Abstract
Pseudomonas putida KT2440 has the ability to colonize the rhizosphere of a wide range of plants and can reach cell densities in the range of 105–106 cfu g soil−1. Using the IVET technology we investigated which KT2440 genes were expressed in the rhizosphere of four different plants: pine, cypress, evergreen oak and rosemary. We identified 39 different transcriptional fusions containing the promoters of annotated genes that were preferentially expressed in the rhizosphere. Six of them were expressed in the rhizosphere of all the plant types tested, 11 were expressed in more than one plant and the remaining 22 fusions were found to be expressed in only one type of plant. Another 40 fusions were found to correspond to likely promoters that encode antisense RNAs of unknown function, some of which were isolated as fusions from the bacteria recovered in the rhizosphere from all of the plants, while others were specific to one or several of the plants. The results obtained in this study suggest that plant-specific signals are sensed by KT2440 in the rhizosphere and that the signals and consequent gene expression are related to the bacteria's successful establishment in this niche.
Introduction
The rhizosphere is a complex ecosystem where a number of dynamic interactions take place between the plant root that exudes a variety of organic molecules and inorganic ions, and prokaryotic and eukaryotic soil microbes and invertebrates. These interactions are also influenced by the physicochemical conditions of the soil. This set of reciprocal interactions causes a network of genetic and physiological responses at the single cell level that have not been fully addressed (Ramos-González et al., 2005; Hartmann et al., 2009). Analyses of bacterial diversity in the rhizosphere using culture-dependent and culture-independent techniques have yielded large amounts of data relating to the microbes that inhabit this niche and their population levels. Bacteria of the genus Pseudomonas are one of the prominent genera of root-associated bacteria, and have been found to be ubiquitous in the rhizosphere of wild and cultivated plants (Haas and Défago, 2005; Uroz et al., 2007; Hartmann et al., 2009; García-Salamanca et al., 2013); however, the responses of these microorganisms to root compounds have not been explored in great detail (Costa et al., 2007; Hartmann et al., 2009).
Pseudomonas putida KT2440 is an efficient root colonizer of a wide range of plants (Molina et al., 2000) and is frequently used as a model organism in studies focusing on rhizoremediation, the removal of pollutants by a microbe when associated with the root of plants (Segura et al., 2009). Evidence for the reciprocal interactions, between the bacterium and the plant roots, comes from the knowledge that this strain exhibits increased catabolic activity in rhizosphere soil when compared with bulk soil (Fernández et al., 2012a). Colonization of roots by KT2440 has been studied in detail using the wild-type strain and a large collection of mutants which had lost their ability to adhere to seeds (Espinosa-Urgel et al., 2000; 2002; Duque et al., 2013), in vivo induced gene expression using IVET technology (Ramos-González et al., 2005) and transcriptional profiling of KT2440 proliferating on the roots of plants grown on vermiculite has been addressed (Matilla et al., 2007).
IVET is essentially a promoter-trapping technique that selects microbial promoters that are active in a specific habitat. These promoters are selected for their ability to drive the expression of a promoterless selection gene marker in vivo, which complements a mutation in an essential gene in the host strain and, therefore enables survival in the tested environment. In contrast to classical approaches by mutagenesis, the advantage of the IVET strategy is the positive selection of genes that are specifically induced by environmental factors (Rediers et al., 2003). The specific IVET system used previously by our group was taken for the identification of P. putida KT2440 in the rhizosphere of maize plants (Ramos-González et al., 2005). In the case of IVET promoters selected in the rhizosphere these promoters are of interest to drive heterologous gene expression such as catabolic genes to remove pollutants or promoters that drive expression of genes involved in biocontrol. The IVET system utilized consists of a plasmid, named pOR1, containing a promoterless ′asd-lacZ reporter cassette, which is used to generate a KT2440 promoter library with DNA fragments in the range of 1000 bp, and a suitable host to select the active transcriptional fusions, P. putida KT2440Δasd which is an auxotroph that requires diaminopimelate (DAP) and three amino acids: lysine, methionine and threonine for growth. The viability of P. putida KT2440Δasd to grow in the rhizosphere is impaired unless the asd gene is expressed in trans from an active promoter cloned in pOR1. In culture medium, in addition to the auxotrophy, activity of the promoters cloned into pOR1 can be measured using the blue/white screening technique on plates with X-gal.
In this study we used the IVET system designed by Ramos-González and colleagues (2005) to perform a comparative study of P. putida KT2440 gene expression in the rhizosphere of four different but representative Mediterranean plants, Pinus halepensis (pine), Quercus ilex (evergreen oak), Cupressus sempervirens (cypress) and Rosemarinus officinalis (rosemary). The aim was to determine which genes were preferentially activated when the strain colonized the rhizosphere and to gain insight into how this bacterium is capable of colonizing the roots of different plants.
Results and discussion
Pseudomonas putida KT2440 has a similar way of colonizing the rhizosphere of different types of plants
We assayed the ability of KT2440 to colonize some of the more common plants on the Mediterranean basin: pine, evergreen oak, cypress and rosemary. With this aim we used seedling trees which were inoculated by dipping the root in a bacterial suspension of KT2440, then the plants were placed in pots with a non-sterilized mixture of peat:sand (1:1) and incubated under controlled growth conditions in a greenhouse (23°C, 12 h:12 h light:dark). In the rhizosphere of the four plants, bacteria survived and bacterial density was between 105 and 106 cfu per gram of rhizosphere soil over a period of more than two months (Fig. 1); these data are in agreement with that reported by Molina and colleagues (2000) of KT2440 colonizing the rhizosphere of corn, broad beans and other herbaceous plants. In the bulk soil of the pots, KT2440 bacterial levels dropped to 103 cfu per gram of soil, which was also in agreement with previous observations (Molina et al., 2000).
Figure 1.
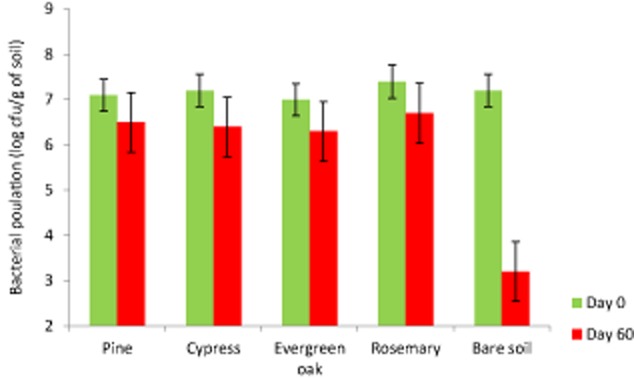
Cell density of P. putida KT2440R populations in the rhizosphere of different plants. Grey bars: population sizes the day of inoculation; dark bars: population size 2 months later. The plant species used are indicated. To facilitate the strain identification by plate counting we used KT2440R, a KT2440 rifampicin-resistant spontaneous mutant. In addition, colonies were confirmed by REPc profile analysis (Aranda-Olmedo et al., 2002). Rhizosphere colonization assays were performed with young trees in a cambisol soil according to our previously published standard protocol (Fernández et al., 2012a). Assays were run in triplicate. Error bars show the standard error.
Unravelling genes/promoters uniquely expressed in the rhizosphere by IVET technology
In this study we aimed to document which genes are specifically expressed in the rhizosphere of the four plants: P. putida Δasd bearing the IVET library was inoculated as above and the plants were placed in pots with peat:sand (1:1) as substrate. We performed two independent rounds of IVET selection, with each plant tested in triplicate on each round. All plants were kept for 2 months under greenhouse conditions before recovering the viable bacterial cells on selective medium. Near 50 000 isolated after spreading serial dilutions in minimal medium agar plates with X-gal, kanamycin, DAP, lysine, methionine and threonine. Around 800 of them (1.6% of the total) formed white or very pale-blue colonies; and were unable to grow in the absence of DAP. This was indicative that the promoter that drove asd expression was expressed in the soil and not in the agar plates. For each clone the pOR1 derivative was recovered (Rainey et al., 1997) and the insert sequenced. Computational analysis of the sequences allowed us to identify a total of 79 different insertions; 39 of which were adjacent to annotated genes (Table 1), whereas the remaining 40 were antisense transcriptional clones (Table S1). From the group of 39 transcriptional fusions, six of them were isolated from the rhizosphere of all of the plants tested, while 11 were isolated from more than one type of plant, and the remaining 22 transcriptional fusions were recovered from the rhizosphere of only one of the four plants tested; specifically, three of them only in cypress, nine in evergreen oak, six in pine and four in rosemary (Fig. 2).
Table 1.
Genes identified as preferentially activated in the rhizosphere by IVET screening
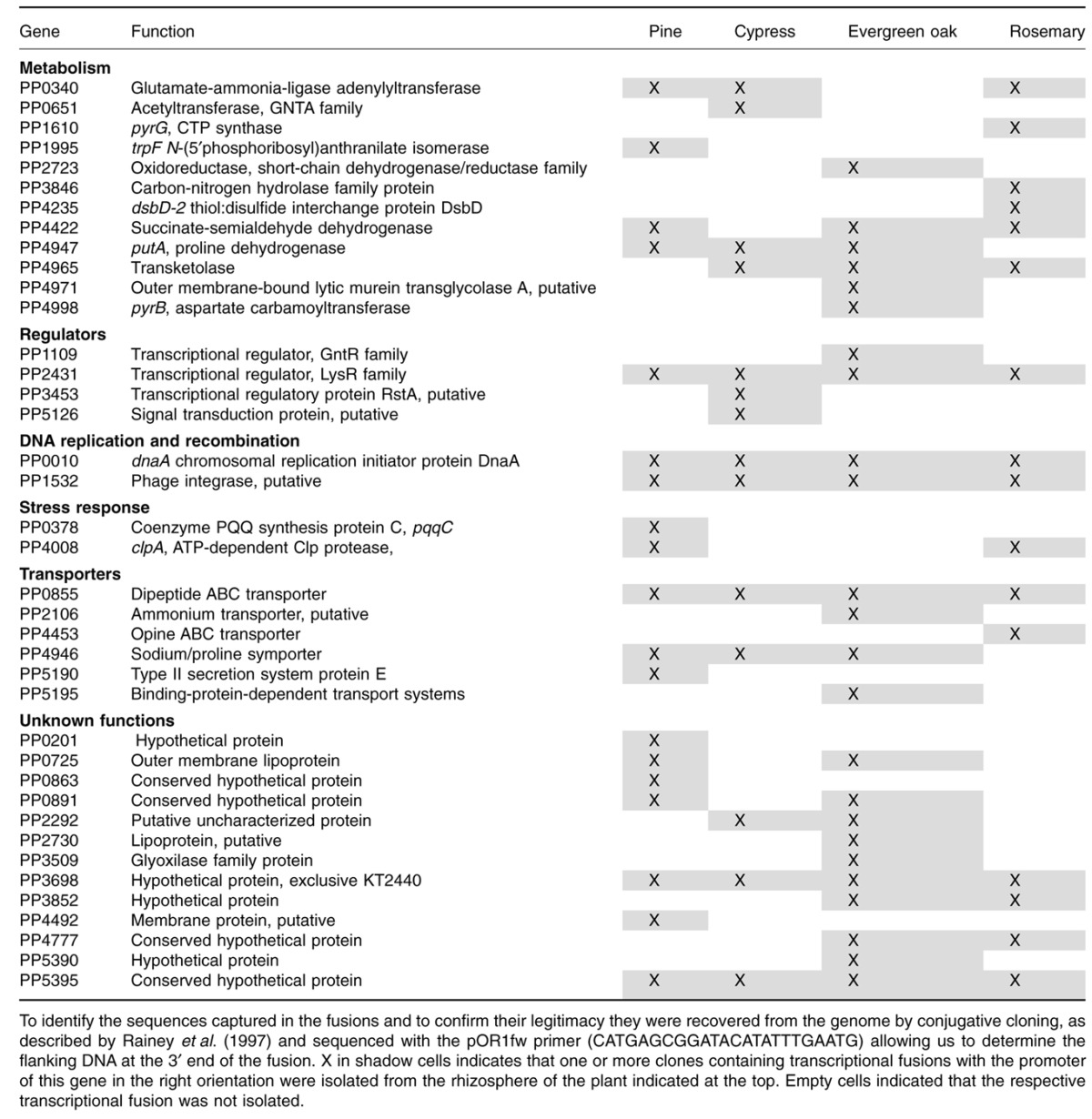 |
Figure 2.
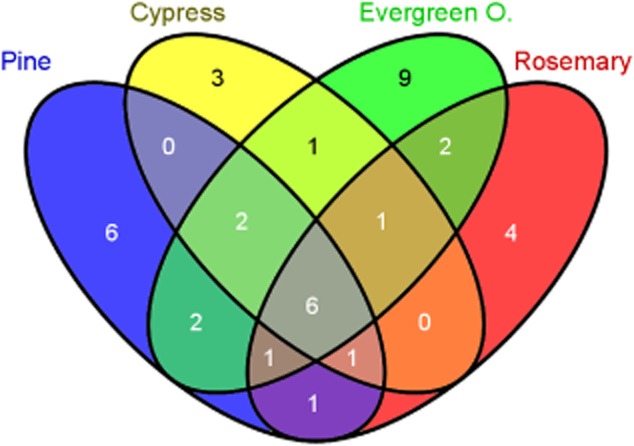
Venn diagram depicting the distribution among the tested plants of the 39 genes identified as preferentially expressed in the rhizosphere during this study. Diagram produced by Venny (Oliveros, 2007).
Although most of the transcriptional fusions were isolated several times from independent samples, we performed an additional set of assays to verify that the isolated clones indeed exhibited asd expression in the rhizosphere from the promoters cloned in the transcriptional fusions. With this aim we randomly chose clones containing a transcriptional fusion found in all plants or only in some of the plants and carried out rhizosphere colonization assays with pure cultures of these clones. The results were compared with those obtained with P. putida Δasd carrying pOR1 (without insert). In all cases, clones with transcriptional fusions kept their population sizes at a level that was at least three orders of magnitude higher than P. putida Δasd with the empty plasmid (Fig. 3). When clones with transcriptional fusions found in only one type of plant were tested in the rhizosphere of another plant, differences in the cell densities were significant, around two orders of magnitude of variation (Table 2).
Figure 3.
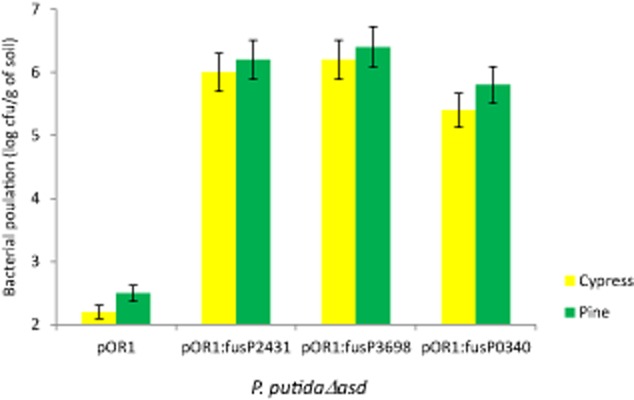
Population size after 2 months in the rhizosphere of P. putida △asd with pOR1 and different transcriptional fusions in pine and cypress. pOR1: plasmid without insert (negative control); pOR1:fusP2431, transcriptional fusion of the asd to the PP2431 promoter, pOR1:fusP3689, transcriptional fusion of the asd to PP3689 promoter, and pOR1:fusP0340, transcriptional fusion the asd with the PP0340 promoter. Population sizes were determined by plate counting in minimal medium supplemented with kanamycin, DAP and amino acids. Experiments were run in triplicate. Transcriptional fusions to verify were chosen at random, error bars show the standard error.
Table 2.
Pseudomonas putida KT2440 population size after 2 months in the rhizosphere of different plants
| cfu per gram of rhizosphere soil | |||
|---|---|---|---|
| pOR1 | pOR1:fusPP4492 | pOR1:fusPP1109 | |
| Pine | 3.6 × 102 ± 8 | 7.9 × 105 ± 27 | 3.9 × 104 ± 57 |
| Evergreen oak | 1.8 × 102 ± 10 | 3.1 × 103 ± 61 | 2.5 × 106 ± 94 |
pOR1: plasmid without insert (negative control); pOR1:fus4492, transcriptional fusion of the asd to the PP4492 promoter; pOR1:fus1109, transcriptional fusion of the asd to the PP1109 promoter. Bacterial densities were determined by plate counting in minimal medium supplemented with kanamycin, DAP and amino acids. Experiments were run in triplicate. Transcriptional fusions to verify were chosen at random.
Specificity of the rhizosphere-activated genes
To further investigate the putative role of the activated genes in rhizosphere adaptation in silico studies were carried out (see below), as well as a detailed analysis of the previously published information on the function of these genes. Functional analysis of the genes expressed from the promoters contained in the transcriptional fusions, revealed six main categories: metabolism, gene regulation, DNA replication and recombination, stress response, transporters, and a number of hypothetical proteins of unknown function (Fig. 4). This reinforces the hypothesis that a strict signalling and a complex network of interactions occur in the rhizosphere enabling bacteria to cope with two general problems: new nutrient sources and significant stress conditions (Hartmann et al., 2009). These categories are in agreement with those described for KT2440 in the rhizosphere of maize grown in hydroponic solution (Ramos-González et al., 2005) and sand (Matilla et al., 2007), although the genes identified in the current study are different.
Figure 4.
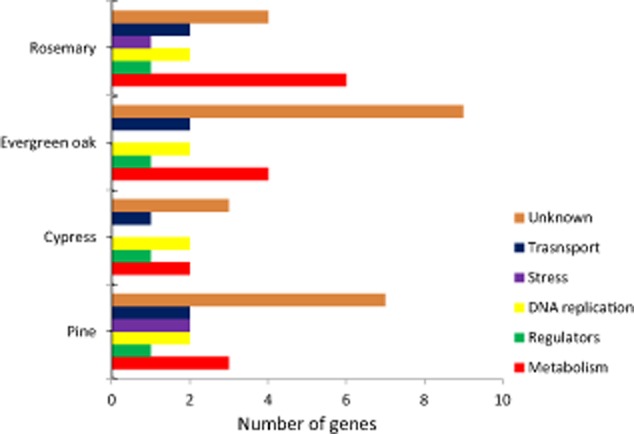
Functional classification of the genes identified as preferentially activated in the rhizosphere by the IVET screening technology. Functional categories have been assigned according to in silico predictions. Only genes whose promoters were trapped in the right sense have been included.
Transcriptional fusions found in the rhizosphere of all or several types of plants
As detailed above, six transcriptional fusions were expressed in the rhizosphere of all of the plants tested. This group includes the promoters of PP0885, encoding a dipeptide ABC transporter which was shown to be upregulated in KT2440 under nitrogen limitation conditions (Hervás et al., 2008). Also induced is the phage integrase encoded by PP1532 that Quesada and colleagues (2012) found to belong to a prophage whose mobilization, mediated by the expression of PP1532, resulted in increased bacterial fitness in the rhizosphere; and that is also activated in response to quorum sensing signals induced by a Pseudomonas aeruginosa strain (Fernández-Piñar et al., 2011). Significant as well is the expression in all of the plants of a transcriptional regulator encoded by PP2431 which can be relevant for cellular adaptation at the genetic level. Also a fusion with dnaA promoter was solely activated in the rhizosphere; this gene is involved in DNA replication, initiation and regulation. Finally, two of the transcriptional fusions found in the four types the plants tested (PP5395 and PP3698) corresponded to genes of unknown function that are exclusively found in the KT2440 strain.
A set of 11 transcriptional fusions were isolated from more than one type of plant; among these transcriptional fusions were genes involved in stress tolerance, such as clpA or PP4422, involved in phenol tolerance in KT2440 (Putrins et al., 2010); genes with a metabolic function, i.e. glnE (PP4965) encodes a transketolase, which was also induced in Streptomyces coelicolor in the presence of plant material (Langlois et al., 2003). In three of the plants we found transcriptional fusions with two genes involved in proline transport and metabolism; the sodium/proline symporter encoded by PP4946, which was also identified as being activated in the rhizosphere of corn (Ramos-González et al., 2005) and putA, involved in proline catabolism (Vílchez et al., 2000). It was previously shown that putA expression is induced in KT2440 in response to maize root exudates (Vílchez et al., 2000), and more recently it has been identified to be induced by a benzoxazinoid, a secondary metabolite involved in plant defence (Neal et al., 2012). Signals for the activation of this group of common genes could be non-specific plant-derived factors or the surrounding microbial signals, without ruling out a combination of both of them together with abiotic elements (Haas and Défago, 2005).
Transcriptional fusions found in only one type of plant
Up to 22 transcriptional fusions were expressed in the rhizosphere of only one of the four tested plants; six of them only in pine, three of them in cypress, nine in evergreen oak and four in rosemary; this group also includes a set of fusions with promoters of genes involved in metabolic processes, transporters, regulators and bacterial stress responses.
Exclusively in the rhizosphere of pine we detected the activation of trpF (PP1995), involved in tryptophan biosynthesis, which in turn is the precursor for indole-3-acetic acid (IAA), a well-characterized phytohormone able to stimulate plant growth and several responses in plants (Cleland, 1990; Hagen, 1990; Patten and Glick, 1996; Spaepen et al., 2007; Roca et al., 2012). A second gene of this group is pqqC (PP0378), involved in the biosynthesis of pyrroloquinoline quinone coenzyme (PQQ), which in Pseudomonas is involved in the stress response (Fernández et al., 2012b), phosphate solubilization (Meyer et al., 2011) and have direct effects on plant growth (Choi et al., 2008).
Two transcriptional fusions with promoters of genes involved in bacterial signalling/regulation processes, PP3453 and PP5126, were isolated only from the cypress rhizosphere. In silico searches using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING 9.0) database (http://string-db.org/) predicted the interaction of the signal transduction protein encoded by PP5126 with four different flagellar motor proteins.
One transcriptional fusion was to the orf PP5390 and it was found in bacteria grown in the evergreen oak rhizosphere, this gene has been previously described as induced by maize root exudates (Ramos-González et al., 2005), and encodes a protein of unknown function.
Among the transcriptionl fusions isolated only in the rhizosphere of rosemary we found the promoter region of PP4453, encoding an opine ABC transporter; since KT2440 carries the set of genes that encodes enzymes for the metabolism of opines, we suggest that induction of this transporter is related to the ability of this bacterium to grow at the expense of plant-produced opines (Nelson et al., 2002).
Antisense transcriptional fusions
Half of the transcriptional fusions isolated in this study lacked a detectable promoter or the captured DNA was oriented in the opposite direction to that necessary for transcription of the promoterless asd gene. Even accepting that some of them could be false positives, the number of such clones and the fact that most of them were recovered on independent occasions and were isolated from some of the tested plants or in all of them, strongly suggested an active role of the fragment trapped in such fusions. Antisense transcriptional fusions are often found in studies using IVET approaches (Silby et al., 2004; Ramos-González et al., 2005; Barr et al., 2008; Silby et al., 2009; Hanin et al., 2010). Authors have speculated on their role as antisense regulatory RNAs; Frank and colleagues (2011) demonstrated the existence of non-coding RNAs in KT2440 in lab conditions, which probably also take place in planta. Also it has been considered that putative errors in in silico gene prediction or even the existence of overlapping protein-coding genes that run in opposite direction to the annotated ORFs (Silby et al., 2004). In any case, there is much to learn about the role of these genes in the bacterial natural environments (Silby et al., 2009). Therefore it is quite possible that we may have detected only a selection of a vast genetic response allowing this bacterium to colonize the rhizosphere of very different types of plants, and these cryptic fusions suggest the existence of genes whose expression in the rhizosphere seems to be independent of the type of plant, and genes whose expression strongly depends on the type of plant.
Root exudates are considered the main factor for changes in bacterial gene expression in the rhizosphere. These exudates are plant-specific and can also vary depending on the plant growth cycle or on the physiological state (Jones et al., 2004; van Veen et al., 2007; Hartmann et al., 2009). Differences in root exudate composition between different plants have also been suggested as the main reason for the changes observed in bacterial rhizospheric communities depending on the type of plant (Hartmann et al., 2009). Mark and colleagues (2005) analysed how the exudates of two varieties of sugar beet influenced the pattern of genetic expression of P. aeruginosa. They found that each exudate provoked a different transcriptional response with only a partial overlap. Such influences of the root exudate composition on bacterial gene expression or on microbial populations contrasts with the fact that while some microbes exhibited a very narrow spectrum of plant roots to colonize, there are others, considered almost ubiquitous in the rhizosphere. Results from this study fully support the influence of plant-specific factors, most probably root exudates, on microbial adaptation to the rhizosphere environment, but possibly in combination with other (still) unknown factors.
In summary, P. putida KT2440 efficiently colonizes the rhizosphere of a wide range of plants and apart from the plasticity of the genome of this strain; successful colonization involves a response to plant-specific signals, which is undoubtedly part of its success in this specific ecological niche.
Acknowledgments
We thank Maribel Ramos-González for IVET libraries and M. Mar Fandila for secretarial assistance.
Conflict of interest
None declared.
Funding Information
Work in our laboratory was supported by Fondo Social Europeo and Fondos Feder from the European Union. Work in this study was also supported by a grant from the Ministry of Ciencia e Innovación BIO2010-17227, Junta de Andalucía CVI-7391, BACSIN and ST-FLOW programmes.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Cryptic transcriptional fusions.
References
- Aranda-Olmedo I, Tobes R, Manzanera M, Ramos JL, Marqués S. Species-specific repetitive extragenic palindromic (REP) sequences in Pseudomonas putida. Nucleic Acids Res. 2002;30:1826–1833. doi: 10.1093/nar/30.8.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M, East AK, Leonard M, Mauchline TH, Poole PS. In vivo expression technology (IVET) selection of genes of Rhizobium leguminosarum biovar viciae A34 expressed in the rhizosphere. FEMS Microbiol Lett. 2008;2:219–227. doi: 10.1111/j.1574-6968.2008.01131.x. [DOI] [PubMed] [Google Scholar]
- Choi O, Kim J, Kim JG, Jeong Y, Moon JS, Park CS, Hwang I. Pyrroloquinoline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiol. 2008;2:657–668. doi: 10.1104/pp.107.112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland RE. Auxin and cell elongation. In: Davies PJ, editor. Plant Hormones and Their Role in Plant Growth and Development. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1990. pp. 132–148. [Google Scholar]
- Costa R, Gomes NC, Krögerrecklenfort E, Opelt K, Berg G, Smalla K. Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ Microbiol. 2007;9:2260–2273. doi: 10.1111/j.1462-2920.2007.01340.x. [DOI] [PubMed] [Google Scholar]
- Duque E, de la Torre J, Bernal P, Molina-Henares MA, Alaminos M, Espinosa-Urgel M, et al. Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ Microbiol. 2013;15:36–48. doi: 10.1111/j.1462-2920.2012.02732.x. [DOI] [PubMed] [Google Scholar]
- Espinosa-Urgel M, Salido A, Ramos JL. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;9:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Urgel M, Kolter R, Ramos JL. Root colonization by Pseudomonas putida: love at first sight. Microbiology. 2002;148:341–343. doi: 10.1099/00221287-148-2-341. [DOI] [PubMed] [Google Scholar]
- Fernández M, Niqui-Arroyo JL, Conde S, Ramos JL, Duque E. Enhanced tolerance to naphthalene and enhanced rhizoremediation performance for Pseudomonas putida KT2440 via the NAH7 catabolic plasmid. Appl Environ Microbiol. 2012a;15:5104–5110. doi: 10.1128/AEM.00619-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández M, Conde S, Torre J, Molina-Santiago C, Ramos JL, Duque E. Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob Agents Chemother. 2012b;2:1001–1009. doi: 10.1128/AAC.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Piñar R, Cámara M, Dubern JF, Ramos JL, Espinosa-Urgel M. The Pseudomonas aeruginosa quinolone quorum sensing signal alters the multicellular behaviour of Pseudomonas putida KT2440. Res Microbiol. 2011;8:773–781. doi: 10.1016/j.resmic.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Frank S, Klockgether J, Hagendorf P, Geffers R, Schöck U, Pohl T, et al. Pseudomonas putida KT2440 genome update by cDNA sequencing and microarray transcriptomics. Environ Microbiol. 2011;5:1309–1326. doi: 10.1111/j.1462-2920.2011.02430.x. [DOI] [PubMed] [Google Scholar]
- García-Salamanca A, Molina-Henares MA, van Dillewijn P, Solano J, Pizarro-Tobías P, Roca A, et al. Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb Biotechnol. 2013;6:36–44. doi: 10.1111/j.1751-7915.2012.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;4:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- Hagen G. The control of gene expression by auxin. In: Davies PJ, editor. Plant Hormones and Their Role in Plant Growth and Development. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1990. pp. 132–148. [Google Scholar]
- Hanin A, Sava I, Bao Y, Huebner J, Hartke A, Auffray Y, Sauvageot N. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PLoS ONE. 2010;7:e11879. doi: 10.1371/journal.pone.0011879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D, Berg G. Plant-driven selection of microbes. Plant Soil. 2009;321:235–257. [Google Scholar]
- Hervás AB, Canosa I, Santero E. Transcriptome analysis of Pseudomonas putida in response to nitrogen availability. J Bacteriol. 2008;1:416–420. doi: 10.1128/JB.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RK, Sun WH, Tang CS, Robert FM. Phytoremediation of petroleum hydrocarbons in tropical coastal soils. II. Microbial response to plant roots and contaminant. Environ Sci Pollut Res Int. 2004;5:340–346. doi: 10.1007/BF02979649. [DOI] [PubMed] [Google Scholar]
- Langlois P, Bourassa S, Poirier GG, Beaulieu C. Identification of Streptomyces coelicolor proteins that are differentially expressed in the presence of plant material. Appl Environ Microbiol. 2003;4:1884–1889. doi: 10.1128/AEM.69.4.1884-1889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark GL, Dow JM, Kiely PD, Higgins H, Haynes J, Baysse C, et al. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe–plant interactions. Proc Natl Acad Sci USA. 2005;48:17454–91745. doi: 10.1073/pnas.0506407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González MI. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 2007;9:R179. doi: 10.1186/gb-2007-8-9-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JB, Frapolli M, Keel C, Maurhofer M. Pyrroloquinoline quinone biosynthesis gene pqqC, a novel molecular marker for studying the phylogeny and diversity of phosphate-solubilizing pseudomonads. Appl Environ Microbiol. 2011;20:7345–7354. doi: 10.1128/AEM.05434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina L, Ramos C, Duque E, Ronchel MC, García JM, Wyke L, Ramos JL. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem. 2000;32:315–321. [Google Scholar]
- Neal AL, Gordon-Weeks R, Ton J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE. 2012;4:e35498. doi: 10.1371/journal.pone.0035498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. 2007. VENNY. An interactive tool for comparing list with Venn Diagrams [WWW document]. URL http:/bioinfogp.cnb.csic.es/tools/venny/index.html.
- Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;3:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- Putrins M, Ilves H, Lilje L, Kivisaar M, Hõrak R. The impact of ColRS two-component system and TtgABC efflux pump on phenol tolerance of Pseudomonas putida becomes evident only in growing bacteria. BMC Microbiol. 2010;10:110. doi: 10.1186/1471-2180-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada JM, Soriano MI, Espinosa-Urgel M. Stability of a Pseudomonas putida KT2440 Bacteriophage-carried genomic island and its impact on rhizosphere fitness. Appl Environ Microbiol. 2012;19:6963–6974. doi: 10.1128/AEM.00901-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey PB, Heithoff DM, Mahan MJ. Single-step conjugative cloning of bacterial gene fusions involved in microbe–host interactions. Mol Gen Genet. 1997;256:84–87. doi: 10.1007/s004380050548. [DOI] [PubMed] [Google Scholar]
- Ramos-González MI, Campos MJ, Ramos JL. Analysis of Pseudomonas putida KT2440 gene expression in the maize rhizosphere: in vivo [corrected] expression technology capture and identification of root-activated promoters. J Bacteriol. 2005;187:4033–4041. doi: 10.1128/JB.187.12.4033-4041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rediers H, Bonnecarrère V, Rainey PB, Hamonts K, Vanderleyden J, De MR. Development and application of a dapB-based in vivo expression technology system to study colonization of rice by the endophytic nitrogen-fixing bacterium Pseudomonas stutzeri A15. Appl Environ Microbiol. 2003;11:6864–6874. doi: 10.1128/AEM.69.11.6864-6874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca A, Pizarro-Tobías P, Udaondo Z, Fernández M, Matilla MA, Molina-Henares A, et al. Analysis of the plant-growth promoting properties encoded by the genome of the rhizobacterium Pseudomonas putida BIRD-1. Environ Microbiol. 2012 doi: 10.1111/1462-2920.12037. doi: 10.1111/1462-2920.12037. [DOI] [PubMed] [Google Scholar]
- Segura A, de Wit P, Preston GM. Life of microbes that interact with plants. Microb Biotechnol. 2009;4:412–415. doi: 10.1111/j.1751-7915.2009.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby MW, Rainey PB, Levy SB. IVET experiments in Pseudomonas fluorescens reveal cryptic promoters at loci associated with recognizable overlapping genes. Microbiology. 2004;150:518–520. doi: 10.1099/mic.0.26871-0. [DOI] [PubMed] [Google Scholar]
- Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;5:R 51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;4:425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- Uroz S, Calvaruso C, Turpault MP, Pierrat JC, Mustin C, Frey-Klett P. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl Environ Microbiol. 2007;73:3019–3027. doi: 10.1128/AEM.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veen JA, Kay E, Vogel TM, Simonet P. Methodological approaches to the study of carbon flow and the associated microbial population dynamics in the rhizosphere. In: Pinton R, Varanini Z, Nannipieri Z, editors. The Rhizosphere: Biochemistry and Organic Substances at the Soil–Plant Interface. 2nd edn. Boca Raton, FL, USA: CRC; 2007. pp. 371–399. [Google Scholar]
- Vílchez S, Manzanera M, Ramos JL. Control of expression of divergent Pseudomonas putida put promoters for proline catabolism. Appl Environ Microbiol. 2000;12:5221–5225. doi: 10.1128/aem.66.12.5221-5225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cryptic transcriptional fusions.


