Summary
There is an increasing need for the use of biocatalysis to obtain enantiopure compounds as chiral building blocks for drug synthesis such as antibiotics. The principal findings of this study are: (i) the complete sequenced genomes of Bacillus cereus ATCC 14579 and Thermoanaerobacter tengcongensis MB4 contain a hitherto undescribed enantioselective and alkaliphilic esterase (BcEST and TtEST respectively) that is specific for the production of (R)‐2‐benzyloxy‐propionic acid ethyl ester, a key intermediate in the synthesis of levofloxacin, a potent antibiotic; and (ii) directed evolution targeted for increased thermostability of BcEST produced two improved variants, but in either case the 3–5°C increase in the apparent melting temperature (Tm) of the mutants over the native BcEST that has a Tm of 50°C was outperformed by TtEST, a naturally occurring homologue with a Tm of 65°C. Protein modelling of BcEST mapped the S148C and K272R mutations at protein surface and the I88T and Q110L mutations at more buried locations. This work expands the repertoire of characterized members of the α/β‐fold hydrolase superfamily. Further, it shows that genome mining is an economical option for new biocatalyst discovery and we provide a rare example of a naturally occurring thermostable biocatalyst that outperforms experimentally evolved homologues that carry out the same hydrolysis.
Introduction
Levofloxacin (LFC) is the biologically active levo‐ or S‐isomer of ofloxacin, a tricyclic fluoroquinolone, a potent antibiotic against Gram‐negative and Gram‐positive bacteria (Hayakawa et al., 1986). The mechanism of action of LFC is through targeting the essential bacterial type II topoisomerases, DNA gyrase and topoisomerase IV that are involved in DNA replication and metabolism. As a third‐generation fluoroquinolone antibiotic, LFC has a broader spectrum of antibacterial activity than the second‐generation ciprofloxacin and related antibiotics (Fu et al., 1992; Ernst et al., 1997). The clinical use of LFC includes long‐term treatment of multidrug‐resistant tuberculosis, community‐acquired pneumonia, acute chronic bronchitis and acute bacterial sinusitis (Mehlhorn and Brown, 2007; Moadebi et al., 2007). Due to increasing resistance of staphylococci for example, new LFC‐containing hybrid chemicals have been synthesized and shown to be potent against these microorganisms (Foroumadil et al., 2006).
There are at least five synthetic routes to LFC that involve numerous chemical steps and solvents (Kang et al., 1996; 1997; Miyadera and Imura, 1999; Kleemann and Engel, 2001). One of the key chemical intermediates is (R)‐2‐benzyloxy‐propionic acid ethyl ester or O‐benzyl lactic acid ethyl ester (BnLAE, Fig. 1). Chemical synthesis of BnLAE would produce a racemic mixture and therefore additional steps for the separation of the isomers are required. The availability of a biocatalyst, an enantioselective esterase capable of resolving the racemic esters, would provide a green and chemo‐enzymatic route towards the production of LFC.
Figure 1.
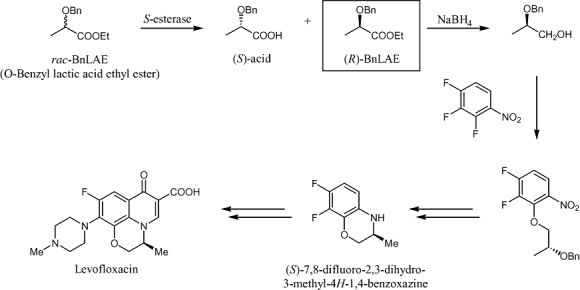
Chemo‐enzymatic scheme for the production of levofloxacin involving the key intermediate (R)‐BnLAE (O‐benzyl lactic acid ethyl ester or 2‐benzyloxy‐propionic acid ethyl ester). The synthesis of (S)‐benzoxazine derivative is also through enzymatic resolution (Miyadera and Imura, 1999).
In this article, we describe the identification, cloning and expression in Escherichia coli of an S‐specific esterase (BcEST) from Bacillus cereus ATCC 14579 that is capable of racemic resolution of BnLAE. Furthermore, we have used directed evolution to attempt improvement the thermostability of this biocatalyst. In addition, a predicted open reading frame (ORF) from a thermophilic microorganism, Thermoanaerobacter tengcongensis, with a presumed hydrolase or acyltransferase function (TTE0556; GenBank NC_003869) was cloned and characterized. Biochemical properties of these biocatalysts are compared and discussed in the context of enhanced thermostability and of a naturally derived biocatalyst versus genetic variants nurtured in the laboratory.
Results
Preliminary microbial screening
Four bacterial strains (Pseudomonas sp. IFO 3925; B. cereus DSC 0007, Microbacterium laevaniformans IFO 14471 and B. cereus ATCC 14579), out of approximately 500 microorganisms screened for the enantioselective hydrolysis reaction using rac‐BnLAE as substrate, were found to have excellent enantiomeric ratio, E, in excess of 100 (not shown). E is the ratio of one enantiomer in a mixture to that of the other (Chen et al., 1982). An initial attempt to purify the desired esterase from Bacillus sp. IFO 3925 proved difficult and therefore attention was turned to strain ATCC 14579 which catalysed the same reaction with an E value > 200.
Cloning and identification of the BcEST‐encoding gene
A 10.5 kb recombinant plasmid, designated pGEMHindIII2, was propagated in E. coli DH5α as a result of cloning a 7.3 kb HindIII genomic fragment of B. cereus ATCC 14579 in plasmid pGEM3zf (see Experimental procedures). Using synthetic primers, the cloned insert was sequenced completely on both strands. This sequence, which encompasses at least eight ORFs, has been deposited in the NCBI GenBank under Accession No. GQ243224. Identification of the coding sequence of BcEST was aided by determining peptide sequences by Edman degradation of the purified enzyme. These sequences consist of the N‐terminal 17 amino acids, and three internal peptide sequences following protease digestion, one of 7 aa and two of 12 aa. In the overall BcEST sequence of 300 aa, the internal peptide sequences were located at positions 139–145, 152–163 and 176–187. Except for one discrepancy (second D in the peptide 139DTDGQPI should be an N), all other peptide sequences are in complete agreement with the DNA‐derived data.
The gene context of BcEST is such that it is flanked by a potential repressor of the MarR‐type (Grkovic et al., 2002; Schumacher and Brennan, 2002) that is divergently transcribed, and downstream where two hypothetical proteins, one being a putative transport protein, are found. This gene organization is in keeping with the annotation in the B. cereus ATCC 14579 genome database (Ivanova et al., 2003).
Plasmid pBcEST was constructed as a subclone of pGEMHindIII2 containing only the est gene in the same orientation as the lac promoter of expression vector pSD80. Cell cultures of E. coli BL21(pBcEST) and DH5α(pGEMHindIII2) were assayed for their enantioselective esterase activity. Both clones produced a pH change causing bromothymol blue to turn yellow when using the substrate (S)‐BnLAE but not the (R)‐substrate. The control cells with only the plasmid vector, IPTG‐induced or non‐induced, did not impart any colour change. The specific activities of the crude enzyme extract prepared from DH5α(pGEMHindIII2), BL21(pBcEST) and the native B. cereus ATCC 14579 were shown to be 0.22, 0.60 and 0.015 U mg−1 protein respectively. This indicated a 15‐ to 40‐fold improvement of activity in the recombinant strains. It is noted that the est gene in pGEMHindIII2 is in the reverse orientation relative to the lac promoter, suggesting that the Bacillus promoter is functional in E. coli.
Characteristics of BcEST and identification of TtEST
The calculated Mr of BcEST of 34 321 is in agreement with that estimated by SDS‐PAGE of an overproduced 32.5 kDa protein prepared from a crude extract of E. coli BL21(pBcEST) (Fig. S1). Gel filtration indicated an apparent Mr of 40 kDa implying a monomeric enzyme (data not shown). The predicted isoelectric point (pI) is 4.8. The NCBI Conserved Domain Search predicted an α/β‐fold hydrolase enzyme of the esterase_lipase superfamily, with the characteristic catalytic triad consisting of S102‐D251‐H279.
A phylogenetic analysis of BcEST is presented in Fig. S2. As expected, the sequence clusters with a number of predicted hydrolases of the α/β‐fold family, invariably annotated as 3‐oxoadipate enol‐hydrolase, that originate from a number of Bacillus spp.; the two closest homologues are from B. cereus B4264 and G9842 whose genomes were completely sequenced as part of the B. cereus group (Joint Genomics Institute). In addition to sequences derived from protozoa, ciliates and cyanobacteria, other notable homologues are from anaerobic dechlorinating microorganisms, e.g. Desulfitobacterium hafniense (41.3% identity) and explosive‐degrading and psychrophilic denitrifiers, such as Shewanella sediminis HAW‐EB3 and S. halifaxensis HAW‐EB4 (42% and 39% identity respectively). Of particular interest is a homologue showing 38% identity, a putative hydrolase or acyltransferase (AAM23832.1; TTE0556) present in the genome of a thermophilic microorganism, T. tengcongensis strain MB4T that originated from a Tengchong freshwater hot spring in Baoshan, Yunnan province, China (Bao et al., 2002). We subsequently cloned this putative gene, designated TtEST, in the IPTG‐inducible vector pET17b (Novogen). Properties of the recombinant 4235 bp plasmid pETTtEST in E. coli BL21 cells are described in a later section.
In the Protein Data Bank (Berman et al., 2000), the structural homologues of BcEST include: an aryl esterase from Pseudomonas fluorescens (PDB 1VA4, 27% identity; Cheeseman et al., 2004); a C–C bond hydrolase (MphC) of the phenylpropionate degradation pathway of E. coli (PDB 1U2E, 25% identity; Dunn et al., 2005); and, a bromoperoxidase A1 from Streptomyces aureofaciens (PDB 1A8Q, 26% identity; Hofmann et al., 1998). The sequence alignment of BcEST, together with TtEST and the three structurally similar enzymes, is given in Fig. 2.
Figure 2.

Multiple sequence alignment of BcEST and TtEST with their closest structural homologues (identified by PBD codes). Identical residues are highlighted black and conserved residues on grey backgrounds. Active‐site catalytic triad residues are marked with a triangle, and mutated residues with a dot. Secondary structure elements are indicated by arrows for β‐strands and by cylinders for α‐helices, as predicted for BcEST and TtEST (shown in light‐grey above the alignment) and observed in the closest structural homologue 1VA4 (shown in dark‐grey below the alignment). For the structural homologues, indicated residue numbers are taken from the respective PDB entries.
Characteristic conserved sequences around the predicted catalytic triad include residues S102 and D251 that are found within the tetrapeptide SMGG and a motif L‐x3‐G‐x2‐D‐x2‐V. The sequence around H279 is less conserved, although BcESt and TtEST share a common (G/A)HS(P/V)FID sequence. In the classification of microbial lipolytic enzymes and carboxylesterases, BcEST and TtEST appear to be members of the Group V family that consists of mostly non‐lipolytic enzymes (Arpigny and Jaeger, 1999; Bornscheuer, 2002).
Evolving the thermostability of BcEST
A preliminary stability assay with crude BcEST enzyme preparation indicated that it is unstable above 40°C as expected of a mesophilic enzyme. We used a thymolblue‐based colorimetric assay to carry out error‐prone PCR (epPCR) with the aim of generating thermostable mutants by screening at two elevated temperatures, first at 50°C and then at 55°C. In the first round, more than 4600 transformants were screened, and the best clone, designated EH5, was subjected to a second round of mutagenesis. The result of screening > 2300 transformants in the second round culminated in the isolation of 2IC12.
DNA sequencing identified three single‐base changes in EH5 that resulted in two amino acid substitutions: S148C due to AGT to TGT; and K272R due to AAG to AGG; the ATT to ATC change at I48 is silent. The 2IC12 mutant was the result of two additional single‐amino‐acid substitutions: I88T (ATT to ACT) and Q110L (CAA to CTA), in addition to S148C and K272R derived from the first round of mutagenesis.
Stability assays for BcEST and the two variants were carried out at 55°C. After a 20 min treatment of the respective crude enzyme, 2IC12 and EH5 retained 80% and 40% of their original activity compared with 25% in the case of the wild‐type BcEST. A 5 min treatment at 60°C resulted in almost total loss of wild‐type activity, whereas 2IC12 retained about 50% and EH5 25% of their activity (Fig. S3).
Properties of purified BcEST, EH5, 2IC12 and TtEST
The four specified esterases were purified to electrophoretic homogeneity (Fig. 3) using a three‐step chromatographic procedure as described in Experimental procedures. By SDS‐PAGE (12%) gel, the proteins appear as single bands with estimated Mr between 35 and 38 kDa, in good agreement with the calculated Mr(s). The 2IC12 and TtEST proteins showed a slightly higher Mr than expected (34 336 and 34 388 respectively). The purified esterases eluted at 36–40 kDa by gel filtration on Superose‐12 indicating monomeric enzymes. Analysis by UV‐Vis showed no other absorbance peak besides that at 280 nm.
Figure 3.
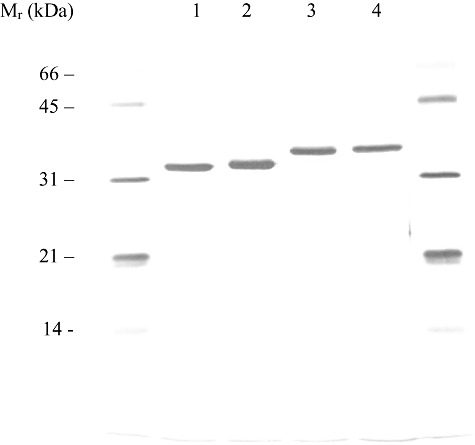
SDS‐PAGE of purified esterases. Lanes 1–4: BcEST, EH5, 2IC12 and TtEST respectively. The following standard proteins were used: lysozyme (14 400), trypsin inhibitor (21 500), carbonic anhydrase (31 000), ovalbumin (45 000) and bovine serum albumin (66 200).
The specific activity of the purified BcEST, EH5, 2IC12 and TtEST towards rac‐BnLAE, assayed at 25°C, was found to be very similar for the former three enzymes (31.5, 25.9 and 32.7 U mg−1) but some three to four times less in the case of TtEST (8.6 U mg−1). Optimal enzyme activity was determined for all enzymes: pH 8.0–9.0 for BcEST and EH5 at 45°C; 2IC12 at 50°C, and at pH 8.0–10 for TtEST at about 60°C.
Kinetic parameters measured under steady‐state condition at 25°C and in Tris‐HCl buffer (pH 8.0) towards the rac‐BnLAE substrate indicated a very similar kcat and Km for BcEST and EH5 (BcEST, 18 s−1, 0.22 mM; EH5, 15 s−1, 0.21 mM). The Km appears to be lower (0.13 mM) for 2IC12 but its kcat (19 s−1) is similar to those of the wild type and EH5 variant. In TtEST, the Km is higher at 0.69 mM but with a lower kcat of 5 s−1. The catalytic efficiency (kcat/Km) is the highest for 2IC12 but the lowest for TtEST (Table 1). By plotting ln kcat versus T−1 and calculating the slopes of the linear regressions, the following activation energies were estimated: BcEST: 6.4 ± 0.3, EH5: 6.3 ± 0.2, 2IC12: 6.1 ± 0.3, and TtEST: 4.8 ± 0.3 kcal mol−1 (Fig. S4A and B). At 25°C the free energy of activation (ΔG‡) was calculated to be 15.7, 15.8, 15.7 and 16.5 kcal mol−1 respectively (BcEST, EH5, IC212 and TtEST).
Table 1.
Summary of selected molecular and kinetic parameters of the esterases.
| Esterase | kcat (s−1) | Km (mM) | kcat /Km (M−1s−1) | Mr (kDa) | Mr (SDS) (kDa) | Tm (°C) |
|---|---|---|---|---|---|---|
| BcEST | 18.0 | 0.22 | 8.2 × 105 | 34.319 | 34.7 | 50 |
| EH5 | 14.8 | 0.21 | 7.0 × 105 | 34.363 | 35.4 | 53 |
| 2IC12 | 18.7 | 0.13 | 1.4 × 106 | 34.336 | 37.9 | 55 |
| TtEST | 4.9 | 0.69 | 7.1 × 104 | 34.388 | 38.2 | 65 |
Although racemic BnLAE was used in the assay, selective cleavage of the (S)‐enantiomer producing pure (R)‐enantiomer was confirmed using HPLC with a chiral column for the four esterases. The enantiomeric excess of (R)‐BnLAE was found to be 99% with high yield (close to 50%) and an E value of > 200.
The specificity of TtEST was evaluated for various 4‐methylumbelliferyl esters, α‐naphthyl esters, indoxyl acetate and p‐nitrophenyl acetate and butyrate (Fig. S6). All the activities were approximately 3–11 times lower than for BnLAE. Methyl cinnamic acid was a poor substrate. No activity was found towards β‐naphthyl caprylate, indicating some regio‐selectivity of TtEST. Tributyrin and esters of fatty acids are not substrates. Similarly, BcEST activities towards α‐naphthylacetate, α‐naphthylbutyrate and α‐indoxylacetate were found to be 8–18 times lower than for the BnLAE substrate (Table S1).
Stability of cloned esterases
Organic solvents such as DMF (up to 30%) and DMSO (up to 50%) did not effect the stability (20 h incubation) nor the activity of the esterases (not shown). After several months of storage as lyophilized powder at room temperature, the enzymes retained full activity. The thermostability, thermodenaturation constant (kd) and half‐life time (t1/2) of these biocatalysts are shown in Fig. 4. TtEST was fully active at 50°C for an extended period of time but lost about 40% of its activity when incubated at 70°C for 20 min. At 50°C, the t1/2 of BcEST was about 40 min, but lost its activity very quickly at higher temperatures. The EH5 and 2IC12 variants have greater t1/2 at 50°C (130 and > 200 min respectively), and also displayed increased stability at 55°C (17 and 59 min).
Figure 4.
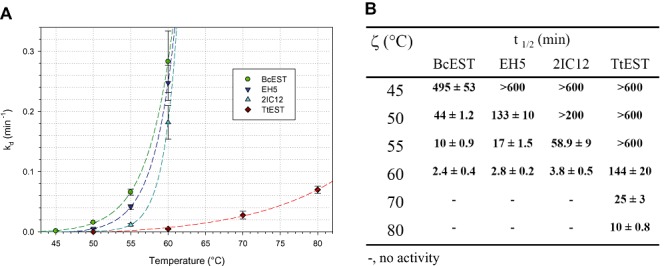
Comparison of (A) denaturation constants (kd) and (B) half‐lives (t1/2) of BcEST, EH5, 2IC12 and TtEST esterases.
According to the Lumry and Eyring model (Lumry and Eyring, 1954; Sanchez‐Ruiz, 1992), the irreversible denaturation of a protein is a two‐step process, in which the native enzyme reversibly unfolds and is then eventually and irreversibly altered to its final non‐native state. Based on the experimentally determined rate constants of denaturation (kd) for the esterases, the transformed Arrhenius equation (similarly to above using kcat) can be used to estimate the activation energy (Ed) of denaturation (unfolding). The Ed was estimated to be 18.6 ± 1.6, 21.0 ± 0.4, 30.0 ± 0.8 and 39.4 ± 3.6 kcal mol−1 for BcEST, EH5, 2IC12 and TtEST respectively.
Circular dichroism spectroscopy
Thermal unfolding experiments monitored by CD spectroscopy revealed apparent melting temperatures (Tm) for BcEST, EH5, 2IC12 and TtEST of 50°C, 53°C, 55°C and 65°C respectively (Fig. S5). The CD spectra of the esterases in the far UV (200–250 nm) revealed typical helical proteins. All four enzymes showed little differences in their far UV spectra and were found to consist of at least 50% α‐helices (not shown). Despite the low sequence identity (38%) between TtEST and BcEST, these proteins share a very similar pattern of secondary structure as predicted by PSIPRED (Jones, 1999) (Fig. 2).
A qualitative interpretation for the enhanced thermostability in BcEST mutants EH1 and 2IC12
In order to find a plausible qualitative explanation for the improved thermostability of the two improved variants of BcEST, we generated a homology model of BcEST using the aforementioned three structural templates. The BcEST homology model displays the α/β‐fold around the catalytic triad, a characteristic of this superfamily of hydrolases (Fig. 5). Three of the four mutated residues are predicted to be found in α‐helices (I88, Q110 and S148) while the fourth one (K272) in an extended β‐strand region (Fig. 5). The S148 and K272 residues mutated in the first round appear surface‐exposed, while the additional two residues, I88 and Q110, from the second round are presumably partially buried. The two mutations in the double mutant, S148C and K272R, are highly conserved. Based on a visual examination of the BcEST homology model one can speculate that the S148C surface mutation would slightly improve hydrophobic packing against neighbouring hydrophobic residues (e.g. I145, V253) owing to a larger, more polarizable S atom compared with the O atom at this position. The K272R surface mutation may lead to some gain in H‐bonding and electrostatic interactions due to a closer approach to neighbouring acidic residues (e.g. E270, D295). However, the net stabilizing effect may not be large since such gains may also incur desolvation and entropic costs. Given the more buried location of the additional I88T and Q110L mutations from the quadruple mutant, the effects of those mutations taken individually may be relatively larger than those of the S148C or K272R surface‐exposed mutations. However, these additional two mutations introduce opposite changes in the physicochemical properties at the site of mutation: I88T increases the polarity while Q110L increases the hydrophobicity. In a plausible scenario one would expect I88T and Q110L to have relatively large opposing effects on protein stability (I88T – destabilizing, Q110L – stabilizing). Hence, the net stabilizing effect of the combined buried mutations I88T and Q110L (large opposing effects) may end up as comparable to that afforded by the combined surface mutations S148C and K272R (small synergistic effects), as observed experimentally.
Figure 5.
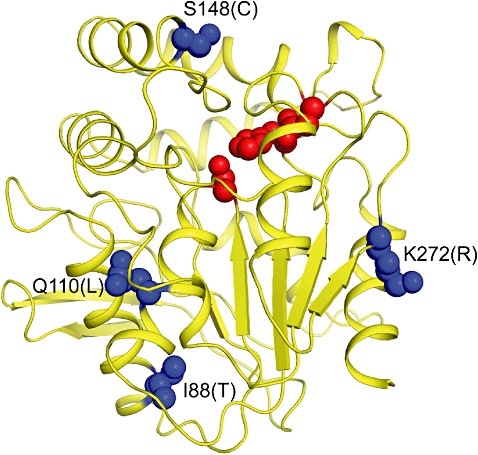
Homology model of wild‐type and mutant BcEST. The overall fold of the enzyme is rendered as a yellow ribbon. The side‐chains of the active‐site catalytic triad residues (S102, D251 and H279) are rendered as red spheres. The side‐chains of mutated residues are rendered as blue spheres.
Discussion
BcEST and TtEST are two new members of the growing superfamily of α/β‐fold hydrolases (Ollis et al., 1992; Nardini and Dijkstra, 1999; Holmquist, 2000). Otherwise annotated as either a 3‐oxoadipate enol‐hydrolase in B. cereus ATCC 14579 (NC_004722; GenBank AE016877) or simply a hydrolase or acyltransferase in the case of T. tengcongensis (NC_003869; AAM 23832.1; Bao et al., 2002), the two enzymes are characterized as alkaliphilic esterases and are remarkable for the racemic resolution of BnLAE to produce R‐BnLAE with an E value > 200. Whereas BcEST has a higher catalytic activity towards BnLAE, TtEST has greatly improved overall thermostability. This pair of enzymes represents the second example of an enzymatic step in the chemoenzymatic synthesis of LFC as shown in Fig. 1, the other example being the resolution of the benzoxazine intermediate by either Bacillus sp. DSC 1012 (Miyadera and Imura, 1999) or Baker's Yeast (Kang et al., 1996).
In the context of comparative genomics, it is noteworthy that BcEST is not an ORF specific to ATCC 14579, unlike two members with unknown function that have the α/β‐hydrolase fold (Mois et al., 2007). On the other hand, the BcEST‐encoding gene and its surrounding sequences that include a putative major facilitator transport protein and a 6‐aminohexanoate‐dimer hydrolase homologue, are very well conserved among several of the completely sequenced genomes of the B. cereus group of bacteria, including the recently described B. cereus Q1 extremophile, isolated from a deep subsurface oil reservoir (GenBank Accession No. CP000227; Xiong et al., 2009). Unfortunately, the physiological substrate and the significance of this potential metabolic pathway are not known at this time. However, one can speculate that the BcEST‐encoding gene is likely controlled by a divergently transcribed repressor of the MarR family, as suggested by the presence of two inverted repeat sequences in the 188 bp intergenic space between the marR homologue and est (Grkovic et al., 2002; Schumacher and Brennan, 2002).
In the T. tengcongensis genome (NC_003869), TtEST (TTE0556) is flanked by a LipA homologue (putative acetyltransferase and hydrolase with an α/β‐hydrolase fold) and a putative FabH2[3‐oxo‐acyl‐(acyl‐carrier protein) synthase III]. We speculate that this pathway is involved in fatty acid metabolism. Previously, Zhang and colleagues (2003) had cloned a thermostable esterase from T. tengcongensis strain MB4T. Although it can hydrolyse tributyrin, it does not hydrolyse olive oil but only hydrolyses short‐chain p‐nitrophenol esters and is therefore not considered to be a true lipase. The enzyme is optimally active at 70°C and at pH 9 (Zhang et al., 2003) similar to TtEST. Indeed, the T. tengcongensis MB4T genome has been used as a source of thermostable proteins, such as tengconglysin, a new serine protease (Koma et al., 2007) and a highly thermostable single‐strand DNA‐binding protein (Olszewski et al., 2008). Characterization of TtEST represents a contribution to the global proteomics of the T. tengcongensis genome by Wang and colleagues (2007).
We modelled BcEST according to three structural homologues in an attempt to explain the added thermostability in the double and quadruple mutants EH5 and 2IC12 respectively. The stabilizing effects observed experimentally are undoubtedly the result of a complex interplay of intramolecular interaction, desolvation and entropy contributions. One interesting hypothesis is that the I88T and Q110L mutations in the quadruple mutant may have actually large opposing effects, which when combined lead to a relatively small stabilization as observed experimentally by a mere 2°C increase of Tm. This leads to the prediction that a triple mutant consisting of Q110L, S148C and K272R would have an improved thermostability compared with the quadruple mutant, a hypothesis that remains to be tested.
A modelled structure of TtEST was also constructed and found to have a very similar structural environment around the catalytic triad as in BcEST and the three template structures (not shown). There appears to be no major structural differences between the modelled TtEST and BcEST structures. As amino acid composition is believed to play an intrinsic role in the stability of proteins (Farias et al., 2004; Zhou et al., 2008), the sequence of TtEST was analysed and compared with the mesophilic BcEST. For TtEST, the E+K/Q+H ratio is 4.8, which falls in the range of thermophiles (3.2–4.6) and hyperthermophiles (> 4.5). This ratio is 1.5 for BcEST that fits the category of mesophiles of < 2.5. However, explaining thermostabilization is more complex since it is known that a multitude of factors are involved. These include additional formation of disulfide bridges, hydrophobic or aromatic interactions, contact order, hydrogen bonding, ion pairing, dimmer–dimer interaction, stability of α‐helices, reduction of the entropy of unfolding etc. (Vieille and Zeikus, 2001; Robinson‐Rechavi and Godzik, 2005; Zhou et al., 2008).
The experimental values of activation energies for denaturation of the BcEST and variants, and TtEST esterases are in good agreement with the respective Tm values. It has been noted that thermophilic enzymes, although adapted to work at high temperatures, are not optimized in terms of activity, whereas their mesophilic counterparts often show optimized catalytic activities but low stability (Vieille and Zeikus, 2001). The BcEST and TtEST described in this study are another example.
Owing to the wide‐ranging use of hydrolases in industry as well as academic interest, protein engineering efforts applied to this family of enzymes have been intense and have utilized a number of approaches (Jaeger et al., 1999; Qian et al., 2007). Using directed evolution and selection at higher temperature we have obtained two thermostable variants. However, the modest increase of 5°C in Tm in one quadruple mutant was less than the 15°C increase in a genome‐mined homologue derived from a naturally occurring thermophilic organism. This suggests that nature has outrun directed evolution in this thermostabilization case, contrary to many examples of directed evolution efforts where bona fide thermophilic counterparts (unlike BcEST–TtEST pair) are not readily available (Woodyer et al., 2004). In any case, judicious genome mining can be a productive and economical way of new biocatalyst discovery. More importantly, functional characterization of the sequence space is key, even though the ‘road’ can be difficult (White, 2006).
Presently, a popular approach to discovering new and improved proteins is through metagenomics, with the marine environment as a rich source for the discovery of novel enzymes (Ferrer et al., 2005; Kennedy et al., 2008). With reference to racemic resolution of building blocks or precursors for pharmaceutical compounds such as antibiotics, Park and colleagues (2007) had reported on a Vibrio‐derived esterase (Vlip509) that is capable of hydrolysing the S‐enantiomer of racemic ofloxacin propyl ester. In a more recent study, two esterases (EstAT1 and EstAT11) from the Arctic sediment metagenome were found to hydrolyse S‐ofloxacin butyl ester (Jeon et al., 2009). In either case, the enantiomeric excess (e.e.) value is modest at 42.9% for Vlip509 and up to 70.3% for EstAT11.
In summary, this study illustrates that despite an enormous database on esterase sequences (ESTHER; http://bioweb.ensam.inra.fr/esther) there is ample room for new findings and it is particularly rewarding for potential industrial applications. There is an increasing demand for the environmentally benign synthesis of enantiopure compounds as chiral building blocks for drug synthesis such as antibiotics (Agranat et al., 2002). Given a multistep synthesis for molecule such as LFC, a chemoenzymatic route appears attractive since it combines proven economics of some unavoidable chemical processes with environmental care through green chemistry.
Experimental procedures
The description of the experimental procedures is contained in Appendix S1 in Supporting information.
Acknowledgments
Financial support by Daiichi Pharmaceuticals is gratefully acknowledged. We thank Michele Loewen for initial help in homology modelling. We are grateful to Allan Matte, Zhizhuang Xiao and Hannes Leisch for suggestions and help with the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Coomassie blue-stained protein gel of E. coli crude extracts separated on SDS-10% PAGE.
Fig. S2. Phylogenetic analysis of BcEST. The GenBank accession numbers for the respective proteins, annotated as either α/β-fold family hydrolase, 3-oxoadipate enol-lactonase, or in some cases, unnamed protein product or conserved hypothetical protein, are as follows: (1) ACK89548.1, (2) AAP27115.1, (3) AAS42215.1, (4) ACM13522.1, (5) AU17266.1, (6) AAT62319.1, (7) ACO28098.1, (8) ABY44194.1, (9) ACK96374.1, (10) BcEST (AAP10252.1), (11) ACK59771.1, (12) ABS22367.1, (13) ABX31253.1, (14) ACB86291.1, (15) ABV38133.1, (16) ABZ77297.1, (17) ACL19790.1, (18) ABX81126.1, (19) TtEST (AAM23832.1), (20) EAS01633.1, (21) EAR92522.1, (22) CAK56695.1, (23) BAH42062.1, (24) BAD39794.1, (25) ACL97306.1, (26) AAK25687.1, (27) ABI78668.1, (28) ABC62566.1, (29) ABS62973.1, (30) ABN96204.1, (31) ABG91905.1, (32) BAH48442.1, (33) ABK68474.1, (34) ABM15056.1, (35) ABP44844.1, (36) ACB02466.1 (1U2E), (37) ACC84816.1, (38) AAB60168.1 (1VA4), (39) AAC43253.1 (1A8Q). The tree was generated using MEGA4 (Kumar et al., 2008).
Fig. S3. Heat inactivation of BcEST and variant enzymes.
Fig. S4. Estimation of free energy of activation in the enzymatic reaction of (A) BcEST and variants and (B) TtEST.
Fig. S5. Tms of the BcEST and variant enzymes, and TtEST.
Fig. S6. Colorimetric assay of esterase activity using Phenol-Red.
A. Time-course of enzyme-induced colour shift of Phenol-Red.
B. Standard curve for the quantification of enzymatic reaction.
C. Linear increase of A435 over time due to esterase activity.
Table S1. Comparison of the substrate specificity of BcEST and TtEST
Appendix S1. Experimental procedures
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agranat I., Caner H., Caldwell J. Putting chirality to work: the strategy of chiral switches. Nat Rev Drug Discov. 2002;1:753–768. doi: 10.1038/nrd915. [DOI] [PubMed] [Google Scholar]
- Arpigny J.L., Jaeger K.E. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- Bao Q., Tian Y., Li W., Xu Z., Xuan Z., Hu S. A complete sequence of the T. tengcongensis genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Bhat T.N., Bourne P.E., Feng Z., Gilliland G., Weissig H., Westbrook J. The protein data base and the challenge of structural genomics. Nat Struct Biol. 2000;7:957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- Bornscheuer U.T. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev. 2002;26:73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- Cheeseman J.D., Tocilj A., Park S., Schrag J.D., Kazlauskas R.J. Structure of an aryl esterase from Pseudomonas fluorescens. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:1237–1243. doi: 10.1107/S0907444904010522. [DOI] [PubMed] [Google Scholar]
- Chen C.‐S., Fujimoto Y., Girdaukas G., Sih C.J. Quantitative analyses of biochemical kinetic resolutions of enantiomers. J Am Chem Soc. 1982;104:7294–7299. [Google Scholar]
- Dunn G., Montgomery M.G., Mohammed F., Coker A., Cooper J.B., Robertson T. The structure of the C–C bond hydrolase MhpC provides insights into its catalytic mechanism. J Mol Biol. 2005;346:253–265. doi: 10.1016/j.jmb.2004.11.033. et al. [DOI] [PubMed] [Google Scholar]
- Ernst M.E., Ernst E.J., Klepser M.E. Levofloxacin and trovalfoxacin: the next generation of fluoroquinolones? Am J Health Syst Pharm. 1997;54:2569–2584. doi: 10.1093/ajhp/54.22.2569. [DOI] [PubMed] [Google Scholar]
- Farias S.T., Van Der Linden M.G., Rego T.G., Araujo D.A., Bonato M.C. Thermo‐search: lifestyle and thermostability analysis. In Silico Biol. 2004;4:30. [PubMed] [Google Scholar]
- Ferrer M., Golyshina O.V., Chernikova T.N., Khachane A.N., Martins dos Santos V.A.P., Yakimov M.M. Microbial enzymes mined from the Urania deep‐sea hypersaline anoxic basin. Chem Biol. 2005;12:895–904. doi: 10.1016/j.chembiol.2005.05.020. et al. [DOI] [PubMed] [Google Scholar]
- Foroumadil A., Mansouri S., Emami S., Mirzaei S., Sorkhi M., Saeid‐Adeli N., Shafieel A. Synthesis and antibacterial activity of nitroaryl thiadiazole‐Levofloxacin hybrids. Arch Pharm Chem Life Sci. 2006;339:621–624. doi: 10.1002/ardp.200600108. [DOI] [PubMed] [Google Scholar]
- Fu K.P., Lafredo S.C., Foleno B., Isaacson D.M., Barrett J.F., Tobia A.J., Rosenthale M.E. In vitro and in vivo antibacterial activities of Levofloxacin (l‐ofloxacin), an optically active ofloxacin. Antimicrob Agents Chemother. 1992;36:860–866. doi: 10.1128/aac.36.4.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S., Brown M.H., Skurray R.A. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa I., Atarashi S., Yokohama S., Imamura M., Sakano K.‐I., Furukawa M. Synthesis and antibacterial activities of optically active ofloxacin. Antimicrob Agents Chemother. 1986;29:163–164. doi: 10.1128/aac.29.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann B., Tolzer S., Pelletier I., Altenbuchner J., Van Pée K.‐H., Hecht H.‐J. Structural investigation of the cofactor‐free chloroperoxidases. J Mol Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]
- Holmquist M. Alpha/beta hydrolase fold enzymes: structures , functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., Kapatral V. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. et al. [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Dijkstra B.W., Reetz M.T. Bacterial biocatalysts: molecular biology, three‐dimensional structures and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- Jeon H.H., Kim J.T., Kang S.G., Lee J.‐H., Kim S.J. Characterization and its potential application of two esterases derived form the Arctic sediment metagenome. Mar Biotechnol. 2009;11:307–316. doi: 10.1007/s10126-008-9145-2. [DOI] [PubMed] [Google Scholar]
- Jones D.T. Protein secondary structure prediction based on position‐specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kang S.B., Ahn E.J., Kim Y. A facile synthesis of (S)‐(‐)‐7,8‐difluoro‐3,4‐dihydro‐3‐methyl‐2H‐1,4‐benzoxazine by zinc chloride assisted Mitsunobu cyclization reaction. Tetrahedron Lett. 1996;37:9317–9320. [Google Scholar]
- Kang S.B., Park S., Kim Y.H., Kim Y. An improved synthesis of levofloxacin. Heterocycles. 1997;45:137–145. [Google Scholar]
- Kennedy J., Marchesi J.R., Dobson A.D.W. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb Cell Factories. 2008;7:27. doi: 10.1186/1475-2859-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann A., Engel J. 4th. Georg Thieme; 2001. [Google Scholar]
- Koma D., Yamanaka H., Moriyoshi K., Ohmoto T., Sakai K. Overexpression and characterization of thermostable serine protease in Escherichia coli encoded by the ORF TTE0824 from Thermoanaerobacter tengcongensis. Extremophiles. 2007;11:769–779. doi: 10.1007/s00792-007-0103-0. [DOI] [PubMed] [Google Scholar]
- Lumry R., Eyring H. Conformational changes in proteins. J Phys Chem. 1954;58:110–120. [Google Scholar]
- Mehlhorn A.J., Brown D.A. Safety concerns with fluoroquinolones. Ann Pharmacother. 2007;41:1859–1866. doi: 10.1345/aph.1K347. [DOI] [PubMed] [Google Scholar]
- Miyadera A., Imura A. Enantioselective synthesis of a key intermediate of Levofloxacin using microbial resolution. Tetrahedron: Asymmetry. 1999;10:119–123. [Google Scholar]
- Moadebi S., Harder C.K., Fitzgerald M.J., Elwood K.R., Marra F. Fluoroquinolones for the treatment of pulmonary tuberculosis. Drugs. 2007;67:2077–2099. doi: 10.2165/00003495-200767140-00007. [DOI] [PubMed] [Google Scholar]
- Mois M., De Been M., Zwietering M.H., Moezelaar R., Abee T. Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ Microbiol. 2007;9:2933–2944. doi: 10.1111/j.1462-2920.2007.01404.x. [DOI] [PubMed] [Google Scholar]
- Nardini M., Dijkstra B.W. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. et al. [DOI] [PubMed] [Google Scholar]
- Olszewski M., Mickiewicz M., Kur J. Two highly thermostable paralogous single‐stranded DNA‐binding proteins from Thermoanaerobacter tengcongensis. Arch Microbiol. 2008;190:79–87. doi: 10.1007/s00203-008-0366-6. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Kim J.T., Kang S.G., Woo J.H., Lee J.‐H., Choi H.‐T., Kim S.‐J. A new esterase showing similarity to putative dienelactone hydrolase from a strict marine bacterium, Vibrio sp. GMD509. Appl Microbiol Biotechnol. 2007;77:107–115. doi: 10.1007/s00253-007-1134-2. [DOI] [PubMed] [Google Scholar]
- Qian Z., Field C.J., Yu Y., Lutz S. Recent progress in engineering α‐β hydrolase‐fold family members. Biotechnol J. 2007;2:192–200. doi: 10.1002/biot.200600186. [DOI] [PubMed] [Google Scholar]
- Robinson‐Rechavi M., Godzik A. Structural genomics of Thermotoga maritima proteins shows that contact order is a major determinant of protein thermostability. Structure. 2005;13:857–860. doi: 10.1016/j.str.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Ruiz J.M. Theoretical analysis of Lumry‐Eyring models in differential scanning calorimetry. Biophys J. 1992;61:921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M.A., Brennan R.G. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- Vieille B.R., Zeikus G.J. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao C., Meng B., Xie J., Zhou C., Chen X. The proteomic alterations of Thermoanaerobacter tengcongensis cultured at different temperatures. Proteomics. 2007;7:1409–1419. doi: 10.1002/pmic.200500226. et al. [DOI] [PubMed] [Google Scholar]
- White R.H. The difficult road from sequence to function. J Bacteriol. 2006;188:3431–3432. doi: 10.1128/JB.188.10.3431-3432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodyer R., Chen W., Zhao H. Outrunning nature: directed evolution of superior biocatalysts. J Chem Educ. 2004;41:81–126. [Google Scholar]
- Xiong Z., Jiang Y., Qi D., Lu H., Yang F., Yang J. The complete genome sequence of the extremophilic Bacillus cereus strain Q1 with industrial applications. J Bacteriol. 2009;191:1120–1121. doi: 10.1128/JB.01629-08. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu J., Zhou J., Ren Y., Dai X., Xiang H. Thermostable esterase from Thermoanaerobacter tengcongensis: high‐level expression, purification and characterization. Biotechnol Lett. 2003;25:1463–1467. doi: 10.1023/a:1025076121700. [DOI] [PubMed] [Google Scholar]
- Zhou X.‐X., Wang Y.‐B., Pan Y.‐J., Li W.‐F. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids. 2008;34:25–33. doi: 10.1007/s00726-007-0589-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Coomassie blue-stained protein gel of E. coli crude extracts separated on SDS-10% PAGE.
Fig. S2. Phylogenetic analysis of BcEST. The GenBank accession numbers for the respective proteins, annotated as either α/β-fold family hydrolase, 3-oxoadipate enol-lactonase, or in some cases, unnamed protein product or conserved hypothetical protein, are as follows: (1) ACK89548.1, (2) AAP27115.1, (3) AAS42215.1, (4) ACM13522.1, (5) AU17266.1, (6) AAT62319.1, (7) ACO28098.1, (8) ABY44194.1, (9) ACK96374.1, (10) BcEST (AAP10252.1), (11) ACK59771.1, (12) ABS22367.1, (13) ABX31253.1, (14) ACB86291.1, (15) ABV38133.1, (16) ABZ77297.1, (17) ACL19790.1, (18) ABX81126.1, (19) TtEST (AAM23832.1), (20) EAS01633.1, (21) EAR92522.1, (22) CAK56695.1, (23) BAH42062.1, (24) BAD39794.1, (25) ACL97306.1, (26) AAK25687.1, (27) ABI78668.1, (28) ABC62566.1, (29) ABS62973.1, (30) ABN96204.1, (31) ABG91905.1, (32) BAH48442.1, (33) ABK68474.1, (34) ABM15056.1, (35) ABP44844.1, (36) ACB02466.1 (1U2E), (37) ACC84816.1, (38) AAB60168.1 (1VA4), (39) AAC43253.1 (1A8Q). The tree was generated using MEGA4 (Kumar et al., 2008).
Fig. S3. Heat inactivation of BcEST and variant enzymes.
Fig. S4. Estimation of free energy of activation in the enzymatic reaction of (A) BcEST and variants and (B) TtEST.
Fig. S5. Tms of the BcEST and variant enzymes, and TtEST.
Fig. S6. Colorimetric assay of esterase activity using Phenol-Red.
A. Time-course of enzyme-induced colour shift of Phenol-Red.
B. Standard curve for the quantification of enzymatic reaction.
C. Linear increase of A435 over time due to esterase activity.
Table S1. Comparison of the substrate specificity of BcEST and TtEST
Appendix S1. Experimental procedures


