Summary
Erwinia amylovora causes fire blight on several plant species such as apple and pear, which produce diverse phytoalexins as defence mechanisms. An evolutionary successful pathogen thus must develop resistance mechanisms towards these toxic compounds. The E. amylovora outer membrane protein, TolC, might mediate phytoalexin resistance through its interaction with the multidrug efflux pump, AcrAB. To prove this, a tolC mutant and an acrB/tolC double mutant were constructed. The minimal inhibitory concentrations of diverse antimicrobials and phytoalexins were determined for these mutants and compared with that of a previously generated acrB mutant. The tolC and arcB/tolC mutants were considerably more susceptible than the wild type but showed similar levels as the acrB mutant. The results clearly indicated that neither TolC nor AcrAB significantly interacted with other transport systems during the efflux of the tested toxic compounds. Survival and virulence assays on inoculated apple plants showed that pathogenicity and the ability of E. amylovora to colonize plant tissue were equally impaired by mutations of tolC and acrB/tolC. Our results allowed the conclusion that TolC plays an important role as a virulence and fitness factor of E. amylovora by mediating resistance towards phytoalexins through its exclusive interaction with AcrAB.
Introduction
Plants produce an array of diverse secondary metabolites that have antimicrobial activities including preformed, so‐called phytanticipants and phytoalexins, which are synthesized in response to pathogen attack. For this purpose, flavonoids, isoprenoids and alkaloids comprise three major classes of secondary metabolites synthesized by higher plants. These compounds are essential for diverse plant defence mechanisms and are furthermore implicated in a broad range of physiological processes (Osbourn, 1996; Hammerschmidt, 1999).
Evolutionary successful pathogens are able to circumvent the toxic effect of antimicrobial compounds by enzymatic inactivation, alteration of the cellular target, reduced uptake of the toxic compounds, or active efflux (Walsh, 2000; VanEtten et al., 2001). In this context, multidrug efflux (MDE) has been suggested for a broad range of structurally unrelated compounds. So far, most emphasis has been devoted to the investigation of MDE phenomena in human pathogens. However, genome analyses of various microorganisms revealed the ubiquitous occurrence of genes encoding MDE pumps (Paulsen et al., 2001). Five well‐characterized families of MDE pump exist: the ATP‐binding cassette (ABC) family, the major facilitator superfamily (MFS), the resistance‐nodulation‐cell division (RND) superfamily, the small multidrug resistance (SMR) family, and the multidrug and toxic compound extrusion (MATE) family (Brown et al., 1999). Efflux by proteins of the MFS, RND, SMR and MATE families is driven by proton (and sodium) motive force while ATP hydrolysis drives efflux in ABC transporters (Moreira et al., 2004; Poole, 2005; Piddock, 2006; Alekshun and Levy, 2007). In Gram‐negative bacteria, RND‐type transporters reside in the inner membrane and function in concert with two types of proteins: the accessory membrane fusion proteins, which are located in the periplasmic space, and outer membrane protein channels (Piddock, 2006; Alekshun and Levy, 2007). These tripartite transport systems enable drug efflux across both membranes (Zgurskaya and Nikaido, 2000).
For plant pathogens, protection against toxic compounds by MDE transporters has particularly been studied in fungal organisms. Mutation of genes encoding ABC transporters of Gibberella pulicaris and Botrytis cinerea resulted in decreased tolerance to phytoalexins and thus led to impaired virulence (Del Sorbo et al., 2000; Schoonbeek et al., 2001; Fleißner et al., 2002). In phytopathogenic and other plant‐associated bacteria so far only few MDE pumps belonging to the RND superfamily have been associated with respective interactions, for example in Agrobacterium tumefaciens (Palumbo et al., 1998; Peng and Nester, 2001), in the nitrogen‐fixing symbiont, Rhizobium etli (Gonzáles‐Pasayo and Martinez‐Romera, 2000), in Pseudomonas syringae (Stoitsova et al., 2008), in Erwinia chrysanthemi (Maggiorani Valecillos et al., 2006) and in Bradyrhizobium japonicum (Krummenacher and Narberhaus, 2000).
One of the economically most important bacterioses represents fire blight on rosaceous plants, which is particularly destructive to apple and pear trees (Eastgate, 2000). The causal agent of this disease is Erwinia amylovora, a member of the Enterobacteriaceae. The initial symptom of fire blight is water soaking followed by wilting and rapid necrosis leaving infected tissue with a scorched, blackened appearance. The spread of the pathogen through the host tissue and xylem into all parts of a plant can lead to loss of entire trees (Vanneste and Eden‐Green, 2000). The commercial implications of fire blight outbreaks are aggravated by the limited effectiveness of current control strategies (Psallidas and Tsiantos, 2000). Thus, a better understanding of the molecular interactions between plant and pathogen can lead to improved design of antibacterial compounds and genetically engineered plants.
In E. amylovora, two essential pathogenicity factors have been identified: the hrp/dsp genes encoding a type III protein secretion system and disease‐specific effector proteins (Steinberger and Beer, 1988; Barny et al., 1990; Vanneste et al., 1990) and the extracellular polysaccharide amylovoran (Bellemann and Geider, 1992). Further studies revealed a number of additional factors that are not directly involved in the induction of disease symptoms but allow the pathogen to survive in planta (Metzger et al., 1994; Aldridge et al., 1997; Bogs and Geider, 2000).
A resistance mechanism to bypass the antimicrobial effects of secondary metabolites of the host defence has been studied for E. amylovora by Burse and colleagues (2004). It was shown that the E. amylovora RND‐family efflux pump AcrAB was essential for resistance against plant‐derived antimicrobial toxins, particularly against the apple phytoalexin, phloretin, and for a successful colonization of plant tissue. Based on that finding, our present study focused on the involvement of the outer membrane protein, TolC, in MDE, its specific interaction with AcrAB, and its involvement in virulence and in planta survival of E. amylovora. TolC homologues have been shown to be involved in resistance against antimicrobial compounds in mammalian pathogens such as Escherichia coli (Fralick and Burns‐Keliher, 1994; Sharff et al., 2001; Bavro et al., 2008), Salmonella enterica (Baucheron et al., 2004; 2005) and Borrelia bugdorferi (Bunikis et al., 2008). Furthermore, TolC has been studied to variable extent in the nitrogen‐fixing organism, Sinorhizobium meliloti (Cosme et al., 2008) and in two plant pathogenic bacteria, Xylella fastidiosa (Reddy et al., 2007) and Dickeya dadantii (formerly known as Erwinia chrysanthemi) (Barabote et al., 2003). Herein, we report on the identification of the gene encoding for TolC in E. amylovora, the generation and resistance analyses of single and double mutants for tolC and acrB/tolC, and the determination of virulence and in planta survival of the respective mutants.
Results
In vitro growth of wild type and mutants of E. amylovora Ea1189
In order to test whether or not individual or combined effects of mutations in tolC and acrB influenced the overall fitness of respective E. amylovora mutants, in vitro growth experiments were conducted. Bacteria were cultured in complex Luria–Bertani (LB) and minimal asparagine minimal medium 2 (AMM2) medium at 28°C. The OD600 was monitored continuously until cultures had entered the late stationary phase (Fig. 1). Growth of wild‐type cultures was generally faster than that of any mutant tested with doubling times of ∼60 min for the wild type; ∼90 min for the acrB mutant; ∼105 min for the tolC mutant and ∼120 min for the acrB/tolC double mutant. Growth of the genetically complemented mutants was similar to that of the wild type proving that growth delays were directly due to lack of tolC and acrB/tolC. Wild type and complemented mutants reached the stationary phase within 7 h with an OD600 of ∼1.5. In contrast, stationary phases were reached at OD600 of ∼1, ∼0.9 and ∼0.8 for the mutants defective in acrB, tolC and acrB/tolC respectively, demonstrating that both, acrB and tolC, contributed to in vitro fitness and that simultaneous mutation of both genes resulted in additive effects. In both media individual growth rates for respective mutants were comparable suggesting that the composition of the growth medium had no effect on particular mutants' phenotypes.
Figure 1.
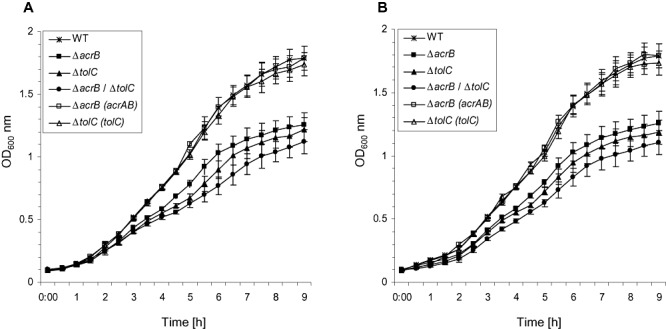
In vitro growth of E. amylovora wild type and mutants in LB (A) and AMM2 (B) medium at 28°C as determined by measurement of the OD600. Data represent the means of three independent cultures ± standard deviation.
Susceptibility of tolC mutants towards phytoalexins and other antimicrobial compounds
Determination of minimal inhibitory concentrations (MICs) of various antibiotics and phytoalexins was used to examine the susceptibility of tolC mutants as compared with the wild‐type strain and the previously studied acrB mutant (Burse et al., 2004) in complex and minimal medium (Tables 1 and 2). As potential substrates of TolC and/or AcrAB different isoflavonoids were analysed, which are known phytoalexins in Rosaceae plants (Harborne, 1999; Grayer and Kokubun, 2001) and which were previously identified and isolated from apple tissue (Treutter, 2001). Additionally, bile salt as a reported substrate for AcrAB in E. coli (Thanassi et al., 1997) and several other antimicrobials were tested in order to determine substrate specificity profiles. In general, differences in MIC values were only considered significant if they were at least fourfold. This cut‐off is consistent with previous publications by others (Stermitz et al., 2000; Andrews, 2001), due to the sensitivity of the used method, and was meant to avoid misinterpretation of data.
Table 1.
Susceptibility of Erwinia amylovora strains to different compounds in medium MHB.
| Compoundsa | MICb (µg ml−1) | |||||
|---|---|---|---|---|---|---|
| Ea1189 | ΔacrB | ΔtolC | ΔacrB/ΔtolC | ΔacrB (acrAB) | ΔtolC (tolC) | |
| Phloretin | 1000 | 125 | 125 | 125 | > 1000 | > 1000 |
| Naringenin | 1000 | 62.5 | 125 | 125 | 1000 | 1000 |
| (+)‐Catechin | > 1000 | 125 | 250 | 250 | > 1000 | > 1000 |
| Quercetin | 1000 | 62.5 | 62.5 | 125 | > 1000 | 1000 |
| Berberine | 1000 | 31.2 | 31.2 | 31.2 | 1000 | 1000 |
| Bile salt | > 1000 | 125 | 125 | 125 | > 1000 | > 1000 |
| Acriflavine | 15.6 | 1.56 | 3.12 | 3.12 | 31.2 | 31.2 |
| Ampicillin | 62.5 | 6.25 | 12.5 | 6.25 | 62.5 | 62.5 |
| Novobiocin | 62.5 | 1.56 | 1.56 | 1.56 | 62.5 | 62.5 |
| Cefoperazone | 12.5 | 3.12 | 3.12 | 3.12 | 12.5 | 12.5 |
| Mitomycin | 6.25 | 0.31 | 0.62 | 0.62 | 12.5 | 12.5 |
| Tetracycline | 6.25 | 0.62 | 0.62 | 0.62 | 12.5 | 6.25 |
| Nalidixic acid | 1.25 | 0.12 | 0.12 | 0.12 | 1.25 | 1.25 |
| Norfloxacin | 0.62 | 0.031 | 0.062 | 0.062 | 0.62 | 0.62 |
| Ciprofloxacin | 0.62 | 0.062 | 0.062 | 0.062 | 0.62 | 0.62 |
| SDS | > 1000 | 100 | 100 | 100 | > 1000 | 1000 |
| Ethidium bromide | 31.2 | 3.12 | 3.12 | 3.12 | 62.5 | 62.5 |
| Crystal violet | 3.12 | 0.62 | 0.62 | 0.62 | 3.12 | 3.12 |
Reserpine and salicylic acid were also tested (data not shown); however, the mutants and the parent strain showed similar MIC values.
Minimal inhibitory concentration determination by the dilution assay was repeated at least three times in each case thereby confirming consistencies of MIC values.
Table 2.
Susceptibility of E. amylovora strains to different compounds in medium AMM2.
| Compoundsa | MICb (µg ml−1) | |||||
|---|---|---|---|---|---|---|
| Ea1189 | ΔacrB | ΔtolC | ΔacrB/ΔtolC | ΔacrB (acrAB) | ΔtolC (tolC) | |
| Phloretin | 1000 | 31.2 | 15.6 | 15.6 | > 1000 | > 1000 |
| Naringenin | 1000 | 15.6 | 31.2 | 31.2 | 1000 | > 1000 |
| (+)‐Catechin | > 1000 | 31.2 | 31.2 | 31.2 | > 1000 | > 1000 |
| Quercetin | 1000 | 31.2 | 62.5 | 62.5 | > 1000 | > 1000 |
| Berberine | 1000 | 15.6 | 31.2 | 31.2 | 1000 | 1000 |
| Bile salt | > 1000 | 31.2 | 62.5 | 31.2 | > 1000 | > 1000 |
| Acriflavine | 15.6 | 3.12 | 3.12 | 3.12 | 31.2 | 31.2 |
| Ampicillin | 31.2 | 1.56 | 0.75 | 0.75 | 31.2 | 62.5 |
| Novobiocin | 125 | 1.56 | 3.12 | 3.12 | 250 | 250 |
| Cefoperazone | 6.25 | 1.56 | 1.56 | 1.56 | 12.5 | 12.5 |
| Mitomycin | 12.5 | 0.31 | 0.31 | 0.31 | 12.5 | 12.5 |
| Tetracycline | 6.25 | 0.62 | 0.31 | 0.62 | 12.5 | 6.25 |
| Nalidixic acid | 2.5 | 0.25 | 0.25 | 0.25 | 2.5 | 2.5 |
| Norfloxacin | 0.31 | 0.031 | 0.062 | 0.062 | 0.62 | 0.62 |
| Ciprofloxacin | 0.31 | 0.031 | 0.015 | 0.015 | 0.62 | 0.62 |
| SDS | 250 | 3.12 | 3.12 | 6.25 | 250 | 500 |
| Ethidium bromide | 62.5 | 3.12 | 1.56 | 1.56 | 62.5 | 62.5 |
| Crystal violet | 0.75 | 0.15 | 0.15 | 0.31 | 3.12 | 1.56 |
Reserpine and salicylic acid were also tested (data not shown); however, the mutants and the parent strain showed similar MIC values.
Minimal inhibitory concentration determination by the dilution assay was repeated at least three times in each case thereby confirming consistencies of MIC values.
In the tolC mutant and the acrB/tolC double mutant MICs for the dihydrochalcone phloretin, the flavonol quercetin, the flavanone naringenin, the monomeric flavanol (+)‐catechin, the isoquinoline alkaloid berberine, and bile salt were similar to that of the acrB mutant in both media and were decreased 8‐fold, 16‐fold, 16‐fold, 8‐fold, 32‐fold and 8‐fold respectively, as compared with the MICs in the wild type. There were no significant (i.e. more than fourfold) differences between the mutants with single tolC knockout or with simultaneous mutations in both, tolC and acrB, suggesting that both gene products were essential for the efflux of the tested compounds and that neither protein significantly interacted with additional third components to accomplish the efflux.
Complementation of the two single mutants with plasmids pNK7 (for the tolC mutant) and pNK8 (for the acrB mutant), respectively, fully restored tolerance towards the tested isoflavonoids thereby confirming that these hydrophobic compounds were substrates of the AcrAB–TolC efflux system in E. amylovora. The results furthermore suggested that lack of TolC resulted in increased susceptibility possibly due to accumulation of antimicrobials in the bacterial cells.
The three tested mutants were susceptible to the polyketides, tetracycline with a fivefold reduction of MIC values, and to hydrophobic quinolon derivatives such as nalidixic acid, norfloxacin and ciprofloxacin, with a 10‐fold reduction of the MICs. In contrast to the wild type, the mutants were sensitive to the beta‐lactam antibiotics, ampicillin and cefoperazone. In addition, the mutants showed increased susceptibility with about 10‐fold reduction of the MIC to the aminocoumarin novobiocin, the aziridine mitomycin, the antiseptic acriflavine, the detergent SDS, and the dyes, crystal violet and ethidium bromide. Again complementation of either single mutant with the respective genetic locus restored wild‐type resistance levels (Tables 1 and 2). Only the MICs of reserpine and salicylic acid did not differ from those determined for the wild type (data not shown) suggesting that neither compound acted as a substrate for AcrAB–TolC in E. amylovora. Just as observed for the in vitro growth, there was no significant (i.e. at least fourfold difference in MIC values) influence of the used growth medium indicating that activity of AcrAB–TolC was not significantly triggered by the physiological state of the cells.
Contribution of TolC to virulence and in planta survival of E. amylovora
First, any potential role for TolC on virulence of the fire blight pathogen was tested by inoculation of apple rootstock MM106 plants with wild type, the tolC mutant, and the acrB/tolC double mutant. The previously reported acrB mutant (Burse et al., 2004) was used as a control. Shoot tips of the plants were inoculated by the so‐called prick technique (May et al., 1997), which mimics the natural infection process and allows for defined numbers of bacterial cells to be inoculated. Observable fire blight symptoms were the typical shepherd's crook‐like bending of the shoot tip after 1 week post inoculation as well as ooze formation and necrosis after 3 weeks post inoculation. Such typical symptoms were induced by the wild type of E. amylovora Ea1189 (Fig. 2). In contrast to this finding and comparable to the previously published results for the acrB mutant of Ea1189, neither the tolC mutant nor the acrB/tolC double mutant exhibited any disease symptoms (Fig. 2). As expected, genetic complementation rendered the mutants virulent with typical symptoms produced. These result suggested that TolC plays an important role in virulence of the fire blight pathogen. As no significant differences in the lack of symptom developments were observed between the tolC, acrB and the acrB/tolC mutants it could be concluded that both proteins, i.e. TolC and AcrB, were fully required for symptom formation.
Figure 2.

Pathogenicity assay on apple plant. Shoot tips were inoculated with 5 µl of 1 × 107 cfu ml−1 bacterial suspensions. Disease symptoms such as the typical ‘shepherd's crook’ formation and wilting developed on plants inoculated with E. amylovora wild type after 1 week of inoculation.
Next, bacterial colonization of the host plant 24 h post inoculation was studied postulating that TolC and AcrB might be likewise important for this initial step of the disease development. Erwinia amylovora Ea1189 and its mutants were inoculated at different cell numbers (103–107 cfu/inoculation sites) and bacterial multiplication was monitored by re‐isolating bacterial cells from infected plant tissue after 24 h (Table 3). In contrast to the wild type, inoculation of all three mutants in low cell numbers (103 and 104 cfu/shoot) yielded in no re‐isolatable colony forming units suggesting that the mutants did not survive. Inoculation of the mutants with cell numbers > 105 cfu/shoot were necessary for re‐isolation of mutant cells. However, the populations decreased approximately 1000‐fold for both single mutants and for the tolC–acrB double mutant indicating that mutants with knockout in tolC were clearly impaired in multiplication in apple tissue. Noteworthy, mutation of tolC in the genomic background of the acrB mutant did not lead to any additive effects in terms of this phenotype. In contrast, the wild type reached an approximately 10‐fold increase in population density at 24 h post inoculation regardless of the initially cell numbers used for inoculation (103–107 cfu/shoot). As observed above, complementation of the mutants with plasmids, pNK7 (for the tolC mutant) and pNK8 (for the acrB mutant), restored the wild‐type phenotype.
Table 3.
Survival of E. amylovora strains in apple rootstock MM106.
| Inoculumsa cfu/shoot | Re‐isolated bacteria cellsb (cfu/shoot) | |||||
|---|---|---|---|---|---|---|
| Ea1189 | ΔacrB | ΔtolC | ΔacrB/ΔtolC | ΔacrB (acrAB) | ΔtolC (tolC) | |
| 1.2 × 103 | 4.4 × 103 ± 2 × 102 | 0 | 0 | 0 | 4.1 × 103 ± 2 × 102 | 4.3 × 103 ± 2 × 102 |
| 1.1 × 104 | 1.6 × 105 ± 2 × 103 | 0 | 0 | 0 | 1.5 × 105 ± 2 × 103 | 1.5 × 105 ± 2 × 103 |
| 1.0 × 105 | 5.4 × 105 ± 1 × 103 | 1.2 × 102 ± 9 × 101 | 1.5 × 102 ± 6 × 101 | 1.9 × 102 ± 6 × 101 | 3.4 × 105 ± 1 × 103 | 3.4 × 105 ± 1 × 103 |
| 1.2 × 106 | 7.1 × 107 ± 1 × 104 | 6.4 × 103 ± 2 × 102 | 8.1 × 103 ± 2 × 102 | 9.9 × 103 ± 2 × 102 | 7.6 × 107 ± 1 × 104 | 7.5 × 107 ± 1 × 104 |
| 1.0 × 107 | 8.8 × 108 ± 2 × 105 | 1.7 × 104 ± 3 × 103 | 1.7 × 104 ± 3 × 103 | 1.8 × 104 ± 3 × 103 | 8.4 × 108 ± 1 × 105 | 8.5 × 108 ± 1 × 105 |
Bacteria were inoculated by prick technique in the shoot tip of plants.
Establishment of a population of E. amylovora strains was determined 24 h after inoculation. Data represent the average cfu numbers from 15 individual plants.
Discussion
Herein the role of the outer membrane protein, TolC, for antibiotics resistance, virulence, and in planta survival of the fire blight pathogen, E. amylovora, was investigated demonstrating its significance for each of these bacterial processes. Burse and colleagues (2004) had previously shown that mutation of acrB caused a clear reduction in resistance towards different antibiotics as well as towards plant isoflavonoids, isoprenoids and alkaloids. Furthermore, mutation of acrB led to an avirulent phenotype and caused a significant reduction in bacterial in planta survival (Burse et al., 2004).
The AcrAB–TolC tripartite system of E. coli mediates resistance towards a wide variety of lipophilic and amphiphilic compounds including bile salt, detergents, dyes and antimicrobial agents and is one of the best‐characterized bacterial RND‐type MDE pumps (Zgurskaya and Nikaido, 2000). AcrAB–TolC complexes were also described for Salmonella enterica (Baucheron et al., 2004; 2005) and Enterobacter cloacae (Perez et al., 2007). However, in E. coli, TolC can also interact with several other transport systems besides AcrAB (Sharff et al., 2001; Ramos et al., 2002; Koronakis et al., 2004) prompting the question whether this is also the case for the enterobacterial fire blight pathogen. In Gram‐negative bacteria other than E. coli, individual TolC‐like outer membrane channels are often unique for a given RND‐type transporter and their genes are consequently co‐expressed in the same gene cluster as the RND‐type pumps, such as in the case of the oprJMN cluster in Pseudomonas aeruginosa (Li et al., 1995; Koehler et al., 1997) or for the outer membrane channel, MtrE, in Neisseria gonorrhoeae (Hagmann et al., 1995). In contrast, in Klebsiella pneumonia AcrAB homologues specifically interact with the outer membrane protein, KocC, but their respective genes are not co‐transcribed (Li et al., 2008). At least in terms of genomic localization of acrAB and tolC, E. amylovora seems to group along with E. coli and some of the other enterobacterial representatives mentioned above. The bicistronic acrAB operon (nucleotide numbers 1120951–1125307) is not co‐transcribed with tolC, which is located at a very distant position (nucleotide numbers 3269783–3271233) within the E. amylovora genome (http://www.sanger.ac.uk/projects/E.amylovora).
The initial finding that the in vitro growth of the acrB/tolC double mutant was more impaired than that of the single mutants seemed to support the idea that AcrAB and TolC of E. amylovora may also interact with other proteins. However, simultaneous mutation of acrB and tolC did not yield in any significant additive or synergistic effects in terms of resistance towards phytoalexins or any other antimicrobial compound suggesting that at least for these functions AcrAB and TolC, respectively, might not recruit other transport partners. For proper in vitro growth in both complex and minimal medium, there might be a particular yet‐to‐be identified additional function, which does not require an exclusive AcrAB–TolC interaction.
In comparison with the wild type, the single tolC mutant and the acrB/tolC double mutant showed significant but similar reductions in resistance towards hydrophobic and amphiphilic antibiotics, dyes and detergents. A wide range of constitutively synthesized isoflavonoids has been described for members of the Rosaceae plant family (Harborne, 1999; Grayer and Kokubun, 2001). Notably, Mayr and colleagues (1995) found that the glycosides, phloretin, phloridzin and quercetin represented the quantitatively most abundant compounds in apple leaves of the cultivar ‘Golden Delicious’. In the current study, the impact of various antibacterial compounds previously described for apple plants (Treutter, 2001) on E. amylovora wild type and its mutants were examined. Phloretin, quercetin, (+)‐catechin and naringenin inhibited growth of the tolC mutant to very similar extent as the growth of the acrB/tolC double mutant and the acrB mutant. Consequently, it can be assumed that these compounds are likely to be exported by the AcrAB–TolC complex but not by AcrAB and TolC respectively, recruiting other components in E. amylovora. A total of six different MDE systems are present in the genome of E. amylovora. A blast search with the genome sequence of E. amylovora Ea237 using the amino acid sequence of AcrB from E. coli K12 strain DH10B (accession number YP‐001729367) as query identified six homologous sequences in the genome of Ea237. At the amino acid sequence level, the respective predicted E. amylovora proteins showed the following identities (similarities given in brackets): AcrB with 83% (92%), AcrD with 78% (89%), MdtB with 81% (90%), MdtC with 73% (86%), and two MdtB‐ and MdtC‐like proteins with 63% (79%) and 56% (73%) respectively (http://www.sanger.ac.uk/projects/E.amylovora).
The increased sensitivity towards diverse antibiotics was comparable to that of the previously described acrB mutant of E. amylovora and was corresponding to the previously reported broad substrate spectrum of AcrAB–TolC from E. coli and other homologous MDE pumps (Poole, 2000; Nishino and Yamaguchi, 2001; Sulavik et al., 2001). However, neither of the tested E. amylovora mutants showed increased sensitivity towards the indole alkaloid, reserpine, or the plant hormone, salicylic acid, which consequently might not belong to the substrate spectrum of AcrAB–TolC of E. amylovora.
In plant‐associated bacteria, TolC was predicted to function in protection against phytoalexins, whose syntheses represent important plant defence mechanisms. For the phytopathogen, Xylella fastidiosa, it was shown that a tolC knockout impaired virulence and increased sensitivity to the grapevine phytoalexin, resveratrol (Reddy et al., 2007). Likewise, in the phytopathogen, Dickeya dadantii (E. chrysanthemi), the importance of tolC for virulence and resistance towards plant‐derived chemicals was shown (Barabote et al., 2003). Cosme and colleagues (2008) demonstrated that mutation of tolC in the nitrogen‐fixing plant symbiont, Sinorhizobium meliloti, strongly affected the resistance against antimicrobial agents and was important for symbiotic functions. Herein, we demonstrated that individual or simultaneous disruption of single components of a MDE pump in E. amylovora resulted in similarly reduced symptom formation and in significantly reduced in planta survival shortly after infection. These findings indicated that the detoxifying activity might be linked to AcrAB–TolC as a complex. Thus, the infected plants were obviously able to block the infection and kill bacterial cells if those lacked the gene products of acrB or tolC. The results were substantiated by genetic complementation of the mutants to wild‐type levels with recombinant acrAB and tolC. It is tempting to speculate that the AcrAB–TolC complex might enable bacterial cells to survive at and outgrow from the local infection site, in which antimicrobial substances of plant defence accumulate during early stages of infection.
Erwinia amylovora wild type and its mutants were additionally inoculated on tobacco leaves to monitor the development of the hypersensitive response (data not shown). Regardless of their reduced virulence, the acrB and tolC mutants elicited typical hypersensitive responses during the incompatible reaction with tobacco plants demonstrating that the hrp type III secretion system (Nissinen et al., 2007) was not influenced by the MDE system. For E. chrysanthemi, the TolC analogue, PrtF, was shown to be required for the secretion of a protease important for virulence (Létofféet al., 1990). Our future studies will therefore focus on the in‐depth analysis of potential function(s) of TolC in protein secretion as well as on the genetic exchange of individual AcrAB–TolC components between E. amylovora and E. coli in order to determine ecological niche‐mediated specializations of either system. Additionally, future work will be directed towards the transcriptional regulation of the tolC gene of E. amylovora in order to determine whether its expression is substrate‐dependent and whether expression of acrAB (Burse et al., 2004) and that of tolC is co‐regulated by any potential substrate(s).
Experimental procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study were listed in Tables 4 and 5. Erwinia amylovora strains were cultured at 28°C on LB medium or AMM2. Asparagine minimal medium 2 has the following composition (per litre of deionized water): fructose, 10 g; l‐asparagine, 4 g; Na2HPO4 × 7 H2O, 12.8 g; K2HPO4, 3 g; NaCl, 3 g; MgSO4 × 7 H2O, 0.2 g; nicotinic acid, 0.25 g; thiamine, 200 µg. Escherichia coli DH5α was used as cloning host. Escherichia coli cells were routinely maintained at 37°C in LB medium. Cultures were supplemented with 50 µg ml−1 ampicillin (Ap), 25 µg ml−1 chloramphenicol (Cm), gentamicin (Gm) at 2 µg ml−1 and 25 µg ml−1 kanamycin (Km) when necessary. Bacterial growth was monitored using a spectrophotometer (OD at 600 nm).
Table 4.
E. coli and E. amylovora strains used in this study.
| Strain | Relevant characteristics | Reverence or source |
|---|---|---|
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi‐1 relA1 | Sambrook and Russel (2001) |
| S17‐1 λ‐pir | λ‐pir lysogen of S17‐1 | Wilson et al. (1995) |
| E. amylovora | ||
| 1189 | Wild type | GSPBa |
| 1189‐3 | Kmr, acrB mutant carrying Kmr cassette in the acrB gene | Burse et al. (2004) |
| 1189‐25 | Gmr, tolC mutant carrying GFP‐Gmr cassette in the tolC gene | This study |
| 1189‐3‐3 | Kmr, Gmr, acrB/tolC mutant carrying GFP‐Gmr cassette in the tolC gene and Kmr cassette in acrB gene | This study |
| 1189‐25‐1 | Gmr, Cmr, complemented tolC mutant carrying pNK7 | This study |
| 1189‐3‐1 | Kmr, Cmr, complemented acrB mutant carrying pNK8 | This study |
GSPB, Göttinger Sammlung phytopathogener Bakterien, Göttingen, Germany.
Table 5.
Plasmids used in this study.
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| pGEM‐T Easy | Apr, ColE1 origin | Promega |
| pNK1 | Apr, contains a 984 bp XhoI PCR fragment that located upstream of tolC. | This study |
| pNK2 | Apr, contains a 912 bp XhoI/BamHI PCR fragment located downstream of tolC | This study |
| pPS858 | Source of the Gm‐GFP cassette | Hoang et al. (1998) |
| pNK3 | Apr, Gmr, 1843 bp Gm‐GFP cassette from pPS858 cloned into BamHI‐linearized pNK2 | This study |
| pNK4 | Apr, Gmr, 2755 bp SpeI‐XhoI GFPGm‐downstream fragment from pNK3 cloned into SpeI‐XhoI‐linearized pNK1 | This study |
| pCAM‐MCS | Apr, pCAM140‐derivative without mini‐Tn5, contains the MCS of pBluescript II SK (+) | Burse et al. (2004) |
| pNK5 | Apr, Gmr, 3739 bp ScaI‐EcoRI upstream‐GFPGm‐downstream fragment cloned into EcoRI‐linearized pCAM‐MCS | This study |
| pNK6 | Apr, contains a 2630 bp PCR fragment carrying the tolC gene, upstream and downstream of the gene | This study |
| pNK7 | Cmr, contains a 2.6 kb fragment carrying tolC of E. amylovora cloned in pBBR1MCS by SpeI and SacII in opposite direction of lacZ | This study |
| pNK8 | Cmr, contains a 4.9 kb fragment carrying acrAB of E. amylovora cloned in pBBR1MCS by SpeI and SacI in opposite direction of lacZ | This study |
Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Cm, chloramphenicol; GFP, green fluorescent protein.
Standard genetic procedures
Restriction digestions, agarose gel electrophoresis, electroporation, PCR, ligation of DNA fragments, plasmid DNA isolation, and other routine molecular methods were performed by standard techniques (Sambrook and Russel, 2001). Purification of PCR products and DNA fragments from agarose gels were performed using Nucleospin columns (Macherey‐Nagel, Dueren, Germany).
Generation of tolC in E. amylovora
Erwinia amylovora tolC‐deficient mutants were generated by marker exchange mutagenesis as follows. Two fragments flanking the tolC gene were PCR‐amplified from strain Ea1189 using the oligonucleotide primer pairs Ea‐tolC‐FwdI, Ea‐tolC‐RevI and Ea‐tolC‐FwdII, Ea‐tolC‐RevII respectively (Table 6). PCR products were cloned into pGEM‐T Easy (Promega, Mannheim, Germany) yielding plasmids pNK1 (3999 bp) and pNK2 (3927 bp). A 1843 bp BamHI fragment containing a green fluorescent protein–gentamicin‐resistance cassette (GFP‐Gmr) was derived from pPS858 (Hoang et al., 1998) and ligated into BamHI‐digested pNK2, yielding the 5767 bp plasmid pNK3. A 984 bp XhoI‐SpeI fragment was derived from pNK1 and ligated into XhoI‐SpeI‐digested pNK3, yielding the 6751 bp plasmid pNK4. A 3739 bp EcoRI fragment was obtained from pNK4 and ligated into EcoRI‐digested suicide plasmid, pCAM‐MCS (Burse et al., 2004), yielding the final tolC replacement plasmid pNK5 (7439 bp). Escherichia coli S17‐1 λ‐pir was used as host for the suicide plasmid. The plasmid was transferred via electroporation to electro‐competent E. amylovora wild type and its acrB mutant, which subsequently were grown at 28°C for 1 h in SOC broth and plated on LB containing gentamicin. Putative mutants were screened for homologous recombination events by checking for antibiotics resistance and GFP fluorescence. To confirm the altered genotypes, PCR amplification of tolC using primers Ea‐tolC‐FwdI and Ea‐tolC‐RevII (Table 6) was carried out, yielding a 3739 bp fragment for the two mutants, as opposed to the presence of wild‐type tolC, forming a 2630 bp PCR fragment in samples of wild type and acrB mutant.
Table 6.
Oligonucleotide primers used in this study.
| Primer | Nucleotide sequence (5′‐3′)a |
|---|---|
| Ea‐tolC‐FwdI | GCTCACCACATGCACAAG |
| Ea‐tolC‐RevI | CTCGAGAGTGTTGCTGTTAGAGCCAC |
| Ea‐tolC‐FwdII | GCCAAATTCAGCCACGCA |
| Ea‐tolC‐RevII | CTCGAGGATCCGCAAGTGAACAGCTCGAAG |
| Ea‐acr‐Com‐Fwd | CGAGAGCTCCGCCAGTGACGTATTAGC |
| Ea‐acr‐Com‐Rev | GATACTAGTCGGTATAGTAAACGTGCG |
Restriction sites incorporated into primers are underlined: CTCGAG, XhoI; GGATCC, BamHI; GAGCTC, SacI; ACTAGT, SpeI.
Complementation of mutants
Genetic complementation of tolC mutants was performed as follows: a 2630 bp fragment including the tolC gene and its upstream and downstream regions was PCR‐amplified from strain Ea1189 using the oligonucleotide primers, Ea‐tolC‐FwdI and Ea‐tolC‐RevII (Table 6), cloned into pGEM‐T Easy yielding the 5645 bp plasmid pNK6. A 2630 bp SpeI‐SacII fragment was derived from pNK6 and ligated into SpeI‐SacII‐digested pBBR1MCS (Kovach et al., 1995) yielding the final tolC complementation plasmid pNK7 (7337 bp). Complementation of the acrB mutant was performed as follows: a 4917 bp fragment including acrAB and its upstream and downstream region was PCR‐amplified from Ea1189 using the oligonucleotide primers, Ea‐acr‐Com‐Fwd and Ea‐acr‐Com‐Rev (Table 6), cloned into SpeI‐SacI‐digested pBBR1MCS (Kovach et al., 1995) yielding the final acrAB complementation plasmid pNK8 (9624 bp). Plasmids pNK7 and pNK8 were transferred to electro‐competent cells of the E. amylovora tolC and acrB mutants respectively, which subsequently were grown at 28°C for 1 h in SOC broth and plated on LB containing appropriate antibiotics. To confirm complementation of the genotypes, plasmids were re‐isolated from the complemented cells and the target gene(s) were PCR‐amplified using appropriate oligonucleotides.
Drug susceptibility tests
The MICs of diverse antibiotics were determined by a dilution assay in Mueller–Hinton broth (MHB) (Becton Dickinson, Heidelberg, Germany) and AMM2 respectively. All tests were done in triplicate following the National Committee for Clinical Laboratory Standards (2000) recommendations. Briefly, testing of MIC was carried out in micro‐titer plates with 96 flat‐bottomed wells. With the exception of those wells used as control, each well received 100 µl of a twofold dilution series of an antibiotic solution in the appropriate medium. Than each well except those used as sterility controls received 100 µl of bacterial suspension at 1 × 106 cfu ml−1. The MIC was defined as the lowest concentration of an antibiotic that completely stopped visible cell growth. Growth of E. amylovora at 28°C was examined by visual and photometric inspection after 24 h of incubation.
Plant materials and pathogenicity assay on apple plants
Apple plants (rootstock Malus MM106) were grown in a light chamber at 20–25°C, 60% humidity, with a 12 h photoperiod (15 000 lux). Erwinia amylovora strains were grown on LB agar plates for 48 h, resuspended in sterile 0.9% NaCl solution and diluted to a cell density of 1 × 107 cfu ml−1 for inoculation. Apple plants were inoculated by the prick technique as described by May and colleagues (1997). Each bacterial strain was inoculated into shoots of five individual plants by placing 5 µl of bacterial suspensions onto each wound on the shoot tip. Plants were monitored for symptom development daily.
To study bacterial survival in plant tissue, 5 µl of bacterial suspensions (102–106 cfu ml−1) was placed onto each wound on the shoot tips of nine individual plants. Sterile 0.9% NaCl solution was used as negative controls. Survival of bacteria in the plant tissue was examined by re‐isolation of bacterial cells 24 h post inoculation. One centimetre of the shoot tip around the inoculation site was cut off. Samples from apple plants were pooled, homogenized in 0.9% NaCl solution, serially diluted, and appropriate dilutions were spread on LB agar plates.
All greenhouse experiments were repeated at least three times to confirm reproducibility and to calculate standard deviations.
Nucleotide sequence accession number
The nucleotide sequence of the tolC gene of E. amylovora 1189 was deposited in GenBank under Accession No. FJ462442.
Acknowledgments
This project was partially funded by a stipend from the Deutscher Akademischer Austauschdienst (DAAD).
References
- Aldridge P., Metzger M., Geider K. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol Gen Genet. 1997;256:611–619. doi: 10.1007/s004380050609. [DOI] [PubMed] [Google Scholar]
- Alekshun M.N., Levy S.B. Molecular mechanism of antimicrobial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Andrews J.M. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Barabote R.D., Johnson O.L., Zetina E., San Francisco S.K., Fralick J.A., San Francisco M.J.D. Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogensis. J Bacteriol. 2003;185:5772–5778. doi: 10.1128/JB.185.19.5772-5778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barny M.A., Guinebretière M.H., Marçais B., Coissac E., Paulin J.‐P., Laurent J. Cloning of a large gene cluster involved in Erwinia amylovora CFBP 1430 virulence. Mol Microbiol. 1990;4:777–786. doi: 10.1111/j.1365-2958.1990.tb00648.x. [DOI] [PubMed] [Google Scholar]
- Baucheron S., Chaslus‐Dancla E., Cloeckaert A. Role of TolC and parC mutation in high‐level fluroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J Antimicrob Chemother. 2004;53:657–659. doi: 10.1093/jac/dkh122. [DOI] [PubMed] [Google Scholar]
- Baucheron S., Mouline C., Praud K., Chaslus‐Dancla E., Cloeckaert A. TolC but not AcrB is essential for multidrug‐resistance Salmonella enterica serotype Typhimurium colonization of chickes. J Antimicrob Chemother. 2005;55:707–712. doi: 10.1093/jac/dki091. [DOI] [PubMed] [Google Scholar]
- Bavro V.N., Pirtras Z., Furnham N., Pérez‐Cano L., Fernández‐Recio J., Pei X.Y. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemann P., Geider K. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. J Gen Microbiol. 1992;138:931–940. doi: 10.1099/00221287-138-5-931. [DOI] [PubMed] [Google Scholar]
- Bogs J., Geider K. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J Bacteriol. 2000;182:5351–5358. doi: 10.1128/jb.182.19.5351-5358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.H., Paulsen I.T., Skurray R.A. The multi‐drug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- Bunikis I., Denker K., Östberg Y., Andersen C., Benz R., Bergström S. An RND‐type efflux system in Borrelia bugdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog. 2008;4:e1000009. doi: 10.1371/journal.ppat.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burse A., Weingart H., Ullrich M. The phytoalexin‐inducible multidrug efflux pump AcrAB contributes virulence in the fire blight pathogen, Erwinia amylovora. Mol Plant Microbe Interact. 2004;17:43–54. doi: 10.1094/MPMI.2004.17.1.43. [DOI] [PubMed] [Google Scholar]
- Cosme A.M., Becker A., Santos M.R., Sharypova L.A., Santos P.M., Moreira L.M. The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol Plant Microbe Interact. 2008;21:947–957. doi: 10.1094/MPMI-21-7-0947. [DOI] [PubMed] [Google Scholar]
- Del Sorbo G., Schoonbeek H.J., De Waard M.A. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet Biol. 2000;30:1–15. doi: 10.1006/fgbi.2000.1206. [DOI] [PubMed] [Google Scholar]
- Eastgate J.A. Erwinia amylovora: the molecular basis of fire blight disease. Mol Plant Pathol. 2000;1:325–329. doi: 10.1046/j.1364-3703.2000.00044.x. [DOI] [PubMed] [Google Scholar]
- Fleißner A., Sopalla C., Weltring K.M. An ATP‐binding cassette multi‐drug‐resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol Plant Microbe Interact. 2002;15:102–108. doi: 10.1094/MPMI.2002.15.2.102. [DOI] [PubMed] [Google Scholar]
- Fralick J.A., Burns‐Keliher L.L. Additive effect of tolC and rfa mutation on the hydrophobic barrier of the outer membrane of Escherichia coli K12. J Bacteriol. 1994;176:6404–6406. doi: 10.1128/jb.176.20.6404-6406.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzáles‐Pasayo R., Martinez‐Romera E. Multiresistance genes of Rhizobium etli CFN42. Mol Plant Microbe Interact. 2000;13:572–577. doi: 10.1094/MPMI.2000.13.5.572. [DOI] [PubMed] [Google Scholar]
- Grayer R.J., Kokubun T. Plant‐fungal interactions: the search for phytoalexins and other antifungal compounds from higher plants. Phytochemistry. 2001;56:253–263. doi: 10.1016/s0031-9422(00)00450-7. [DOI] [PubMed] [Google Scholar]
- Hagmann K.E., Pan W., Spratt B.G., Balthazar J.T., Judd R.C., Schafer W.M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. Phytoalexins: what we have learned after 60 years? Annu Revs Phytopathol. 1999;37:285–306. doi: 10.1146/annurev.phyto.37.1.285. [DOI] [PubMed] [Google Scholar]
- Harborne J.B. The comparative biochemistry of phytoalexin induction in plants. Biochem System Ecol. 1999;27:335–367. [Google Scholar]
- Hoang T.T., Karkhoff‐Schweizer R.R., Kutchma A.J., Schweizer H.P. A broad‐host‐range Flp‐FRT recombination system for site‐specific excision of chromosomally‐located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Koehler T., Michea‐Hamzehpour M., Henze U., Gotoh N., Curty L.K., Pechere J.C. Characterization of MexE‐MexF‐OprN, a positively regulated multi‐drug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Eswaran J., Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drug. Annu Rev Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., Peterson K.M. Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Krummenacher P., Narberhaus F. Two genes encoding a putative multi‐drug efflux pump of the RND/MFP family are cotranscribed with an rpoH gene in Bradyrhizobium japonicum. Gene. 2000;241:247–254. doi: 10.1016/s0378-1119(99)00490-4. [DOI] [PubMed] [Google Scholar]
- Li D.W., Onishi M., Kishino T., Matsuo T., Ogawa W., Kuroda T., Tsuchiya T. Properties and expression of a multidrug efflux pump AcrAB‐KocC from Klebsiella pneumoniae. Biol Pharm Bull. 2008;31:577–582. doi: 10.1248/bpb.31.577. [DOI] [PubMed] [Google Scholar]
- Li X.‐Z., Nikaido H., Poole K. Role of MexA‐MexB‐OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létoffé S., Delepelaire P., Wandersman C. Protease secretion by Erwinia chrysanthemi: the specific secretion functions are analogous to those of Escherichia coliα‐haemolysin. EMBO J. 1990;9:1375–1382. doi: 10.1002/j.1460-2075.1990.tb08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiorani Valecillos A.M., Palenzuela P.R., López‐Solanilla E. The role of several multidrug resistance systems in Erwinia chrysanthemi pathogenesis. Mol Plant Microbe Interact. 2006;19:607–613. doi: 10.1094/MPMI-19-0607. [DOI] [PubMed] [Google Scholar]
- May R., Völksch B., Kampmann G. Antagonistic activities of epiphytic bacteria from soybean leaves against Pseudomonas syringae pv. glycinea in vitro and in planta. Microb Ecol. 1997;34:118–124. doi: 10.1007/s002489900041. [DOI] [PubMed] [Google Scholar]
- Mayr U., Treutter D., Santos‐Buelga C., Bauer H., Feucht W. Developmental changes in the phenol concentrations of, Golden Delicious' apple fruits and leaves. Phytochemistry. 1995;38:1151–1155. doi: 10.1016/0031-9422(94)00760-q. [DOI] [PubMed] [Google Scholar]
- Metzger M., Bellemann P., Bugert P., Geider K. Genetics of galactose metabolism of Erwinia amylovora and its influence on polysaccharide synthesis and virulence of the fire blight pathogen. J Bacteriol. 1994;176:450–459. doi: 10.1128/jb.176.2.450-459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira M.A.S., De Souza E.C., De Moraes C.A. Multidrug efflux system in gram negative bacteria. Braz J Microbiol. 2004;35:19–28. [Google Scholar]
- National Committee for Clinical Laboratory Standards. 2000. [PubMed]
- Nishino K., Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissinen R.M., Ytterberg A.J., Bogdanove A.J., Van Wijk K.J., Beer S. Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel typeIII secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol Plant Pathol. 2007;8:55–67. doi: 10.1111/j.1364-3703.2006.00370.x. [DOI] [PubMed] [Google Scholar]
- Osbourn A.E. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo J.D., Kado C.I., Phillips D.A. An isoflavonoid‐inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol. 1998;180:3107–3113. doi: 10.1128/jb.180.12.3107-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I.T., Cen J., Nelson K.E., Saier M.H., Jr Comparative genomics of microbial drug efflux systems. J Mol Microbiol Biotechnol. 2001;3:145–150. [PubMed] [Google Scholar]
- Peng W.T., Nester E.W. Characterization of a putative RND‐type efflux system in Agrobacterium tumefaciens. Gene. 2001;270:245–252. doi: 10.1016/s0378-1119(01)00468-1. [DOI] [PubMed] [Google Scholar]
- Perez A., Canle D., Latasa C., Poza M., Beceiro A., Tomas M.M. Cloning, nucleotide sequencing, and analysis of the AcrAB‐TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolates. Antimicrob Agent Chemother. 2007;51:3247–3253. doi: 10.1128/AAC.00072-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L.J.V. Multidrug resistance efflux pumps‐not just for resistance. Microbiology. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Poole K. Efflux‐mediated resistance to fluoroquinolones in gram‐negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux‐mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Psallidas P.G., Tsiantos J. Chemical control of fire blight. In: Vanneste J.L., editor. CABI Publishing; 2000. pp. 199–234. , and . In Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora (ed.). Oxon: , pp. [Google Scholar]
- Ramos J.L., Duque E., Gallegos M.T., Godoy P., Ramos‐González M.I., Rojas A. Mechanism of solvent tolerance in gram‐negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. et al. [DOI] [PubMed] [Google Scholar]
- Reddy J.D., Reddy S.L., Hopkins D.L., Gabriel D.W. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera Grapevines. Mol Plant Microbe Interact. 2007;20:403–410. doi: 10.1094/MPMI-20-4-0403. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. Cold Spring Habor Laboratory Press; 2001. [Google Scholar]
- Schoonbeek H., Del Sorbo G., De Waard M.A. The ABC transporter BcatrB affects the sensitivity of Botrytis cinerea to the phytoalexin resveratrol and the fungicide fenpiclonil. Mol Plant Microbe Interact. 2001;14:562–571. doi: 10.1094/MPMI.2001.14.4.562. [DOI] [PubMed] [Google Scholar]
- Sharff A., Fanutii C., Shi J., Calladine C., Luisi P. The role of the TolC family in protein transport and multidrug efflux from stereochemical certainty to mechanistic hypothesis. Eur J Biochem. 2001;268:5011–5026. doi: 10.1046/j.0014-2956.2001.02442.x. [DOI] [PubMed] [Google Scholar]
- Steinberger E.M., Beer S.V. Creation and complementation of pathogenicity mutants of Erwinia amylovora. Mol Plant Microbe Interact. 1988;1:135–144. [Google Scholar]
- Stermitz F.R., Lorenz P., Tawara J.N., Zenewicz L.A., Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′‐methyoxy‐hydrocarpin, a multidrug pump inhibitor. Proc Nat Acad Sci. 2000;97:1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoitsova S.O., Braun Y., Ullrich M.S., Weingart H. Characterization of the RND‐type multidrug efflux pump MexAB‐OprM of the plant pathogen Pseudomonas syringae. Appl Environ Microbiol. 2008;74:3387–3393. doi: 10.1128/AEM.02866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulavik M.C., Houseweart C., Cramer C., Jiwani N., Murgolo N., Greene J. Antibiotic susceptibility profiles of Escherichia coli strains lacking multi‐drug efflux pump genes. Antimicrob Agents Chemother. 2001;45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi D.G., Cheng L.W., Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D. Biosynthesis of phenolic compounds and its regulation in apple. Plant Grow Regul. 2001;34:71–89. [Google Scholar]
- VanEtten H., Temporine E., Wasmann C. Phytoalexin (and Phytoanthicipin) tolerance as a virulence trait: why is it not required by all pathogens? Physiol Mol Plant Pathol. 2001;59:83–93. [Google Scholar]
- Vanneste J.L., Eden‐Green S. Migration of Erwinia amylovora in host plant tissue. In: Vanneste J.L., editor. CABI Publishing; 2000. pp. 9–36. , and . In Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora (ed.). Oxon: , pp. [Google Scholar]
- Vanneste J.L., Paulin J.‐P., Expert D. Bacteriophage Mu as a genetic tool to study Erwinia amylovora pathogenicity and hypersensitive reaction on tobacco. J Bacteriol. 1990;172:932–941. doi: 10.1128/jb.172.2.932-941.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- Wilson K.J., Sessitsch A., Corbo J.C., Giller K.E., Akkermans A.D.L., Jefferson R.A. Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram‐negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- Zgurskaya H., Nikaido H. Multidrug resistance mechanisms: drug efflux across two membranes. Mol Microbiol. 2000;37:219–225. doi: 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]


