
- Thimann K.V., Mahadevan S. Enzymatic hydrolysis of indoleacetonitrile. Nature. 1958;181:1466–1467. doi: 10.1038/1811466a0. [DOI] [PubMed] [Google Scholar]
- Vannesland B., Conn E.E., Knowles C.J., Westley J., Wissing F. Academic Press; 1981. [Google Scholar]
- Vetter J. Plant cyanogenic glycosides. Toxicon. 2000;38:11–36. doi: 10.1016/s0041-0101(99)00128-2. [DOI] [PubMed] [Google Scholar]
- Vorwerk S., Biernacki S., Hillebrand H., Janzik I., Muller A., Weiler E.W., Piotrowski M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3‐gene cluster. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- Wang P., VanEtten H.D. Cloning and properties of a cyanide hydratase gene from the phytopathogenic fungus Gloeocercospora sorghi. Biochem Biophys Res Commun. 1992;187:1048–1054. doi: 10.1016/0006-291x(92)91303-8. [DOI] [PubMed] [Google Scholar]
- Wang P., Matthews D.E., VanEtten H.D. Purification and characterization of cyanide hydratase from the phytopathogenic fungus Gloeocercospora sorghi. Arch Biochem Biophys. 1992;298:569–575. doi: 10.1016/0003-9861(92)90451-2. [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K., Adachi K., Nishijima M., Takadera T., Tamaki S., Harada K. Beta‐cyanoalanine production by marine bacteria on cyanide‐free medium and its specific inhibitory activity toward cyanobacteria. Appl Environ Microbiol. 2000;66:718–722. doi: 10.1128/aem.66.2.718-722.2000. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


