Abstract
Poly-γ-glutamic acid (γ-PGA) is a promising environmental-friendly material with outstanding water solubility, biocompatibility and degradability. However, it is tough to determine the relationship between functional synthetic enzyme and the strains' yield or substrate dependency. We cloned γ-PGA synthetase genes pgsBCA and glutamate racemase gene racE from both L-glutamate-dependent γ-PGA-producing Bacillus licheniformis NK-03 and L-glutamate-independent B. amyloliquefaciens LL3 strains. The deduced RacE and PgsA from the two strains shared the identity of 84.5% and 78.53%, while PgsB and PgsC possessed greater similarity with 93.13% and 93.96%. The induced co-expression of pgsBCA and racE showed that the engineered Escherichia coli strains had the capacity of synthesizing γ-PGA, and LL3 derived PgsBCA had higher catalytic activity and enhanced productivity than NK-03 in Luria–Bertani medium containing glucose or L-glutamate. However, the differential effect was weakened when providing sufficient immediateness L-glutamate substrate, that is, the supply of substrate could be served as the ascendance upon γ-PGA production. Furthermore, RacE integration could enhance γ-PGA yield through improving the preferred d-glutamate content. This is the first report about co-expression of pgsBCA and racE from the two Bacillus strains, which will be of great value for the determination of the biosynthetic mechanism of γ-PGA.
Introduction
Poly-γ-glutamic acid (γ-PGA) is a naturally occurring polyanionic polypeptide that consists of repeating units of D- and L-glutamic acid via amide linkages between α-amino and γ-carboxyl groups (Ashiuchi and Misono, 2002). The molecular weight of microbial γ-PGA varies from 10k to 10 000k and the stereochemical structure includes three types: a homopolymer of d-glutamic acid, a homopolymer of L-glutamic acid, and copolymer of random combinations of D-/L-glutamic acid (γ-DL-PGA) (Ashiuchi and Misono, 2003). With the chiral centre existing in glutamate unit and the abundant active sites of carboxylic groups present in the main chain, γ-PGA and its derivatives were endowed with outstanding water solubility, biocompatibility and degradability, and have been successfully utilized in hydrogels, humectants, flocculants, thickeners, dispersants, cryoprotectants, drug carriers, and cosmetic and biological food additives (Shih and Van, 2001; Sung et al., 2005).
The microorganisms, capable of producing γ-PGA and drawing the molecules as capsule protective components and extracellular nutritious secretion, mainly belong to Gram-positive bacteria, such as Bacillus genus, Staphylococcus epidermidis (Kocianova et al., 2005), Archaeobacteria species Natronococcus occultus (Niemetz et al., 1997) and Natrialba aegyptiaca (Hezayen et al., 2000). However, Candela and colleagues (2009) classified the first Gram-negative bacterium, Fusobacterium nucleatum, which demonstrated to produce γ-PGA as interaction factor for dental plaque formation. γ-PGA-producing strains are divided into two types: one produces γ-PGA in the presence of L-glutamate in medium, and the other does not (Ito et al., 1996). Most of the known γ-PGA producers, such as B. subtilis (natto) IFO 3335 (Goto and Kunioka, 1992), B. subtilis chungkookjang (Ashiuchi et al., 2001), B. licheniformis ATCC9945a (Gardner and Troy, 1979) and B. licheniformis NK-03 (Cao et al., 2010), belong to the former group, whereas for the latter group, just a few strains have been characterized, including B. subtilis TAM-4 (Ito et al., 1996), B. licheniformis A35 (Cheng et al., 1989) and B. amyloliquefaciens LL3 (Cao et al., 2011). Although γ-PGA producers with high productivity and industrial applications are mostly L-glutamic acid-dependent strains, it is of great interest to choose L-glutamic acid-independent producers for the studies on γ-PGA biosynthesis mechanism and its development potential because of the lower cost and simplified downstream processing.
To date, heterologous expression of γ-PGA synthetase genes (pgsBCA) from L-glutamic acid-dependent strains have been achieved in various organisms (Ashiuchi et al., 1999; 2001; Jiang et al., 2006). It was previously reported that tobacco leaf cells (Tarui et al., 2005) and the glutamate-producing Corynebacterium glutamicum strains harbouring pgsBCA expression vectors could display γ-PGA synthetase and accumulate extracellular γ-PGA without L-glutamate (Sung et al., 2003; Cao et al., 2010). Cao et al. (2011) reported the successful expression of pgsBCA genes from B. amyloliquefaciens LL3 in Escherichia coli JM109, resulting in the production of γ-PGA without L-glutamate. However, it would be more significant if the differential expression of pgsBCA genes from these two strains was demonstrated. Furthermore, glutamate racemase (racE) can be overexpressed to control the stereochemical composition of D-/L-glutamate leading to an increase in the production of γ-PGA, as well as the proportion of d-glutamate present in the polymer. It was revealed that overexpression of glr (racE) gene not only increased the production of γ-PGA but also increased the proportion of d-glutamate in γ-PGA (Ashiuchi et al., 1999; Jiang et al., 2011).
In this study, the pgsBCA genes and glutamate racemase gene (racE) from L-glutamic acid-dependent strain B. licheniformis NK-03 and L-glutamic acid-independent strain B. amyloliquefaciens LL3 were cloned and expressed in E. coli JM109. The differential expression was carried out in the medium containing either L-glutamate or glucose as carbon source. The extraction products were characterized by the weighing yields and fraction ratios of D-/L-isomer of γ-PGA monomer using reversed-phase high-performance liquid chromatography (reversed-phase HPLC). The results herein can supply the clue to the comparable functional structure of synthetase and be used to elucidate the molecular catalytic mechanism and the stereochemical modulation in γ-PGA biosynthesis.
Results and discussion
Cloning and alignment of pgsBCA genes from Bacillus strains
As expected for representative γ-PGA-producing strains, B. licheniformis NK-03 and B. amyloliquefaciens LL3 were capable of synthetizing γ-PGA with different molecular weight in the presence or absence L-glutamate, respectively. It is known that the γ-PGA synthetase complex consists of three functional subunits (PgsB, PgsC and PgsA) and is responsible for catalysing glutamate to synthesize γ-PGA with the type of membranous adenosine triphosphate (ATP)-dependent amide-ligase (Ashiuchi et al., 2004; Wang et al., 2011). Therefore, the cloning and sequence alignment of pgsBCA genes will be fundamental to understand the molecular mechanism of γ-PGA synthesis.
The sequence of pgsBCA was amplified from genome DNA of NK-03 and LL3 strains as before (Cao et al., 2010; 2011), which was about 3000 bp and contained three open-reading frames (ORFs), designated as pgsB, pgsC and pgsA. After alignment by BLAST (Basic Local Alignment Search Tool) and DNAMAN, the synthetase constituents PgsB and PgsC showed great similarity above 93%, while the PgsA showed only about 78.5% homology (Table S1). Based on the consensus of PgsBCA components and their amino acids sequences (Fig. 1), the feature of each element have been analysed and validated as that reported (Ashiuchi and Misono, 2003). It was revealed that PgsB conserves the consensus sequences found in enzymes of an amide ligase superfamily (Eveland et al., 1997). It harbours an ATP-binding motif at the N-teminal residues of 37–42 (GIRGKS), which is responsible for catalysing the hydrolysis of essential ATP, providing the energy for γ-PGA synthesis. PgsC, the most conserved component of PgsBCA complex, is a hydrophobic and a membrane-bound protein containing four transmembrane helices regions. The active site of PgsBCA, which was supposed to be constituted by PgsB and PgsC, was assumed to display ATPase activity and assimilate both isomers of γ-DL-PGA as the substrate (Urushibata et al., 2002). As the core structure for membrane integration and the transporter of γ-PGA, PgsA consisted mainly of hydrophobic and cationic amino acid residues, and showed the most variable part of synthetase to generate the differential elongation (molecular weight) of γ-PGA (Candela and Fouet, 2006). However, to elucidate the precise function of each membrane-associated component in γ-PGA polymerization and transportation, it should be recurred to three-dimensional modelling and crystal structure resolution.
Figure 1.
Sequence alignment of amino acid sequences of γ-PGA synthetase complex (PgsBCA) and glutamate racemase (RacE) from B. licheniformis NK-03 and B. amyloliquefaciens LL3. The residues with identity are represented by lower case beneath the sequences.
Characterization of racE gene from Bacillus strains
RacE, the glutamate racemase, was universally acknowledged as the primary enzyme of d-glutamate conversion from its enantiomer L-glutamate in the dynamic kinetic resolution of γ-PGA synthesis (Ashiuchi et al., 2001). The racE gene was cloned into pMD 19-Simple T vector and sequenced; in addition, the deduced amino acid sequence was aligned using DNAMAN software. It was revealed that the ORFs of racE gene from NK-03 and LL3 has size of 816 bp, and were deposited into GenBank accession nos. GQ375411 and GQ375412, respectively. The racE gene encodes 271 amino acids, the initial codon of which was TTG but not ATG. According to the deduced amino acid analysis, the RacE was estimated to have a molecular mass of 30 kDa, and the 12 amino acid sequence (MEQPIGVIDSGV) in its N-terminal region was determined to be Edman degradation region (Fig. 1). Furthermore, the regions surrounding the two cysteine residues (Cys-73 and Cys-184) are highly conserved; glutamate racemase reactions are proposed to proceed through a two-base mechanism involving the two essential cysteine residues (Gallo et al., 1993; Doublet et al., 1996; Ashiuchi et al., 1998). As it was reported that racemase from Bacillus was inactivated by 2-nitro-5-thiocyanatobenzoate, the two conserved cysteine residues are deemed to play an important role in the catalysis (Ashiuchi et al., 1998).
As shown in Fig. 1, the homology score of the glutamate racemase between B. licheniformis NK-03 and B. amyloliquefaciens LL3 was about 84.5%, and it was completely identical at the N-terminal region, composed by 55 amino acids. Because of the γ-PGA products from the two strains had the similar low content of d-glutamate (less than 2%), we hypothesized that the N-terminal sequence may act as the binding domain of L-glutamate accumulated in cells and the enhancer of the racemization. In addition, the initial codon of TTG presents low translation efficiency, which brought about the low activity of RacE and then the low content of d-glutamate in γ-PGA products. Ashiuchi and colleagues (1998) cloned the glr (racE) gene from B. subtilis IFO 3336 and overproduced it in the soluble fraction of the E. coli clone cells with the initial codon substitution of ATG for TTG. The cloned enzyme showed similar properties to those of the racemase from B. subtilis IFO 3336, suggesting that the enzyme spontaneously and effectively folds to become active in the E. coli overproducer. Therefore, the initial TTG of racE gene from NK-03 and LL3 was replaced with ATG to produce a large amount of the gene products from the recombinant cells of E. coli in the following co-expression of pgsBCA and racE.
Construction of co-expression pTrcNRP and pTrcLRP vectors
The 3.0 kb size of pgsBCA genes from B. licheniformis NK-03 and B. amyloliquefaciens LL3 were fused between BamHI-HindIII sites of pTrc99A (size of 4.2 kb), constructing recombinant plasmid that were named pTrcNpgs and pTrcLgs (size of 7.2 kb), respectively. Subsequently, the 0.8 kb racE gene holding KpnI-BamHI restriction sites was inserted into the foregoing site of pgsBCA located after the trc promoter, incorporating co-expression vectors pTrcNRP and pTrcLRP (size of 8.0 kb). Escherichia coli JM109 clones that harboured pTrc99A-pgsBCA-racE were selected by the methods of colony polymerase chain reaction (PCR) and recombinant plasmid enzyme digestion.
Judging from the bands migration distance compared with Marker III (Tiangen) in 0.8% agarose gel electrophoresis shown in Fig. 2, it was revealed that the fusion expression vectors pTrcLRP and pTrcNRP were successfully introduced into E. coli JM109. To our knowledge, the PgsBCA synthetase and glutamate racemase (RacE) genes in recombinant strains that could synthesize γ-PGA were only from B. subtilis strains (Ashiuchi et al., 1999). Therefore, the related work about pgsBCA and racE cloned from B. licheniformis and B. amyloliquefaciens strains and transformed into E. coli will enrich the co-expression systems and supply the clue for differential synthesis of γ-PGA as well.
Figure 2.
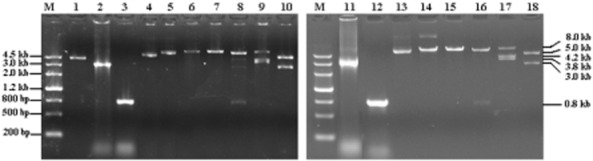
Electrophoresis analysis profile of recombinant plasmid pTrcLRP and pTrcNRP in E. coli JM109. Lane 1–10, test of pTrc99A harbouring LL3 pgsBCA and racE genes; Lane 11–18, test of pTrc99A harbouring NK-03 pgsBCA and racE genes.
Note: Lane M: Tiangen Marker III (Tiangen); Lane 1: pTrc99A/BamHI (4.2 kb); Lane 2: LL3 pgsBCA (3.0 kb); Lane 3: LL3 racE (0.8 kb); Lane 4: pTrcLRP (Supercoil, 8.0 kb); Lane 5: pTrcLpgs/BamHI (7.2 kb); Lane 6: pTrcLRP/KpnI (8.0 kb); Lane 7: pTrcLRP/BamHI (8.0 kb); Lane 8: pTrcLRP/KpnI+BamHI (7.2 and 0.8 kb); Lane 9: pTrcLRP/KpnI+HindIII (4.2 and 3.8 kb); Lane 10: pTrcLRP/BamHI+HindIII (5.0 and 3.0 kb); Lane 11: NK-03 pgsBCA (3.0 kb); Lane 12: NK-03 racE (0.8 kb); Lane 13: pTrcNpgs/BamHI (7.2 kb); Lane 14: pTrcNRP/KpnI (8.0 kb); Lane 15: pTrcNRP/BamHI (8.0 kb); Lane 16: pTrcNRP/KpnI+BamHI (7.2 and 0.8 kb); Lane 17: pTrcNRP/KpnI+HindIII (4.2 and 3.8 kb); Lane 18: pTrcNRP/BamHI+HindIII (5.0 and 3.0 kb).
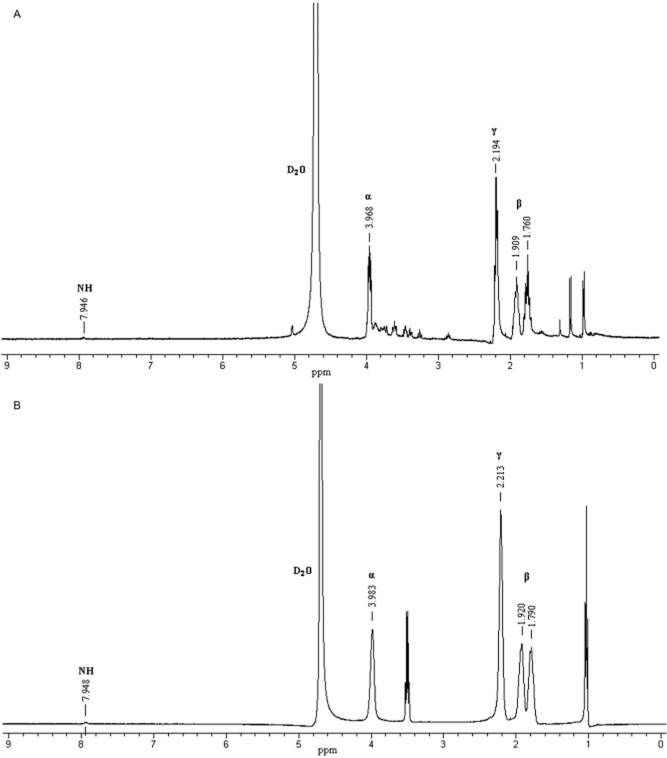
Determination of co-expression of racE and pgsBCA genes in E. coli
The recombinant E. coli JM109 strains, harbouring pTrcLpgs, pTrcNpgs, pTrcLRP and pTrcNRP were designated as 109LP, 109NP, 109LRP and109NRP, respectively (Table 1). The flask culture of Luria–Bertani (LB) plus 2% carbon source, metal ions (Mg2+ and Mn2+) suggested that the recombinant strains had the capacity of synthesizing γ-PGA. The 1H NMR spectrum of purified products revealed that the chemical shifts of α-CH, β-CH2, γ-CH2 and N-H are nearly overlapped with that of Sigma γ-PGA standard and the origin strains of LL3 and NK-03 (Cao et al., 2010; 2011). As shown in Fig. 3, the peak positions of γ-PGA from 109LRP and 109NRP were displayed as follows: α-CH (3.968/3.983 ppm), β-CH2 (1.760/1.790 ppm and 1.909/1.920 ppm), γ-CH2 (2.194/2.213 ppm) and N-H (7.946/7.948 ppm). In addition, the products were further determined for amino acid analysis by reversed phase HPLC and thin layer chromatography (data not shown). The results suggest that the fusion of racE and pgsBCA genes were successfully expressed, and γ-PGA was the primary product of the four engineered strains. However, it was surprising that there was only an obscure band corresponding to a molecular mass of 30 kDa (RacE), yet no clear band of target protein with molecular mass of 43 kDa (PgsB), 16 kDa (PgsC) and 42 kDa (PgsA) observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown). As we know, the PgsB, PgsC and PgsA parts constitute the active form of the synthetase complex, which is generally anchored into Bacillus cell membrane. It is conjectured that the different hydrophobicity and permeability of cell membrane structure between E. coli and Bacillus would bring about the inaccurate location and incapable function of PgsBCA, then following the scarce quantity for electrophoresis analysis. Considering there was no reports about the enzymatic purification and characterization of PgsBCA complex, this study will be employed to interpret the instability of membranous complex and supply a promising approach to obtain the crystallized amount of PgsBCA.
Table 1.
Strains and plasmids used in this study
| Stain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Plasmids | ||
| pMD 19-Simple T vector | T/A-cloning vector; lacZ; Ampr | TaKaRa (Dalian) |
| pTrc99A | Cloning vector, ori from pBR322, trc promoter, Ampr | Amann and colleagues (1988) |
| pTrcLpgs | pTrc99A carrying pgsBCA genes from LL3 strain | This study |
| pTrcNpgs | pTrc99A carrying pgsBCA genes from NK-03 strain | This study |
| pTrcLRP | pTrc99A carrying racE and pgsBCA genes from LL3 strain | This study |
| pTrcNRP | pTrc99A carrying racE and pgsBCA genes from NK-03 strain | This study |
| Strains | ||
| B. licheniformis NK-03 | Wild type, L-glutamic acid dependent producer of γ-PGA | Laboratory stock |
| B. amyloliquefaciens LL3 | Wild type, L-glutamic acid independent producer of γ-PGA | Laboratory stock |
| E. coli strains | ||
| JM109 | recA supE44 endA1 hsdR17 gyrA96relA1 thi △(lac-proAB) F′ (traD36 proAB+ lacIqlacZ△M15) | TaKaRa (Dalian) |
| 109LP | JM109 harbouring vector pTrcLpgs | This study |
| 109NP | JM109 harbouring vector pTrcNpgs | This study |
| 109LRP | JM109 harbouring vector pTrcLRP | This study |
| 109NRP | JM109 harbouring vector pTrcNRP | This study |
Figure 3.
1H NMR spectrum of γ-PGA produced from engineered 109LRP and 109NRP.
A. 1H NMR spectrum of γ-PGA from 109LRP product in D2O. The chemical shifts of α-CH (3.968 ppm), β-CH2 (1.76 and 1.909 ppm), γ-CH2 (2.194 ppm) and N-H (7.946) were labelled in the peaks location.
B. 1H NMR spectrum of γ-PGA from 109NRP product in D2O. The chemical shifts of α-CH (3.983 ppm), β-CH2 (1.790 and 1.920 ppm), γ-CH2 (2.213 ppm) and N-H (7.948) were labelled in the peaks location.
Fermentation and characterization of γ-PGA produced by engineered strains
A previous study showed that the synthetase encoded by pgsBCA from B. amyloliquefaciens LL3 could produce γ-PGA in engineered E. coli with the de novo pathway from glucose, and the yield was resembled to that of the recombinant C. glutamicum strain harbouring pgsBCA from B. licheniformis NK-03 (Cao et al., 2011). However, the PgsBCA activity and productivity differences between NK-03 and LL3, following with the possible molecular catalysis mechanism of L-glutamic acid-dependent and -independent γ-PGA-producing strains, remained undetermined. Furthermore, addition of Mn2+, cooperated with Mg2+, to the polymer-synthesis medium of E. coli cells co-expression genetic system harbouring pgsBCA and racE genes could increase the yield and d-glutamate content of γ-PGA, which could be adopted in the fermentation process of γ-PGA (Ashiuchi et al., 1999; Jiang et al., 2011).
The resultant cultivation showed that the four engineered strains grew almost synchronously and could produce γ-PGA in both glucose and L-glutamate medium after 24 h of induction. It was interesting that regardless of the harbouring vector, the yield of γ-PGA produced by LL3 pgsBCA was higher than that of NK-03; in other words, the LL3-derived PgsBCA appeared to have greater catalytic activity. As shown in Table 2, 109LP and 109LRP could synthesize 22% and 48% higher yields of γ-PGA than 109NP (0.308 ± 0.025 g l−1) and 109NRP (0.349 ± 0.016 g l−1), respectively, in glucose medium, while it was not so distinctive when the strains cultivated in L-glutamate medium. However, the molecular weight of γ-PGA produced by 109LP and 109LRP in both glucose and L-glutamate medium possessed lower Mw than 109NP and 109NRP, which had almost the same tendency as that of the original LL3 (Mw = 470 kDa) and NK03 (Mw = 1360 kDa). These observations suggested that the catalytic efficiency of PgsBCA from LL3 could really outperform that from NK-03, while the elongation dynamics was affected by the separate subunit of the synthetase complex. In addition, the activity bias could be offset by supplying enough substrate of L-glutamate, i.e. the divergence of PgsBCA did not contribute very well to the glutamic acid-independent γ-PGA-producing strains, while the glutamate synthase likely played an important part in γ-PGA synthesis.
Table 2.
Characterization of γ-PGA produced by the four E. coli recombinant strains
| Strains | Yield (g l−1) | D-glutamate content (%) | Molecular weight (× 104Da) |
|---|---|---|---|
| E. coli-pTrcLpgs (109LP)a | 0.376 ± 0.021 | 2.46 ± 0.32 | 3.23 ± 0.26 |
| E. coli-pTrcNpgs (109NP)a | 0.308 ± 0.025 | 3.28 ± 0.27 | 3.74 ± 0.38 |
| E. coli-pTrcLRP (109LRP)a | 0.517 ± 0.027 | 8.96 ± 0.53 | 5.09 ± 0.33 |
| E. coli-pTrcNRP (109NRP)a | 0.349 ± 0.016 | 9.33 ± 0.47 | 5.83 ± 0.42 |
| E. coli-pTrcLpgs (109LP)b | 0.558 ± 0.018 | 3.01 ± 0.45 | 5.35 ± 0.28 |
| E. coli-pTrcNpgs (109NP)b | 0.533 ± 0.022 | 5.94 ± 0.66 | 5.89 ± 0.55 |
| E. coli-pTrcLRP (109LRP)b | 0.645 ± 0.016 | 18.13 ± 1.36 | 6.23 ± 0.46 |
| E. coli-pTrcNRP (109NRP)b | 0.603 ± 0.033 | 19.59 ± 0.98 | 6.95 ± 0.79 |
Carbon source of glucose.
Substrate of L-glutamate.
As the glutamate racemase could supply the preferred d-glutamate substrate for γ-PGA polymerization, we estimated that the d-monomer content and productivity of γ-PGA should be improved significantly in 109LRP and 109NRP versus that of the parent strains. The results from the γ-PGA yield and d-glutamate content measurement using reversed phase HPLC (Fig. S1) showed great consistency with our hypothesis in either glucose or L-glutamate medium. When using L-glutamate as substrate, the d-monomer content in recombinant 109LRP (18.13%) and 109NRP (19.45%) strains was much higher than that of the 109LP (3.01%), 109NP (5.94%), and the original LL3 (1.5%) and NK-03 (2.0%) Bacillus strains, which was due to the regulation of RacE caused by the initial codon substitution of ATG for TTG. Meanwhile, the racemization capacity of RacE from LL3 was conjectured to be better than that of NK-03, owing to the elevated racemization level of RacE of sixfold and threefold, respectively. The similar result of d-monomer content of γ-PGA was acquired in glucose medium (Table 2), while the improvement of γ-PGA yield from 109LRP was much higher than that from 109NRP, which may be attributed to the stronger coordination ability of L-glutamate synthesis and RacE catalysis reaction. Furthermore, the molecular weight was increased as the yield and d-monomer content of γ-PGA, which was evidently affected by the synergy between RacE and PgsBCA. At present, the detailed works on stereochemical composition control of Bacillus γ-PGA regulated by RacE are effectively carried out through the combinatorial test and overexpression, including the directed-sites mutation of racE gene, which would supply the two species with the needs of different applications.
Although the co-expression of racE and pgsBCA had successfully presented an appreciable productivity in glucose medium (without L-glutamate), compared with that of other reported engineered E. coli strains harbouring only pgsBCA genes (Ashiuchi et al., 1999; Jiang et al., 2011; Wang et al., 2011), the optimized production of γ-PGA by the candidate 109LRP in flask culture was just 1.26 g l−1, which was still not as high as the wild type of Bacillus strains (Cao et al., 2010; 2011). Therefore, lots of works will be done to regulate the composition and the biosynthesis of γ-PGA, including the synergism and combinational performances of the concentration of substrate, the stability and permeability of cell membrane, and the activities of glutamate synthase, glutamate dehydrogenase, glutamate racemase, γ-PGA synthetase and depolymerase. Worth to be mentioned, the genome sequence of B. amyloliquefaciens LL3 has been completed and deposited into GenBank with accession no. CP002634 (Geng et al., 2011); the genome of B. licheniformis NK-03 is under sequencing. The future focus will be on systematical analysis of functional genes and regulatory elements screened from the grand data of proteomics and metabolomics. Furthermore, the ongoing crystal structure resolution, and rational designs of glutamate and γ-PGA metabolic pathways' enzymes will provide us in-depth understanding of the biochemical and molecular mechanism of γ-PGA synthesis.
Experimental procedures
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Bacillus and E. coli JM109 as the hosts for cloning and expression were routinely cultivated aerobically on LB (10 g tryptone, 5 g yeast extract, 10 g NaCl per litre of water) solid or liquid medium at 37°C. The media were supplemented with the following carbon sources, metal ions and antibiotics when required: 2% L-glutamate or glucose, 0.5% MgCl2, 0.05% MnSO4 and 60 μg ml−1 of filter sterilized ampicillin (Amp). Samples were regularly taken for measurement of optical density at 600 nm (OD600) and γ-PGA.
Reagents and kits
The DNA polymerases (rTaq, Ex-Taq and LA-Taq), restriction enzymes and DNA ligase solution were supplied by TaKaRa Biotech (Dalian, China). All oligonucletide primers used in this study (Table 3) were ordered from BGI Genomics (Shenzhen, China), and DNA sequencing was performed by Tianyi Huiyuan (Beijing, China). Extraction and purification of DNA samples were accomplished by Tiangen DNA gel purification kit, plasmid extraction kit and DNA Marker III (Beijing, China). All chemicals and standards were purchased from Sigma (Shanghai, China).
Table 3.
Primers used in this study
| Primer | Sequence (5′-3′)a | Function |
|---|---|---|
| Npgs-F | CGCGGATCCAAGGAGATGTCGAAAGCAATGT | NK-03 pgsBCA cloning |
| Npgs-R | CCCCAAGCTTCATCTTTATCACTCCGTTTATT | |
| Lpgs-F | CGCGGATCCAGAAGGAGATGTCAAAAATCAATG | LL3 pgsBCA cloning |
| Lpgs-R | CCCAAGCTTGATTTTCATTTGTTTTTCACTCCGC | |
| NracE-F | CGGGGTACCGAGGCGATTTTGATGGAAC | NK-03 racE cloning |
| NracE-R | CGCGGATCCACTATCTTTTAATCGGTTCTT | |
| LracE-F | CGGGGTACCGAGGCGATTTTGATGGAAC | LL3 racE cloning |
| LracE-R | CGCGGATCCAACAGCGGGTTTTTTGATTTAT |
The restriction sites are underlined.
Cloning and alignment of genes encoding γ-PGA synthetase and glutamate racemase
Routine DNA manipulations were carried out as described by Sambrook and Russell (2001). The pgsBCA genes responsible for γ-PGA synthesis were amplified from chromosomal DNA of B. licheniformis NK-03 and B. amyloliquefaciens LL3 using the restriction sites (forward: BamHI, reverse: HindIII) incorporated primers Npgs-F/Npgs-R and Lpgs-F/Lpgs-R. TA-cloning was processed with the purified PCR product and pMD 19-Simple T vector (TaKaRa), the positive clones of which were screened by colony PCR and enzyme digestion. Subsequently, the inserted fragments from positive clones were sequenced, and the ORFs were identified using the National Center for Biotechnology Information ORF finder tool and DNASTAR software (DNASTAR, Inc., Madison, WI, USA). The alignments of nucleotides and amino acid sequences between NK-03 and LL3 were accomplished by using BlastN and DNAMAN software (Lynnon Co., Pointe-Claire, QC, Canada). Likewise, the previous manipulations of pgsBCA could be employed to perform the cloning and intercomparsion of the glutamate racemase gene racE and the related coding enzyme. However, the difference was that the restriction sites were KpnI in forward primer (NracE-F and LracE-F) and BamHI in reverse primer (NracE-R and LracE-R), respectively.
Plasmid construction for expression of racE and pgsBCA genes
Based on the deduced ORFs, the sequenced fragments of pgsBCA from L-glutamic acid-dependent producer NK-03 or L-glutamic acid-independent producer LL3 were digested with BamHI-HindIII from T-vector and then subcloned into the same sites of pTrc99A to generate the PgsBCA expression vector pTrcNpgs or pTrcLpgs, respectively. In succession, the racE gene was excised from T-vector by KpnI-BamHI and fused in the foregoing site of pgsBCA, i.e. in the direction of trc promoter of pTrc99A, incorporating PgsBCA and RacE co-expression vectors pTrcNRP and pTrcLRP. The positive clones were tested by colony PCR and recombinant plasmid enzyme digestion.
Co-expression of racE and pgsBCA in E. coli
Strains 109LP, 109NP, 109LRP and109NRP were obtained by transforming E. coli JM109 competent cells with pTrcNpgs, pTrcLpgs, pTrcNRP and pTrcLRP, respectively (Table 1). The recombinant cells were selected on LB plates supplemented with Amp (100 μg ml−1) and were subsequently inoculated into 5 ml LB broth containing Amp (60 μg ml−1). After overnight shaking cultivation at 37°C (200 rpm), 1% (v/v) inoculum was transferred into 500 ml conical flasks containing 100 ml LB medium plus 2% L-glutamate or glucose, 0.5% MgCl2, 0.05% MnSO4 and 60 μg ml−1 of Amp. The optimal expression conditions of trc promoter for fusion genes of racE and pgsBCA were obtained when mid-log-phase cells (OD600 = 1.0) were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 28°C for 24 h with stirring.
The resulting broth was separated by centrifugation for 15 min at 10 000 g. The supernatant was used for γ-PGA separation and purification, while the harvested cells were gathered for protein analysis by SDS-PAGE with linear gradient of gel concentration from 8∼12%. The cells were resuspended in 0.1 M sodium phosphate buffer (pH 7.4) and disrupted by sonication in ice-cold buffer of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10 mM dithiothreitol. The supernatant liquid and sedimentation pellets were collected selectively and boiled for denaturation with the electrophoresis detection of potential RacE and PgsBCA.
Purification and characterization of γ-PGA
γ-PGA was recovered and purified by a modified precipitation method reported previously (Cao et al., 2011). The supernatant of IPTG-induced broth was poured into fourfold volumes of cold anhydrous ethanol; then, the precipitate was centrifuged at 20 000 g (20 min, 4°C) and dissolved in deionized water. The γ-PGA-containing solution was dialysed against ice-cold Milli-Q water in a dialysis membrane bag (molecular weight cut-off = 8000–14 000) for 24 h at 4°C, and the remain was lyophilized (−50°C) to obtain the purified biopolymer.
The 1H NMR spectroscopy was carried out to identify products of γ-PGA, using the heavy water (D2O)-dissolved samples scanned in nuclear magnetic resonance spectrometer (Varian Infinity plus 400, Varian Inc., Palo Alto, CA, USA), and the fingerprints were compared with a standard γ-PGA (Sigma). To determine the ratios of L- and d-isomer of γ-PGA hydrolysate, reversed phase HPLC was employed according to the FDAA (Marfey's reagent) precolumn derivation method with an Alltech GRACE C18 column (Alltech Associates, Inc., Deerfield, IL, USA) (Cao et al., 2010). The molecular weight of the products was measured by gel permeation chromatography using the eluent of 0.25 M NaNO3 in Alltech system controller equipped with a Shodex KW804 column (Showa Denko KK, Tokyo, Japan) and Schambeck SFD refractive index detector (Schambeck SFD GmbH, Bad Honnef, Germany) (Cao et al., 2011).
Acknowledgments
This study was financially supported by the National key Basic Research Program of China (‘973’ Program) 2012CB725204, the National High Technology Research and Development Program of China (‘863’ Program) 2012AA021505, and the Natural Science Foundation of China Grant Nos. 31070039, 31170030 and 51073081. We thank to Dr. Miguel Suastegui from Iowa State University for the manuscript modification.
Conflict of interest
None declared.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. d-glutamate content measurement of γ-PGA produced by the four E. coli recombinant strains using the reversed phase HPLC method.
Table S1. The comparison of PgsBCA synthetase complex and RacE from NK-03 and LL3.
References
- Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Misono H. Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl Microbiol Biotechnol. 2002;59:9–14. doi: 10.1007/s00253-002-0984-x. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Misono H. Poly-γ-glutamate synthetase of Bacillus subtilis. J Mol Catal B. 2003;23:101–106. [Google Scholar]
- Ashiuchi M, Tani K, Soda K, Misono H. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-γ-glutamate. J Biochem. 1998;123:1156–1163. doi: 10.1093/oxfordjournals.jbchem.a022055. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Soda K, Misono H. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem Biophys Res Commun. 1999;263:6–12. doi: 10.1006/bbrc.1999.1298. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Kamei T, Baek DH, Shin SY, Sung MH, Soda K, et al. Isolation of Bacillus subtilis (chungkookjang), a poly-γ-glutamate producer with high genetic competence. Appl Microbiol Biotechnol. 2001;57:764–769. doi: 10.1007/s00253-001-0848-9. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Shimanouchi K, Nakamura H, Kamei T, Soda K, Park C, et al. Enzymatic synthesis of high-molecular-mass poly-γ-glutamate and regulation of its stereochemistry. Appl Environ Microbiol. 2004;70:4249–4255. doi: 10.1128/AEM.70.7.4249-4255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela T, Fouet A. Poly-gamma-glutamate in bacteria. Mol Microbiol. 2006;60:1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x. [DOI] [PubMed] [Google Scholar]
- Candela T, Moya M, Haustant M, Haustant M, Fouet A. Fusobacterium nucleatum, the first Gram-negative bacterium demonstrated to produce polyglutamate. Can J Microbiol. 2009;55:627–632. doi: 10.1139/w09-003. [DOI] [PubMed] [Google Scholar]
- Cao MF, Song CJ, Jin YH, Liu L, Liu J, Xie H, et al. Synthesis of poly(γ-glutamic acid) and heterologous expression of pgsBCA genes. J Mol Catal B. 2010;67:111–116. [Google Scholar]
- Cao MF, Geng WT, Liu L, Song CJ, Xie H, Guo WB, et al. Glutamic acid independent production of poly-γ-glutamic acid by Bacillus amyloliquefaciens LL3 and cloning of pgsBCA genes. Bioresour Technol. 2011;102:4251–4257. doi: 10.1016/j.biortech.2010.12.065. [DOI] [PubMed] [Google Scholar]
- Cheng C, Asada Y, Aida T. Production of γ-polyglutamic acid by Bacillus licheniformis A35 under denitrifying conditions. Agric Biol Chem. 1989;53:2369–2375. [Google Scholar]
- Doublet P, van Heijenoort J, Mengin-Lecreulx D. Regulation of the glutamate racemase of Escherichia coli investigated by site-directed mutagenesis. Microb Drug Resist. 1996;2:43–49. doi: 10.1089/mdr.1996.2.43. [DOI] [PubMed] [Google Scholar]
- Eveland SS, Pompliano DL, Anderson MS. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-gamma-glutamate ligases: identification of a ligase superfamily. Biochemistry. 1997;36:6223–6229. doi: 10.1021/bi9701078. [DOI] [PubMed] [Google Scholar]
- Gallo KA, Tanner ME, Knowles JR. Mechanism of the reaction catalyzed by glutamate racemase. Biochemistry. 1993;32:3991–3997. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- Gardner JM, Troy FA. Chemistry and biosynthesis of the poly (gamma-D-glutamyl) capsule in Bacillus licheniformis. Activation, racemization, and polymerization of glutamic acid by a membranous polyglutamyl synthetase complex. J Biol Chem. 1979;254:6262–6269. [PubMed] [Google Scholar]
- Geng WT, Cao MF, Song CJ, Xie H, Liu L, Yang C, et al. Complete genome sequence of Bacillus amyloliquefaciens LL3, which exhibits glutamic acid-independent production of poly-γ-glutamic acid. J Bacteriol. 2011;193:3393–3394. doi: 10.1128/JB.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Kunioka M. Biosynthesis and hydrolysis of poly (γ-glutamic acid) from Bacillus subtilis IFO 3335. Biosci Biotech Biochem. 1992;56:1031–1035. doi: 10.1271/bbb.56.1031. [DOI] [PubMed] [Google Scholar]
- Hezayen FF, Rehm BHA, Eberhardt R, Steinbüchel A. Polymer production by two newly isolated extremely halophilic Archaea: application of a novel corrosion-resistant bioreactor. Appl Microbiol Biotechnol. 2000;54:319–325. doi: 10.1007/s002530000394. [DOI] [PubMed] [Google Scholar]
- Ito Y, Tanaka T, Ohmachi T, Asada Y. Glutamic acid independent production of poly(γ-glutamic acid) by Bacillus subtilis TAM-4. Biosic Biotech Biochem. 1996;60:1239–1242. [Google Scholar]
- Jiang F, Qi GF, Ji ZX, Zhang SL, Liu J, Ma X, Chen SW. Expression of glr gene encoding glutamate racemase in Bacillus licheniformis WX-02 and its regulatory effects on synthesis of poly-γ-glutamic acid. Biotechnol Lett. 2011;33:1837–1840. doi: 10.1007/s10529-011-0631-7. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shang LA, Yoon SH, Lee SY, Yu ZN. Optimal production of poly-γ-glutamic acid by metabolically engineered Escherichia coli. Biotechnol Lett. 2006;28:1241–1246. doi: 10.1007/s10529-006-9080-0. [DOI] [PubMed] [Google Scholar]
- Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemetz R, Kärcher U, Kandler O, Tindall BJ, König H. The cell wall polymer of the extremely halophillic arxhaeon Natronococcus occultus. Eur J Biochem. 1997;249:905–911. doi: 10.1111/j.1432-1033.1997.00905.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratry Manual. 3rd edn. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shih IL, Van YT. The production of poly(γ-glutamic acid) from microorganisms and its various applications. Bioresour Technol. 2001;79:207–225. doi: 10.1016/s0960-8524(01)00074-8. [DOI] [PubMed] [Google Scholar]
- Sung MH, Park C, Kim KS, Park JH, Choi YH, Kim HB, et al. 2003. Method for producing poly-γ-glutamate using glutamate producing Coryne-bacteria. KR/10-0062761, 08, 09.
- Sung MH, Park C, Kim CJ, Poo H, Soda K, Ashiuchi M. Natural and edible biopolymer poly-g-glutamic acid: synthesis, production, and applications. Chem Rec. 2005;5:352–366. doi: 10.1002/tcr.20061. [DOI] [PubMed] [Google Scholar]
- Tarui Y, Iida H, Ono E, Miki W, Hirasawa E, Fujita K, et al. Biosynthesis of poly-γ-glutamic acid in plants: transient expression of poly-γ-glutamate synthetase complex in tobacco leaves. J Biosci Bioeng. 2005;100:443–448. doi: 10.1263/jbb.100.443. [DOI] [PubMed] [Google Scholar]
- Urushibata Y, Tokuyama S, Tahara Y. Characterization of the Bacillus subtilis ywsC gene, involved in γ-polyglutamic acid production. J Bacteriol. 2002;184:337–343. doi: 10.1128/JB.184.2.337-343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang G, Che C, Liu Y. Heterogenous expression of poly-γ-glutamic acid synthetase complex gene of Bacillus licheniformis WBL-3. Appl Biochem Microbiol. 2011;47:381–385. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.