Summary
The endophytic phase of Pseudomonas savastanoi pv. savastanoi in olive stems and the structural and ultrastructural histogenesis of olive knots have been studied. Construction of a stable plasmid vector expressing the green fluorescent protein, in combination with the use of in vitro olive plants, allowed real‐time monitoring of P. savastanoi pv. savastanoi infection. The infection process was also examined by bright field and epifluorescence microscopy as well as by scanning and transmission electron microscopy. Hypertrophy of the stem tissue was concomitant with the formation of bacterial aggregates, microcolonies and multilayer biofilms, over the cell surfaces and the interior of plasmolysed cells facing the air‐tissue interface of internal opened fissures, and was followed by invasion of the outer layers of the hypertrophied tissue. Pathogenic invasion of the internal lumen of newly formed xylem vessels, which were connected with the stem vascular system, was also observed in late stages of infection. Ultrastructural analysis of knot sections showed the release of outer membrane vesicles from the pathogen surface, a phenomenon not described before for bacterial phytopathogens during host infection. This is the first real‐time monitoring of P. savastanoi disease development and the first illustrated description of the ultrastructure of P. savastanoi‐induced knots.
Introduction
Pseudomonas savastanoi pvs. savastanoi, fraxini and nerii (Gardan et al., 1992) cause knots and galls on members of the various genera of the Oleaceae (including olive, ash, jasmine and Phyllyrea spp.) and oleander (Nerium oleander L.). Symptoms of infected trees include overgrowths (tumorous galls or knots) on the stems and branches of the host plant, occasionally occurring on the leaves and fruits. At present, the only P. savastanoi determinants known to be involved in knot development are the phytohormones indoleacetic acid and the cytokinins (Smidt and Kosuge, 1978; Comai and Kosuge, 1980; Surico et al., 1985; Rodríguez‐Moreno et al., 2008), and the biosynthesis of a functional type III secretion system (TTSS), encoded by the hrp/hrc gene clusters (Sisto et al., 2004). Recently, a global genomic analysis of P. savastanoi pv. savastanoi plasmids allowed for the identification of several putative virulence factors in this olive pathogen, including several TTSS protein effectors and a variety of genes encoding known P. syringae virulence determinants (Pérez‐Martínez et al., 2008).
Besides from knots, P. savastanoi pv. savastanoi has been isolated from the phyllosphere of diseased and healthy olive leaves and stems where the pathogen has a resident epiphytic phase. The seasonal dynamics of P. savastanoi pv. savastanoi epiphytic populations, as well as the composition of the bacterial community found on olive tree leaves, has been studied in detail using isolation (Ercolani, 1978; Ercolani, 1985; Ercolani, 1991), molecular detection (Penyalver et al., 2000; Quesada et al., 2007) and scanning electron microscopy (Surico, 1993a,b). However, although knot histology has been examined by light microscopy for olive (Smith, 1920; Surico, 1977), oleander (Wilson and Magie, 1964; Wilson, 1965) and, more recently, for buckthorn (Temsah et al., 2007a) and myrtle (Temsah et al., 2007b), data concerning the lifestyle of P. savastanoi cells during its endopathogenic phase are scarce. Anatomical studies of disease development in oleander showed that P. savastanoi pv. nerii systematically invades the host through laticifers and, less frequently, through xylem vessels (Wilson and Magie, 1964). In contrast, it is not clear whether P. savastanoi pv. savastanoi cells invade the xylem and migrate through olive tissues (Wilson and Magie, 1964; Penyalver et al., 2006).
A valuable tool for analysing the behaviour of microbes in their natural environments, whether they are in soil, living plants or animal hosts, is the use of autofluorescent reporter proteins. The vast majority of studies utilizing this technology to examine bacterial–plant interactions lifestyle in association with plants have used modified forms of green fluorescent protein (GFP) (Chalfie et al., 1994) that confer high levels of fluorescence, e.g. GFPmut3 (Cormack et al., 1996). GFP in combination with epifluorescence microscopy techniques has been used broadly for the visualization of Pseudomonas cells in the rhizosphere (e.g. Bloemberg et al., 2000; Ramos et al., 2000; Boldt et al., 2004), on leaf surfaces (Normander et al., 1998; Sabaratnam and Beattie, 2003; Wang et al., 2007), and within infected leaf tissues of herbaceous plants (Badel et al., 2002; Boureau et al., 2004; Wang et al., 2007). However, the broad and efficient application of GFP to the investigation of bacterial interactions with woody plants has presented two main restrictions. First, in order to detect GFP in planta, its level of expression should be high enough to overcome the interference by high levels of autofluorescence emitted by secondary wall thickenings. Second, the GFP gene must be stable in the labelled bacterial cells throughout the process of infection.
We have recently shown that olive plants micropropagated in vitro can serve as a valuable model system for evaluating the pathogenicity and virulence of P. savastanoi pvs. savastanoi and nerii (Rodríguez‐Moreno et al., 2008). In this study, we report the construction of a stable, broad‐host‐range plasmid vector, pLRM1‐GFP, which expresses GFPmut3 at high levels from a constitutive promoter in Pseudomonas (PA1/04/03). GFP tagging of P. savastanoi pv. savastanoi strain NCPPB 3335 allows the real‐time monitoring of bacterial disease development at the whole‐tumour level by epifluorescence microscopy, as well as the monitoring of bacterial localization inside knots at the single‐cell level by scanning confocal electron microscopy (SCLM). Additionally, scanning and transmission electron microscopy were used for a detailed ultrastructural analysis of tumour histology, as well as for the visualization of the P. savastanoi pv. savastanoi lifestyle within knot tissues.
Results
Plasmid pLRM1‐GFP is stable in P. savastanoi pv. savastanoi during growth in vitro and in planta
The stability of plasmid pLRM1‐GFP (Fig. 1) in P. savastanoi pv. savastanoi NCPPB 3335‐GFP was examined in vitro. As judged by resistance to the appropriate antibiotics, the strain stably maintained the plasmid (between 90% and 100% of the cells) for at least 40 generations (Fig. 2A). Furthermore, 100% of the colonies isolated at the end of the experiment from media not containing antibiotics exhibited uniform green fluorescence. The stability of the plasmid was also assessed in planta by infecting in vitro olive plants with NCPPB 3335‐GFP. During the 28 days of the experiment, estimated colony‐forming units (cfu) counts in media with and without antibiotics were similar (Fig. 2B). Moreover, 100% of the colonies isolated at 28 days post‐inoculation (dpi) were fluorescent green, further confirming the stability of the plasmid in P. savastanoi pv. savastanoi during bacterial growth in olive plants.
Figure 1.
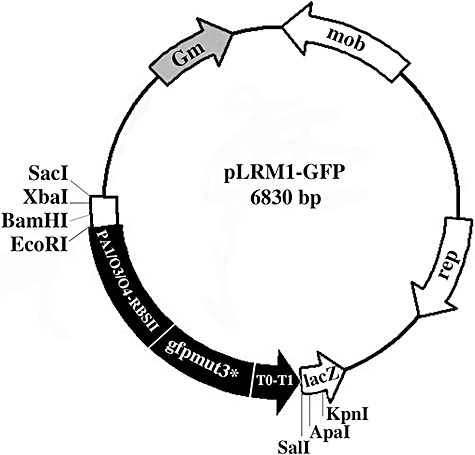
Schematic drawing of plasmid pLRM1‐GFP. The GFP expression cassette from plasmid pJBA29 (Andersen et al., 1998), shown in black, was cloned into SalI/EcoRI‐digested plasmid pBBR1‐MCS5 (Kovach et al., 1995). pLRM1‐GFP consists of a 2.05 kb fragment carrying a fusion of the IPTG‐inducible promoter PA1/04/03(Lanzer and Bujard, 1988) to the gfpmut3* gene, encoding GFPmut3 (Cormack et al., 1996), a synthetic ribosome binding site (RBSII), translational stop codons in all three reading frames, and two strong transcriptional terminators, T0 (derived from phage λ) and T1 (derived from the rrnB operon of E. coli).
Figure 2.
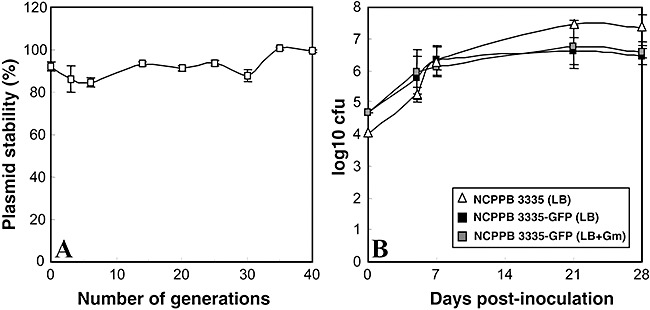
(A) Stability of plasmid pLRM1‐GFP in P. savastanoi pv. savastanoi NCPPB 3335. The transformant strain was grown in the absence of antibiotic pressure for 40 generations. Cultures were plated on LB‐Nf with and without Gm to score the presence of pLRM1‐GFP and to determine cell titres respectively. (B) Growth curves of Pseudomonas savastanoi pv. savastanoi NCPPB 3335‐GFP in in vitro olive plants compared to its parental strain. Colony‐forming unit counts were estimated by dilution and plating in the indicated media. Data represent the average of three independent experiments; error bars indicate the standard deviation from the average.
GFP expression does not affect in planta growth and pathogenicity of P. savastanoi pv. savastanoi
Similar disease symptoms developed from 1 to 120 dpi for the GFP‐labelled and the wild‐type strains. As previously reported for NCPPB 3335 (Rodríguez‐Moreno et al., 2008), swelling of the stem tissue observed in plants inoculated with NCPPB 3335‐GFP was followed by the formation of hyperplasic knots already visible at 9 dpi. Size of the overgrowths slightly increased over time from 9 dpi to approximately 28 dpi after which necrosis developed, which covered all of the hypertrophied tissue at 120 dpi (Fig. 3A, not shown for the wild‐type strain). Both bacterial strains were able to multiply inside the olive tissue during the first week post‐inoculation, reaching around 106 cfu at 7 dpi. In both cases, bacterial counts increased slightly, from 14 to 21 dpi, and then remained steady at this level until 28 dpi (106–107 cfu). Although average cfu counts were slightly higher for the wild‐type strain than for the GFP‐tagged strain, no significant differences between the strains were observed at any time post‐inoculation (Fig. 2B). Taken together, these results show that GFP expression from plasmid pLRM1‐GFP in the tested P. savastanoi strain did not significantly affect its in planta growth or pathogenicity.
Figure 3.
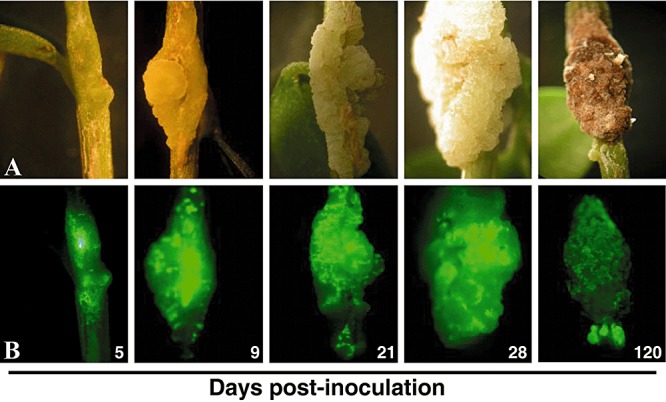
Real‐time monitoring of P. savastanoi pv. savastanoi NCPPB 3335‐GFP infection of in vitro olive plants. Evolution of knot symptoms (A) and epifluorescence microscopy images of whole tumours (B) were recorded at different times after inoculating 104 cfu of the pathogen.
Real‐time monitoring of the P. savastanoi pv. savastanoi infection of olive plants
Whole knots induced on in vitro olive plants by NCPPB 3335‐GFP were visualized at different post‐inoculation times using an epifluorescense stereoscopic microscope. A green fluorescent spot covering the inoculation point was already visible in the stem at 1 dpi (data not shown) and then GFP fluorescence spread both upwards and downwards from the inoculation point. Green fluorescent clusters were clearly seen in the swollen stem at 5 dpi. From 9 to 28 dpi, the number and size of green fluorescence spots increased over time and expanded by the entire knot surface. After approximately 30 dpi, and concomitant with the progressive necrosis of the knot tissue, green fluorescence intensity decreased with time. At 120 dpi, with the exception of a few bright GFP spots corresponding to small overgrowths of new formations located below the main tumour, the necrotized tissue showed diffused spots of green fluorescence not distinguishable from the level of background green autofluorescence shown by plants inoculated with the wild‐type strain (Fig. 3B, not shown for the wild‐type strain).
Vascular bundles of new formation in olive knots are connected with the stem vascular cylinder
In order to determine whether knots induced by P. savastanoi pv. savastanoi NCPPB 3335‐GFP on in vitro olive plants exhibited similar histological structures to those previously reported for older olive plants, semi‐thin and fixed‐frozen transverse sections of knots collected at different times after pathogen inoculation (for details see Fig. 4) were stained with toluidine blue or methylene blue‐picrofuchsin, respectively, and visualized by light microscopy. As previously reported for olive knots developed under natural conditions, a panoramic view of knot sections stained with toluidine blue clearly showed two structurally different regions: a vascular cylinder similar to that of non‐infected stems, composed of the pith parenchyma, xylem, cambium, phloem and epidermis; and expanding from this area, a hypertrophied parenchymatic tissue (Fig. 4A). Hypertrophied tissue was predominantly made up of disorganized cells and showed internal open fissures surrounded by plasmolysed cells and clusters of primary cell walls stained in intense dark blue (Fig. 4A and B). A closer view of this area also showed the existence of longitudinal xylem vessels running out of the stem vascular system towards the hypertrophied knot tissue (Fig. 4B). In addition, magnification of the knot outer layers allowed visualization of both hypertrophic and hyperplasic cells, which frequently showed irregular thickenings of the primary wall. Hyperplasic cells were clearly distinguishable by the formation of a slim middle lamella separating two nuclei (Fig. 4C). Knot sections stained with methylene blue‐picrofuchsin showed newly formed bundles of spiral xylem vessels inside the hypertrophied area (Fig. 4D and E). Interestingly, a connection between the primary vascular cylinder and newly formed spiral vessels was clearly observed in longitudinal sections of knots (Fig. 4F and G). Identical results were obtained from sections of knots collected from olive plants inoculated with the wild‐type strain NCPPB 3335 (data not shown).
Figure 4.
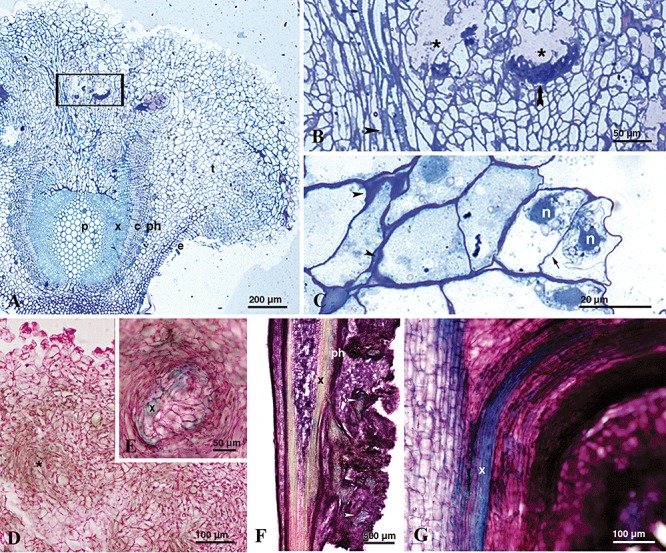
Light microscopy images of cross‐sections of knots induced by P. savastanoi pv. savastanoi NCPPB‐3335‐GFP on in vitro olive plants. A–C. Cross‐semi‐thin sections of knots induced at 35 dpi, stained with toluidine blue. (A) Pith parenchyma (p), secondary xylem (x), vascular cambium (c), secondary phloem (ph), epidermis (e) and hypertrophied parenchyma (t). Stained primary and secondary walls show dark and light blue colour respectively. (B) Detail of a hypertrophied area of the tissue showing internal cavities surrounded by collapsed host cells (asterisks), longitudinal xylem vessels running out of the stem vascular system towards the knot tissue (black arrowhead) and intensively stained cell wall accumulations (black arrow). (C) Detail of hyperplasic cells showing two nuclei (n) separated by a slim middle lamella (black arrow). Irregular thickenings of the primary cell wall surrounding parenchymatic‐like cells of outer knot layers (arrowheads). D and E. Cross‐sections of knots, collected at 28 dpi, stained with methylene blue‐picrofuchsin. (D) Groups of parenchymatous‐like cells, showing a slight blue‐green stain of the cell walls due to the formation of secondary walls during differentiation (asterisks). (E) Detail of a newly formed bundle of xylem vessels (x). F. Panoramic view of a longitudinal section of a knot, collected at 21 dpi, stained with methylene blue‐picrofuchsin. Bundles of xylem are distinguished inside the hypertrophied area (white arrowheads). G. Detail of a vascular connection between the vascular cylinder and the hypertrophied parenchymatic tissue observed in a longitudinal section of a knot collected at 21 dpi.
Spatial distribution of P. savastanoi pv. savastanoi inside olive knots
Autofluorescence emitted by plant tissues after excitation with blue fluorescence was used to differentiate different histological structures: parenchymatic cells (red), xylem and epidermis (yellow) and sclereids (dark‐green). The levels of background green autofluorescence emitted by the stem tissues of non‐inoculated plants (Fig. 5A), and by the knots induced by the wild‐type strain (data not shown), were clearly reduced in comparison to that of NCPPB 3335‐GFP cells inside olive knots. Sections of knots collected at 5 dpi showed only a few green fluorescent spots located near the inoculation point (Fig. 5B and C). At 9 dpi (Fig. 5D), the infection sites of NCPPB 3335‐GFP inside olive knots were clearly visible as numerous bright green spots, which increased over time in size and number. Figure 5E and F, Fig. 5G and Fig. 5H and I correspond to sections of knots collected at 14, 21 and 120 dpi respectively. As previously described for olive (Smith, 1920; Surico, 1977), oleander (Wilson and Magie, 1963), buckthorn (Temsah et al., 2007a) and myrtle (Temsah et al., 2007b), P. savastanoi cells were localized inside internal cavities of the hypertrophied knot tissue (Fig. 5E–H). Moreover, GFP tagging of the pathogen allowed its identification in other histological structures, such as the outer layers of the hypertrophied tissue (Fig. 5E and G). In spite of the low green fluorescence level emitted by whole tumours at 120 dpi (Fig. 3B), bright green fluorescent spots were clearly visible in transverse sections of these tumours, not only inside internal cavities or in outer layers of the knot, but also surrounding the stem vascular cylinder (Fig. 5H), and filling the internal lumen of newly formed xylem vessels (Fig. 5I).
Figure 5.
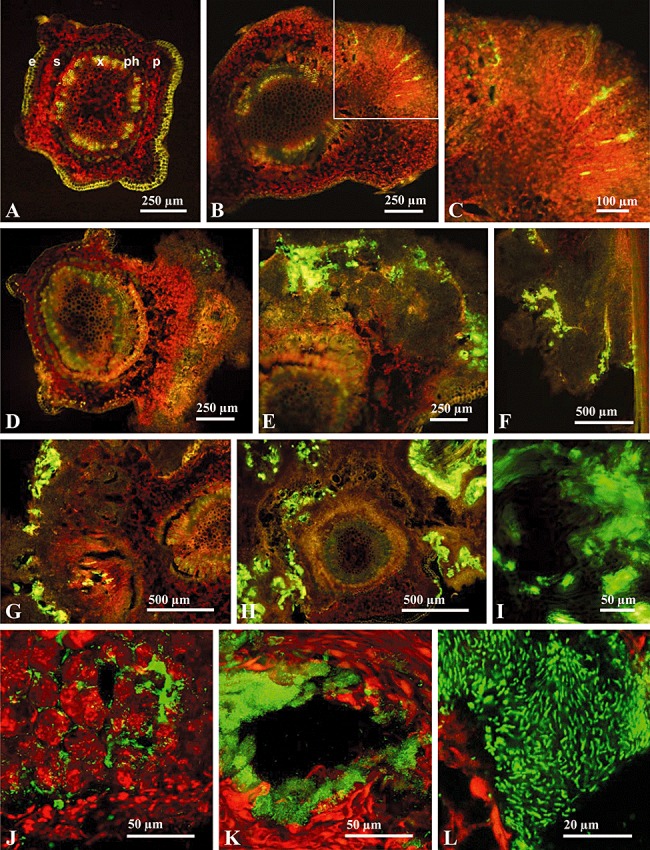
In situ visualization of P. savastanoi pv. savastanoi NCPPB 3335‐GFP cells by epifluorescence microscopy (A–I) and SCLM (J–L). (A) Cross‐section of a non‐infected olive stem. Autofluorescence emitted by plant tissues allowed differentiation of different histological structures; epidermis (e), schlereids (s), xylem (x), phloem (ph) and parenchyma (p). Epifluorescence images of cross‐sections of knots induced by NCPPB 3335‐GFP at 5 dpi (B and C), 9 dpi (D), 14 dpi (E and F), 21 dpi (G) and 120 dpi (H). (C) Detail of the area indicated in (B). (I) Detail of (H) showing the emission of green fluorescence within the lumen of xylem vessels localized within the hypertrophied tissue. (J–L) SCLM images of cross‐sections of olive knots. The green fluorescence emitted by NCPPB 3335‐GFP cells and the red autofluorescence emitted by the plant tissue was recorded. (J) GFP‐tagged P. savastanoi pv. savastanoi cells colonizing the intercellular spaces of the host tissue at 5 dpi. (K) Internal open fissure of a knot colonized by a biofilm of GFP‐tagged cells at 21 dpi. (L) SCLM visualization of GFP‐tagged cells forming a biofilm at 21 dpi in an internal knot cavity.
To analyse the distribution of P. savastanoi pv. savastanoi cells inside olive knots in detail, sections of knots induced by NCPPB 3335‐GFP were examined by SCLM. Microcolonies of GFP‐tagged cells were visualized filling the intercellular spaces of the parenchymatic tissue (Fig. 5J). Moreover, GFP‐tagged cells organized in biofilms profusely colonized the internal cavities of the hypertrophied tissues (Fig. 5K and L). A closer view of the cavities filled with bacteria allowed visualization of NCPPB 3335‐GFP at the single cell level (Fig. 5L).
Organization of P. savastanoi pv. savastanoi cell clusters inside olive knots
To visualize the organization of P. savastanoi pv. savastanoi cell clusters during their endopathogenic phase in greater detail, cross‐sections of olive knots induced by NCPPB 3335‐GFP at 35 dpi were examined by scanning electron microscopy. Small groups (Fig. 6A and B) and microcolonies (Fig. 6C) of rod‐shaped bacterial cells, about 0.5 µm wide and 1–2 µm long, were visualized attached to each other and to the surface of hypertrophied host cells by an extracellular fibrillar matrix. However, the majority of the bacterial cells were found forming biofilms composed of a multilayer of bacterial cells. Biofims were observed colonizing the surface (Fig. 6D) and interior of plasmolysed host cells (Fig. 6E).
Figure 6.
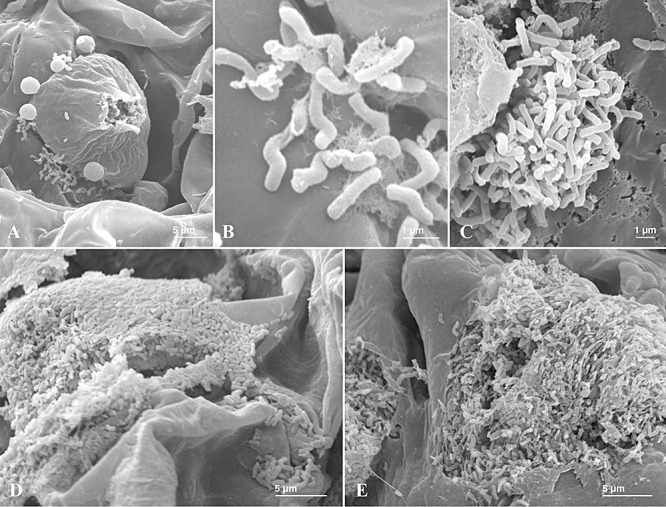
Scanning electron micrographs of knot sections induced by P. savastanoi pv. savastanoi NCPPB 3335‐GFP at 35 dpi. A. A small group of bacterial cells is seen attached to the cell wall of a plasmolysed host cell. B. Detail of (A), rod‐shaped bacterial cells attached to each other and to the cell walls by a fibrillar matrix. C. Bacterial microcolony attached to a host cell. D. Bacterial biofilm colonizing the surface of plasmolysed host cells. E. Mass of bacterial cells colonizing the interior of a plasmolysed host cell.
Ultrastructural analysis of olive knots induced by P. savastanoi pv. savastanoi
Ultrathin knot sections of olive knots induced by NCPPB 3335‐GFP at 35 dpi were also analysed using high‐resolution transmission electron microscopy. Proliferation of P. savastanoi pv. savastanoi cells inside olive induced clearly visible modifications of the parenchymatous tissue, such as abnormal cell wall thickenings and intercellular spaces showing a degraded middle lamella. Moreover, bacterial cells were detected colonizing the intercellular spaces (Fig. 7A) and were in close contact to the degraded middle lamella (Fig. 7B) and primary cell wall (Fig. 7C), suggesting pathogen‐mediated degradation of these host cell structures. Parenchymatic‐like cells in contact with the pathogen usually showed a fibrillar and electrodense cytoplasm, probably due to the degeneration of cytoplasmatic organelles and plasma membranes (Fig. 7D and E).
Figure 7.
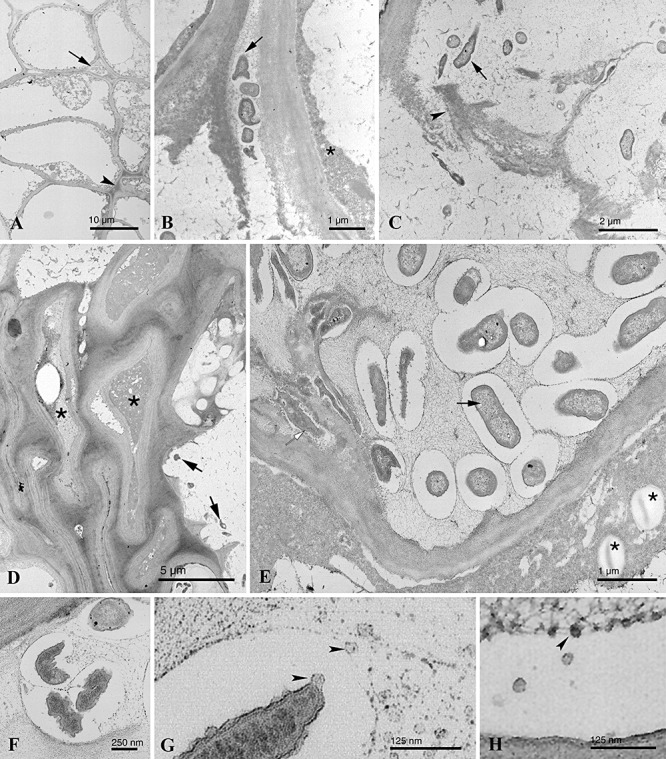
Transmission electron micrographs of ultrathin sections of knots induced by P. savastanoi pv. savastanoi NCPPB‐3335‐GFP at 35 dpi on in vitro olive plants. A. Ultrastructure of knot tissue showing parenchymatic‐like cells containing a fibrillar cytoplasm and irregular cell wall thickenings (arrowhead). Bacterial cells were visualized at intercellular spaces (black arrow). B. Pseudomonas savastanoi pv. savastanoi cells (black arrow) localized at the intercellular space of two host cells. A condensed cytoplasm (asterisk) and a degraded middle lamella are shown. C. Bacterial cells (black arrow) in contact with a primary cell wall in the process of degradation (arrowhead). D. Abnormal cell wall accumulations of host cells in close contact with degenerated cytoplasm (asterisks) and pathogen cells (black arrow). E. Rod‐shaped (black arrow) and irregular (open arrow) bacterial cells colonizing the extracellular space of a host cell showing degenerated organelles (asterisks). F. Group of P. savastanoi pv. savastanoi cells within a degenerated host cell. Bacterial cells are seen surrounded by an electrolucent halo and immersed in a fibroreticular matrix. G. Detail of (F), high‐magnification image of the bacterial surface releasing outer‐membrane vesicles (arrowheads). H. Detail of the fusion of outer‐membrane vesicles (arrowhead) to the fibroreticular matrix.
Pseudomonas savastanoi pv. savastanoi cells colonizing extracellular spaces (Fig. 7E) and the interior of plasmolysed host cells (Fig. 7F) were frequently seen surrounded by an electrolucent halo and immersed in a fibroreticular matrix. Interestingly, a closer view of these bacterial cells revealed the release of outer membrane vesicles (OMVs), filled by an electrodense material, from the bacterial surface (Fig. 7G and H). These vesicles, which have not been previously visualized for any other bacterial phytopathogen during the infection of host plants, were observed both being released into the electrolucent halo that covered the bacterial surface and fusing with the fibroreticular matrix, where the bacterial cells were immersed (Fig. 7G and H). Besides the characteristic rod‐shaped morphology showed by P. savastanoi pv. savastanoi cells, bacterial cells showing an irregular shape were also frequently found inside olive knots (Fig. 7E).
Discussion
Research on diseases caused by Pseudomonas in herbaceous plants has progressed rapidly after the complete genome sequencing of three P. syringae pathovars and the comparison of their repertoire of virulence‐associated genes (Lindeberg et al., 2008). However, knowledge on the infection of woody plants by strains belonging to the genus Pseudomonas lags far behind. Research on P. savastanoi strains affecting woody hosts is now ready to move forward, due to the development of molecular detection methods for identifying P. savastanoi pathovars (Penyalver et al., 2000; Bertolini et al., 2003), the selection of virulent strains and the improvement of gene transfer for genetic manipulation of P. savastanoi isolated from olive knots (Pérez‐Martínez et al., 2007), the establishment of the in vitro olive plant as model system for studying the pathogenicity and virulence of P. savastanoi pathovars (Rodríguez‐Moreno et al., 2008), the identification of several putative virulence determinants by global genomic analysis of their plasmids (Zhao et al., 2005; Pérez‐Martínez et al., 2008) and DNA microarray‐based comparative genomic hybridization (Sarkar et al., 2006). Future investigations on the function of P. savastanoi genes that undertake a phenotypic analysis of mutants would first require a broader understanding of the biology of the wild‐type bacterium for comparison. For this purpose, P. savastanoi pv. savastanoi strain NCPPB 3335‐GFP was constructed. GFP tagging of P. savastanoi pv. savastanoi NCPPB 3335, using a mini‐Tn5 transposon encoding GFPmut3, has been previously reported (Pérez‐Martínez et al., 2007); however, GFP emission by this strain was not as strong as that observed in this study for NCPPB 3335 containing plasmid pLRM1‐GFP, which was found to be stable in this strain both in vitro and in planta (Fig. 2A and B). NCPPB 3335‐GFP performed like the wild‐type in all features tested, including growth in olive and virulence (Fig. 2B and Fig. 3), thus allowing real‐time monitoring of the bacterial infection and for an in planta analysis of its endophytic phase.
Histological modifications observed in in vitro olive plants after infection of P. savastanoi pv. savastanoi NCPPB 3335‐GFP (Fig. 4) were very similar to those previously observed in older olive plants (Smith, 1920; Surico, 1977), oleander (Wilson and Magie, 1963; Wilson, 1965), buckthorn (Temsah et al., 2007a) and myrtle (Temsah et al., 2007b), further confirming the suitability of this model system to analyse P. savastanoi pathogenicity. A connection between the primary vascular cylinder and newly formed spiral vessels was found in stem knots induced by P. savastanoi in in vitro olive plants (Fig. 4F and G). Although neovascularization is a general requirement for the growth of plant and animal tumours (Abbott et al., 1977; Ullrich and Aloni, 2000), the existence of an interconnection between new‐formed bundles and the vascular system of the host plant has only been reported before in Agrobacterium turmefaciens‐induced crown galls (Aloni et al., 1995). The existence of this network of vascular tissues in olive knots could be related to the supply of nutrients and water necessary to reach their final size. These results encourage the future utilization of in vitro olive plants for elucidating the mechanisms of phytohormone‐dependent vascular development in plants.
Pseudomonas savastanoi cells have been previously observed to fill the internal cavities of hypertrophied knot tissues that developed on several plants (Smith, 1920; Wilson, 1965; Surico, 1977; Temsah et al., 2007a,b); however, xylem invasion of olive knots and systemic movement of the pathogen in olive plants is a controversial point of discussion (Wilson and Magie, 1964; Penyalver et al., 2006). GFP tagging of P. savastanoi pv. savastanoi and examination of knot sections by epifluorescence microscopy allowed us to demonstrate invasion of newly formed xylem vessels by NCPPB 3335‐GFP cells at 120 dpi (Fig. 5I). These results, together with the observed vascular network of knots mentioned above, suggest a possible systemic movement of the pathogen through the vascular system; however, real‐time monitoring of whole plants infected with P. savastanoi pv. savastanoi NCPPB 3335‐GFP did not detect the pathogen outside the knot area in 120 days post inoculation. Using epifluorescence microscopy to examine olive stem sections above and below the knots induced by this strain might help to clarify this issue. Pseudomonas savastanoi pv. savastanoi cells were also observed colonizing the outer layers of olive knots (Fig. 5G). Perhaps the tangential division of parenchymatic host cells moved the pathogen forward in this direction, or the pathogen actively moved to this site due to oxygen requirements. This position of P. savastanoi pv. savastanoi cells might also be related to the release of pathogen cells through knot exudates, which could serve as new inoculum source.
Visualization of olive knot sections by SCLM (Fig. 5K and L) and electron microscopy (Fig. 6C and D) allowed us to analyse, for the first time, the organization of P. savastanoi cell clusters (microcolonies and biofilms) inside the host tissue. A previous study reported the ability of several P. syringae and P. savastanoi strains, including NCPPB 3335, to form air‐liquid biofilms over abiotic surfaces; however, the formation of biofilm structures has not been described before for P. savastanoi during its internal colonization of host tissues.
In vitro activity of cell wall degrading enzymes has been reported for P. savastanoi (Magie, 1963) and is hypothesized to be involved in the formation of internal fissures in olive and oleander knots (Wilson, 1965). Transmission electron microscopy images clearly showed degradation of the primary cell wall and middle lamella of host cells in close contact with the pathogen (Fig. 7B and C). Moreover, vesicles released from the envelope of P. savastanoi pv. savastanoi cells were observed during the infection of host tissues. Bacterial release of OMVs, well studied in Gram‐negative animal pathogens but, to the best of our knowledge, not reported before for any other bacterial phytopathogen during plant infection, is known to play roles in establishing a colonization niche, carrying and transmitting virulence factors into host cells, and modulating host defence and response (Kuehn and Kesty, 2005). Recently, a proteome analysis of OMVs released in vitro by the pathogenic plant bacterium Xanthomonas campestris identified several virulence‐associated proteins in the OMV fraction (Sidhu et al., 2008). As OMV release from P. savastanoi pv. savastanoi cells was visualized both in extracellular spaces (Fig. 7E) and in the interior of plasmolysed host cells (Fig. 7F and G), it is difficult to attribute a synthetic or degrading function to them in this pathogen. Further investigations will be necessary to clarify this matter, including visualization of OMVs released from a wild‐type P. savastanoi pv. savastanoi strain not expressing GFP.
The results shown here establish the basis for the future utilization of plasmid pLRM1‐GFP, in combination with in vitro olive plants, for the real‐time monitoring of P. savastanoi pv. savastanoi systemic movement and for the in vivo analysis of virulence mutants in comparison to wild‐type strains. In addition, these results encourage the in planta analysis of transient (real‐time) gene expression in P. savastanoi using promoter fusions to genes encoding unstable GFP proteins (Andersen et al., 1998). Additional lines of research are also opened by this study, such as the determination of the role of OMVs in bacterial phytopathogens.
Experimental procedures
Bacterial strains and growth conditions
Pseudomonas savastanoi pv. savastanoi strain NCPPB 3335 and its GFP‐tagged derivative NCPPB 3335‐GFP, containing the plasmid pLRMl‐GFP, were routinely cultured in Luria–Bertani (LB) broth (Sambrook and Russell., 2001) at 28°C. When appropriate, gentamicin (Gm), kanamycin (Km) and nitrofurantoin (Nf) were added to the culture medium to a final concentration of 10 µg ml−1. Escherichia coli strains S17‐λpir [pJBA29] and DH5α were routinely cultured in LB broth and, when appropriate, supplemented with Km or Gm to a final concentration of 50 and 10 µg ml−1 respectively.
Construction of plasmid pLRM1‐GFP
Digestion of plasmid pJBA29 (Andersen et al., 1998) with SalI/EcoRI yielded a 2.05 kb fragment containing a fusion of the synthetic isopropyl‐β‐d‐galactopyranoside (IPTG)‐inducible promoter PA1/04/03(Lanzer and Bujard, 1988) to the gfpmut3* gene, encoding GFPmut3 (Cormack et al., 1996), a synthetic ribosome binding site (RBSII), translational stop codons in all three reading frames, and two strong transcriptional terminators, T0 (derived from phage λ) and T1 (derived from the rrnB operon of E. coli). This fragment (PA1/04/03‐RBSII‐gfpmut3*‐T0‐T1 cassette) was subsequently inserted into the plasmid pBBR1‐MCS5 (Kovach et al., 1995), which had been digested with SalI/EcoRI. The resulting plasmid, pLRM1‐GFP (6.83 kb) (Fig. 1), was maintained in E. coli DH5α. The pLRM1‐GFP plasmid was isolated from E. coli using the NucleoSpin plasmidic DNA purification kit (Macherey‐Nagel GmbH and Co. KG, Düren, Germany) and transformed into electrocompetent P. savastanoi pv. savastanoi NCPPB 3335 as previously described (Pérez‐Martínez et al., 2007). Green fluorescent transformants, containing pLRM1‐GFP (NCPPB 3335‐GFP), were selected on LB plates containing Gm and identified by epifluorescence microscopy using a stereoscopic microscope (Leica MZFLIII).
Plant inoculation and isolation of bacteria from olive knots
Micropropagated olive (Olea europaea L.) plants were derived from an in vitro germinated seed, collected from a cv. Arbequina plant, established in Driver–Kuniyuki walnut medium (Driver and Kuniyuki, 1984) and supplemented with sucrose at 20 g l−1 and myo‐inositol at 1 g l−1 as the carbon source. Plant micropropagation and rooting was performed as previously reported (Rodríguez‐Moreno et al., 2008).
In vitro olive plants were wounded by the excision of an intermediate leaf and inoculated with bacterial cells (approximately 104 cfu) as previously reported (Rodríguez‐Moreno et al., 2008). Plants were incubated in a growth chamber at 25 ± 1°C with a 16‐hour photoperiod. After different periods of time, P. savastanoi pv. savastanoi cells were recovered from inoculated explants as follows. Stem fragments sampled from 1 cm above and 1 cm below the inoculation point were crushed in a mortar with 1 ml of 10 mM MgCl2. Serial dilutions of the obtained suspensions were spotted onto LB‐Gm plates and incubated at 28°C for 3 days. Population densities were calculated from at least three replicates.
Symptoms induced on inoculated olive plants were visualized with a stereoscopic microscope (Leica MZ FLIII). Images were captured with a high‐resolution digital camera (Nikon DXM 1200) and processed for display using Adobe Photoshop CS software.
Plasmid pLRM1‐GFP stability assays in vitro and in planta
Stability of pLRM1‐GFP was assessed in vitro as follows. NCPPB 3335‐GFP, carrying pLRM1‐GFP, was grown overnight at 28°C in liquid LB‐Gm. The cultures were then diluted to an OD600 of 0.01 into fresh LB‐Nf with and without Gm, grown to stationary phase and then diluted again into the same medium as described above. This cycle was repeated until the strains were grown for 40 generations. At the time of each dilution, the cultures were plated onto LB‐Nf with and without Gm to score the presence of pLRM1‐GFP and to determine cell titres respectively.
In vitro olive plants were inoculated with NCPPB 3335‐GFP and, after different periods of time, bacteria were recovered from developed knots as described above. Serial dilutions of the obtained suspensions were spotted onto LB‐Nf with and without Gm to score the presence of pLRM1‐GFP and to determine cell titres respectively. All experiments were repeated at least three times.
Epifluorescence microscopy and confocal laser scanning microscopy
To visualize bacterial infection at real time in tumours, whole knots were directly examined with a stereoscopic fluorescence microscope (Leica MZ FLIII) equipped with a 100 W mercury lamp and FITC filter. Images were captured with a high‐resolution digital camera (Nikon DXM 1200) and processed for display using Adobe Photoshop CS software.
To visualize bacterial infection inside olive tumours, knots were sampled at different days post inoculation, 1 cm above and 1 cm below the inoculation point. Knot samples were fixed and included in agarose as described by Alvarez and colleagues (2006). Samples were fixed overnight in 2.5% paraformaldehyde (PFA), pH 7.4, at 4°C. Fixed samples were then transferred into 2.5% PFA with an ascending gradient of 10%, 20% and 30% sucrose for 10, 20 and 30 min respectively. Finally, samples were included into 7% low melting point agarose and cooled down to 4°C. Sections of 40 and 60 µm in thickness were obtained from olive knot samples with a freezing microtome (Leica CM1325). Green fluorescence emitted by bacterial cells inside olive knot sections was visualized by epifluorescence microscopy, using a Nikon Microphot‐FXA microscope. For confocal microscopy, an inverted confocal laser scanning microscope (model TCS‐NT, Leica, Germany), equipped with detectors and filters set that simultaneously monitor GFP and red fluorescence, was used. Image acquisitions of green fluorescence were carried out using excitation at 488 nm and by collecting emitted light from 500 to 560 nm. Images were obtained by sequential scan analyses; image projections were processed using Adobe PhotoShop 6 software (Adobe, Mountain View, CA, USA).
Methylene blue‐picrofuchsin stain
Olive knot samples, sectioned and fixed as described above, were stained for 10 s in 1% methylene blue. Then they were washed in ethanol (96%), followed by distilled water and finally stained for 5 min in picrofuchsin. Picrofuchsin contained 0.1% acid fuchsin in a saturated picric acid solution. Stained sections were dehydrated, mounted on slides with Canadian balsam and visualized with a Nikon Eclipse 800 light microscope.
Scanning electron microscopy
Knot samples, obtained as described above, were cut into cross‐sections of approximately 1–2 mm in thickness with a blade. Sections were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, overnight at 4°C. After dehydration in a gradient of alcohol and critical point drying, tissue blocks were coated with ionized gold and scanned with an electron microscope JEOL JSM‐840 using a secondary electron detector.
Transmission electron microscopy
Knot cross‐sections of 1–2 mm in thickness were obtained with a blade, immersed into 3% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 2 h at room temperature and then at 4°C overnight. Post‐fixation took place in cold 1% osmium tetroxide for 2 h at 4°C. Fixed sections were dehydrated and embedded in Araldite 502. Semi‐thin (1 µm thick) and ultrathin (50–70 nm thick) sections were obtained with an ultramicrotome (model Ultracut E, Leica, Germany). Semi‐thin sections were mounted on glass slides and stained with 1% toluidine blue. Ultrathin sections were mounted in grids, stained using Reynold's lead citrate solution and uranyl acetate and visualized using a Philips CM100 electron microscope.
Acknowledgments
This project was supported by Spanish MEC grants, numbers AGL2005‐02090 and AGL2008‐05311‐C02‐02, a Junta de Andalucía grant CVI‐264, and co‐financed by FEDER. We thank D. Navas for assistance with confocal microscopy, as well as G. Martín and A. Martínez for help with scanning and transmission electron microscopy respectively. A. Rivera and M. Reina are thanked for help with preparation of olive knot samples for epifluorescence microscopy. We are grateful to M. Duarte for excellent technical assistance and to A. Barceló and I. Vidoy for the micropropagation of olive plants.
References
- Abbott J.G., Bornman C.H., Noel A.R.A. Nicotiana tabacun callus studies. V Ontogeny and structure of phloem. Z Pflanzenphysiologie. 1977;83:1–10. [Google Scholar]
- Aloni R., Pradal K.S., Ullrich C.I. The three‐dimensional structure of vascular tissues in Agrobacterium tumefaciens‐induced crown galls and in the host stems of Ricinus communis L. Planta. 1995;196:597–605. [Google Scholar]
- Alvarez J.P., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. Endogenous and synthetic MicroRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.B., Sternberg C., Poulsen L.K., Bjørn S.P., Givskov M., Molin S. New unestable of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel J.L., Charkowski A.O., Deng W.L., Collmer A. A gene in the Pseudomonas syringae pv. tomato Hrp pathogenicity island conserved effector locus, hopPtoA1, contributes to efficient formation of bacterial colonies in planta and is duplicated elsewhere in the genome. Mol Plant Microbe In. 2002;15:1014–1024. doi: 10.1094/MPMI.2002.15.10.1014. [DOI] [PubMed] [Google Scholar]
- Bertolini E., Penyalver R., García A., Olmos A., Quesada J.M., Cambra M., López M.M. Highly sensitive detection of Pseudomonas savastanoi pv. savastanoi in asymptomatic olive plants by nested PCR in a single closed tube. J Microbiol Meth. 2003;52:261–266. doi: 10.1016/s0167-7012(02)00163-x. [DOI] [PubMed] [Google Scholar]
- Bloemberg G.V., Wijfjes A.H.M., Lamers G.E.M., Stuurman N. Simultaneous imaging of Pseudomonas fluorescens WCS365 population expressing three different autofluorescent proteins in the rizhosphere: new perspectives for studying microbial communities. Mol Plant Microbe In. 2000;13:1170–1176. doi: 10.1094/MPMI.2000.13.11.1170. [DOI] [PubMed] [Google Scholar]
- Boldt T.S., Sørensen J., Karlson U., Molin S., Ramos C. Combined use of different Gfp reporters for monitoring single‐cell activities of a genetically modified PCB degrader in the rhizosphere of alfalfa. FEMS Microbiol Ecol. 2004;48:139–148. doi: 10.1016/j.femsec.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Boureau T., Jacques M.A., Berruyer R., Dessaux Y., Dominguez H., Morris C.E. Comparison of the phenotypes and genotypes of biofilm and solitary epiphytic bacterial populations on broad‐leaved endive. Microb Ecol. 2004;47:87–95. doi: 10.1007/s00248-002-0106-4. [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Eushirchen G., Ward W.W., Prasher D.C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Comai L., Kosuge T. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J Bacteriol. 1980;143:950–957. doi: 10.1128/jb.143.2.950-957.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia R.H., Falkow S. FACS optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Driver J.A., Kuniyuki A. In vitro propagation of paradox walnut rootstock. HortScience. 1984;19:507–509. [Google Scholar]
- Ercolani G.L. Pseudomonas savastanoi and other bacteria colonising the surface of olive leaves in the field. J Gen Microbiol. 1978;109:245–257. [Google Scholar]
- Ercolani G.L. Factor analysis of fluctuation in populations of Pseudomonas syrinage pv. savastanoi on the phylloplane of the olive. Microb Ecol. 1985;11:41–49. doi: 10.1007/BF02015107. [DOI] [PubMed] [Google Scholar]
- Ercolani G.L. Distribution of epiphytic bacteria on olive leaves and the influence of leafage and sampling time. Microb Ecol. 1991;21:35–48. doi: 10.1007/BF02539143. [DOI] [PubMed] [Google Scholar]
- Gardan L., Bollet C., Abu‐Ghorrah M., Grimont F., Grimont P.A.D. DNA relatedness among the patovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int J Syst Bacteriol. 1992;42:606–612. [Google Scholar]
- Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., Peterson K.M. Four new derivatives of the broad host‐range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Kuehn M.J., Kesty N.C. Bacterial outer membrane vesicles and the host‐pathogen interaction. Gene Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M., Myers C.R., Collmer A., Schneider D.J. Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol Plant Microbe In. 2008;21:685–700. doi: 10.1094/MPMI-21-6-0685. [DOI] [PubMed] [Google Scholar]
- Magie E.E. University of California; 1963. Physiological factors involved in tumor production by the oleander knot pathogen. [Google Scholar]
- Normander B., Christensen B.B., Molin S., Kroer N. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris. Appl Environ Microbiol. 1998;64:1902–1909. doi: 10.1128/aem.64.5.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penyalver R., García A., Ferrer A., Bertolini E., López M.M. Detection of Pseudomonas savastanoi pv. savastanoi in olive plants by enrichment and PCR. Appl Environ Microbiol. 2000;66:2673–2677. doi: 10.1128/aem.66.6.2673-2677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penyalver R., Garcia A., Ferrer A., Bertolini E., Quesada J.M., Salcedo C.I. Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopahology. 2006;96:313–319. doi: 10.1094/PHYTO-96-0313. et al. [DOI] [PubMed] [Google Scholar]
- Pérez‐Martínez I., Rodríguez‐Moreno L., Matas I.M., Ramos C. Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Res Microbiol. 2007;158:60–69. doi: 10.1016/j.resmic.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Pérez‐Martínez I., Zhao Y., Murillo J., Sundin G.W., Ramos C. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. J Bacteriol. 2008;190:625–635. doi: 10.1128/JB.01067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada J.M., García A., Bertolini E., López M.M., Penyalver R. Recovery of Pseudomonas savastanoi pv. savastanoi from symptomless shoots of naturally infected olive trees. Int Microbiol. 2007;10:77–84. [PubMed] [Google Scholar]
- Ramos C., Mølbak L., Molin S. Bacterial activity in the rhizosphere at the single‐cell level by monitoring ribosome contents and synthesis rates. Appl Environ Microbiol. 2000;66:801–809. doi: 10.1128/aem.66.2.801-809.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Moreno L., Barceló‐Muñoz A., Ramos C. In vitro analysis of the interaction of Pseudomonas savastanoi pvs. savastanoi and nerii with micropropagated olive plants. Phytopathology. 2008;98:815–822. doi: 10.1094/PHYTO-98-7-0815. [DOI] [PubMed] [Google Scholar]
- Sabaratnam S., Beattie G.A. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl Environ Microbiol. 2003;69:1220–1228. doi: 10.1128/AEM.69.2.1220-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. 3rd. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sarkar S.F., Gordon J.S., Martin G.B., Guttman D.S. Comparative genomics of host‐specific virulence in Pseudomonas syringae. Genetics. 2006;174:1041–1056. doi: 10.1534/genetics.106.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu V.K., Vorhölter F.J., Niehaus K., Watt S.A. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008;8:87. doi: 10.1186/1471-2180-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisto A., Cipriani M.G., Morea M. Knot formation caused by Pseudomonas savastanoi subsp. savastanoi on olive plants is hrp/hrc dependent. Phytopathology. 2004;94:484–489. doi: 10.1094/PHYTO.2004.94.5.484. [DOI] [PubMed] [Google Scholar]
- Smidt M., Kosuge T. The role of indole‐3‐acetic acid accumulation by alpha‐methyl tryptophan‐resistant mutants of Pseudomonas savastanoi in gall formation in oleander. Physiol Plant Pathol. 1978;13:203–214. [Google Scholar]
- Smith E.F. W.B. Saunders Company; 1920. [Google Scholar]
- Surico G. Osservazioni istologiche sui tubercoli della ‘ogna’ dell'Olivo. Phytopathol Mediterr. 1977;16:109–125. [Google Scholar]
- Surico G. Scanning electron microscopy of olive and oleander leaves colonized by Pseudomonas syringae subsp. savastanoi. J Phytopathol. 1993a;138:31–40. [Google Scholar]
- Surico G. Symptom development in olive and oleander leaves inoculated with Pseudomonas syringae subsp. savastanoi and scanning electron microscopy on infections. Petria. 1993b;3:117–127. [Google Scholar]
- Surico G., Iacobelis N.S., Sisto S. Studies on the role of indole‐3‐acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol Plant Pathol. 1985;26:309–320. [Google Scholar]
- Temsah M., Hanna L., Saad A.T. Anatomical observations of Pseudomonas savastanoi on Rhamnus alaternus. Forest Pathol. 2007a;37:64–72. [Google Scholar]
- Temsah M., Hanna L., Saad A.T. Histological Pathogenesis of Pseudomonas savastanoi on Myrtus communis. J Plant Pathol. 2007b;89:241–249. [Google Scholar]
- Ullrich C.I., Aloni R. Vascularization is a general requirement for growth of plant and animal tumours. J Exp Bot. 2000;51:1951–1960. doi: 10.1093/jexbot/51.353.1951. [DOI] [PubMed] [Google Scholar]
- Wang K., Kang L., Anand A., Lazarovits G., Mysore K.S. Monitoring in planta bacterial infection at both cellular and whole‐plant levels using the green fluorescent protein variant GFPuv. New Phytol. 2007;174:212–223. doi: 10.1111/j.1469-8137.2007.01999.x. [DOI] [PubMed] [Google Scholar]
- Wilson E.E. Pathological histogenesis in oleander tumors induced by Pseudomonas savastanoi. Phytopathology. 1965;55:1244–1249. [Google Scholar]
- Wilson E.E., Magie A.R. Pathological histology of tumors induced in oleander by Pseudomonas savastanoi. Phytopathology. 1963;53:893. [Google Scholar]
- Wilson E.E., Magie A.R. Systemic invasion of the host plant by the tumor inducing bacterium, Pseudomonas savastanoi. Phytopathology. 1964;54:577–579. [Google Scholar]
- Zhao Y., Ma Z., Sundin G.W. Comparative genomic analysis of the pPT23A plasmid family of Pseudomonas syringae. J Bacteriol. 2005;187:2113–2126. doi: 10.1128/JB.187.6.2113-2126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


