Summary
The major biological pesticide for the control of insect infestations of crops, Bacillus thuringiensis was found to be present naturally within cotton plants from fields that had never been treated with commercial formulations of this bacterium. The ability of B. thuringiensis to colonize plants as an endophyte was further established by the introduction of a strain marked by production of green fluorescent protein (GFP). After inoculation of this preparation close to the roots of cotton and cabbage seedlings, GFP‐marked bacteria could be re‐isolated from all parts of the plant, having entered the roots and migrated through the xylem. Leaves taken from the treated plants were able to cause toxicity when fed to the Lepidoptera Spodoptera frugiperda (cotton) and Plutella xylostella (cabbage). These results open up new horizons for understanding the natural ecology and evolution of B. thuringiensis and use of B. thuringiensis in insect control.
Introduction
Bacillus thuringiensis (Bt) is a naturally occurring Gram positive spore‐forming bacterium commonly used in the biological control of insects. The biologically active components of Bt are its crystal proteins synthesized by the bacterium upon sporulation and then released into a crystal upon spore lysis. Despite its extensive use for decades, the natural ecology of Bt remains poorly understood.
Bacillus thuringiensis can be successfully isolated from the soil, dead insects and the phylloplane (Schnepf et al., 1998; Damgaard, 2000). Although Bt can be found on the phylloplane and is most typically used in the control of insects that live there, a great mystery is how the bacterium ends up on plants. Spores can reach the lower levels of plants by rain splash (Pedersen et al., 1995), but in general there has been little work to demonstrate how the bacterium is able to colonize plants so successfully in order to reach insects on the phylloplane.
Studies indicate that the Bt species found in the soil and phylloplane may be the same, and it has been suggested in one study that Bt found on cabbage foliage was somehow transported there from the soil (Damgaard, 2000). More recently, a different study observed that Bt inoculated into soil sown with clover seeds could end up in the leaves after the seeds germinated and grew (Bizzarri and Bishop, 2007). However, how the Bt might have made its way from the soil to the leaves was not explained (e.g. it could have been brought up on sprouts) nor could the results explain the observation that Bt is present in stable populations on the leaves of deciduous trees (Smith and Barry, 1998). These data all point to the enigma of how Bt is able to stably colonize the phylloplane, how it evolved to kill insects that live there, and how Bt is able to, presumably, cycle between the phylloplane and soil.
Here we discover that Bt can naturally be found inside plants. Furthermore, Bt introduced into the soil can be taken up by plants and translocated throughout the plant. Such translocated Bt living inside plants have insecticidal activity. These results open up new horizons for understanding the natural ecology and evolution of Bt and use of Bt in insect control.
Results
Live Bt can be isolated from field plants
Ninety‐day‐old cotton plants from a field site close to Brasilia, which had never been the site of Bt application, were collected, surface sterilized, sectioned and passed through several steps to select for Bt that might be present specifically inside the plants. Seven colonies were isolated from throughout the plants and showed Bt‐like morphology under light microscopy. Further analysis by SDS‐PAGE indicated the presence of approximately 130 and 70 kDa proteins in the spore preparation of one strain, named S1942. PCR screening and toxicity assays produced no amplicons for the other six strains, which also showed no toxicity against the insects tested. In contrast, S1942 produced amplicons indicating the presence of cry1Aa, cry1Ab, cry1Ac, cry1B and cry2A genes, consistent with the production of ∼130 and 70 kDa protoxins as seen by SDS‐PAGE (not shown). This strain was also toxic to lepidopteran insects (Spodoptera frugiperda, Anticarsia gemmatalis and Plutella xylostella) but was not toxic to the other tested insects. The protein and cry gene profiles indicated that this endophytic strain S1942 was similar to Bt serotype kurstaki HD1 (Monnerat et al., 2007).
Bacillus thuringiensis introduced into soil is taken up by plants
The above results demonstrated that Bt could be found inside living plants. We hypothesized that the Bt might be taken up from the plant roots. To test this hypothesis, we used the standard Bt strain, Btk HD1, transformed with a construct to allow expression of green fluorescent protein (GFP), thereby permitting us to identify the inoculated strain unambiguously and, therefore, rule out contamination with other bacteria. This strain (Btk::GFP) was inoculated into soil containing 28‐day‐old cotton or cabbage plants. Sample plants were uprooted weekly and processed as above to detect the internal presence of Bt. Confirmation that the Bt found in the plants was the same as that inoculated into the soil was based on: selection on penicillin; the ability to sporulate; production of GFP; and the detection of appropriate crystal toxin genes for Btk (cry1Aa, cry1Ab, cry1Ac, cry2). These results (Table 1, cotton plants; identical results were obtained for cabbage plants) show that Btk::GFP introduced into the soil was taken up by the plant and could make its way into any part of the plant. The bacterium was able to persist in plant tissues for at least 7 weeks following an inoculation (Table 1, column ‘S’). Fluorescent bacteria were never recovered from control plants that had not been inoculated, thereby excluding contamination as a source of fluorescent Bt in the experimental plants.
Table 1.
Detection of Btk::GFP in different cotton tissues.
| Weeks | Soil | Root | Stem | Petiole | Leaf | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | S | W | C | S | W | C | S | W | C | S | W | C | S | W | |
| 0 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 2 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 3 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 4 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 5 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 6 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 7 | − | + | + | − | + | + | − | + | + | − | + | + | − | + | + |
| 8 | − | + | + | − | + | + | − | − | + | − | + | + | − | + | + |
Presence (+) or absence (−) of Btk::GFP is indicated in the 8 weeks following the initial inoculations: after single application, (S); after weekly applications, (W); and in the un‐inoculated control, (C).
To investigate the presence of bacteria directly inside the plant, fluorescence microscopy was employed to visualize bacteria in plant sections (using a 530–550 nm filter to exclude other fluorescence). Individual bacteria, fluorescent with GFP and motile, could be seen inside the plant in such areas as the cotton petiole (representative image shown in Fig. 1) (similar results were seen in cabbage plants). We are confident that the bacteria seen were from inside the plant and not from external contamination (a potential issue with Bt) because the plants were surface sterilized and the bacteria were clearly and consistently seen inside the xylem. The association of the bacteria with the lumen of the xylem suggests that they are being translocated through the plant in the ascending water column.
Figure 1.
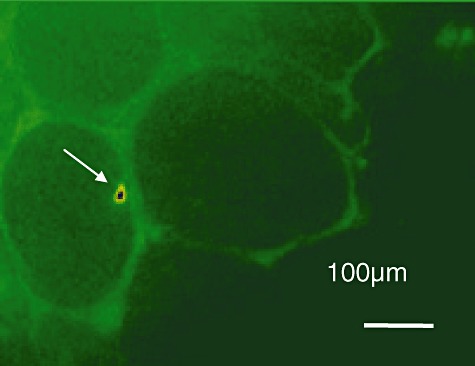
Localization of Btk::GFP in petiole. The arrow indicates the presence of Btk::GFP visible in the Xylem of the plant. Fluorescence signal as well as its movement in the xylem was used to ascertain the presence of the bacterium. Many Btk::GFP were visible in the samples although only a few were seen at any time at high resolution due to the narrow focal plane.
To confirm uptake of Bt into plants independent of GFP, accumulation of Bt in the leaves was studied by autoradiography following inoculation of plants with radiolabelled bacteria (Fig. 2). The radioactivity could be detected in all parts of inoculated plants. In contrast, the radioactivity was not detected in plants inoculated with the final wash solution used to remove residual, unincorporated 35S‐methionine from the preparation and thus providing a control demonstrating that the label observed on addition of Bt was not due to unincorporated radiolabel.
Figure 2.
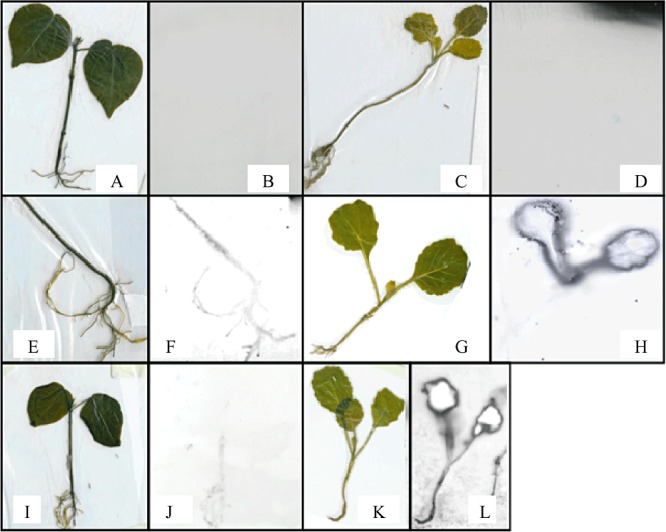
Plants inoculated with radiolabelled Btk 35SMet. A and C. Control plants of cotton and cabbage respectively, inoculated with supernatant of the final wash of Bt culture. B and D. Autoradiography of control plants of cotton and cabbage respectively. E and I. Cotton inoculated once with 2.0 ml radiolabelled Btk 35SMet. F and J. Autoradiography of cotton inoculated once with 2.0 ml radiolabelled Btk 35SMet; G. Cabbage inoculated once with 2.0 ml radiolabelled Btk 35SMet. H. Autoradiography of cabbage inoculated once with 2.0 ml radiolabelled Btk 35SMet. K. Cabbage inoculated five times with 2.0 ml radiolabelled Btk 35SMet. L. Autoradiography of cabbage inoculated five times with 2.0 ml radiolabelled Btk 35SMet. The plants were exposed for autoradiography for 7 days.
To get a better idea of the distribution of the bacterium, we looked at the presence of GFP in plants using an anti‐GFP antibody and laser confocal microscopy. The ability of the method to label GFP within non‐fixed bacteria was first confirmed using bacteria alone (Fig. 3). Analysis of control plants inoculated with non‐labelled bacteria showed only parenchyma (the cellular surface of the leaves) and guard‐cell autofluorescence in the tissue and no fluorescence signal in the veins (Fig. 4A–I). In contrast, observation of the leaves of inoculated plants showed a major presence of Btk::GFP in the veins, probably attached to the walls and almost no fluorescent signal in the parenchyma (Fig. 4). However, some labelling could sometimes be found in this tissue adjacent to the end of the veins (Fig. 4), indicating that the bacteria can reach the parenchyma in these specific regions.
Figure 3.
Confocal microscopy images of the Btk::GFP, showing (A) control bacteria, (B) Btk::GFP labelled with Alexa 594 in the bacteria.
Figure 4.
Confocal microscopy images of cabbage leaves. Leaves were taken from cabbage plants 48 h after root inoculation with sterile NYSM medium (controls: A–C) or Btk::GFP (D–I). Images show: autofluorescence of plant tissue alone at 515 nm (A, D, G,); fluorescence at 594 nm due to anti‐GFP antibody (B, E, H); merged images of autofluorescence and antibody fluorescence (C, F, I). The following structures are labelled: c, chloroplasts; p, parenchyma; v, vein.
Plant tissues containing endophytic Bt are toxic to insects
Because Bt are taken up by plants and because we found an insecticidal Bt naturally occurring in plants, we hypothesized that Bt in plants might actually have insecticidal activity. To test this hypothesis, leaves from Btk::GFP‐treated plants were fed to S. frugiperda and P. xylostella larvae. Although there was significant variation among experimental treatments, mortality was always observed with both the single and weekly inoculated plants, typically at a level of 10–20%, and never observed with non‐treated plants. Single inoculation gave maximal mortality around 3–5 weeks post inoculum. When leaves were taken from plants that had been inoculated at weekly intervals, insect mortality levels appeared slightly higher (10–25%). To confirm that lethality was coming from the bacteria inoculated into the plants, bacteria were re‐isolated from the guts of both dead and living larvae. No Bt were produced from control larvae from untreated plants while many Bt colonies were isolated from larvae from treated plants. The GFP expression was observed by fluorescence microscopy for all of these colonies, confirming the origin of these isolates from the experimental inoculation.
To analyse the effects on insects in detail, the midguts of S. frugiperda fed for 24 h on leaves from Bt‐inoculated cotton plants and in artificial diet were dissected out and analysed using anti‐GFP antibodies. We found most bacteria in the insect midgut (Fig. 5). Other organs like adipose and tracheal systems were infected as well (not shown). Tissues from insects fed on control plants showed structural integrity and no fluorescence signal (Fig. 5).
Figure 5.
Confocal microscopy images of S. frugiperda larvae. Larvae were fed on leaves of cabbage plants inoculated with sterile NYSM medium (controls: A–D) or with Btk::GFP for 24 h (I–L) and larvae fed on diet containing 1.3 × 103 cells to Btk::GFP for 24 h. (E–H) Images show: fluorescence due to DAPI blue (A, E, I). Fluorescence at 488 nm due to GFP protein (C, G, K). Fluorescence at 594 nm due to anti‐GFP antibody (B, F, J); merged images of DAPI blue, GFP fluorescence and antibody fluorescence (D, H, L). The following structures are labelled: av, apical vesicles; bl, basal lamina; n, nucleus; sb, striated border.
Discussion
Here we demonstrate for the first time the uptake of live Bt from soil, up through the plant, and into leaves. Our evidence is as follows. Live GFP‐labelled Bt inoculated into soil near roots of two different plants can be recovered in all tissues of the plant; radiolabelled Bt can be taken up and detected in all tissues of the plants; bacteria can be seen inside the plant, in particular in the xylem (suggesting a means by which it is transported); and insects fed upon the leaves of such plants can be intoxicated by the Bt and the labelled Bt can be recovered from live and dead insects. We have taken great care to ensure that the Bt is not being introduced from outside sources by labelling the bacteria introduced into the soil to follow it unambiguously; by covering the soil to prevent aerosolylated Bt from contaminating the phylloplane; by extensive surface sterilization of plant tissues; and by showing that neighbouring control pots of plants, in which Bt was not introduced, always showed a complete lack of Bt.
Recent reports (Bizzarri and Bishop, 2007; Maduell et al., 2007) have described the isolation from leaves of Bt strains that were inoculated along with seeds. However, in these reports it was not possible to establish whether the presence of Bt on aerial parts of the plants was due to direct surface contact of emerging plant tissues with bacteria in the soil or on the surface of the seed. The experiments described here represent the first unequivocal demonstration of the systemic endophytic migration of Bt in seedlings from root to leaf.
There are two dramatic implications of this work. First is for understanding the evolution of Bt. Bacillus thuringiensis is an insect pathogen and tends to target insects that live in the phylloplane. Yet an important reservoir of this organism resides in the soil. How can the bacteria move between these localities? Our results here provide a rational explanation. Bacillus thuringiensis in the soil can be taken up into the plant and deposited into the leaves, where it can intoxicate insects. Either via insect droppings (Bizzarri and Bishop, 2007) or via occasional dead insects falling into the soil, the bacterium can recycle back to the soil reservoir. This then allows the host–pathogen interaction to evolve over time in an arms‐race leading to different susceptibilities to Bt toxins. Perhaps insects are in contact with various pathogenic Bt over a long period of time (via persistence of Bt in the phylloplane from constant root uptake) and thus can develop resistance, which in turn selects for new Bt variants that newly target the same insect or different insect populations.
The second area that this work impacts is biological control. Although high levels of lethality were not seen here, our experiments suggest that Bt applied directly into the soil can serve as a source for biological control from pests. As such Bt would be shielded from UV in sunlight (which damages the crystals), it could serve as a protected source of biological control over periods of time. Our methodology implied here would represent a powerful new method for crop protection that offers the possibility of serial inoculation of plants with multiple Bt strains at different stages in a flexible response to combat a range of pest insects that may infest crops at different stages of growth or from one season to the next. For instance, cotton may be subject to serial infestation with the Lepdioptera Alabama argilacea, S. frugiperda and the Coleopteran Anthonomus grandis. The cotton crop is regularly fertilized close to the soil and during this procedure the appropriate Bt strains that target all of these different insects could be applied either simultaneously or serially. The levels of insect toxicity reported herein were achieved without optimization and it is likely that an understanding of the interactions between Bt and host plants will allow specific adaptation of methodologies to suit both the Bt strain deployed and the particular plant host, to attain viable levels of control for plant protection in the field.
Experimental procedures
Isolation of Bt from field plants
On each of two trips to field sites that had never been used in the application of Bt, five cotton plants, including roots, were dug up from soil, brought to the laboratory, washed extensively with water, and dried in a sterile hood, where all subsequent procedures were carried out. The root was surface sterilized by immersion in 5% hypochlorite for 5 min followed by 90% ethanol for 5 min and allowed to dry for a few minutes. Using a sterile scalpel, the ends of one of the roots were cut on a sterile surface and the newly exposed end pressed on NYSM medium agar (Myers and Yousten, 1978) plates in three different places on a single plate (see below). This procedure was repeated two to three more times with different roots. A similar procedure was then carried out working our way up the rest of the plant, namely stems, branches, petioles (leaf stalks) and leaves. The plates were then incubated at 30°C for 72 h. Each colony was then incubated in NYSM liquid medium at 30°C for 72 h to allow for sporulation. Then 1 ml of each sample was heat‐shocked at 80°C for 12 min. Using a sterile loop, an aliquot from each sample was spread onto NYSM medium agar plates containing 10 µg ml−1 penicillin to select for the growth of Bt (Silva et al., 2002).
To determine the identity of the bacteria that grew, the colonies were examined under phase contrast microscopy to look for spores and crystals diagnostic of Bt. These samples were further analysed for the presence of crystal proteins by (i) SDS‐PAGE, (ii) crystal toxin gene profiles and (iii) toxicity bioassays against a variety of insect larvae as follows. (i) For SDS‐PAGE bacterial proteins were extracted from the culture according to Lecadet and colleagues (1991). Samples of 15 µl were separated by electrophoresis on 10% polyacrylamide gels before staining with 0.1% (w/v) Coomassie blue. (ii) For crystal toxin gene profiles, Bt isolates identified in (i) were grown on NYSM agar for 14–15 h, at 30°C and DNA was extracted as described by Sambrook and colleagues (1989). Molecular characterization through PCR was performed to identify the toxin‐coding genes, by using a variety specific primers for the following genes/gene families: cry1, cry2, cry3, cry4, cry5, cry7, cry8, cry9, cry10, cry11, cry12, cry13, cry14, cry17, cry19, cry21, cry24, cry25, cry27, cry29, cry30, cry32, cry39, cry40, cyt1 and cyt2 (Ceron et al., 1995; Bravo et al., 1998; Ibarra et al., 2003). (iii) For insect bioassays, insects were reared in the laboratory, at 26 ± 2°C, 70 ± 10% relative humidity, and a photoperiod of 14:10 (L:D) (Schmidt et al., 2001). Qualitative bioassays were performed in order to identify those isolates capable of killing 100% of the larvae tested at a single dose. The bioassays were performed as previously described against larvae of S. frugiperda (Lepidoptera: Noctuidae), P. xylostella (Lepidoptera: Plutellidae), Anticarsia gemmatalis (Lepidoptera: Noctuidae) (Tabashnik et al., 1990; Monnerat et al., 1999), Culex quinquefasciatus (Diptera: Culicidae), Aedes aegypti (Diptera: Culicidae) (Monnerat et al., 2005) and Anthonomus grandis (Coleoptera: Curculionidae) (Martins et al., 2007).
Systemic uptake of Bt into plants
To follow the uptake of Bt into plants, a GFP marked Bt strain was constructed. Bacillus thuringiensis kurstaki strain HD1 (obtained from the Collection of Entomopathogenic bacilli at the Institut Pasteur, Paris, France) was transformed by eletroporation (Bone and Ellar, 1989) using standard Bt protocols with plasmid pGFP304, which expresses GFP during the vegetative phase (a kind gift from Professor Lin Li, Huazhong Agricultural University, Wuhan, China). This GFP can be detected in growing Bt cells and more faintly in spores. The resulting strain, designated Btk::GFP, was tested along with its parental Btk strain in bioassays using S. frugiperda and P. xylostella. Both strains had the same toxicity on both insects, demonstrating that GFP expression did not affect the functionality of Btk (results not shown).
(i) Uptake of GFP bacteria: To test Bt uptake into plants by root inoculation, cotton (variety Delta‐Opal) and cabbage (hybrid Matsukaze Sakata) plants were grown in unautoclaved soil in plastic pots with four plants per pot. After ∼1 week, when the seedlings emerged, the soil was covered in plastic (with the plant rising through it) for the duration of the experiment to prevent any possible transference from the soil to the plant except via root uptake. During watering or inoculations, the plastic was temporarily and carefully lifted to allow access to the soil. After 28 days, these seedlings were split into three groups of 30 seedlings. Thirty ‘control plants’ were watered daily with 20 ml water per plant. Thirty ‘single inoculation plants’ were given a single inoculation of 5 ml of a 108 cells ml−1 suspension of Btk::GFP applied carefully with a pipette near the roots of each plant in the pot. These plants were watered thereafter as for the control group. Thirty ‘weekly inoculation plants’ were subjected to weekly administration of 5 ml of a 108 cells ml−1 suspension of Btk::GFP for an 8 week period. Every week, three plants randomly selected from each group were collected and surface sterilized before sectioning and plating for bacteria as described above for field cotton and cabbage plants.
To confirm the identity of the bacteria that grew on the penicillin selection plates, a sample of each colony was taken 12 h after inoculation on the plate to detect the presence of GFP in vegetative cells using a Zeiss Axiophot microscope and a 40× 0.75 NA Plan Neofluor lens. In addition, 2 days later, a sample of each colony was examined under phase contrast for the presence of spores. Furthermore, one colony from each week's sample was examined by PCR for the presence of all applicable crystal toxin genes. All tests carried out on all the colonies confirmed that each colony was Btk::GFP. For the image in Fig. 1, plant tissue was sectioned by hand and observed in Zeiss Axiophot microscope. These experiments were repeated twice for cotton and three times for cabbage, always with the same result. The data in Table 1 are from one of these experiments in cotton.
(ii) Uptake of radiolabelled bacteria. Systemic uptake of Bt by cotton and cabbage plants was also studied using radiolabelled Bt. Bacillus thuringiensis kurstaki strain HD1 (non‐GFP) was grown for 24 h in 10 ml NYSM medium supplemented with 35S‐methionine (final 1 µCi ml−1). The resulting spores and vegetative cells were washed three times with 10 ml phosphate‐buffered saline (PBS). The final wash solution was kept for use as a control (see below) and the cell pellet was resuspended in 10 ml PBS and used to inoculate 28‐day‐old cabbage and cotton plants as above with 4 × 106 cells ml−1. The experiment was repeated twice with one pot of cotton and one pot of cabbage per experiment (four plants per pot). Each plant received five inoculations of 2 ml of bacterial suspension applied at the roots at 5 h intervals. Control plants received inoculations with the last wash that was performed to remove radioactivity to control for any residual radioactivity in the inoculum. Two hours after the final inoculation, all the plants were taken from the pots and the roots washed with water. The plants were dried in a hood on paper for 2 h, wrapped in plastic, and then subjected to autoradiography. Typical results are shown in Fig. 2.
Detection of bacteria in plants using indirect immunofluorescence and confocal laser scanning microscopy
In order to confirm that detection of GFP in non‐fixed bacteria was possible using the anti‐GFP antibody Alexa Fluor 594 method, a preliminary assay was performed according to the manufacturer's protocol (Invitrogen).
Twenty‐eight‐day‐old cabbage plants were inoculated as above by root application of 1.5 × 106 cells of Btk::GFP grown in NYSM medium for 24 h. Control plants were inoculated with 5 × 106 cells of non‐labelled Btk. After 2 days, leaves were cut from the petioles of both sets of plants while immersed in water to avoid the entrance of air and formation of bubbles that could block the entrance of the subsequent solutions. The antibody labelling was conducted by immersing the leaves from both control and Btk::GFP‐inoculated plants in PBS with 5.0 µg ml−1 anti‐GFP antibody (goat anti‐GFP IgG Alexa Fluor 594 from Invitrogen, Cat. No. A21312) and incubating overnight to optimize the absorption of the antibodies by the plant. The following day, the leaves were immersed in PBS for 4 h to wash away unbound antibody. Non‐fixed/embedded leaf tissue (∼1.5 cm2) immersed in PBS was mounted on regular glass slides and analysed using a Zeiss LSM 410 confocal fluorescence microscope equipped with Ar (Ex 488 nm) and He/Ne (Ex 543 nm) lasers. Digital images were acquired using LP 515 filter for 488 nm and BP 590‐610 filter for 543 nm. The antibody conjugate (Alexa Fluor 594) and excitation filter used were chosen to minimize autofluorescence from the plants and insects. Insect tissue autofluorescence ranges around the green spectrum (∼500–550 nm) when excited under blue light (488 nm), and plant autofluorescence, mostly from chlorophyll, ranges around the red spectrum (∼600–650 nm) under blue light. The conjugate antibody was exposed to the 543 nm laser and under this wavelength only the antibody fluorescence could be observed.
Insect assays and confocal microscopy
Single leaves (approximately 12 cm2) from treated or control plants were collected into sterile Petri dishes prior to the addition of 10 second‐instar S. frugiperda for cotton leaves and P. xylostella larvae for cabbage leaves. Each treatment was carried out in triplicate and larval mortality was assessed after 7 days. In contrast to some S. frugiperda colonies, the insects used in this study are highly susceptible to Btk HD1 (Monnerat et al., 2007).
To test for the presence of bacteria in the insects that fed on the leaves, both living and dead insects were surface sterilized by washing in 5% hypochlorite, before homogenization and plating onto NYSM agar containing 10 µg ml−1 penicillin for selection. After 24 h incubation at 30°C, the colonies were observed by fluorescence microscopy to confirm the presence of GFP in the isolates.
To localize the bacteria in the insects, 28‐day‐old cabbage plants were inoculated as above by root application of 1.5 × 106 cells of Btk::GFP grown in NYSM medium for 24 h. After 2 days, leaves were cut from the petioles and offered to second‐instar S. frugiperda larvae. Control plants were inoculated with 1.5 × 106 cells of unlabelled Btk. In addition second‐instar S. frugiperda larvae were fed in S. frugiperda artificial diet (Monnerat et al., 2007) inoculated with 1.5 × 106 cells of Btk::GFP grown in NYSM medium for 24 h. Twenty‐four hours latter, the larvae (from diet and plant) were dissected with a dorsal incision to facilitate the entrance of reagents. After incision, the whole larvae were fixed in Zamboni's Fixative (Stefanini et al., 1967) for 18 h at 4°C and then washed three times with Sorensen Phosphate Buffer 0.2 M. The larvae were dehydrated in increasing concentrations of ethanol (70%, 95% and 100% twice) for 15 min each. After dehydration, the paraplast® (Fisherbrand) embedding procedure was initiated with ethanol 100% and xylol (1:1) for 1 h at room temperature; xylol and paraplast (1:1) for 1 h at 58°C; paraplast alone at 58°C for 16 h and then two paraplast exchanges for 2 h each. Microtomy was performed 48 h after embedding and the sections (5 µm thick) produced were mounted in poly‐l‐lysine covered slides (Superfrost Plus®, Erviegas). Sections were deparaffinated by immersing the slides in absolute xylol, ethanol 100% and xylol (1:1) and decreasing concentrations of ethanol (90%, 70%, 50% and 15%) for 15 min each, in this order, followed by a 5 min immersion in distilled water. The slides were dried at room temperature. The antibody labelling was conducted according to the manufacturer's instructions (Invitrogen). The slides were washed in PBS 1× for 1 min followed by addition of Block solution (10% BSA in PBS 1× pH 7.4) and incubation for 1 h. The slides were then washed twice in PBS 1× for 1 min each time. The antibody was diluted to a final concentration of 5.0 µg ml−1 and incubated on the slide for 1 h at room temperature. The slides were incubated with 200 ng ml−1 of DAPI blue (Invitrogen) for 30 min. The solution was then removed and the slides washed twice in PBS for 1 min each time. Finally, the solution was removed and the slides washed twice in PBS for 2 min each time. The slides were analysed in a Leika SP5. Digital images were acquired using laser excitation wavelengths of 488 and 590 nm.
References
- Bizzarri M.F., Bishop A.H. The recovery of Bacillus thuringiensis in vegetative form from the phylloplane of clover (Trifolium hybridum) during a growing season. J Invertebr Pathol. 2007;94:38–47. doi: 10.1016/j.jip.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Bone E.J., Ellar D.J. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett. 1989;58:171–178. doi: 10.1016/0378-1097(89)90033-5. [DOI] [PubMed] [Google Scholar]
- Bravo A., Sarabia S., Lopez L., Ontiveros H., Abarca C., Ortiz A. Characterization of cry genes in Mexican B. thuringiensis strain collection. Appl Environ Microbiol. 1998;64:4965–4972. doi: 10.1128/aem.64.12.4965-4972.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceron J., Ortiz A., Quintero R., Guereca L., Bravo A. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl Environ Microbiol. 1995;61:3826–3831. doi: 10.1128/aem.61.11.3826-3831.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard P.H. Natural occurrence and dispersal of Bacillus thuringiensis in the environment. In: Charles J.‐F., Delécluse A., Nielsen‐LeRoux C., editors. Kluwer Academic Publishers; 2000. pp. 23–40. [Google Scholar]
- Ibarra J., Rincon C., Ordúz S., Benintende G., Monnerat R., Regis L. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquitoes species. Appl Environ Microbiol. 2003;69:5269–5274. doi: 10.1128/AEM.69.9.5269-5274.2003. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecadet M.M., Chaufaux J., Ribier J., Lereclus D. Construction of novel Bacillus thuringiensis strains with different insecticidal activities by transduction and transformation. Appl Environ Microbiol. 1991;58:840–849. doi: 10.1128/aem.58.3.840-849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduell P., Armengol G., Llagostera M., Lindow S., Orduz S. Immigration of Bacillus thuringiensis to bean leaves from soil inoculum or distal plant parts. J App Microbiol. 2007;103:2593–2600. doi: 10.1111/j.1365-2672.2007.03509.x. [DOI] [PubMed] [Google Scholar]
- Martins E., Praça L.B., Dumas V.F., Silva‐Werneck J.O., Sone E.H., Waga I.C. Characterization of Bacillus thuringiensis isolates toxic to cotton boll weevil (Anthonomus grandis. Biol Control. 2007;40:65–68. et al. [Google Scholar]
- Monnerat R.G., Masson L., Brousseau R., Pusztai‐Carey M., Bordat D., Frutos R. Differential activity and activation of Bacillus thuringiensis insecticidal proteins in Diamondback moth, Plutella xylostella. Curr Microbiol. 1999;39:159–162. doi: 10.1007/s002849900438. [DOI] [PubMed] [Google Scholar]
- Monnerat R.G., Dias D., Silva S., Martins E., Berry C., Falcão R. Screening of Bacillus thuringiensis strains effective against mosquitoes. PAB. 2005;40:103–106. et al. [Google Scholar]
- Monnerat R.G., Batista A.C., Medeiros P., Martins E., Melatti V.M., Praça L., Berry C. Screening of Brazilian Bacillus thuringiensis isolates active against Spodoptera frugiperda, Plutella xylostella and Anticarsia gemmatalis. Biol Control. 2007;41:291–295. [Google Scholar]
- Myers P., Yousten A. Toxic activity of Bacillus sphaericus SSII‐1 for mosquito larvae. Infect Immun. 1978;19:1047–1053. doi: 10.1128/iai.19.3.1047-1053.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.C., Damgaard P.H., Eilenberg J., Hansen B.M. Dispersal of Bacillus thuringiensis kurstaki. Can J Microbiol. 1995;41:118–125. [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. 2nd. CSHL; 1989. [Google Scholar]
- Schmidt F.G.V., Monnerat R.G., Borges M., Carvalho R. 2001.
- Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S.F., Dias J.M.C.S., Monnerat R.G. 2002.
- Smith R.A., Barry J.W. Environmental persistence of Bacillus thuringiensis spores following aerial application. J Invertebr Pathol. 1998;71:263–267. doi: 10.1006/jipa.1997.4738. [DOI] [PubMed] [Google Scholar]
- Stefanini M., De Martino C., Zamboni L. Fixation of ejaculated spermatozoa for electron microscopy. Nature. 1967;216:173–176. doi: 10.1038/216173a0. [DOI] [PubMed] [Google Scholar]
- Tabashnik B.E., Cushing N.L., Finson N., Johnson M.W. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1990;83:1671–1676. [Google Scholar]


