Summary
Light control over enzyme function represents a novel and exciting field of biocatalysis research. Blue‐light photoreceptors of the Light, Oxygen, Voltage (LOV) family have recently been investigated for their applicability as photoactive switches. We discuss here the primary photochemical events leading to light activation of LOV domains as well as the proposed signal propagation mechanism to the respective effector domain. Furthermore, we describe the construction of LOV fusions to different effector domains, namely a dihydrofolate reductase from Escherichia coli and a lipase from Bacillus subtilis. Both fusion partners retained functionality, and alteration of enzyme activity by light was also demonstrated. Hence, it appears that fusion of LOV photoreceptors to functional enzyme target sites via appropriate linker structures may represent a straightforward strategy to design light controllable biocatalysts.
Introduction
The remote temporal and spatial control of protein functions within living cells is a major goal long wished for in life sciences. It is only in the last couple of years that significant progress has been made in this exciting field of research by developing different methods to allow for the light‐dependent control of protein and cellular functions. Light as a trigger for cellular functions is of outstanding interest as it provides a non‐invasive method for (photo)stimulation which thus limits cellular damage to a minimum. Furthermore, light‐induced manipulations allow for high temporal and spatial resolution.
At an early stage, photolabile caged compounds, e.g. caged ions, and neurotransmitters such as Ca2+ and glutamate were used to non‐invasively stimulate neuronal activity (Zemelman et al., 2003; Shoham et al., 2005). Generally, such systems rely on the irreversible photolysis of a photolabile caged complex that releases a signalling molecule after photostimulation. The photochromic azobenzene system (Sadovski et al., 2009) provides an attractive alternative to caged compounds because it can reversibly be switched between two states by using two different wavelengths of excitation allowing for a reversible and repeatable on/off switching of protein function achieved by coupling of a receptor ligand to an azobenzene derivative. Illumination of the azobenzene‐coupled ligand results in a cis/trans isomerization of the azobenze moiety thus placing the ligand in a position that allows binding to the respective receptor and hence its activation. To this end, different cellular receptors and channels such as the nicotinic acetylcholine receptor and several ionotrophic glutamate receptors could be light‐dependently controlled (Lester et al., 1980; Volgraf et al., 2007). More direct control over protein functions can be exerted by attaching azobenzene derivatives to a protein backbone by cross‐linking to a position that influences the respective protein function. In this way, photo‐control over skeletal muscle myosin (Umeki et al., 2004) and a kinesin ATPase (Yamada et al., 2007) could be obtained in vitro. Hence, reversible and irreversible photocaged ligands present powerful tools to achieve a remote control over various cellular functions (Goeldner and Givens 2005; Banghart et al., 2006; Gorostiza and Isacoff, 2008). Nevertheless, their use is still limited by several drawbacks: (i) freely diffusible photocaged ligands and cross‐linked azobenzenes can never be completely selective in vivo for just a single protein target, and (ii) the photo‐released ligand will diffuse freely through the cell thus limiting spatial resolution.
Photochromic proteins for control of protein functions
Genetically encoded photochromic proteins represent a promising alternative to photocaged or coupled ligands. Retinal is a well‐known naturally occurring photochromic switch which functions as light‐sensitive chromophore of visual opsins (rhodopsins) found in the mammalian eye (Yoshizawa, 1984). In bacteria, retinal photoswitches occur as light‐driven proton pumps (bacteriorhodopsins) (Lozier et al., 1975). Here, the retinal molecule (much like the above‐mentioned azobenzene derivatives) isomerizes between the cis and the trans isomer in a light‐dependent manner. Recently, light‐gated retinal proteins such as the algal channelrhodopsin‐2 (Nagel et al., 2003), engineered variants of the same (Lin et al., 2009), as well as light‐driven chloride pumps of the halorhodopsin family (Schobert and Lanyi, 1982) have found their way into the toolbox of neurobiologists (Zhang et al., 2007a). In particular, channelrhodopsins are nowadays widely employed in neurosciences to selectively and non‐invasively control the firing of specific neurones (Boyden et al., 2005; Nagel et al., 2005; Zhang et al., 2007b; Pulver et al., 2009). Other naturally occurring light‐sensitive proteins can also be employed including the photoactivated adenylyl cyclases (PACs) of the unicellular flagellate Euglena gracilis (Iseki et al., 2002) which represent potent alternative systems for the photocontrol of neuronal activity (Nagahama et al., 2007; Schröder‐Lang et al., 2007; Bucher and Buchner, 2009). Photoactivated adenylyl cyclases are blue light‐sensitive photoreceptors that bind a flavin adeninedinucleotide (FAD) molecule as chromophore in two so‐called BLUF (sensors of Blue‐Light Using FAD) domains (Gomelsky and Klug, 2002). These BLUF domains are assumed to undergo a light‐dependent conformational rearrangement that triggers the catalytic conversion of adenosine triphosphate to the secondary messenger molecule cyclic adenosine‐monophosphate (cAMP) (Ntefidou et al., 2006) which in turn regulates various cellular functions (Yoshikawa et al., 2005).
These naturally occurring light‐gated systems attenuate the problem of spatial control over the cellular response, as they can be selectively expressed, e.g. in specific types of neurones. However, the problem remains that the function of a specific cellular protein cannot be probed selectively in vivo. Both channelrhodopsin‐mediated synaptic membrane depolarization and PAC‐triggered control of the intracellular cAMP levels will affect cellular control mechanisms at different levels and initiate different physiological responses at the same time. Thus, a generally applicable strategy to engineer optical control into any type of target protein in any type of organism including bacteria, plants and animals still represents a tempting goal.
Light, Oxygen, Voltage (LOV) photoreceptors for control of protein functions
Light, Oxygen, Voltage (LOV) blue‐light photoreceptor proteins (Christie et al., 1999) exert a light‐triggered control mechanism present in the three kingdoms of life. LOV domains belong to the Per, Arndt, Sim (PAS) family of sensor domains (Taylor and Zhulin, 1999) which is one of the most widely spread sensor module in nature found in all three kingdoms of life. The family includes apart from blue‐light photoreceptors, other ligand‐binding sensors suitable to detect changes in oxygen levels and redox potential (Taylor and Zhulin, 1999). LOV photoreceptors, like PAS sensor domains themselves, are found in the Archaea, the Bacteria, in lower and higher plants as well as in fungi (U. Krauss, B.Q. Minh, A. Losi, W. Gärtner, T. Eggert, A. von Haeseler and K.‐E. Jaeger, in preparation). Recently, even the possibility of LOV signalling modules to occur in animals including humans was highlighted (van der Horst et al., 2007).
In nature, LOV photoreceptors are fused mostly N‐terminally to a wide array of different effector domains (Crosson et al., 2003; Losi, 2006). In plants, fusion partners include Ser/Thr kinases where a light‐dependent autophosphorylation is initiated in response to blue‐light illumination (Huala et al., 1997). In the bacterial kingdom the LOV sensor module is found in two‐ to multi‐component systems together with other PAS domains, His‐kinases, anti‐sigma factors, helix–turn–helix DNA‐binding domains, phosphatases, phosphodiesterases and guanylate cyclases (Losi, 2004; Losi and Gärtner, 2008). This broad spectrum of coupled effector domains which exert an array of different in vivo functionalities already suggests that sensor signal transduction mechanisms may be successfully transferred to many structurally and functionally different protein effector domains.
The functional engineering of a light‐activated protein requires (i) an understanding of the primary photochemical events that lead to effector domain activation, (ii) knowledge of the signal propagation mechanism from the site of photon capture through the sensor core to the effector domain, and (iii) an effector domain whose activity can be controlled by a structural perturbation transmitted from the coupled sensor domain. For LOV domains, issues (i) and (ii) are well resolved. Thus, the question arises whether LOV can be employed as a universal light switch to control protein functions. The fundamental principle underlying this approach is illustrated in Fig. 1.
Figure 1.
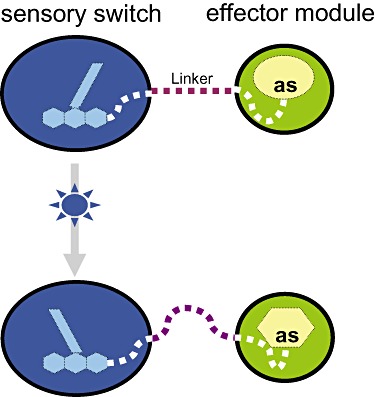
Principle of engineering a genetically encoded photoswitchable enzyme. The chimeric protein must consist of a photoreceptor (or sensor) domain (in blue) harbouring a chromophore which functions as the light‐sensitive ‘switch’. Upon illumination with light of appropriate wavelength the chromophore absorbs light energy and undergoes electronic excitation which is accompanied by a conformational rearrangement in the sensor domain protein backbone around the chromophore. This change is further transmitted via relay effects (e.g. via a linker polypeptide) into an ‘effector’ domain (in green) possessing enzymatic activity. Hence, the light signal propagates to the effectors' active site (as) causing a structural pertubation which may in turn result in increased or decreased activity, altered substrate specificity or enantioselectivity.
LOV photocycle and signal transduction mechanisms
Photon capture within the light‐sensitive FMN chromophore bound non‐covalently in the LOV domain dark state (LOV447) results in the formation of the singlet exited state of FMN on a picosecond timescale. The singlet excited state in part relaxes back to the ground state by emitting photons in form of fluorescence and decays via intersystem crossing to the FMN triplet state (LOV660). The triplet in turn gives rise to the so‐called signalling state (LOV390) on a microsecond timescale. In the signalling state, a covalent bond is formed between a highly conserved cysteine residue in the protein core and the carbon (C4) – atom of the flavin isoalloxazine ring (Fig. 2). From this longest living species in the LOV photocycle the protein returns to the dark state (LOV447) within seconds, hours or days depending on the type of LOV protein (Losi, 2007). Presumably, the signalling state must trigger a conformational change in the LOV domain which then relays the light signal to fused effector domains. However, the molecular mechanism of this signal propagation is still somewhat controversial as crystal structures solved for the dark and light state of the protein did not display major differences (Crosson and Moffat, 2001; 2002; Fedorov et al., 2003; Halavaty and Moffat, 2007). Recently, solution NMR spectroscopy was used to analyse a plant LOV2 sensor domain responsible for phototropin activation. Hereby, Gardner's group suggested that the unfolding or dissociation of a helical‐segment located C‐terminally to the LOV core and termed Jα‐helix triggers phototropin autophosphorylation (Harper et al., 2003; 2004; Yao et al., 2008). Different possible mechanisms of activation were suggested with the LOV domain acting either as a dark‐state inhibitor or as a light‐state activator (Harper et al., 2004). For bacterial LOV proteins, no light‐dependent unfolding of helices could be observed so far (Buttani et al., 2007; Möglich and Moffat, 2007), although the respective bacterial LOV proteins apparently possess C‐terminally fused helical polypeptides (Krauss et al., 2005; Buttani et al., 2007; Möglich and Moffat, 2007). Recently, structural and in vitro functional data suggested that the signal propagation in some bacterial LOV proteins might occur via a rotational movement of the C‐terminal Jα‐helices (Möglich and Moffat, 2007; Möglich et al., 2009). Thus, the mechanisms of effector domain activation by LOVs currently appear to be different in both plant and bacterial systems. However, in both bacterial and plant LOV systems, photon capture in the LOV domain results in a structural perturbation that transmits conformational changes through the LOV sensor core, either via unfolding or via rotation of the Jα‐helical linker located at the C‐terminus of the LOV domain. This structural perturbation is then relayed into the effector domain resulting in changes or alterations of cellular activities like phosphorylation, protein–protein interaction or DNA binding.
Figure 2.
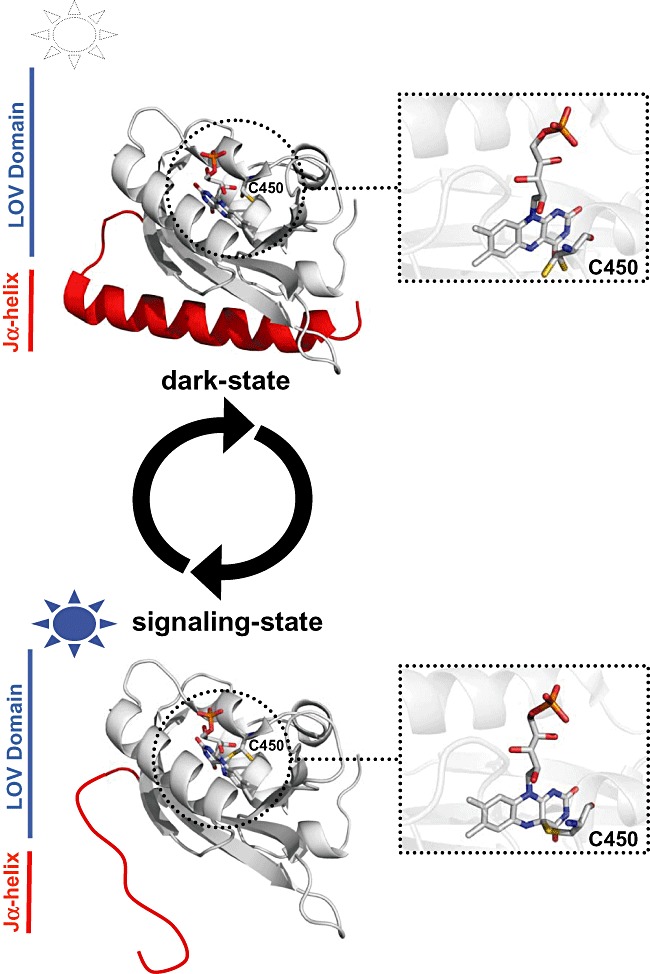
LOV photocycle and signal propagation. The figure shows the LOV core structure of the Avena sativa phototropin1 LOV2 domain (AsLOV2, pdb entry: 2v1a) in the dark state (upper part) with the light‐sensitive flavin chromophore in stick representation. The inset shows a close‐up view of the FMN binding pocket in dark state, with a non‐covalently bound FMN molecule in close vicinity of the photoactive cystein (C450). The lower part of the figure visualizes the conformational change suggested to occur in the signalling state after blue‐light illumination. A covalent bond is formed between the photoactive cysteine (C450) (see inset, lower part of the figure) and the flavin isoalloxazine ring. This in turn triggers unfolding (or dissociation) of the C‐terminally located Jα‐helix which results in the activation of plant phototropin (Harper et al., 2003; 2004). Please note that the mechanistic model is based on NMR data of AsLOV2 whereas no unfolding of the Jα‐helix is observed in the light‐state crystal structure of AsLOV2. All structural changes are completely reversible in the dark thus concluding the LOV photocycle.
Chimeric light‐controlled proteins
Natural allosteric systems
Do we understand this signal transduction mechanism well enough to engineer light‐switching abilities into any target effector protein using LOV sensor domains as the light trigger? The first such system described used a plant LOV domain fused to the well‐characterized Escherichia coli tryptophane repressor protein (TrpR) (Strickland et al., 2008). The corresponding chimeric construct was designated LovTAP. An allosterically regulated ‘lever‐arm’ formed from shared helices on both the LOV domain (Jα‐helix) and the TrpR side allowed for light‐dependent control over DNA binding to the TrpR helix–turn–helix DNA binding site (Strickland et al., 2008). More recently, a similar design strategy was used to construct a light‐controlled histidine kinase by joining the His‐kinase domain of the PAS sensor FixL of Bradyrhizobium japonicum with the LOV domain of the Bacillus subtilis YtvA photoreceptor protein (Möglich et al., 2009). Again, shared helical linkers were employed as signal propagating elements between the sensor core and the respective effector domain.
Both studies demonstrate proof‐of‐principle; however, they closely mimic naturally existing sensor‐effector systems. FixL contains a haem‐binding PAS sensor domain that structurally resembles the FMN‐binding LOV domain of YtvA (Möglich and Moffat, 2007). For the LovTAP construct, a helix–turn–helix DNA‐binding domain was employed that is found in nature fused to PAS sensors (Aravind et al., 2005) including LOV domains (Losi, 2004).
Artificial allosteric systems
Thus far, just a single example exists which demonstrates light‐dependent control over a completely sensor‐unrelated protein. Here, the extensively characterized enzyme, dihydrofolate reductase (DHFR) from E. coli, was used as target effector module to be engineered with a plant LOV domain allowing for allosteric control over DHFR biocatalytic activity (Lee et al., 2008). DHFR is an enzyme that catalyses the reduction of 5,6‐dihydrofolate (DHF) to 5,6,7,8‐tetrahydrofolate (THF) using NADPH as a cofactor. The enzyme plays a central role to maintain the cellular levels of tetrahydrofolate and its derivatives which are essential for purine and thymidylate biosynthesis. Therefore, this enzyme has been an important target for the development of antibacterial agents, anti‐cancer drugs and other therapeutics (Volpato and Pelletier, 2009). It also serves as an important system for understanding the mechanism of enzyme catalysis. Extensive kinetic studies of E. coli DHFR and various mutants along with X‐ray crystallography, NMR spectroscopy and computational studies have revealed the correlation between protein dynamics and catalytic function (Sawaya and Kraut, 1997; Schnell et al., 2004; Hammes‐Schiffer and Benkovic, 2006) and provided evidence for a network of coupled motions. Figure 3A depicts the structure of DHFR showing the ligand binding site as well as the location of important loop regions that are involved in enzyme catalysis.
Figure 3.
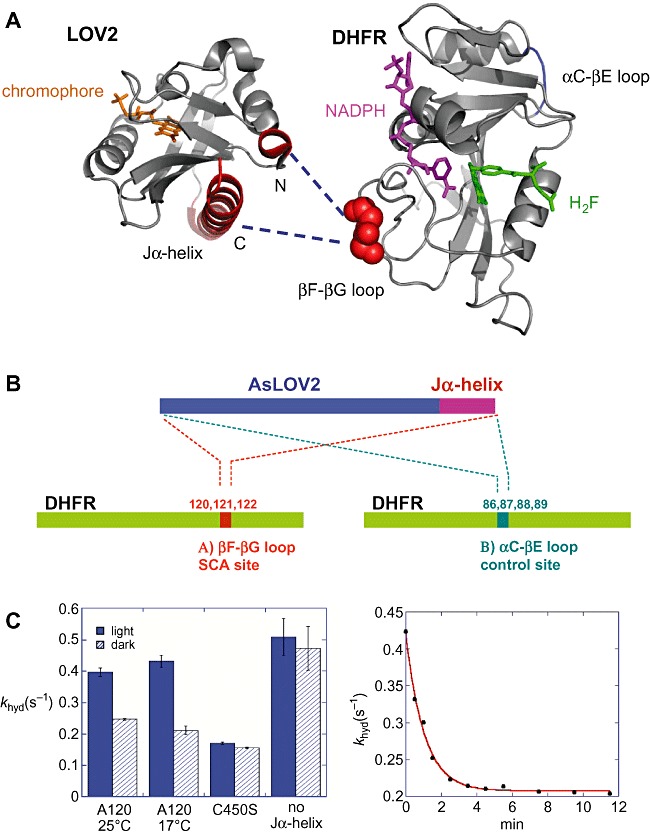
A. Structures of AsLOV2 (pdb entry: 2v0u) and DHFR (pdb entry: 1rx2), illustrating surface‐exposed SCA sites (red), the chromophore FMN (orange), NADPH (magenta) and folate (green). The SCA method identifies networks of statistically correlated amino acid residues in the LOV domain as well as in DHFR. The network links the FMN binding pocket of the LOV domain to N‐ and C‐terminal helices (shown in red) that undergo significant conformational changes upon light activation (see Fig. 3). The design strategy was to insert the light‐sensing domain into one of the surface positions (βF–βG loop, in red) in the DHFR network so that the light signal can be transmitted to the DHFR active site though the network. B. Schematic representation of the LOV2–DHFR chimeric constructs. To functionally couple the light‐induced signal with the DHFR catalysis, sets of chimeric proteins were generated. Site A (amino acids 120–122) is an allosteric site within the coupled network of DHFR, while site B (amino acids 86–89) is a control site that is not correlated with the active site. C. Light‐dependent enzyme activity in the LOV2–DHFR chimera. Hydride transfer rates (khyd) were measured by a stopped‐flow kinetic method under dark (shown as hatched bars) and light‐exposed conditions (blue bars). Among the chimeras, light dependence was observed only when the LOV domain is attached to a specific position (A120). This chimera exhibited a twofold increase of hydride transfer rate in the light condition. The construct with a point mutation (C450S) that locks the LOV domain into the dark state or a chimera lacking the Jα‐helix did not show the light dependence. The kinetics of the dark recovery of the hydride transfer rate exhibits a single exponential decay (right part of C), which is comparable to the kinetics of the FMN‐thiol covalent adduct decay.
An evolutionary approach using statistical coupling analysis (SCA) also has identified networks of co‐evolving amino acids in various proteins (Hatley et al., 2003; Peterson et al., 2004). Statistical coupling analysis for a multiple sequence alignment of 1104 PAS domains (Hefti et al., 2004) (the general folding class LOV domains belong to) revealed a spatially contiguous network of amino acids that linked the chromophore binding site to surface‐exposed residues at the N‐ and C‐terminal regions of the domain. Statistical coupling analysis of 418 members of the DHFR family also probed for network residues within DHFR and identified the βF–βG loop, one of the functional loops in the DHFR‐coupled network, as a potential surface allosteric site (Lee et al., 2008). The strategy for engineering a new allosteric protein then was to connect the networks of these two protein modules through a covalent linkage joining their surface sites such that light can modulate the DHFR activity through the response of a light‐sensing LOV domain from plant phototropin (Avena sativa LOV2) (Salomon et al., 2000; Halavaty and Moffat, 2007). In summary, the computational SCA method enabled the prediction of allosteric surface sites where the perturbation of a regulatory element (input domain) will control protein function at the remote site (output domain).
Several sets of chimeric proteins were generated in which the LOV2 domain is inserted via its N‐ and C‐terminal helical extensions into DHFR at two different surface sites (sites A and B, Fig. 3B). Site A chimeras have the LOV domain insertion at an allosteric site within the coupled network (A120–A122, βF–βG loop), and site B chimeras served as a control with another surface site (B86–B89, αC–βE loop) that is similarly distant but statistically uncorrelated with the active site. The engineered LOV2–DHFR chimera exhibited light‐dependent enzyme activity based on the functional communication between the light‐sensing protein module and DHFR. Dark recovery kinetics based on a 447 nm absorbance and enzyme activity showed that the intrinsic features of each domain are structurally and functionally intact in the LOV2–DHFR chimera. The light switch worked only when the light‐sensing domain is attached to a specific position within the network of DHFR. In the presence of light, the hydride transfer rate measured by a stopped‐flow kinetic method exhibited a twofold increase of enzyme activity compared with the dark condition. The photocycle between light and dark state generated a reversible change in the enzyme activity and the single exponential measure of the enzyme activity (khyd) from the light‐excited chimera to the dark species matched the rate of thermal relaxation of the FMN.
In the case of LOV2–DHFR, the kinetic results showed that the signal initiated by light absorption by the FMN chromophore within the hydrophobic binding pocket of the LOV domain is propagated through the structure to cause conformational changes at the opposite site of the sensor domain, particularly by the destabilization of the C‐terminal Jα‐helix, that in the natural phototropin system is implicated in the signal‐progation mechanism (Harper et al., 2003). In turn these conformational changes act to affect the DHFR conformation, leading to an enzyme activity altered by the light and dark state of the LOV domain.
The construct lacking the Jα‐helix or a variant with a C450S mutation in the LOV2 domain did not show this light dependence. Hereby, the mutation of C450 abolishes the LOV photocycle and thus traps the sensor in a dark state conformation (Salomon et al., 2000). In addition, the switch did not operate when the LOV domain was connected to the other functional loop (αC–βE loop) that is not in the network, clearly suggesting the presence of unique network(s) in proteins through which the signal is transmitted. Obviously, these results demonstrate that the light‐dependent activity of the LOV2–DHFR results from the allosteric communication between light‐sensing domain and DHFR. This novel approach of connecting SCA implicated sectors from two different proteins may furnish a general tool for the creation of a new allosteric proteins from any sensor‐unrelated protein.
Future perspectives: de novo design of light‐dependent control over protein function
One of the remaining open questions is how to modulate different levels of control over protein function by using a light signal. The detected effect in the LOV2–DHFR construct was only twofold; theoretically it can be as high as hundreds fold (Yao et al., 2008). This chimera also showed a fast transition from the light‐induced state to the dark state, which may not maximize the active catalytic population within the experimental time limit. Since bacterial LOV proteins are many times slower in their dark recovery kinetics compared with the plant phot‐LOV domains (Losi et al., 2002; Cao et al., 2007), the engineering of new bacterial LOV domains with wide ranges of recovery kinetics as well as optimizing the linker composition should guide us to more optimal light‐activated allosteric protein switches. To this end we are currently investigating several bacterial LOV domains as sensor switches for the control of DHFR.
Furthermore, we have constructed fusions of bacterial as well as plant LOV domains with a non‐allosteric regulated protein module, namely a lipase from B. subtilis (Eggert et al., 2001). Chimeric LOV–lipase constructs indeed retained both functionalities, namely blue‐light activation of the LOV domain and enzymatic activity (Fig. 4). Preliminary data obtained with the LOV–lipase fusion protein suggest that lipase activity can also be controlled by light. Furthermore, we expect that subtle structural perturbations in the lipase, i.e. propagated from the sensor domain, may result also in an alteration of enzyme properties such as substrate specificity and (enantio)selectivity.
Figure 4.
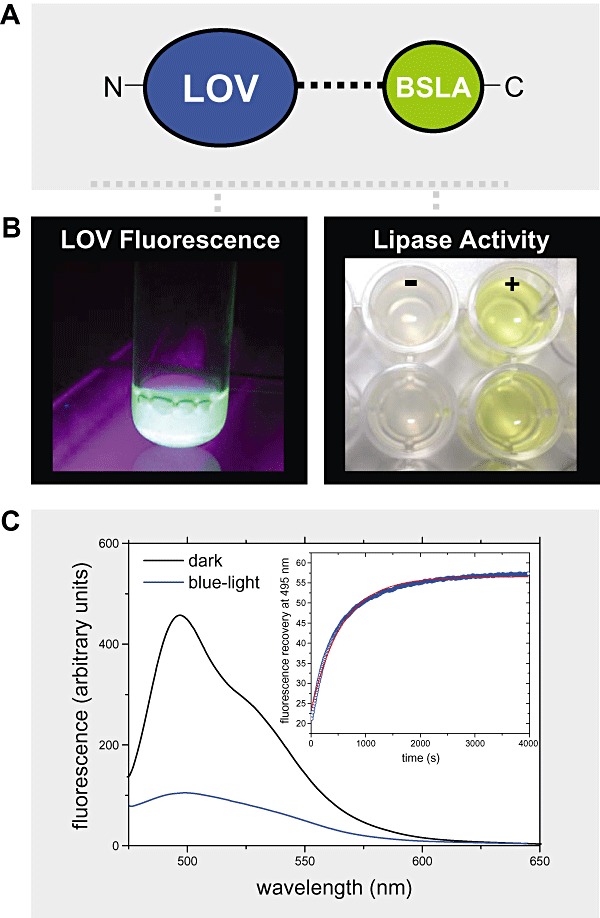
A and B. (A) Illustration of the fusion strategy: a bacterial LOV domain (here of the B. subtilis YtvA protein) was fused N‐terminally to the lipase A of B. subtilis (BSLA) resulting in the construct nLOV‐BSLA. The two protein parts are linked via a helical linker polypeptide naturally present in B. subtilis YtvA. The fusion construct can be readily expressed in soluble form and retains both chromophore binding capacity in the LOV domain as evidenced by flavin fluorescence (left part of B) and lipolytic activity of the lipase (right part of B). Here shown is the result of a standard lipase activity assay using p‐nitrophenylpalmitate (pNPP) as substrate (+). The corresponding negative control (−) contained no enzyme. The two rows of wells depict the results of a duplicate measurement. The lipase cleaves the ester‐bond of pNPP resulting in the formation of the yellow‐coloured p‐nitrophenylate anion indicative for enzymatic activity. C. Observed light sensitivity of nLOV‐BSLA: Flavin‐fluorescence was recorded in the proteins dark state (black line) as well as 30 s after blue‐light illumination (blue line). In the dark, the protein displays a bimodal fluorescence with maxima occurring at around 495 and 520 nm. Upon formation of the signalling state, this fluorescence is effectively quenched by the formation of the covalent flavin‐cysteinyl adduct. The inset shows the dark‐recovery time trace of the FMN fluorescence peak recorded at 495 nm.
The key question is whether a general strategy can be devised that allows one to choose any target enzyme, link it to a light activating domain at predetermined amino acid residues and thus create a light‐controlled enzyme. One would hope to minimize the trial and error aspects of a combinatorial approach and instead rely on SCA or other computational methods to predict the residues to be used for successful attachment. In the future, one can imagine light control encompassing not only biological regulation processes but also biocatalytic reactions and even complex reaction systems.
Acknowledgments
Part of this work has been funded by the Deutsche Forschungsgemeinschaft (DFG) (Forschergruppe ‘Blue‐Light Photoreceptors’, FOR526). J.L. and S.J.B. acknowledge support by the Defense Advanced Research Projects Agency (GMO‐700123)
References
- Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. The many faces of the helix–turn–helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Banghart M.R., Volgraf M., Trauner D. Engineering light‐gated ion channels. Biochemistry. 2006;45:15129–15141. doi: 10.1021/bi0618058. [DOI] [PubMed] [Google Scholar]
- Boyden E.S., Zhang F., Bamberg E., Nagel G., Deisseroth K. Millisecond‐timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Bucher D., Buchner E. Stimulating PACalpha increases miniature excitatory junction potential frequency at the Drosophila neuromuscular junction. J Neurogenet. 2009;23:220–224. doi: 10.1080/01677060802441356. [DOI] [PubMed] [Google Scholar]
- Buttani V., Losi A., Eggert T., Krauss U., Jaeger K.E., Cao Z., Gärtner W. Conformational analysis of the blue‐light sensing protein YtvA reveals a competitive interface for LOV‐LOV dimerization and interdomain interactions. Photochem Photobiol Sci. 2007;6:41–49. doi: 10.1039/b610375h. [DOI] [PubMed] [Google Scholar]
- Cao Z., Buttani V., Losi A., Gärtner W. A blue light inducible two component signal transduction system in the plant pathogen Pseudomonas syringae pv. tomato. Biophys J. 2007;94:897–905. doi: 10.1529/biophysj.107.108977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M., Salomon M., Nozue K., Wada M., Briggs W.R. LOV (light, oxygen, or voltage) domains of the blue‐light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S., Moffat K. Structure of a flavin‐binding plant photoreceptor domain: insights into light‐mediated signal transduction. Proc Natl Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S., Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light‐driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S., Rajagopal S., Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- Eggert T., Van Pouderoyen G., Dijkstra B.W., Jaeger K.E. Lipolytic enzymes LipA and LipB from Bacillus subtilis differ in regulation of gene expression, biochemical properties, and three‐dimensional structure. FEBS Lett. 2001;502:89–92. doi: 10.1016/s0014-5793(01)02665-5. [DOI] [PubMed] [Google Scholar]
- Fedorov R., Schlichting I., Hartmann E., Domratcheva T., Fuhrmann M., Hegemann P. Crystal structures and molecular mechanism of a light‐induced signaling switch: the Phot‐LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeldner M., Givens R. Wiley‐VCH; 2005. [Google Scholar]
- Gomelsky M., Klug G. BLUF: a novel FAD‐binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- Gorostiza P., Isacoff E.Y. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halavaty A.S., Moffat K. N‐ and C‐terminal flanking regions modulate light‐induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- Hammes‐Schiffer S., Benkovic S.J. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- Harper S.M., Neil L.C., Gardner K.H. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Harper S.M., Christie J.M., Gardner K.H. Disruption of the LOV‐Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- Hatley M.E., Lockless S.W., Gibson S.K., Gilman A.G., Ranganathan R. Allosteric determinants in guanine nucleotide‐binding proteins. Proc Natl Acad Sci USA. 2003;100:14445–14450. doi: 10.1073/pnas.1835919100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti M.H., Francoijs K.J., De Vries S.C., Dixon R., Vervoort J. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur J Biochem. 2004;271:1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x. [DOI] [PubMed] [Google Scholar]
- Van Der Horst M.A., Key J., Hellingwerf K.J. Photosensing in chemotrophic, non‐phototrophic bacteria: let there be light sensing too. Trends Microbiol. 2007;15:554–562. doi: 10.1016/j.tim.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Huala E., Oeller P.W., Liscum E., Han I.S., Larsen E., Briggs W.R. Arabidopsis NPH1: a protein kinase with a putative redox‐sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K. A blue‐light‐activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. et al. [DOI] [PubMed] [Google Scholar]
- Krauss U., Losi A., Gärtner W., Jaeger K.E., Eggert T. Initial characterization of a blue‐light sensing, phototropin‐related protein from Pseudomonas putida: a paradigm for an extended LOV construct. Phys Chem Chem Phys. 2005;7:2804–2811. doi: 10.1039/b504554a. [DOI] [PubMed] [Google Scholar]
- Lee J., Natarajan M., Nashine V.C., Socolich M., Vo T., Russ W.P. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester H.A., Krouse M.E., Nass M.M., Wassermann N.H., Erlanger B.F. A covalently bound photoisomerizable agonist: comparison with reversibly bound agonists at Electrophorus electroplaques. J Gen Physiol. 1980;75:207–232. doi: 10.1085/jgp.75.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Lin M.Z., Steinbach P., Tsien R.Y. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi A. The bacterial counterparts of plants phototropins. Photochem Photobiol Sci. 2004;3:566–574. doi: 10.1039/b400728j. [DOI] [PubMed] [Google Scholar]
- Losi A. Flavin‐based photoreceptors in bacteria. In: Silva E., Edwards A.M., editors. The Royal Society of Chemistry; 2006. pp. 223–276. [Google Scholar]
- Losi A. Flavin‐based blue‐light photosensors: a photobiophysics update. Photochem Photobiol. 2007;6:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- Losi A., Gärtner W. Bacterial bilin‐ and flavin‐binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- Losi A., Polverini E., Quest B., Gärtner W. First evidence for phototropin‐related blue‐light receptors in prokaryotes. Biophys J. 2002;82:2627–2634. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozier R.H., Bogomolni R.A., Stoeckenius W. Bacteriorhodopsin: a light‐driven proton pump in Halobacterium halobium. Biophys J. 1975;15:955–962. doi: 10.1016/S0006-3495(75)85875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Moffat K. Structural basis for light‐dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Ayers R.A., Moffat K. Design and signaling mechanism of light‐regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama T., Suzuki T., Yoshikawa S., Iseki M. Functional transplant of photoactivated adenylyl cyclase (PAC) into Aplysia sensory neurons. Neurosci Res. 2007;59:81–88. doi: 10.1016/j.neures.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P. Channelrhodopsin‐2, a directly light‐gated cation‐selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G., Brauner M., Liewald J.F., Adeishvili N., Bamberg E., Gottschalk A. Light activation of channelrhodopsin‐2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Ntefidou M., Ludtke T., Ahmad M., Hader D.P. Heterologous expression of photoactivated adenylyl cyclase (PAC) genes from the flagellate Euglena gracilis in insect cells. Photochem Photobiol. 2006;82:1601–1605. doi: 10.1562/2006-04-06-RA-867. [DOI] [PubMed] [Google Scholar]
- Peterson F.C., Penkert R.R., Volkman B.F., Prehoda K.E. Cdc42 regulates the Par‐6 PDZ domain through an allosteric CRIB‐PDZ transition. Mol Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- Pulver S.R., Pashkovski S.L., Hornstein N.J., Garrity P.A., Griffith L.C. Temporal dynamics of neuronal activation by Channelrhodopsin‐2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovski O., Beharry A.A., Zhang F., Woolley G.A. Spectral tuning of azobenzene photoswitches for biological applications. Angew Chem Int Ed Engl. 2009;48:1484–1486. doi: 10.1002/anie.200805013. [DOI] [PubMed] [Google Scholar]
- Salomon M., Christie J.M., Knieb E., Lempert U., Briggs W.R. Photochemical and mutational analysis of the FMN‐binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Sawaya M.R., Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- Schnell J.R., Dyson H.J., Wright P.E. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- Schobert B., Lanyi J.K. Halorhodopsin is a light‐driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- Schröder‐Lang S., Schwärzel M., Seifert R., Strunker T., Kateriya S., Looser J. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4:39–42. doi: 10.1038/nmeth975. et al. [DOI] [PubMed] [Google Scholar]
- Shoham S., O'Connor D.H., Sarkisov D.V., Wang S.S. Rapid neurotransmitter uncaging in spatially defined patterns. Nat Methods. 2005;2:837–843. doi: 10.1038/nmeth793. [DOI] [PubMed] [Google Scholar]
- Strickland D., Moffat K., Sosnick T.R. Light‐activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.L., Zhulin I.B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeki N., Yoshizawa T., Sugimoto Y., Mitsui T., Wakabayashi K., Maruta S. Incorporation of an azobenzene derivative into the energy transducing site of skeletal muscle myosin results in photo‐induced conformational changes. J Biochem. 2004;136:839–846. doi: 10.1093/jb/mvh194. [DOI] [PubMed] [Google Scholar]
- Volgraf M., Gorostiza P., Szobota S., Helix M.R., Isacoff E.Y., Trauner D. Reversibly caged glutamate: a photochromic agonist of ionotropic glutamate receptors. J Am Chem Soc. 2007;129:260–261. doi: 10.1021/ja067269o. [DOI] [PubMed] [Google Scholar]
- Volpato J.P., Pelletier J.N. Mutational ‘hot‐spots’ in mammalian, bacterial and protozoal dihydrofolate reductases associated with antifolate resistance: sequence and structural comparison. Drug Resist Updat. 2009;12:28–41. doi: 10.1016/j.drup.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Yamada M.D., Nakajima Y., Maeda H., Maruta S. Photocontrol of kinesin ATPase activity using an azobenzene derivative. J Biochem. 2007;142:691–698. doi: 10.1093/jb/mvm183. [DOI] [PubMed] [Google Scholar]
- Yao X., Rosen M.K., Gardner K.H. Estimation of the available free energy in a LOV2‐J alpha photoswitch. Nat Chem Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S., Suzuki T., Watanabe M., Iseki M. Kinetic analysis of the activation of photoactivated adenylyl cyclase (PAC), a blue‐light receptor for photomovements of Euglena. Photochem Photobiol Sci. 2005;4:727–731. doi: 10.1039/b417212d. [DOI] [PubMed] [Google Scholar]
- Yoshizawa T. Photophysiological functions of visual pigments. Adv Biophys. 1984;17:5–67. doi: 10.1016/0065-227x(84)90024-8. [DOI] [PubMed] [Google Scholar]
- Zemelman B.V., Nesnas N., Lee G.A., Miesenbock G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci USA. 2003;100:1352–1357. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Aravanis A.M., Adamantidis A., De Lecea L., Deisseroth K. Circuit‐breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007a;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang L.P., Brauner M., Liewald J.F., Kay K., Watzke N. Multimodal fast optical interrogation of neural circuitry. Nature. 2007b;446:633–639. doi: 10.1038/nature05744. et al. [DOI] [PubMed] [Google Scholar]


