Abstract
Ras activation is critical for T-cell development and function, but the specific roles of the different Ras isoforms in T-lymphocyte function are poorly understood. We recently reported T-cell receptor (TCR) activation of ectopically expressed H-Ras on the the Golgi apparatus of T cells. Here we studied the isoform and subcellular compartment specificity of Ras signaling in Jurkat T cells. H-Ras was expressed at much lower levels than the other Ras isoforms in Jurkat and several other T-cell lines. Glutathione S-transferase-Ras-binding domain (RBD) pulldown assays revealed that, although high-grade TCR stimulation and phorbol ester activated both N-Ras and K-Ras, low-grade stimulation of the TCR resulted in specific activation of N-Ras. Surprisingly, whereas ectopically expressed H-Ras cocapped with the TCRs in lipid microdomains of the Jurkat plasma membrane, N-Ras did not. Live-cell imaging of Jurkat cells expressing green fluorescent protein-RBD, a fluorescent reporter of GTP-bound Ras, revealed that N-Ras activation occurs exclusively on the Golgi apparatus in a phospholipase Cγ- and RasGRP1-dependent fashion. The specificity of N-Ras signaling downstream of low-grade TCR stimulation was dependent on the monoacylation of the hypervariable membrane targeting sequence. Our data show that, in contrast to fibroblasts stimulated with growth factors in which all three Ras isoforms become activated and signaling occurs at both the plasma membrane and Golgi apparatus, Golgi-associated N-Ras is the critical Ras isoform and intracellular pool for low-grade TCR signaling in Jurkat T cells.
Ras proteins are small, guanine-nucleotide-binding proteins that are implicated in the regulation of multiple cellular functions, including proliferation, differentiation, and apoptosis (26). Mutations that render Ras constitutively active occur in many human malignancies (5, 33). Ras proteins cycle between inactive GDP-bound and active GTP-bound states. The two main groups of molecules involved in the regulation of Ras proteins are the guanine nucleotide exchange factors that promote the transition from the inactive GDP-bound to the active GTP-bound state and the GTPase-activating proteins (GAPs) that inhibit Ras by stimulating its GTPase activity.
Mammals have three functional ras genes, H-ras, K-ras, and N-ras, the products of which have very similar structures (26). The K-ras gene contains two alternative fourth coding exons, giving rise to two splice variants, K-Ras4A and K-Ras4B. Since the K-Ras4A alternative accounts for less than 10% of total K-ras mRNA, we will refer to K-Ras4B as the K-Ras isoform. At the amino acid level, Ras isoforms are identical for the first 80 amino acids, exhibit 85% identity for the next 80 residues, and display only 15% amino acid conservation within the C-terminal 25 amino acids (3, 6). The C-terminal hypervariable region directs the posttranslational modifications of the primary ras gene products that determine their subcellular localization (18, 19).
Ras proteins play an important role in the signaling pathways that activate cytokine gene induction and in the control of T-cell development (17). Since the activation of Ras upon T-cell stimulation was first demonstrated (12), a critical role of Ras in antigen receptor signaling in lymphocytes has been appreciated (1, 16, 20). In fact, the loss of Ras function prevents the proliferation, cytokine production, and lymphocyte development induced by the recognition of the antigen (39, 43).
A number of functional differences between the Ras isoforms have been reported (26). For example, different ras genes have been found mutated in different tumor types (5, 33), and mice deficient in the different Ras isoforms exhibit different developmental phenotypes (14, 21, 24, 44). Despite these differences, the specific function(s), if any, of the various Ras isoforms is poorly understood. However, a number of studies have pointed out the importance of N-Ras in T-cell function. Firstly, activating mutations of N-Ras are frequently found in human and mouse hematopoietic tumors (5, 27, 33, 38, 42). More recently, by using an N-Ras-deficient mouse model, we have shown that N-Ras is an important component of the T-cell signaling network and its function (29). The functional consequences of the absence of N-Ras in T cells include deficient CD8+ selection, a decreased thymocyte proliferation, a significant reduction in the production of interleukin-2 upon thymocyte activation, and an increased sensitivity to influenza infection in vivo.
The purpose of this work was to determine the mechanism(s) underlying the specific role of N-Ras in T-cell function. Our results show that, although all three Ras isoforms are expressed in human T cells, N-Ras is the only isoform activated following low-grade stimulation of the T-cell receptors (TCR) in Jurkat T cells. Moreover, N-Ras activation takes place exclusively on the Golgi apparatus as a consequence of signaling through phospholipase Cγ1 (PLCγ1) and RasGRP1.
MATERIALS AND METHODS
Cells and transfection assays.
Jurkat T leukemia cells (clone E6-1) and the PLCγ1-deficient mutant (J gamma 1) are derived from a human acute T-cell leukemia and were obtained from the American Type Culture Collection. CEM (CCRF-CEM) and Karpas (KARPAS-299) cell lines are derived from a human T-cell acute lymphoblastic leukemia and a human T-cell non-Hodgkin lymphoma, respectively. HEK293 cells, which are a permanent line of primary human embryonic kidney cells, were also obtained from the American Type Culture Collection. All the cells were kept at logarithmic growth in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and 100 U of penicillin G and streptomycin each per ml.
The majority of the transfection assays were performed by lipofection with DMRIE-C (Gibco BRL) (for Jurkat) or Superfect (Gibco BRL) (for COS-1) and the conditions recommended by the manufacturer. In the indicated cases, Amaxa technology was used to transfect Jurkat T cells according to the manufacturer's recommendations.
Plasmids, antibodies, and reagents.
Yellow fluorescent protein (YFP)-Ras-binding domain (RBD), untagged Ras, green fluorescent protein (GFP)-Ras, and cyan fluorescent protein (CFP)-Ras vectors were previously described (8, 9). CFP-H-RasC184L and CFP-N-RasL184C palmitoylation mutants were generated by using a QuikChange site-directed mutagenesis kit (Stratagene). Human RasGRP cDNAs were amplified by PCR (primer sequences available upon request) and cloned in frame into the mammalian expression vectors pYFP-N1 (Clontech) and pcDNA3.1(+)/Neo (Invitrogen). All plasmids were verified by bidirectional sequencing. Antibodies used for Ras detection included agarose-conjugated anti-pan-Ras Y13-259 (Oncogene Research) and monoclonal antibodies for mouse N-Ras (F155), H-Ras (F235), and K-Ras (F234) (Santa Cruz Biotechnology). Mouse anti-human CD3 (UCHT1) and CD28 (5D10) (Ancell) were used for TCR-dependent activation, whereas phorbol 12-myristate 13-acetate (PMA) plus ionomycin (Sigma-Aldrich) was used for TCR-independent activation of Jurkat cells.
Cell stimulation and imaging.
For TCR-dependent stimulation, Jurkat cells were incubated with high (5 μg/ml) or low (1 μg/ml) doses of both mouse anti-human CD3 plus anti-CD28 antibodies. PMA (100 ng/ml) plus ionomycin (500 ng/ml) was used for TCR-independent stimulation. For the specific microlocalization of Ras proteins in the plasma membranes of T cells, Jurkat cells were first incubated with anti-CD3 and anti-CD28, washed with phosphate-buffered saline, and then incubated with Texas red-conjugated donkey anti-mouse immunoglobulin G (heavy plus light chains) (Jackson ImmunoResearch Laboratories, Inc). The stimulation of COS-1 cells was performed by adding 40 ng of epidermal growth factor (EGF)/ml to the media. For examination by fluorescence microscopy, the cells were plated in 35-mm dishes containing a glass coverslip-covered 15-mm cutout (MatTek). By using a Harvard Apparatus microincubator and dual confocal microscopy, the images were captured with a Zeiss 510 laser-scanning confocal microscope (LSM) with the manufacturer-specified filter sets for single and dual emissions. TIFF images were processed with Adobe Photoshop 6.0.
Protein expression and Ras activation.
For the detection of Ras variants, the proteins were extracted in 200 μl of lysis buffer (10% glycerol, 1% Nonidet P-40, 50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin, 10 μg of soybean trypsin inhibitor per ml, 0.1 μM aprotinin). Lysates were cleared by centrifugation, and the protein concentration was quantified by the Bradford method. Ras proteins from Jurkat cells (1 mg per sample) were also immunopurified by using an anti-pan-Ras immunoaffinity column. In all cases, the proteins were fractionated in triplicate in sodium dodecyl sulfate-15% polyacrylamide gels, and Ras variants were detected by using antibodies specific for each isoform.
For Ras activation experiments, 2 × 107 Jurkat T cells were serum starved at 37°C for 2 h and then incubated in the presence or absence of anti-CD3 plus anti-CD28 or PMA plus ionomycin for 10 min. To detect GTP-bound Ras, 75% of the cells were quickly sedimented and lysed in 400 μl of lysis buffer. Lysates were cleared by centrifugation, incubated for 2 h at 4°C with glutathione S-transferase (GST)-Raf-RBD fusion protein (a gift of J. L. Bos, Utrech University, Utrech, The Netherlands) coupled to glutathione agarose beads, and washed four times with lysis buffer. To detect total Ras, the remaining cells (25%) were lysed in 100 μl of Laemmli buffer. Finally, GTP-bound and total Ras proteins were detected by Western blotting with isoform-specific antibodies.
Active ERK1/2 proteins were detected in the primary lysates by using phospho-specific antibodies according to the manufacturer's recommendations (Promega). To normalize the activation of endogenous ERK1/2, lysates were also probed with anti-ERK1/2.
In all cases, the immunoblots were revealed by using Supersignal West Femto maximum sensitivity substrate (Pierce). Scanned TIFF images were processed and bands were quantified with the Quantity One software (Bio-Rad).
Indirect immunofluorescence assays.
Jurkat T cells were fixed with 4% paraformaldehyde and permeabilized and blocked with 0.5% Triton X-100-5% bovine serum albumin in phosphate-buffered saline. The cells were dual stained with monoclonal antibodies to N-Ras (F155; Santa Cruz Biotechnology) or RasGRP1 (M133; a gift of James C. Stone, University of Alberta, Alberta, Canada) and a polyclonal antiserum against human giantin (PRB-114; Covance Research) (each diluted 1:250), followed by Texas red-conjugated horse anti-mouse combined with fluorescein isothiocyanate-conjugated goat anti-rabbit antisera (Jackson ImmunoResearch). The cells were mounted with photobleach retardant medium (DAKO Corp.), and they were imaged with a Zeiss 510 inverted LSM.
RESULTS
N-Ras mediates TCR signaling in Jurkat T cells upon low-grade stimulation.
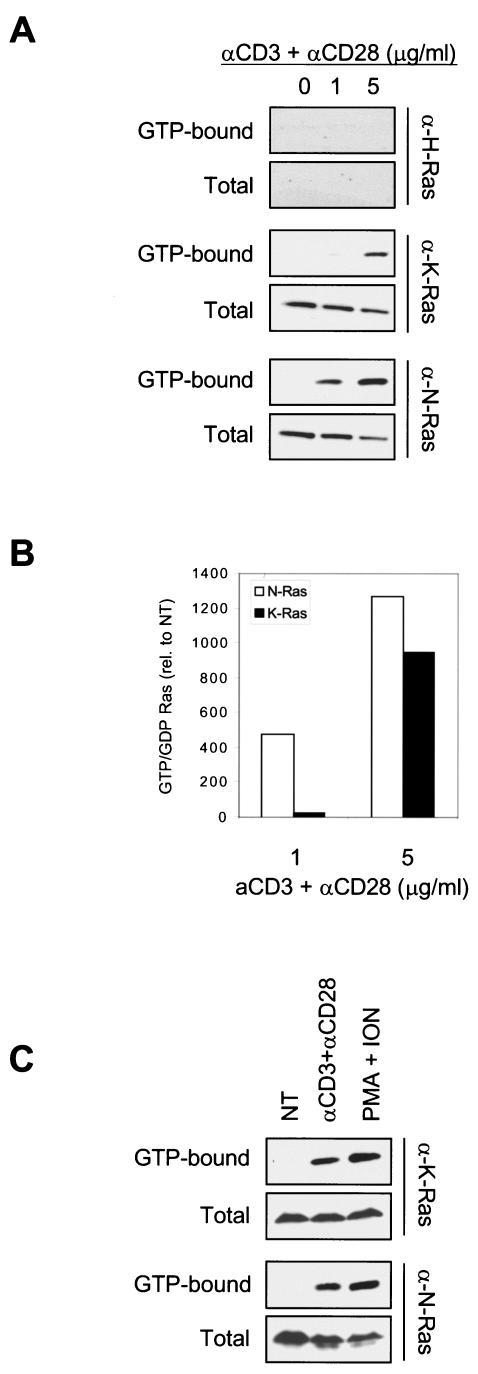
To determine the isoform specificity of Ras activation downstream of the TCR, we stimulated Jurkat T cells by cross-linking the TCR with anti-CD3 and anti-CD28 antisera, and we analyzed cell lysates for GTP-bound Ras by using the GST-RBD pulldown method (10) combined with isoform-specific immunoblotting. As shown in Fig. 1A, K-Ras and N-Ras, but not H-Ras, were activated following TCR cross-linking. Interestingly, whereas the stimulation of Jurkat T cells with 5 μg of anti-CD3 plus anti-CD28/ml showed similar activation of both N-Ras and K-Ras, low-grade TCR stimulation (1 μg of anti-CD3 plus anti-CD28/ml) induced activation only on N-Ras (Fig. 1B). Since the majority of evidence supports that a weak-moderate signal results in T-cell positive selection (22, 36), our data indicate that N-Ras should be the Ras isoform involved in transducing the TCR-dependent signals that mediate cell survival and differentiation into mature T cells.
FIG.1.
N-Ras-specific activation in Jurkat cells following low-grade TCR stimulation. (A) Jurkat cells (2 × 107 per point) were serum starved for 2 h and incubated with or without the indicated amounts of anti-CD3 plus anti-CD28. Proteins from stimulated and unstimulated cells were used to collect GTP-bound and total Ras as described in Materials and Methods. (B) To quantify Ras activation, GDP-bound and GTP-bound bands were quantified by using Quantity One software. The graph shows GTP/GDP ratios relative to the nontreated cells (NT). (C) Both N-Ras and K-Ras are equally activated by strong TCR-dependent and TCR-independent stimuli. Cells were kept untreated or activated with the indicated mitogens and processed as described for panel A. ION, ionomycin.
Since our results showed no activation of endogenous H-Ras upon TCR stimulation, we tested the capacity of the GST-RBD pulldown method to detect GTP-bound H-Ras. As expected, when a constitutively active H-Ras mutant (H-RasL61) was transiently overexpressed in Jurkat T cells, GTP-bound H-Ras was affinity purified from Jurkat cell lysates by using GST-RBD (data not shown), validating the ability of this assay to detect GTP-bound H-Ras.
The apparent differential activation of N-Ras and K-Ras upon low-grade TCR stimulation may be due to different sensitivities of the method toward the different isoforms. However, two factors argue against this possibility. First, there is no evidence that GST-RBD shows any Ras isoform preference. Second, the K-Ras- versus N-Ras-specific antibodies used to analyze the pulldown were equally efficient at recognizing their cognate proteins (data not shown). Nevertheless, we directly tested the possibility that the difference was due to different sensitivities of detection by activating Jurkat T cells with either a high concentration (10 μg/ml) of anti-CD3 and anti-CD28 or a strong TCR-independent stimulus (PMA plus ionomycin). In both cases, K-Ras activation was readily detected in this cell type, and no differences were detected between its activation levels and those of N-Ras (Fig. 1C). Thus, the preferential activation of N-Ras upon low-grade TCR stimulation is not due to differential sensitivities of the assay but rather to an intrinsic difference among the isoforms that is likely to have physiologic consequences.
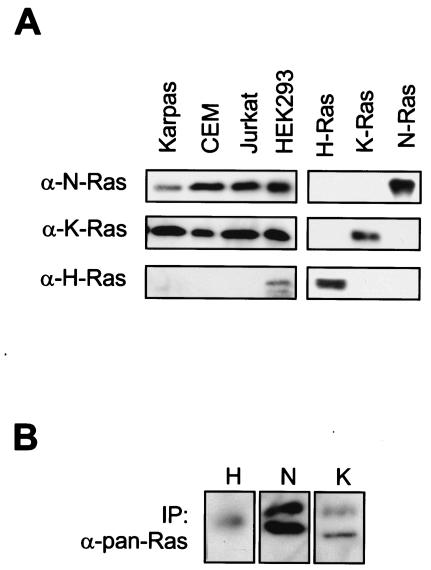
Although mammalian ras genes are expressed in all cell lineages and organs, some differences have been detected in the levels of expression of each of these genes in embryonic development and in various adult tissues (15, 25). For example, human leukemia cells and mouse primary thymocytes express substantially more N-Ras and K-Ras than H-Ras (25, 37). Since differential expression of Ras isoforms could explain why only N-Ras was activated in Jurkat cells upon TCR engagement, we determined the levels of expression of N-, K-, and H-Ras in human T cells. First, we determined the relative affinities of the isoform-specific anti-Ras antisera and discovered that, whereas the anti-K-Ras and anti-N-Ras reagents had very similar affinities for their cognate antigens, the anti-H-Ras antiserum was 10-fold more efficient (data not shown). Ras expression in Jurkat cells, two other human T-cell lines (Karpas and CEM), and a human epithelial cell line (HEK293) was analyzed by immunoblotting with these isoform-specific antibodies (40). HEK293 cells expressed all three Ras isoforms (Fig. 2A). In contrast, whereas all three T-cell lines expressed relatively high levels of K- and N-Ras, we did not detect H-Ras expression in these cells, despite the higher sensitivity afforded by the anti-H-Ras antibody (Fig. 2A). To increase the sensitivity of the assay, we immunoprecipitated all cellular Ras with a pan-Ras antibody and then probed the immunoprecipitates with isoform-specific antisera. By using this method, H-Ras was detected in Jurkat cells, although at a much lower level than were K-Ras and N-Ras (Fig. 2B).
FIG. 2.
Expression levels of the three Ras isoforms in different human cell lines. (A) Jurkat, Karpas, and CEM T cells and HEK293 epithelial cells were analyzed for Ras isoform levels by immunoblotting as described in Materials and Methods (left panels). Recombinant proteins for each Ras isoform were used as positive controls (right panels). (B) For Jurkat cells, total Ras proteins were also immunopurified (IP) by using an anti-pan-Ras immunoaffinity column, and Ras isoforms were detected by using antibodies specific for each isoform. As has been previously reported (40), a doublet was detected for K-Ras and N-Ras.
N-Ras does not colocalize with TCR complexes upon CD3-plus-CD28 stimulation.
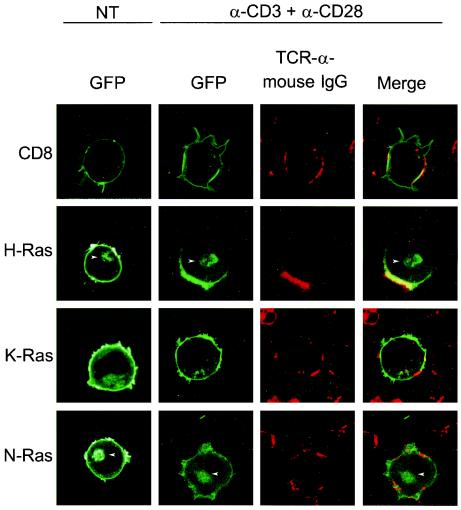
The distribution of Ras isoforms within subdomains of the plasma membrane may influence the activation and signaling of the GTPase (31). The TCR is thought to partition into lipid rafts. Indeed, cross-linking of TCR complexes in T cells induces the coalescence of lipid raft microdomains thought to organize signaling components (23). To examine the microlocalization at the plasma membrane of each Ras isoform during TCR activation, we transfected Jurkat cells with GFP-tagged Ras proteins and observed their subcellular localization in living cells by using LSM before and after activating Jurkat cells by cross-linking TCR complexes. To augment patching and to directly visualize patched lipid rafts, we followed anti-CD3 and anti-CD28 antibodies with Texas red-conjugated goat anti-mouse antiserum (Fig. 3). As a positive control, we utilized GFP-tagged CD8, which is known to associate with the TCR complexes in lipid rafts (2). As expected, CD8-GFP was localized homogeneously along the plasma membrane in untreated cells but was redistributed along with TCR to membrane patches after cross-linking. The steady-state localization of GFP-H-Ras, GFP-K-Ras, and GFP-N-Ras in untreated Jurkat cells paralleled that observed in other cell types (9). Whereas GFP-K-Ras was expressed predominantly on the plasma membrane, GFP-N-Ras and GFP-H-Ras were expressed on both the plasma membrane and Golgi apparatus. GFP-H-Ras behaved like CD8-GFP, colocalizing in patches with the TCR after cross-linking. In contrast, neither GFP-K-Ras nor GFP-N-Ras was enriched in these membrane patches following TCR cross-linking. Paradoxically, N-Ras, the isoform preferentially activated downstream of the TCR upon low-grade stimulation, was not enriched along with the TCR in lipid rafts.
FIG. 3.
N-Ras does not colocalize with TCR complexes in TCR-dependent activation of Jurkat cells. Jurkat cells were transfected with expression vectors encoding CD8-GFP, GFP-H-Ras, GFP-K-Ras, and GFP-N-Ras and were imaged alive 48 h later by LSM. Untreated, transfected Jurkat cells (NT) revealed the intrinsic steady-state localization of each fusion protein that included the plasma membrane and, in the case of GFP-N-Ras and GFP-H-Ras, the Golgi apparatus (arrowheads). Jurkat cells were incubated with anti-CD3 plus anti-CD28, and then TCR complexes were visualized by adding a Texas red-conjugated goat anti-mouse antibody. Jurkat T cells activated in this fashion showed a characteristic TCR patching revealed by red fluorescence. The overlay reveals areas of colocalization (yellow) between patched TCR (red) and GFP-tagged molecules (green).
Low-grade TCR stimulation in Jurkat cells activates N-Ras on Golgi apparatus.
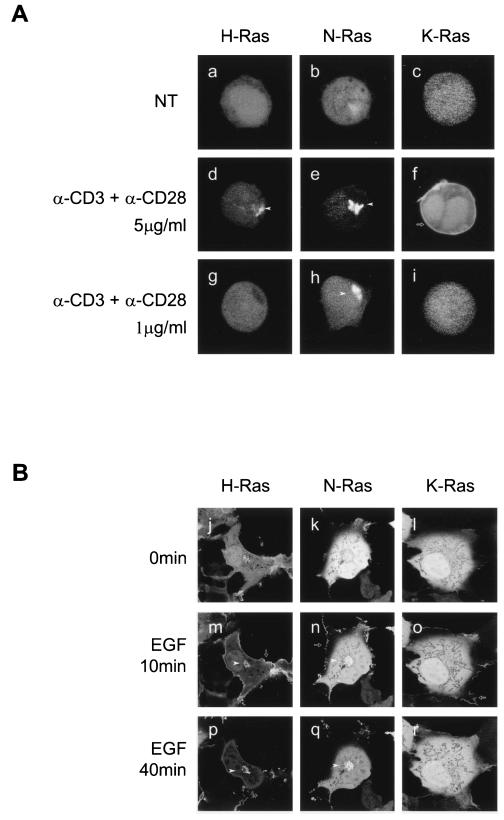
By using GFP-RBD, an in vivo probe for GTP-bound Ras, we have recently demonstrated that, although overexpressed H-Ras is present on both the plasma membrane and Golgi apparatus of Jurkat cells, high-level TCR stimulation activated only the pool on the Golgi (4). To extend this analysis to the other Ras isoforms and to study low-level TCR stimulation, we cotransfected Jurkat cells with H-Ras, N-Ras, or K-Ras together with the YFP-RBD. As in fibroblasts, Jurkat cells transfected only with YFP-RBD and then stimulated by cross-linking the TCR showed no recruitment of the probe to any membrane compartment (data not shown), demonstrating that endogenous levels of Ras are insufficient to generate a signal and confirming that the assay, as utilized, reports the activation state of only the ectopically expressed GTPase, permitting isoform-specific analysis. Cells expressing YFP-RBD and CFP-tagged forms of the three Ras isoforms were incubated with different amounts of anti-CD3 plus anti-CD28 to generate either low- or high-grade TCR stimulation, and the cells were imaged alive every 30 s for 20 min (Fig. 4A). Consistent with previous observations for COS-1 cells (8), in resting Jurkat cells, YFP-RBD was expressed homogenously in both the nucleoplasm and cytosol (Fig. 4A, a to c). High-grade stimulation of the TCR elicited recruitment of YFP-RBD by both N-Ras and H-Ras exclusively to a paranuclear structure consistent with the Golgi apparatus (Fig. 4A, d and e). This pattern of activation was observed in the vast majority of cotransfected cells. YFP-RBD recruitment to the plasma membrane was never observed in cells transfected with N-Ras or H-Ras. In contrast, low-grade stimulation of the TCR stimulated activation of N-Ras but not H-Ras (Fig. 4A, g and h). As with high-grade stimulation, the pool of N-Ras that was activated appeared exclusively on the Golgi apparatus (Fig. 4A, h). The kinetics of N-Ras activation on the Golgi apparatus in response to TCR stimulation was relatively rapid: 30, 50, and 90% of transfected Jurkat cells showed activation 2, 5, and 10 min after stimulation. Activation was transient: the number of positive cells returned to unstimulated levels after 20 min (data not shown). In contrast to the cells cotransfected with YFP-RBD and H-Ras or N-Ras, the majority of cells cotransfected with YFP-RBD and K-Ras showed little membrane recruitment of the reporter upon high-grade TCR stimulation (Fig. 4A, c, f, and i). However, in those cells that showed a redistribution of the probe (∼20%), recruitment was exclusively to the plasma membrane (Fig. 4A, f). Importantly, no membrane recruitment to any compartment was observed in cells coexpressing YFP-RBD and K-Ras following low-grade TCR stimulation (Fig. 4A, i). Thus, results with YFP-RBD membrane recruitment (Fig. 4A) were concordant with those obtained by GST-RBD pulldown (Fig. 1) and confirmed that, under conditions of low-intensity TCR stimulation, N-Ras was preferentially activated. Moreover, the latter assay demonstrated that the pool of activated N-Ras was restricted to the Golgi apparatus.
FIG. 4.
N-Ras is the only Ras isoform activated on the Golgi apparatus upon low-grade TCR engagement in Jurkat T cells. Jurkat (A) and COS-1 (B) cells were cotransfected with YFP-Raf1-RBD plus either H-Ras, N-Ras, or K-Ras tagged with CFP. Forty-eight hours later, cells were serum starved and either left untreated (a to c and j to l), stimulated with the indicated amounts of anti-CD3 plus anti-CD28 (d to i), or stimulated with EGF (m to r) at the times indicated. Cells were imaged alive by LSM, and those expressing equivalent amounts of H-Ras, K-Ras, or N-Ras were selected to analyze YFP-Raf1-RBD distribution. Arrowheads and arrows indicate Raf1-RBD redistribution to the Golgi apparatus and plasma membrane, respectively. NT, nontreated cells.
Because the lack of Ras activation at the plasma membrane of Jurkat cells was unexpected, we confirmed that our probe was capable of reporting the activation of all Ras isoforms on the plasma membranes of fibroblasts stimulated with growth factors. As previously reported (8), in COS-1 cells overexpressing H-Ras, YFP-RBD was transiently recruited in response to EGF to the plasma membrane and subsequently to the Golgi apparatus (Fig. 4B, j, m, and p). An identical pattern was observed in COS-1 cells expressing N-Ras (Fig. 4B, k, n, and q). Interestingly, whereas Ras activation on the Golgi apparatus of Jurkat cells was observed within 5 min of TCR stimulation, activation on the Golgi apparatus of COS-1 cells in response to EGF was relatively delayed (peak at 40 min). COS-1 cells expressing K-Ras showed only transient activation on the plasma membrane in response to growth factor (Fig. 4B, l, o, and r). Thus, Ras activation in Jurkat T cells differs from that in COS-1 cells in several respects. First, N-Ras is preferentially activated in response to low-grade TCR stimulation. Second, N-Ras activation takes place exclusively on the Golgi apparatus. Finally, activation on the Golgi apparatus downstream of TCR stimulation is rapid relative to that on the Golgi apparatus of fibroblasts stimulated by growth factors.
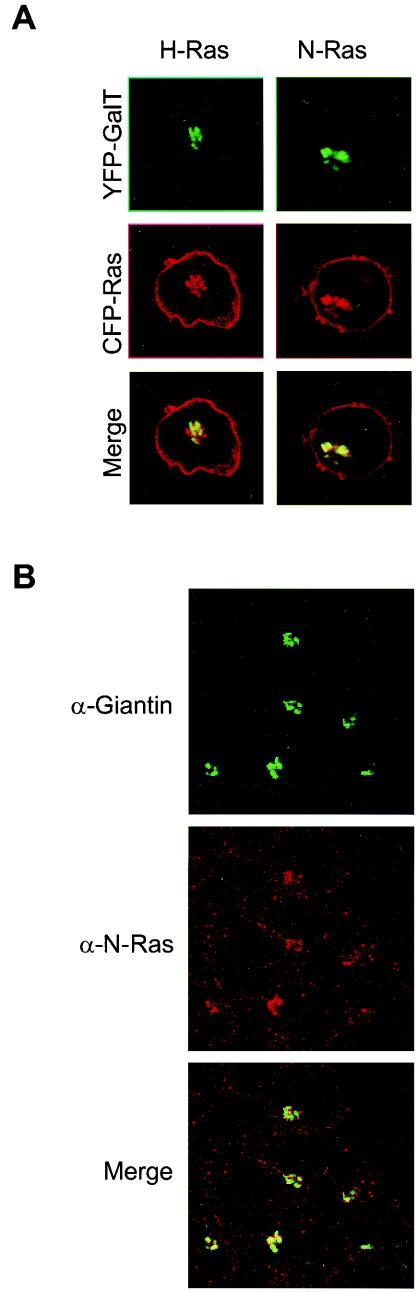
To unambiguously identify the Golgi apparatus as the paranuclear structure on which N-Ras and H-Ras are activated upon TCR engagement, we used Golgi-specific markers in both the live-cell imaging and indirect immunofluorescence of fixed cells (Fig. 5). We cotransfected Jurkat cells with CFP-tagged N-Ras or H-Ras plus the Golgi marker galactosyl transferase (GalT) tagged with YFP. The paranuclear pool of N-Ras and H-Ras colocalized with GalT (Fig. 5A), confirming the Golgi localization of both Ras isoforms. We also analyzed the localization of endogenous N-Ras by indirect immunofluorescence with an N-Ras isoform-specific antibody and anti-giantin as a Golgi marker. Interestingly, the staining for endogenous N-Ras was stronger on the Golgi apparatus than on the plasma membrane (Fig. 5B). These data demonstrate that the Golgi localization of CFP-N-Ras reflects the localization of the endogenous protein in Jurkat cells.
FIG.5.
N-Ras and H-Ras are expressed on the Golgi apparatus of Jurkat cells. (A) CFP-H-Ras and CFP-N-Ras vectors were cotransfected in Jurkat cells together with a vector expressing the Golgi marker GalT tagged with YFP. CFP-Ras and YFP-GalT distributions were determined by LSM in living cells. (B) The localization of endogenous N-Ras in the Golgi apparatus was determined by immunofluorescence. As described in Materials and Methods, endogenous N-Ras and giantin (a Golgi-specific marker) were detected in fixed Jurkat T cells.
Mono- versus dipalmitoylation of H-Ras and N-Ras affects TCR-mediated activation on the Golgi apparatus of Jurkat cells.
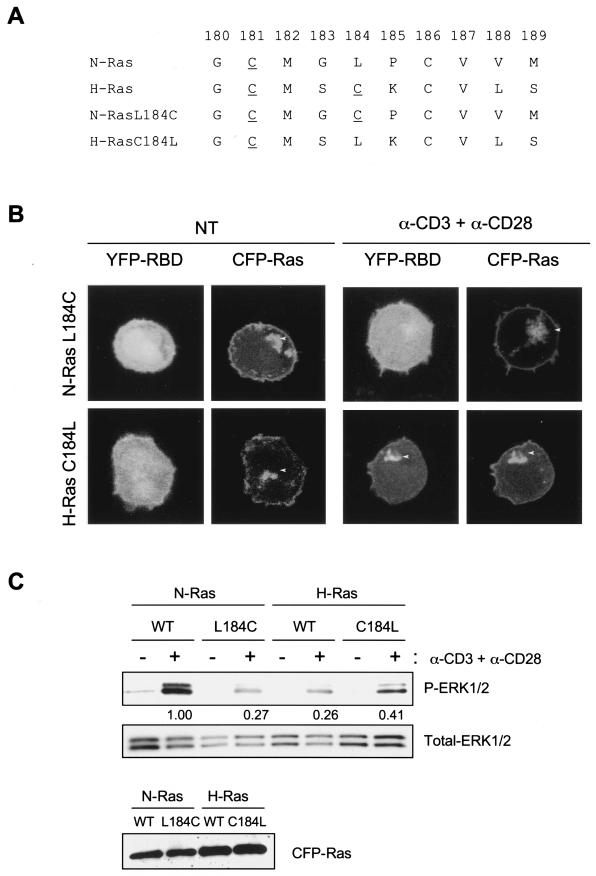
To investigate the molecular basis for the specificity of N-Ras over H-Ras signaling on the Golgi apparatus in response to low-grade TCR stimulation (Fig. 4), we analyzed the hypervariable regions of these isoforms. H-Ras differs from N-Ras in that it contains two (rather than one) palmitoylation sites upstream of its CAAX motif (Fig. 6A). We introduced a second palmitoylation site into N-Ras at codon 184 (N-RasL184C). Conversely, we removed the second palmitoylation site in H-Ras (H-RasC184L). Each construct, when tagged with CFP, localized like wild-type N-Ras and H-Ras on the plasma membrane and the Golgi apparatus (Fig. 6B). When these constructs were expressed along with YFP-RBD in Jurkat cells, low-grade cross-linking of the TCR induced recruitment of the probe to H-RasC184L, but not N-RasL184C, on the Golgi apparatus (Fig. 6B). This result suggests that monoacylation mediates the specificity of Ras activation on the Golgi apparatus.
FIG.6.
A single palmitoylation event in the C-terminal region of N-Ras is crucial for its specific role in TCR-dependent signaling. (A) Wild-type and mutant H-Ras and N-Ras protein sequences corresponding to the membrane targeting domain. Cysteine palmitoylation sites are underlined. (B) Jurkat cells were cotransfected with YFP-Raf1-RBD and either CFP-N-RasL184C or CFP-H-RasC184L mutants. Forty-eight hours later, cells were serum starved and either left untreated (NT) or activated with 1 μg of anti-CD3 plus anti-CD28/ml. Ras localization and RBD redistribution were analyzed in live cells by LSM and compared with those found for wild-type H-Ras and N-Ras isoforms (Fig. 3 and Fig. 4, g to i). Arrowheads indicate the Golgi apparatus. (C) Jurkat cells were transfected by using Amaxa technology with either CFP-N-Ras, CFP-N-RasL184C, CFP-H-Ras, or CFP-H-RasC184L vectors. Forty-eight hours later, cells were serum starved and either left untreated (−) or activated with 1 μg of anti-CD3 plus anti-CD28/ml (+). ERK1/2-activated proteins were detected with phospho-specific antibodies. Values between the upper panels indicate ERK1/2 activation relative to the CFP-N-Ras transfected cells and were determined by quantification of phospho-ERK1/2 bands (P-ERK1/2) and subsequent normalization with total ERK1/2 (middle panel). The bottom panel shows the levels of CFP-Ras proteins (∼52 kDa) detected for each construct. WT, wild type.
To extend these studies downstream of Ras activation, we studied TCR-stimulated Erk1/2 activation in Jurkat cells expressing N-Ras, H-Ras, or the palmitoylation mutants described above (Fig. 6C). Low-grade TCR stimulation induced Erk1/2 activation in N-Ras-overexpressing cells to a much higher degree than that observed in cells expressing similar levels of H-Ras. The addition of a second palmitoylation site in the hypervariable region of N-Ras (L184C) resulted in a markedly diminished ability to support Erk1/2 activation. Conversely, removal of one of the two acylation sites in the hypervariable region of H-Ras (C184L) resulted in a more robust activation of Erk1/2 in response to low-grade TCR stimulation. Thus, the differential activation of N-Ras versus H-Ras on the Golgi apparatus that is controlled by the acylation state of the Ras protein translates into downstream signaling.
TCR stimulation in Jurkat cells activates N-Ras via PLCγ and RasGRP1.
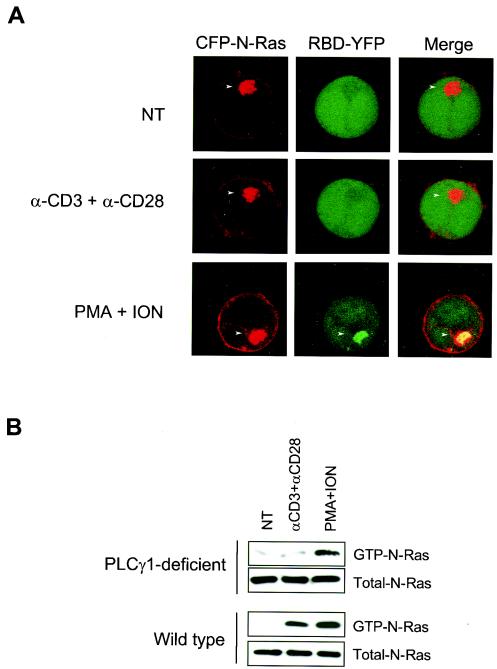
It has recently been demonstrated that H-Ras activation on the Golgi apparatus of fibroblasts is mediated by a pathway dependent on PLCγ and RasGRP1 (4, 7). Similar results were obtained in Jurkat cells overexpressing H-Ras (4). Because the in vitro studies described above and previous in vivo studies of lymphocytes from N-Ras-deficient mice (29) suggest that Ras signaling in lymphocytes may preferentially involve N-Ras, we sought to extend the studies of PLCγ and RasGRP1 to N-Ras. Whereas N-Ras was expressed on both the plasma membrane and the Golgi apparatus in Jurkat cells deficient in PLCγ, stimulation of the TCR failed to induce the activation of N-Ras on any compartment (Fig. 7A). Similar results were obtained when wild-type Jurkat cells were pretreated with the PLCγ inhibitor U73122 (4). In contrast, when the TCR was bypassed by stimulation with PMA and ionomycin, YFP-RBD was recruited to the Golgi apparatus in PLCγ-deficient cells (Fig. 7A). The spatially resolved results obtained with YFP-RBD recruitment were recapitulated by GST-RBD pulldown assays. Low-grade stimulation of the TCR activated N-Ras in wild-type, but not PLCγ-deficient, Jurkat cells (Fig. 7B). In contrast, bypassing the TCR by stimulation with PMA plus ionomycin activated N-Ras in both types of cells (Fig. 7B). Thus, the activation of N-Ras on the Golgi apparatus of Jurkat cells following TCR stimulation depends on PLCγ.
FIG. 7.
N-Ras activation upon TCR engagement in Jurkat cells is PLCγ1-dependent. (A) J gamma 1 cells, which are deficient in PLCγ1, were cotransfected with CFP-N-Ras and YFP-Raf1-RBD and, 48 h later, stimulated as indicated. Colocalization of CFP-N-Ras (red) and YFP-Raf1-RBD (green) is shown in yellow. (B) Wild-type and PLCγ1-deficient Jurkat cells (2 × 107 per point) were serum starved for 2 h and left untreated (NT) or incubated with 1 μg of anti-CD3 plus anti-CD28/ml (αCD3+αCD28) or PMA plus ionomycin (PMA+ION). Proteins from stimulated and unstimulated cells were used to collect and detect GTP-bound and total N-Ras as described in Materials and Methods.
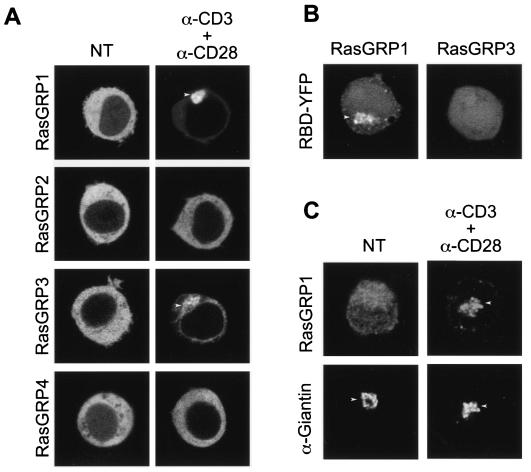
RasGRP1 has been shown to be critical for T-cell activation in vivo (11), and we have shown that it is required to activate overexpressed H-Ras on the Golgi apparatus of both fibroblasts and Jurkat cells (4). To determine if RasGRP1, among the four RasGRP family members, has a particular preference for targeting to the Golgi apparatus in Jurkat cells, we tagged each member of the family with GFP and studied their subcellular localization before and after low-grade TCR stimulation (Fig. 8A). In serum-starved, unstimulated cells, each RasGRP family member was distributed homogeneously in the cytosol. Upon TCR stimulation, RasGRP1 and RasGRP3 translocated to endomembranes but not to the plasma membrane. No relocalization of RasGRP2 or RasGRP4 was observed. Whereas RasGRP3 relocalized in stimulated cells to both the endoplasmic reticulum and Golgi apparatus, RasGRP1 translocation was specific for the Golgi apparatus. To validate the translocation of GFP-RasGRP1, we studied the translocation of the endogenous protein in Jurkat cells by indirect immunofluorescence. Low-grade TCR stimulation induced translocation of endogenous RasGRP1 to the Golgi apparatus, as indicated by colocalization with giantin (Fig. 8C). Because both RasGRP1 and RasGRP3 were observed to translocate to the Golgi apparatus, we sought to determine whether either or both could activate N-Ras on that compartment. We coexpressed N-Ras with each RasGRP, along with YFP-RBD, and observed recruitment of the reporter to the Golgi apparatus as a consequence of the overexpression of RasGRP1 but not RasGRP3 (Fig. 8B). These data are consistent both with the RasGRP1 knockout studies (11) and with RasGRP1 activation of H-Ras on the Golgi apparatus (4, 7). Taken together, our data suggest that low-grade TCR stimulation of Jurkat cells promotes the activation of N-Ras on the Golgi apparatus via a PLCγ/Ca2+ plus diacylglycerol/RasGRP1-dependent pathway.
FIG. 8.
RasGRP1 mediates TCR-dependent activation of N-Ras on the Golgi apparatus of Jurkat cells. (A) Wild-type Jurkat cells were transfected with YFP-tagged RasGRP proteins. Forty-eight hours posttransfection, cells were serum-starved for 2 h and stimulated as described for Fig. 6B. Panels show the localization of RasGRP1 to RasGRP4 proteins before (left) and after (right) TCR stimulation. (B) To assess the role of RasGRP1 and RasGRP3 in N-Ras activation on the Golgi apparatus, wild-type Jurkat cells were cotransfected with untagged expression vectors of each RasGRP protein, CFP-N-Ras, and YFP-RBD. Forty-eight hours later, cells were serum starved, and RBD redistribution was analyzed by LSM. Arrowheads indicate the Golgi apparatus. (C) Recruitment of endogenous RasGRP1 to the Golgi apparatus upon TCR engagement was analyzed by immunofluorescence. As described in Materials and Methods, endogenous RasGRP1 and giantin (a Golgi-specific marker) were detected in nontreated (NT) and TCR-stimulated (5 μg of anti-CD3/ml plus 5 μg of anti-CD28/ml) Jurkat T cells.
DISCUSSION
Ras activation is critical for T-cell function (17). Recently, we analyzed the role of N-Ras in T-cell signaling by using mice deficient in N-Ras (29). Compared to wild-type littermates, mice lacking N-Ras were more sensitive to low-titer influenza virus infection. Moreover, N-Ras-deficient mice exhibited a defect in the selection of CD8-positive T cells, a process that is regulated by low-grade TCR signaling (45). In the present work, we have complemented these in vivo genetic experiments with a cell biological analysis of Ras isoform expression and signaling in T cells. Our study demonstrates that, under conditions of low-grade TCR stimulation, N-Ras is preferentially activated in Jurkat T cells. This observation explains why other Ras isoforms cannot substitute for N-Ras in deficient animals and demonstrates a critical role for N-Ras in T-cell function.
Until recently, the three Ras isoforms have been considered redundant, and few biochemical differences have been described. The embryonic lethality of K-Ras deficiency (24), but not of N-Ras (44) or H-Ras (14) deficiencies, demonstrates conclusively that the functions of all Ras proteins are not entirely overlapping. Differential membrane trafficking of the various Ras isoforms is firmly established (9) and has led to a search for cell biological rather than biochemical differences among the isoforms. Recent studies have focused on isoform differences in localization in plasma membrane microdomains (32, 34) and in endomembrane signaling (8) as possible explanations.
TCR are believed to signal from plasma membrane microdomains known as lipid rafts that are enriched in signaling molecules, including the adaptor protein LAT and enzymes such as Lck and PLCγ (23). The enrichment of Ras in lipid rafts is somewhat controversial (31). In two recent studies of nonlymphoid cells, H-Ras was enriched in lipid rafts but K-Ras was excluded (32, 34). Our failure to observe K-Ras in clustered rafts on Jurkat cells is consistent with these studies. Interestingly, whereas inactive H-Ras was found to be enriched in lipid rafts, activated, GTP-bound H-Ras was excluded, suggesting a dynamic interaction with the microdomain (32). Our observation that H-Ras is not activated by TCR stimulation but that it is nevertheless colocalized with the receptor in lipid rafts is consistent with the lipid raft association of inactive H-Ras. In support of this interpretation, when GFP-H-Ras61L, a constitutively active mutant, was substituted in our system for GFP-H-Ras, the GTP-bound H-Ras protein failed to cocap with the TCR (I. Pérez de Castro, T. G. Bivona, A. Pellicer, and M. R. Philips, unpublished observation). Although in MDCK cells N-Ras has been colocalized with H-Ras in lipid rafts (28), its localization had not been previously analyzed in T cells. Our results demonstrate that N-Ras behaves like K-Ras in failing to partition into the T-cell membrane microdomains defined by TCR and CD8. From these data, we conclude that diacylation is required for Ras proteins to partition into lipid rafts of Jurkat T cells. Since the diacylated form of Ras, H-Ras, is expressed at very low levels in T cells and, even when overexpressed, is not activated downstream of the TCR, we further conclude that the lipid rafts of T-cell plasma membranes do not participate directly in Ras activation and cannot explain the preference for N-Ras over K-Ras in activation following low-grade TCR stimulation.
Having failed to explain the isoform preference of Ras signaling in T cells on plasma membrane microdomains, we next investigated subcellular compartment-specific signaling. It has recently been demonstrated that, although H-Ras expressed ectopically in Jurkat cells was present on both the plasma membrane and Golgi apparatus, the signaling in response to high-grade TCR activation was restricted to the Golgi apparatus and was dependent on PLCγ and RasGRP1 (4). Moreover, we showed that the Ca2+-activated Ras GAP CAPRI blocked H-Ras activation on the plasma membrane (4). We have now shown that, like H-Ras, N-Ras was activated only on the Golgi apparatus following TCR stimulation. Unlike H-Ras, low-grade TCR stimulation was sufficient to activate N-Ras on the Golgi apparatus. N-Ras activation on the Golgi apparatus was also dependent on both PLCγ and RasGRP1. These results suggest that plasma membrane-associated Ras exchange factors such as Grb2/Sos are counterbalanced in T cells by CAPRI. These data are consistent with the finding that the mutation of the phospho-tyrosine docking sites for Grb2/Sos on LAT does not inhibit TCR-mediated Ras activation (46). However, since we observed the activation of K-Ras during high-grade stimulation of TCR, our results are consistent with a role for Grb2/Sos in the activation of K-Ras on the plasma membrane. Thus, the intensity of TCR stimulation controls not only the Ras isoform utilization but also the subcellular compartment from which the Ras signal is propagated.
Our analysis of palmitoylation mutants of N-Ras and H-Ras demonstrates that monoacylation regulates the ability of Golgi-associated Ras to become activated in response to low-grade TCR stimulation. One model that may explain this result is that in which monoacylation is required for the relevant Ras protein to partition into the proper microdomain of the trans-Golgi network membrane to be acted upon by RasGRP1. Alternatively, mono- versus diacylation may specify interactions between Ras and various guanine nucleotide exchange factors or GAPs. Indeed, the posttranslational modification of Ras influences not only subcellular localization but also interaction with regulators (35, 41).
Because working with primary lymphocytes presents obstacles, several T-cell lines have been extensively used for studying T-cell signaling and function. Although the results obtained with these cell lines require validation in primary T cells, increasing evidence supports the utility of Jurkat and other T-cell lines in elucidating signaling pathways. For example, the requirement for RasGRP1 in the activation of N-Ras on the Golgi apparatus of Jurkat cells is consistent with the severe impairment in Ras signaling observed in murine T cells deficient in this exchange factor (11). Indeed, RasGRP1 has been strongly associated with Ras activation in T cells, thymocyte development, and TCR signaling (11, 13). Importantly, it was recently reported that RasGRP1 plays a critical role in T-cell development, homeostasis, and differentiation by transducing low-grade TCR signals (30). Moreover, N-Ras-deficient mice are also defective in some T-cell functions mediated by low-grade stimuli (29). Thus, the striking similarities between the T-cell phenotypes of RasGRP1- and N-Ras-deficient mice can be explained by the elimination of elements of a common pathway. These data strongly support the idea that the TCR/PLCγ/RasGRP1/N-Ras pathway plays a pivotal role in low-grade TCR signaling.
Acknowledgments
We are grateful to David J. McKean, James C. Stone, Konstantina Alexandropoulos, and Johannes Bos for providing plasmids.
This work was supported by grants AI36224 and GM55279 (to M.R.P.), grants CA36327 and CA50434 (to A.P.) from the National Institutes of Health, the New York State Breast Cancer Research Program, and the Burroughs Welcome Fund (to M.R.P.), and by a General Clinical Research Center grant from NIH NCRR (M01RR00096) awarded to the New York University School of Medicine.
REFERENCES
- 1.Alberola-Ila, J., K. A. Forbush, R. Seger, E. G. Krebs, and R. M. Perlmutter. 1995. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature 373:620-623. [DOI] [PubMed] [Google Scholar]
- 2.Arcaro, A., C. Gregoire, N. Boucheron, S. Stotz, E. Palmer, B. Malissen, and I. F. Luescher. 2000. Essential role of CD8 palmitoylation in CD8 coreceptor function. J. Immunol. 165:2068-2076. [DOI] [PubMed] [Google Scholar]
- 3.Barbacid, M. 1987. ras genes. Annu. Rev. Biochem. 56:779-827. [DOI] [PubMed] [Google Scholar]
- 4.Bivona, T. G., I. Perez De Castro, I. M. Ahearn, T. M. Grana, V. K. Chiu, P. J. Lockyer, P. J. Cullen, A. Pellicer, A. D. Cox, and M. R. Philips. 2003. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424:694-698. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 6.Brandt-Rauf, P. W., R. P. Carty, J. Chen, M. Avitable, J. Lubowsky, and M. R. Pincus. 1988. Structure of the carboxyl terminus of the RAS gene-encoded P21 proteins. Proc. Natl. Acad. Sci. USA 85:5869-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caloca, M. J., J. L. Zugaza, and X. R. Bustelo. 2003. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 278:33465-33473. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson II, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed]
- 9.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 10.de Rooij, J., and J. L. Bos. 1997. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene 14:623-625. [DOI] [PubMed] [Google Scholar]
- 11.Dower, N. A., S. L. Stang, D. A. Bottorff, J. O. Ebinu, P. Dickie, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317-321. [DOI] [PubMed] [Google Scholar]
- 12.Downward, J., J. D. Graves, P. H. Warne, S. Rayter, and D. A. Cantrell. 1990. Stimulation of p21ras upon T-cell activation. Nature 346:719-723. [DOI] [PubMed] [Google Scholar]
- 13.Ebinu, J. O., S. L. Stang, C. Teixeira, D. A. Bottorff, J. Hooton, P. M. Blumberg, M. Barry, R. C. Bleakley, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP links T-cell receptor signaling to Ras. Blood 95:3199-3203. [PubMed] [Google Scholar]
- 14.Esteban, L. M., C. Vicario-Abejon, P. Fernandez-Salguero, A. Fernandez-Medarde, N. Swaminathan, K. Yienger, E. Lopez, M. Malumbres, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furth, M. E., T. H. Aldrich, and C. Cordon-Cardo. 1987. Expression of ras proto-oncogene proteins in normal human tissues. Oncogene 1:47-58. [PubMed] [Google Scholar]
- 16.Gartner, F., F. W. Alt, R. Monroe, M. Chu, B. P. Sleckman, L. Davidson, and W. Swat. 1999. Immature thymocytes employ distinct signaling pathways for allelic exclusion versus differentiation and expansion. Immunity 10:537-546. [DOI] [PubMed] [Google Scholar]
- 17.Genot, E., and D. A. Cantrell. 2000. Ras regulation and function in lymphocytes. Curr. Opin. Immunol. 12:289-294. [DOI] [PubMed] [Google Scholar]
- 18.Grand, R. J., and D. Owen. 1991. The biochemistry of ras p21. Biochem. J. 279:609-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 20.Iritani, B. M., J. Alberola-Ila, K. A. Forbush, and R. M. Perimutter. 1999. Distinct signals mediate maturation and allelic exclusion in lymphocyte progenitors. Immunity 10:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ise, K., K. Nakamura, K. Nakao, S. Shimizu, H. Harada, T. Ichise, J. Miyoshi, Y. Gondo, T. Ishikawa, A. Aiba, and M. Katsuki. 2000. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene 19:2951-2956. [DOI] [PubMed] [Google Scholar]
- 22.Jameson, S. C., K. A. Hogquist, and M. J. Bevan. 1995. Positive selection of thymocytes. Annu. Rev. Immunol. 13:93-126. [DOI] [PubMed] [Google Scholar]
- 23.Janes, P. W., S. C. Ley, A. I. Magee, and P. S. Kabouridis. 2000. The role of lipid rafts in T cell antigen receptor (TCR) signalling. Semin. Immunol. 12:23-34. [DOI] [PubMed] [Google Scholar]
- 24.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene 15:1151-1159. [DOI] [PubMed] [Google Scholar]
- 25.Leon, J., I. Guerrero, and A. Pellicer. 1987. Differential expression of the ras gene family in mice. Mol. Cell. Biol. 7:1535-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malumbres, M., and A. Pellicer. 1998. RAS pathways to cell cycle control and cell transformation. Front. Biosci. 3:d887-d912. [DOI] [PubMed] [Google Scholar]
- 27.Mangues, R., W. F. Symmans, S. Lu, S. Schwartz, and A. Pellicer. 1996. Activated N-ras oncogene and N-ras proto-oncogene act through the same pathway for in vivo tumorigenesis. Oncogene 13:1053-1063. [PubMed] [Google Scholar]
- 28.Matallanas, D., I. Arozarena, M. T. Berciano, D. S. Aaronson, A. Pellicer, M. Lafarga, and P. Crespo. 2003. Differences on the inhibitory specificities of H-Ras, K-Ras, and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J. Biol. Chem. 278:4572-4581. [DOI] [PubMed] [Google Scholar]
- 29.Perez de Castro, I., R. Diaz, M. Malumbres, M. I. Hernandez, J. Jagirdar, M. Jimenez, D. Ahn, and A. Pellicer. 2003. Mice deficient for N-ras: impaired antiviral immune response and T-cell function. Cancer Res. 63:1615-1622. [PubMed] [Google Scholar]
- 30.Priatel, J. J., S. J. Teh, N. A. Dower, J. C. Stone, and H. S. Teh. 2002. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity 17:617-627. [DOI] [PubMed] [Google Scholar]
- 31.Prior, I. A., and J. F. Hancock. 2001. Compartmentalization of Ras proteins. J. Cell Sci. 114:1603-1608. [DOI] [PubMed] [Google Scholar]
- 32.Prior, I. A., A. Harding, J. Yan, J. Sluimer, R. G. Parton, and J. F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 33.Rodenhius, S. 1992. ras and human tumors. Semin. Cancer Biol. 3:241-247. [PubMed] [Google Scholar]
- 34.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 35.Rubio, I., U. Wittig, C. Meyer, R. Heinze, D. Kadereit, H. Waldmann, J. Downward, and R. Wetzker. 1999. Farnesylation of Ras is important for the interaction with phosphoinositide 3-kinase gamma. Eur. J. Biochem. 266:70-82. [DOI] [PubMed] [Google Scholar]
- 36.Sebzda, E., S. Mariathasan, T. Ohteki, R. Jones, M. F. Bachmann, and P. S. Ohashi. 1999. Selection of the T cell repertoire. Annu. Rev. Immunol. 17:829-874. [DOI] [PubMed] [Google Scholar]
- 37.Shen, W. P., T. H. Aldrich, G. Venta-Perez, B. R. Franza, Jr., and M. E. Furth. 1987. Expression of normal and mutant ras proteins in human acute leukemia. Oncogene 1:157-165. [PubMed] [Google Scholar]
- 38.Sinn, E., W. Muller, P. Pattengale, I. Tepler, R. Wallace, and P. Leder. 1987. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell 49:465-475. [DOI] [PubMed] [Google Scholar]
- 39.Swan, K. A., J. Alberola-Ila, J. A. Gross, M. W. Appleby, K. A. Forbush, J. F. Thomas, and R. M. Perlmutter. 1995. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 14:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor, S. J., R. J. Resnick, and D. Shalloway. 2001. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Methods Enzymol. 333:333-342. [DOI] [PubMed] [Google Scholar]
- 41.Thissen, J. A., J. M. Gross, K. Subramanian, T. Meyer, and P. J. Casey. 1997. Prenylation-dependent association of Ki-Ras with microtubules. Evidence for a role in subcellular trafficking. J. Biol. Chem. 272:30362-30370. [DOI] [PubMed] [Google Scholar]
- 42.Tremblay, P. J., F. Pothier, T. Hoang, G. Tremblay, S. Brownstein, A. Liszauer, and P. Jolicoeur. 1989. Transgenic mice carrying the mouse mammary tumor virus ras fusion gene: distinct effects in various tissues. Mol. Cell. Biol. 9:854-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, H., and D. A. Cantrell. 1997. Distinct Ras effector pathways are involved in Fc epsilon R1 regulation of the transcriptional activity of Elk-1 and NFAT in mast cells. J. Exp. Med. 185:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umanoff, H., W. Edelmann, A. Pellicer, and R. Kucherlapati. 1995. The murine N-ras gene is not essential for growth and development. Proc. Natl. Acad. Sci. USA 92:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe, N., H. Arase, M. Onodera, P. S. Ohashi, and T. Saito. 2000. The quantity of TCR signal determines positive selection and lineage commitment of T cells. J. Immunol. 165:6252-6261. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W., R. P. Trible, M. Zhu, S. K. Liu, C. J. McGlade, and L. E. Samelson. 2000. Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell antigen receptor-mediated signaling. J. Biol. Chem. 275:23355-23361. [DOI] [PubMed] [Google Scholar]