Abstract
Modern machine learning algorithms are increasingly being used in neuroimaging studies, such as the prediction of Alzheimer’s disease (AD) from structural MRI. However, finding a good representation for multivariate brain MRI features in which their essential structure is revealed and easily extractable has been difficult. We report a successful application of a machine learning framework that significantly improved the use of brain MRI for predictions. Specifically, we used the unsupervised learning algorithm of locally linear embedding (LLE) to transform multivariate MRI data of regional brain volume and cortical thickness to a locally linear space with fewer dimensions, while also utilizing the global nonlinear data structure. The embedded brain features were then used to train a classifier for predicting future conversion to AD based on a baseline MRI. We tested the approach on 413 individuals from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) who had baseline MRI scans and complete clinical follow-ups over 3 years with following diagnoses: Cognitive normal (CN; n= 137), stable mild cognitive impairment (s-MCI; n=93), MCI converters to AD (c-MCI, n=97), and AD (n=86). We found classifications using embedded MRI features generally outperformed (p < 0.05) classifications using the original features directly. Moreover, the improvement from LLE was not limited to a particular classifier but worked equally well for regularized logistic regressions, support vector machines, and linear discriminant analysis. Most strikingly, using LLE significantly improved (p = 0.007) predictions of MCI subjects who converted to AD and those who remained stable (accuracy/sensitivity/specificity: = 0.68/0.80/0.56). In contrast, predictions using the original features performed not better than by chance (accuracy/sensitivity/specificity: = 0.56/0.65/0.46). In conclusion, LLE is a very effective tool for classification studies of AD using multivariate MRI data. The improvement in predicting conversion to AD in MCI could have important implications for health management and for powering therapeutic trials by targeting non-demented subjects who later convert to AD.
Keywords: Alzheimer’s disease, locally linear embedding, statistical learning, classification of AD, MRI
1. Introduction
Machine learning methods have attracted considerable interest in recent years for analyzing neuroimaging data. The multivariate nature of machine learning algorithms overcomes limitations of traditional methods by using available information simultaneously to understand how variables jointly characterize data structures. One area of neuroimaging research where the use of machine learning has been growing rapidly is the prediction and early detection of Alzheimer’s disease (AD), the most frequent cause of age-related dementias (Brookmeyer et al., 2007). Machine learning has been explored for its potential to uncover subtle patterns of distributed brain tissue loss in AD that traditional methods may fail to detect (Stonnington et al., 2010; Vemuri et al., 2008; Fan et al., 2008; Filipovych and Davatzikos, 2011; Casanova et al., 2011; Davatzikos, 2004). In a typical example of machine learning applied to brain MRI, an algorithm is trained on a set of MRI features, such as regional volumes and cortical thickness, to create a classifier which predicts the correct diagnostic outcome for new observations. To reduce the high dimensionality of the MRI features and to stabilize predictions, shrinkage methods, such as principal components analysis or partial least squares have sometimes been added prior to the training step (Franke et al., 2010; Pelaez-Coca et al., 2011; Teipel et al., 2007; Phan et al., 2010). MRI data pose an especially challenging problem for predictions because of the difficulty in finding a good representation of brain features that makes them easily extractable and reveals their essential structure. The problem is perhaps most evident when it comes to choosing the optimal kernel function, or avoiding kernels all together, when transforming the data for feature extraction. Unless the transformation for a good representation of the brain MRI features is known a-priori, any arbitrary choice of remapping could potentially result in a loss of information and in inferior results.
We propose embedding the data into a system of linear coordinates of fewer dimensions prior to training a classifier to alleviate the problem of finding a good data representation for MRI based predictions. To relax assumptions of conventional approaches for reducing dimensionality, such as global linearity in principal components analysis (PCA) or particular kernel shapes in supervised learning algorithms (Westman et al., 2011; Koikkalainen et al., 2011; Eskildsen et al., 2013), we tested the use of local linear embedding (LLE) (Roweis and Saul, 2000). LLE neither relies on linear transformations nor requires supervised learning. The algorithm can transform the brain MRI data into a linear space of fewer dimensions by capturing local linear symmetries in the data while learning from the global nonlinear data structure. We also predict that accuracy will become less dependent on the particular choice of classifiers once the data are linearly embedded. Since LLE was introduced in 2000, the algorithm has been used successfully in a wide range of applications, including image classification (Ridder and Duin, 2002; Zheng and Jie, 2005), feature fusion (Sun et al., 2010), remote sensing (Ma et al., 2010), and face recognition (Sun et al., 2010; Chang and Yeung, 2006; Chen et al., 2007; Geng et al., 2005). More recently, LLE has also been used in MRI, e.g. for shape analysis of the hippocampus in AD (Yang et al., 2011), breast lesion segmentation (Akhbardeh and Jacobs, 2012), functional MRI (Chaillou et al., 2008; Mannfolk et al., 2010) and diffusion tensor imaging (Goh and Vidal, 2008). The overall goal of our study was to explore whether LLE benefits the classification of individuals with various levels of cognitive deficits, including AD, based on multivariate brain MRI data. Specifically, using baseline brain MRI data we aimed to determine the extent to which LLE improves the differentiation between subjects with mild cognitive impairments (MCI) who later decline to AD (within 3 years of baseline MRI) and subjects who remain stable in comparison to traditional methods that do not use LLE.
The rest of this paper is organized as follows. In Section 2, we describe the experimental data and also explain the theoretical basis of LLE in greater detail. In Section 3, we compare classifications using embedded MRI features of either regional brain volume, cortical thickness, or volume and thickness together, with classifications using the features directly. We also compare the classification performance from an LLE for different standard classifiers, i.e. elastic nets (EN) regularized regressions, support vector machine (SVM), and linear discriminant analysis (LDA). In Section 4, we interpret the findings and discuss limitations of the study.
2. Methods
2.1. Subjects and MRI
Subjects
Data used in the preparation of this article were obtained from the Alzheimers Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu).1
For the purpose of this study, we included all subjects recruited between 2005 and 2008 from the ADNI database who had 1.5T MRI scans, had successful evaluations of their MRIs using FreeSurfer (version 4.4) (Reuter et al., 2012), and also had a diagnosis over 3 years consistent with either stable cognitive normal (CN), stable mild cognitive impairment (s-MCI), MCI converters to AD (c-MCI), or AD. Subjects were excluded if their diagnosis reverted, e.g. MCI reversion to CN or AD reversion to MCI, or their MRI data was of insufficient quality for FreeSurfer processing. The final sample included 413 subjects total of which 137 were CN, 93 s-MCI, 97 c-MCI and 86 AD. The study codes of the subjects and other relevant information are provided as supplementary material to give readers the opportunity for direct comparisons with other methods. The subjects also had a battery of clinical and cognitive assessments each time they had their MRI scans. Each subject’s cognitive evaluation included: 1) The minimental state examination (MMSE) (Folstein et al., 1975), which provides a global measure of mental status and 2) the Alzheimer’s disease assessment scale-cognitive subscale (ADAS-Cog) with 11 test items, which provides a summary measure of disease severity by evaluating the most characteristic AD symptoms, such as disturbances in memory, language, attention and judgment (Mohs et al., 1997). We also used the clinical dementia rating sum of boxes (CDR-SOB) scale as a measure of cognitive and functional impairment (Morris, 1993). The CDR-SOB is a well validated instrument that assesses three domains of cognition (memory, orientation, judgement) and three domains of function (community affairs, home/hobbies, personal care) using structured interviews. More details about the tests can be found on the ADNI website www.loni.ucla.edu/ADNI. The average time of MCI conversion to AD was 19±8 months from baseline visit. A summary of the demographic and neurocognitive data of each group is provided in Table 1.
Table 1.
Group demographics and clinical summary
| CN | s-MCI | c-MCI | AD | p-value a | p-value b | |
|---|---|---|---|---|---|---|
| No. of subjects | 138 | 93 | 97 | 86 | ||
| Male (%) | 59.7 | 69.1 | 61.8 | 53.5 | 0.01c | 0.01c |
| APOE e4 carrier (%) | 28.3 | 48.4 | 68.0 | 68.6 | < 0.001 c | 0.14c |
| Baseline | ||||||
| Age (years) | 76±5 | 75±7 | 75±7 | 75±8 | 0.6 | 0.78 |
| MMSEd | 29.2±1 | 27.4±2 | 26.7±2 | 23.1±2 | < 0.001 | 0.02 |
| ADAS-Cog11e | 6±3 | 10±4 | 13±4 | 19±6 | < 0.001 | < 0.001 |
| LIMMTOTAL f | 14±3 | 8±3 | 7± 3 | 3±3 | < 0.001 | 0.02 |
| LDELTOTAL g | 13±3 | 4±3 | 3±3 | 1±2 | < 0.001 | < 0.001 |
| CDR-SOB h | 0±0 | 1±1 | 2±1 | 4±1 | < 0.001 | 0.001 |
| Annual rate of change | ||||||
| MMSE(×10−3) | −0.1±1 | −1±3 | −5±4 | −5±7 | < 0.001 | < 0.001 |
| ADAS-Cog11 | 0±3 | 1±6 | 5±8 | 7±9 | < 0.001 | < 0.001 |
| LIMMTOTAL | 1±2 | 0±2 | −1±1 | 0±1 | < 0.001 | < 0.001 |
| LDELTOTAL | 1±2 | 1±2 | −1±1 | 0±1 | < 0.001 | < 0.001 |
| CDR-SOB(×10−3) | 0±1 | 1±1 | 2±2 | 3±4 | < 0.001 | < 0.001 |
p-values indicate effects across the groups.
p-value indicate differences between s-MCI and c-MCI. The p-values for all pair-wise comparisons are provided in Supplementary Material.
Using Fisher exact test; all other tests using analysis of variance (ANOVA) (Fisher, 1925);
MMSE: Mini-mental state examination score (Folstein et al., 1975); total score range 0–30 with lower scores indicate greater cognitive impairment.
ADAS-Cog11: Alzheimer’s Disease Assessment Scale-cognitive subscale with 11 items (Mohs et al., 1997); total score range 0–70 with larger scores indicate greater impairment.
LIMMTOTAL: Memory score of immediate recall, maximal range 0–24, lower score indicate greater impairment (Wechsler, 2008).
LDELTOTAL: Memory score of delay recall, maximal range 0–25, lower score indicate greater impairment.
CDR-SOB: Clinical dementia rating sum of boxes, maximal range 0–18, higher score indicate greater impairment (Morris, 1993).
MRI acquisition and brain morphometry
The subjects underwent at each site the standardized 1.5 T MRI protocol of ADNI as described in http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml. Briefly, the protocol includes T1-weighted MRI based on a sagittal volumetric magnetization prepared rapid gradient echo (MP-RAGE) sequence (echo time/repetition time TE/TR= 4/9ms, 8° flip angle, 0.94×0.94×1.2 mm nominal resolution). Image quality and pre-processing was performed at a designated MRI center, as described in (Jack et al., 2008). The images were intensity-normalized, aligned to a brain atlas, skull-stripped, and segmented into regional volume using freely available FreeSurfer software, version 4.4 (http://surfer.nmr.mgh.harvard.edu/). For a more complete description of the FreeSurfer processing steps, see (Fischl et al., 2002; Fischl, 2004; Reuter et al., 2012). The outcome measures of the FreeSurfer work flow were 94 automatically labeled anatomical regions including cortical gyri and subcortical structures, yielding the volume of 94 brain regions and average cortical thickness of 68 regions for each subject. Accuracy of anatomical labeling was visually rated by experienced staff. If quality criteria were not met, the data were excluded from the analysis.
2.2. Locally Linear Embedding
Consider the region volume and cortical thickness across brain regions and subjects are arranged in matrix format X̂ ∈ Rn×D, where n is the number of subjects and D is the number of brain features. Because high-dimensional features often bear many redundancies and correlations that hide important relationships, we seek a more compact representation of X̂ prior to the classification of the subjects. In more detail, we use LLE to map the high-dimensional manifold X̂ ∈ Rn×D to one of lower dimensions Y = {y1, y2,…,yn} ∈ Rn×d, where d < D. The dimension reduction is accomplished in LLE based on the the principle idea that each point in a manifold and its nearest neighbors lie on or close to a patch whose local geometry is approximately linear even if the manifold is globally nonlinear. By recovering each point from its neighbors, LLE yields low-dimensional, neighborhood-preserving embeddings of the high-dimensional inputs, while learning from global relationships.
The LLE algorithm consists of three main steps:
Assign K neighbors to each data point x̂i = x̂i1, x̂i2,…,x̂iD. In the context of our study, each point represents a particular subject who is characterized by her/his individual features of brain regional volume and/or cortical thickness. Most commonly, nearest neighbors are defined using Euclidean distance or normalized dot products. We use the nearest neighbors in Euclidean space in our study.
- Compute the weights Wi that best recover linearly the subject’s data x̂i from the nearest neighbors by minimizing the error function:
In order to linearly reconstruct data point x̂i from its nearest neighbors, there are several constraints on the weights: Σk wik = 1, and wik = 0 if k ∉ N(i).N represents nearest neighbors.(1) - Compute the low-dimensional embeddings yi that best estimate the local data ỹi given the weights Wi:
with respect to yi ∈ Rd. The cost function can be rewritten as:(2)
where M = (I – W)T(I – W) and with the following two constraints on Y. First, YTY = I, so that the solution is of rank d. Second, Σi Yi = 0, to ensure that the embedding is centered on the local origin of the original feature space coordinates. The third step can be reduced to an optimization problem according to:(3) (4)
This is an eigenvalue problem, and also the step of LLE learning the global structure of the data. All the eigenvectors of M are solutions of Y, but the eigenvectors corresponding to the smallest eigenvalues minimize the cost function in Eq. 2. After discarding the smallest eigenvalue, which will always be zero, the next n – 1 smallest eigenvalues are used as dimensions of the transformed data (the smallest given the first dimension for each point in the data, the next smallest the 2nd dimension and so on).
After LLE, three different classifiers were compared for performing pair-wise classifications. We trained regularized logistic regressions with elastic nets (EN)(Hastie et al., 2009), support vector machines (SVM) (Cortes and Vapnik, 1995) as well as a linear discriminant analysis (LDA) to predict future diagnosis (within 3 years) from baseline MRI scans. Since LLE provides embedded features globally in linear coordinates, we limited the tests to linear classifiers, including SVM with a linear kernel only. We implemented the EN classifier using the MATLAB toolbox Glmnet (Friedman et al., 2010) and SVM and LDA using the respective MATLAB build-in functions. The optimal thresholding algorithm by Otsu (Otsu, 1979) was used to obtain binary classification results. Although automated methods for the optimal numbers of neighbors K and corresponding dimensionality D have been proposed (Kayo, 2006), in this study we choose to tune K and D by cross-validation in combination with cross-validations of the parameters of the linear classifiers. We used leave-one-out (LOO) cross validation and evaluated classification performance in terms of a receiver operator characteristic (ROC) analysis for parameter optimization. Measures of sensitivity and specificity, followed by computation of overall classification accuracy were obtained. Note, LLE can remain outside the LOO loop, since the algorithm performs dimension reduction unsupervised, i.e. without the requirement of labeling a training set. LOO was performed exactly once for each subject and per comparison hence no bias was introduced at the subject level. For each classification problem, the cross-validated results were then used to build a ROC curve using the pROC package in R and augmented by a stratified bootstrap with 2000 repetitions to obtain a distribution.
3. Experimental Results
The demographic and clinical characteristics of the study groups are shown in Table 1. Differences between the groups were tested using t-tests or Chi-squared tests, as appropriate. Age and gender variations across the groups were well matched. As expected, the groups differed significantly in their respective levels of cognitive deficits at baseline as well as in the annual rates of cognitive decline, based on MMSE and ADAS-Cog11 scores.
Visualization
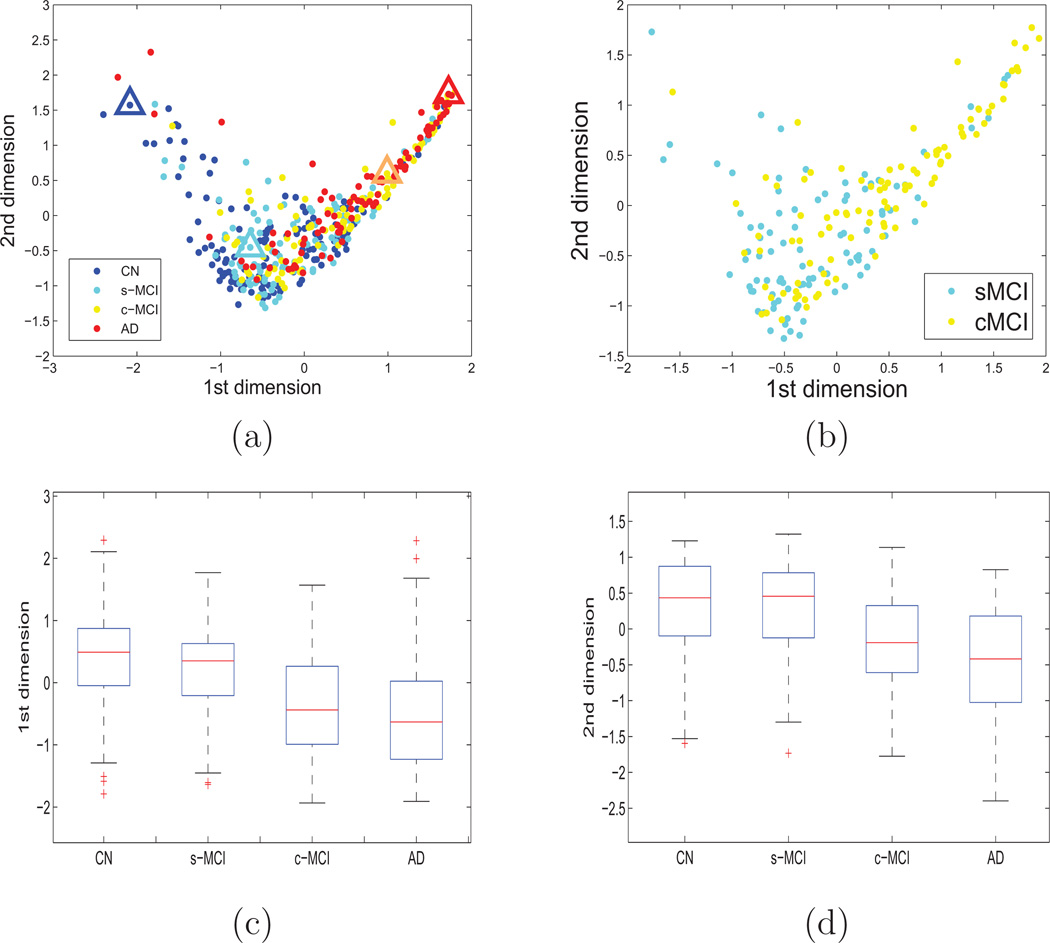
Figure 1(a) shows the locations of all 413 subjects in an embedded feature space of 162 brain features, which include 94 regional brain volume and 68 cortical thickness values, reduced to two dimensions. The figure demonstrates a distinct clustering of the healthy subjects shown in blue and the AD patients shown in red. Furthermore, the c-MCI subjects (yellow) appear on average localized near the AD patients, whereas s-MCI subjects (cyan) appear closer to the CN subjects. The separation between s-MCI and c-MCI subjects in LLE space is depicted in more detail in Figure 1(b). In addition, the distributions of the subject locations along the first and second LLE dimension are depicted in Figure 1(c) and (d), respectively and separately by group.
Figure 1.
Visualization of all 413 subjects in LLE space based on brain volume and cortical thickness. Only the first two dimensions of the LLE space are shown for visualization purpose. Color indicates diagnosis (blue = normal control, cyan = s-MCI, yellow = c-MCI, and red = AD). Part (a) shows all four goups’ subjects in the embedded space, (b) depicts the separation between s-MCI and c-MCI in greater detail; (c) and (d) show the distribution of subject locations by group along the first and second LLE dimension, respectively.
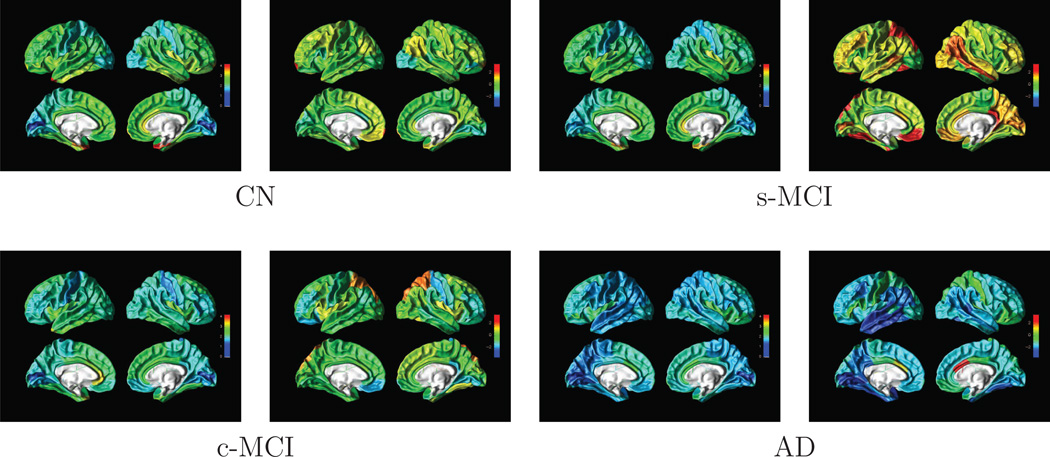
To demonstrate benefit of LLE for data visualization, we selected an example subject from each group based on the subject’s LLE coordinates and then compared the patterns of regional brain volume and cortical thickness. The example subjects were selected based on their extreme location in the embedded space within each population cluster to visualize the variability in MRI features for the classification problem. The triangles in Figure 1 indicate the locations of these example subjects in LLE space. The patterns of cortical thickness and regional brain volume of each of these subjects are depicted on surface rendered brain plots in Figure 2. Note, however, that from the locations in LLE space alone it remains uncertain whether the differences are related to the subjects’ diagnosis or individual variability in atrophy. Cooler colors indicate a thinner than average cortical thickness or smaller than average volume whereas warmer colors indiate a thicker than average cortical thickness or larger than average volume. Together, Figures 1–2 indicate that the locations of the subjects in LLE space reflect generally their differences in cortical thickness and regional brain volumes. Specifically, comparing the surface rendered plots of the control subject and AD patient shown in Figure 2(a), we can see a substantial thinning in parietal lobe, temporal lobe and temporal lobe in the AD patient compared to the control subject. Similarly, comparing the plot of the c-MCI subject in Figure 2, whose location in LLE space is closer to that of the AD patient than to the CN subject as well as to s-MCI subject, has in general a thinner cortex than the CN and s-MCI subjects but still a thicker cortex than the AD patient. In particular, this c-MCI subject has a thicker temporal cortex than the AD patient, while differences in other regions between the two are more subtle. Also of note in these plots from 4 individual subjects is the level of individual variability in atrophy.
Figure 2.
Surface rendered illustrations of cortical thickness and z-score normalized regional volumes from four example subjects, one from each group. The location of each subject in LLE space is indicated by a color coded triangle in Figure 1. For each subject, the left panel illustrates cortical thickness and the right panel illustrates normalized regional volumes. Cooler colors indicate a thinner cortex and a smaller regional volume than average; whereas warmer colors indicate a thicker cortex and a larger regional volume than average.
Classifications
Tables 2– 4 summarize group classifications with and without LLE embedding, separately for the three different classifiers. Classifications differentiating s-MCI from c-MCI patients based on baseline MRI are highlighted. Overall, classifications using LLE features performed significantly better (p<0.05) or at least as good as classifications that used the features directly. Most importantly, almost all predictions of the MCI subjects who later converted to AD and those who remained stable became reliable with the use of LLE (exception SVM+LLE using thickness, see Table 3 in comparison to the same classifications without LLE that performed not better than by chance. The improvement in classification was driven by the LLE transformations of brain volume features and to a lesser extent by cortical thickness. Although it may seem a surprise that the classification between AD and CN based on linear SVM and brain volumes was not better than by chance (see Table 3), one needs to note that we deliberately chose a linear kernel and accepted irrelevant features in the SVM for a fair comparison with LLE. Together, these steps may have sacrificed SVM performance.
Table 2.
Accuracy of classifications based on elastic net regression with and without LLE feature embeddings
| CN vs. s-MCI | CN vs. c-MCI | CN vs. AD | s-MCI vs. c-MCI | s-MCI vs. AD | c-MCI vs. AD | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ||
| Volumes | EN | 0.56a | 0.59 | 0.54 | 0.78 | 0.79 | 0.77 | 0.82 | 0.85 | 0.80 | 0.46a | 0.48 | 0.44 | 0.59a | 0.70 | 0.48 | 0.50a | 0.45 | 0.54 |
| EN+LLE | 0.64 | 0.53 | 0.72 | 0.81 | 0.80 | 0.82 | 0.87 | 0.80 | 0.91 | 0.69 | 0.66 | 0.72 | 0.77 | 0.85 | 0.69 | 0.51a | 0.52 | 0.51 | |
| p-valueb | 0.09 | 0.02 | 0.01 | <0.001 | <0.001 | 0.62 | |||||||||||||
| Cortical thickness |
EN | 0.55a | 0.53 | 0.57 | 0.75 | 0.69 | 0.80 | 0.83 | 0.78 | 0.86 | 0.56a | 0.53 | 0.59 | 0.67 | 0.79 | 0.56 | 0.59a | 0.69 | 0.51 |
| EN+LLE | 0.62 | 0.57 | 0.66 | 0.78 | 0.79 | 0.78 | 0.85 | 0.79 | 0.89 | 0.61 | 0.48 | 0.74 | 0.71 | 0.78 | 0.65 | 0.58a | 0.70 | 0.48 | |
| p-valueb | 0.09 | 0.03 | 0.09 | 0.54 | 0.37 | 0.39 | |||||||||||||
| Thickness +volumes |
EN | 0.55a | 0.53 | 0.57 | 0.75 | 0.67 | 0.80 | 0.80 | 0.73 | 0.84 | 0.56a | 0.65 | 0.46 | 0.60 | 0.59 | 0.61 | 0.48a | 0.67 | 0.30 |
| EN+LLE | 0.64 | 0.65 | 0.63 | 0.82 | 0.81 | 0.82 | 0.90 | 0.86 | 0.93 | 0.68 | 0.80 | 0.56 | 0.75 | 0.77 | 0.73 | 0.56a | 0.51 | 0.61 | |
| p-valueb | 0.01 | <0.001 | <0.001 | 0.007 | <0.001 | 0.02 | |||||||||||||
indicates classification is not better than by chance (p>0.05), whereas no superscripts indicate a significant classification;
p-value of improvement with LLE compared to non-LLE.
Table 4.
Accuracy of classifications based on linear discriminant analysis with and without LLE feature embeddings
| CN vs. s-MCI | CN vs. c-MCI | CN vs. AD | s-MCI vs. c-MCI | s-MCI vs. AD | c-MCI vs. AD | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ||
| Volumes | LDA | 0.56a | 0.60 | 0.54 | 0.77 | 0.75 | 0.78 | 0.83 | 0.85 | 0.82 | 0.46a | 0.48 | 0.44 | 0.58a | 0.66 | 0.51 | 0.49a | 0.45 | 0.53 |
| LDA+LLE | 0.59 | 0.56 | 0.61 | 0.81 | 0.79 | 0.83 | 0.84 | 0.77 | 0.89 | 0.61 | 0.70 | 0.62 | 0.67 | 0.72 | 0.62 | 0.45a | 0.47 | 0.43 | |
| p-valueb | 0.59 | 0.01 | 0.14 | 0.08 | 0.05 | 0.88 | |||||||||||||
| Cortical thickness |
LDA | 0.55a | 0.54 | 0.57 | 0.73 | 0.66 | 0.78 | 0.84 | 0.81 | 0.86 | 0.57a | 0.54 | 0.60 | 0.66 | 0.80 | 0.53 | 0.56a | 0.58 | 0.55 |
| LDA+LLE | 0.69 | 0.67 | 0.71 | 0.78 | 0.79 | 0.77 | 0.85 | 0.79 | 0.77 | 0.65 | 0.52 | 0.56 | 0.66 | 0.71 | 0.61 | 0.58a | 0.70 | 0.48 | |
| p-valueb | 0.002 | 0.07 | 0.61 | 0.04 | 0.79 | 0.66 | |||||||||||||
| Thickness +volume |
LDA | 0.55a | 0.56 | 0.54 | 0.67 | 0.69 | 0.65 | 0.69 | 0.62 | 0.73 | 0.51a | 0.57 | 0.45 | 0.58a | 0.63 | 0.54 | 0.45a | 0.43 | 0.46 |
| LDA+LLE | 0.51a | 0.48 | 0.49 | 0.75 | 0.69 | 0.80 | 0.89 | 0.86 | 0.91 | 0.68 | 0.82 | 0.53 | 0.65 | 0.70 | 0.60 | 0.57a | 0.51 | 0.63 | |
| p-valueb | 0.89 | <0.001 | <0.001 | 0.01 | 0.03 | 0.15 | |||||||||||||
indicates classification is not better than by chance (p>0.05), whereas no superscripts indicate a significant classification;
p-value of improvement with LLE compared to non-LLE.
Table 3.
Accuracy of classifications based on support vector machine with and without LLE feature embeddings
| CN vs. s-MCI | CN vs. c-MCI | CN vs. AD | s-MCI vs. c-MCI | s-MCI vs. AD | c-MCI vs. AD | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ACR | SEN | SPE | ||
| Volume | SVM | 0.55a | 0.60 | 0.52 | 0.52a | 0.40 | 0.60 | 0.53a | 0.49 | 0.55 | 0.51a | 0.54 | 0.48 | 0.50a | 0.56 | 0.45 | 0.49a | 0.38 | 0.58 |
| SVM+LLE | 0.64 | 0.52 | 0.72 | 0.81 | 0.80 | 0.82 | 0.85 | 0.86 | 0.85 | 0.67 | 0.74 | 0.59 | 0.76 | 0.86 | 0.67 | 0.54a | 0.86 | 0.67 | |
| p-valueb | 0.08 | 0.01 | 0.03 | 0.001 | <0.001 | 0.67 | |||||||||||||
| Cortical thickness |
SVM | 0.57a | 0.46 | 0.64 | 0.72 | 0.66 | 0.77 | 0.86 | 0.79 | 0.91 | 0.56a | 0.52 | 0.61 | 0.72 | 0.78 | 0.66 | 0.60 | 0.62 | 0.58 |
| SVM+LLE | 0.62 | 0.53 | 0.68 | 0.78 | 0.82 | 0.74 | 0.84 | 0.78 | 0.88 | 0.56a | 0.45 | 0.58 | 0.69 | 0.71 | 0.67 | 0.56a | 0.72 | 0.41 | |
| p-valueb | 0.1 | 0.03 | 0.85 | 0.85 | 0.5 | 0.34 | |||||||||||||
| Thickness +volume |
SVM | 0.45a | 0.65 | 0.32 | 0.52a | 0.41 | 0.60 | 0.50a | 0.48 | 0.51 | 0.53a | 0.66 | 0.49 | 0.57a | 0.48 | 0.51 | 0.57a | 0.48 | 0.66 |
| SVM+LLE | 0.64 | 0.58 | 0.67 | 0.80 | 0.72 | 0.86 | 0.90 | 0.87 | 0.92 | 0.66 | 0.75 | 0.56 | 0.73 | 0.73 | 0.72 | 0.57a | 0.48 | 0.65 | |
| p-valueb | 0.01 | <0.001 | <0.001 | 0.008 | <0.001 | 0.90 | |||||||||||||
indicates classification is not better than by chance (p>0.05), whereas no superscripts indicate a significant classification
p-value of improvement with LLE compared to non-LLE.
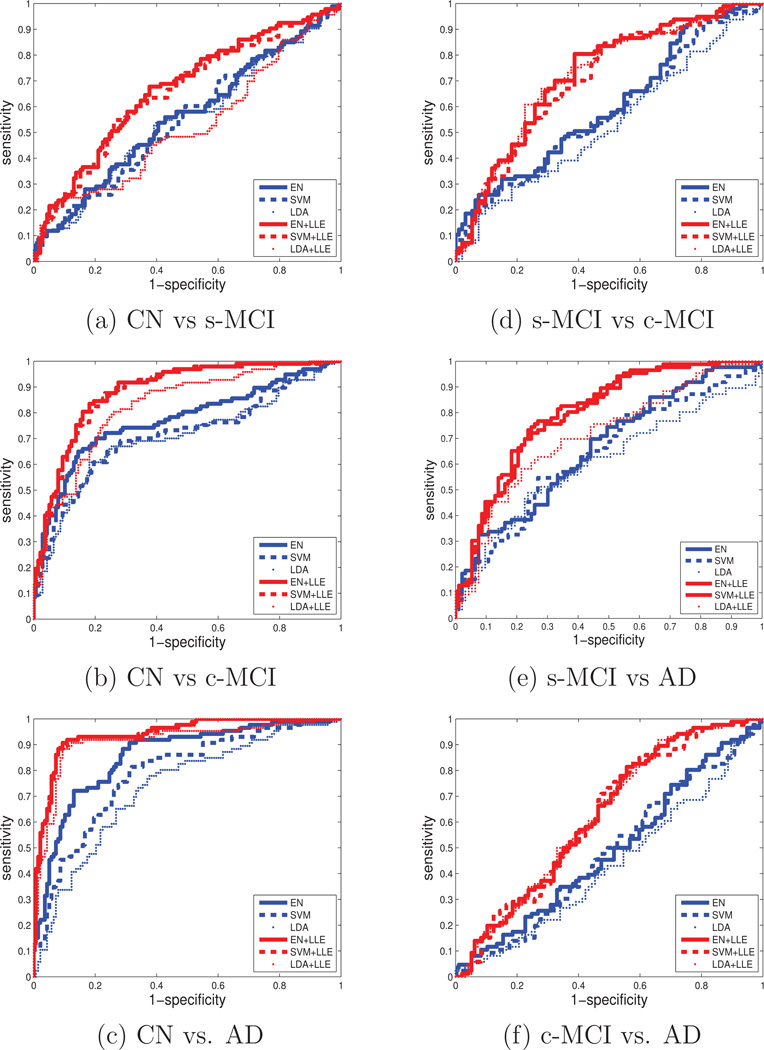
A summary illustration of the improvements with LLE is shown in Figure 3, separately for each pairwise classification. ROC curves are shown from cross-validated classifications, separately for each classifier (EN, SVM, or LDA) when both regional volume and cortical thickness features were used together with and without LLE. The figures indicate classifications using LLE improved across the board. The figure also indicates that the improvements were achieved regardless of the type of classifier.
Figure 3.
ROC curves of pairwise classifications with and without feature embeddings by LLE and separately using as classifier either elastic net (EN) regularized linear regression, a support vector machine (SVM) with a linear kernel, or linear discriminant analysis (LDA). The features are brain regional volume and cortical thickness used together. Accuracy of the classifications generally improves significantly with LLE features, regardless which classifier EN, SVM or LDA is used. FPR stands for false positive rate, and TPR stands for true positive rate.
Since manifold learning can be sensitive to sample size, we also tested the robustness of LLE with fewer samples. Specifically, we repeated the predictions of MCI conversion with 20% and 50% fewer samples by removing subjects at random while still maintaining the group balance. We performed the tests using the EN classifier with or without LLE. Accuracy (and similarly AUC) decreased progressively from 0.68 at 100% of the sample to as low as 0.60 at 50% (AUC decrease: 0.72 at 100% to 0.63 at 50%), as expected. However, the decline was not worse and still remained better in comparison to predictions without LLE with fewer samples (accuracy change: 0.51 at 100% and 0.57 at 50%; AUC change: 0.53 at 100% and 0.61 at 50%).These results indicate that LLE does not exacerbate the sensitivity of manifold learning to sample size.
Lastly, we compared the benefit of LLE for predicting MCI conversion with traditional MRI and clinical methods in terms of accuracy and AUC values. The MRI methods included feature reduction by PCA, a nonlinear SVM using a Gaussian kernel, and selection of region-of-interest (ROI), e.g. the hippocampus or entorhinal cortex. The clinical measures included MMSE and ADAS-Cog11 scores. The results are summarized in Table 5 and ranked from best to worst accuracy. The outcome shows that LLE performance ranked at the top.
Table 5.
Comparison of MRI and clinical measures for prediction of MCI conversion to AD
| Method | Measures | Classifier | ACR | AUC | p-value | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MRI: whole brain | volume+thickness | LLE+LDA | 0.68 | 0.72 | – | ||||||||||||||
| MRI: ROI | hippocampus | 0.66 | 0.69 | 0.19 | |||||||||||||||
| hippocampus +entorhinal corex | LDA | 0.65 | 0.69 | 0.26 | |||||||||||||||
| entorhinal cortex | 0.61 | 0.62 | 0.08 | ||||||||||||||||
| MRI: whole brain | volume+thickness | SVM (Gaussian) | 0.61 | 0.62 | < 0.001 | ||||||||||||||
| neuropsychological scores | ADAS-Cog11+LDEL+CDR+MMSE | 0.62 | 0.69 | 0.27 | |||||||||||||||
| ADAS-Cog11 | 0.59 | 0.67 | 0.14 | ||||||||||||||||
| LDEL | LDA | 0.60 | 0.61 | 0.01 | |||||||||||||||
| CDR | 0.56 | 0.52 | < 0.001 | ||||||||||||||||
| MMSE | 0.57 | 0.54 | < 0.001 | ||||||||||||||||
| MRI: whole brain | volume+thickness | SVM (linear) | 0.53 | 0.58 | 0.002 | ||||||||||||||
| PCA+LDA | 0.51 | 0.53 | < 0.001 | ||||||||||||||||
The results are ranked by accuracy values from high to low.
p-values indicate prediction performance compared with LLE.
All predications used a LDA classifier. ROI = region of interest; PCA = principal component analysis; all other abbreviations are explained in the text.
4. Discussion
In this study, we presented a new approach for classification which transformed MRI features to a linear space of fewer dimensions using LLE, prior to training a classifier. We applied this approach to high-dimensional classifications, especially for the differentation between MCI subjects who remain stable and those who decline to AD based on brain volume and cortical thickness measurements at baseline. The three main findings were: First, LLE generally improved classifications, implying that conventional classification methods that exploited original features without embedding, were not optimal. Second, the improvements in classifications with LLE worked equally well for three types of frequently used linear classifiers: EN, SVM and LDA. This result implied that the benefit of LLE was generally valid and not confined to a particular classifier class. Third, and perhaps most important clinically, was that LLE significantly improved the differentiation between MCI subjects who converted to AD within 3 years from baseline MRI and those who remained stable. In contrast, the same differentiation performed without LLE achieved outcomes that were not better than by chance. Taken together, the results suggested that machine learning methods of AD classification using brain MRI should incorporate LLE.
The finding of a general improvement in high-dimensional classifications of MCI and AD when embeddings of the features are used implies that alterations of regional brain volume and cortical thickness as seen on MRI can involve inherently nonlinear feature structures. Training linear classifiers directly on MRI features may therefore under-utilize the classification power, since these classifiers cannot capture nonlinearity. One might argue that even nonlinear classifiers may sacrifice classification power, if the nonlinear feature structure is not adequately modeled. LLE, on the other hand, generally benefits classifications not only by reducing the dimensionality of features but also by embedding them into linear coordinates while learning their potential nonlinear characteristics. In the context of brain imaging, linear embedding is consistent with the reasonable assumption that subjects who are clinically similar will also have similar distributions of brain features even if the features across all subjects is globally nonlinear. LLE is therefore crucial for maximizing classifications based on structural MRI data. This point is further supported by our finding that LLE outperformed popular traditional MRI methods, including PCA and nonlinear SVM.
However, LLE is not the only algorithm for linear embedding. Various other techniques have been proposed over the years, including high-dimensional scaling (Lespinats et al., 2007), maximum variance unfolding (Weinberger and Saul, 2004), Isomap (Tenenbaum et al., 2000), Laplacian eigenmaps (Belkin and Niyogi, 2001), and nonlinear PCA (Scholz et al., 2005). These algorithms can differ in performance, depending on the data dimensionality, noise, and sampling uniformity among other factors. The extent to which LLE performs better or worse than other embedding algorithms will be a subject of future research. In this study, we choose LLE for two reasons. First, there is only one parameter which needs to be tuned in LLE: the number of nearest neighbors. Although algorithms for automatic optimal tuning this parameter are available (Kayo, 2006), we performed optimization by cross-validation to demonstrate the robustness of LLE. Second, LLE, being rooted in the concept of unsupervised statistical learning, has a non-iterative solution, thus avoiding the convergence to local minima that often plaque iterative techniques (Roweis and Saul, 2000). In addition, we verified that LLE does not exacerbate the sensitivity of manifold learning to sample size.
Compared to other structural MRI studies predicting conversion to AD, LLE performance was among the best (Chupin et al., 2009; Querbes et al., 2009; Davatzikos et al., 2011; Westman et al., 2011; Wolz et al., 2011; Cho et al., 2012; Eskildsen et al., 2013; Suplber et al., 2010). Findings of conversion to AD over a period of at least 30 months are summarized in Table 6. It shows that our LLE approach was only surpassed by studies which had either a much smaller sample size (Ferrarini et al., 2009; Plant et al., 2010) or a much smaller ratio of non-converters to converters (Misra et al., 2009; Koikkalainen et al., 2011). Although comparisons between studies are limited since the number of subjects and processing methods vary, we like to point out that in our study in contrast to most others - the ratio of converters to non-converters was much more balanced. A greater balance is expected to yield better measures of accuracy, since an unbalance can induce bias toward the minority group (Imam et al., 2006). Let alone, we used a much larger sample size (93+97=190), compared with the other three studies. The accuracy with LLE was also surpassed by two MRI studies that used hippocampal shape as predictor for conversion to AD. Although one of the studies (Ferrarini et al., 2009) had a much smaller sample size compared to our study and the other (Costafreda et al., 2011) had only 12 months follow-up, their findings suggest that shape is potentially an addition feature to volume and thickness for LLE. The only other MRI reports (not listed in Table 6) which outperformed LLE were studies that used MRI together with either other imaging modalities (Zhang and Shen, 2012), or cognitive scores, or CSF biomarkers (Hinrichs et al., 2011; Zhang et al., 2011; Vemuri et al., 2008).
Table 6.
Other MRI studies predicting MCI conversion beyond 30 months.
| Study | Data open source | Data type | Region | sMCI/cMCI | Follow-up (months) | ACR | SEN | SPE |
|---|---|---|---|---|---|---|---|---|
| Misra et al. (2009) | ADNI | VBM (GM and WM) | Whole Brain | 76/27 | 0–36 | 82% | - | - |
| Ferrarini et al. (2009) | N/A | Shape | Hippocampus | 15/15 | 0–33 | 80% | 90% | 80% |
| Plant et al. (2010) | N/A | VBM | Whole Brain | 15/9 | 0–30 | 75% | 56% | 87% |
| Koikkalainen et al. (2011) | ADNI | TBM | Whole Brain | 215/154 | 0–36 | 72% | 77% | 71% |
| Our Study | ADNI | VBM | Whole Brain | 93/97 | 0–36 | 69% | 66% | 72% |
| Wolz et al. (2011) | ADNI | TBM+CTH+VBM+CSF | Whole Brain | 238/167 | 0–48 | 68% | 67% | 69% |
| Suplber et al. (2010) | AddNeuroMed+ADNI | CTH+VBM | Whole Brain | 261/173 | 0–36 | 68% | 70% | 67% |
| Eskildsen et al. (2013) | ADNI | CTH | Whole Brain | 227/161 | 0–36 | 67% | 66% | 68% |
AddNeuroMed is an Integrated Project funded by the European Union Sixth Framework program.
VBM represents voxel based morphometry, GM: gray matter, WM: white matter, TBM: tensor based morphometry, CTH: cortical thickness, CSF: cerebrospinal fluid.
The comparison of LLE performance with ROI methods is also interesting and worth a discussion. Some MRI studies have questioned the benefit of machine learning methods over ROI methods for predicting AD (Cuingnet et al., 2011; Chu et al., 2012). Our results are generally in agreement with these studies in that using the volume of the hippocampus (or the entorhinal cortex) alone for AD prediction remained competitive to using LLE. On the other hand, the classification accuracy for AD patients decreased substantially in our data using the hippocampus alone compared to classifications using LLE. The result suggests that ROI methods may not work always well across various disease stages. For intermediate disease stages, the use of multiple ROIs has been proposed as alternative to using the hippocampus alone (Cuingnet et al., 2011). Here, however, LLE has a conceptual advantage over multiple ROI methods, since the algorithm learns unsupervised global MRI features and can therefore detect changes in the brain without specific input how the disease spreads.
Aside from the advantage of LLE for MRI based predictions, LLE also outperformed predictions based on the clinical MMSE and ADAS-Cog11 scores. The result further supports the benefit of LLE for predicting conversion and also justifies the use of this sophisticated methodology as substitute for simpler clinical tests. Whether or not the use of LLE helps to improve such predictions over longer periods than 36 months, when disease progression is slower and MRI effects are presumably more subtle, needs to be determined.
Another observation requiring further investigation is that LLE improved classification accuracy unequally for volume and cortical thickness with volume often outperforming cortical thickness in classifications. The reason for this outcome is not clear but there are several explanations. One explanation is that there is much less global structure in the thickness data than in volumes to regulate the learning step in LLE according to Equation 4. This can result in the diminished separation of subjects who otherwise differ in cortical volume.
A potentially practical benefit of LLE is that it can help readers to perceive similarities in high-dimensional data from visualization. In the context of this study and as illustrated in Fig. 1, individuals can be localized relative to others in an embedded feature space of few dimensions. As seen in Fig. 1 the c-MCI subjects appear on average at locations close to AD patients, whereas s-MCI subjects appear at locations closer to controls. The visualization of the features in a low-dimensional embedded space can also help to design more efficient classification schemes by keeping only the most important dimensions, e.g. the ones that hold the most information to identify subjects.
One limitation of the study is lack of confirmed diagnosis of AD. Another limitation is that Freesurfer was used for MRI pre-processing, which involved excluding data of substandard quality. The extent to which LLE performs better or worse in combination with other pre-processing software that has less stringent quality requirements than Freesurfer warrants further tests. In addition, LLE sacrifices simple means to track which brain features contributed to the classifications. Recovering this critical information is in principle feasible by exploring the distribution of the weights in Eq. 1, but this requires further investigations and it is beyond the scope of the present work. In addition, the dimensionality of features was restricted to the number of anatomical brain regions that FreeSurfer provides. Potentially, all MRI voxels or vertices could be used for a more comprehensive classification analysis. The performance of LLE as dimensionality increases is also a subject for future investigations. Although the LLE approach is promising, more studies are warranted to understand in greater detail the susceptibility of LLE to outliers, skewed distributions, and noise in MRI data. Improved LLE versions that are more robust to noise and outliers have been proposed (Yin et al., 2007; Wang et al., 2006, 2011), but their benefit for MRI needs to be determined in future studies. It should be noted that the stable MCIs and MCI converters differed already at baseline in major clinical measures and not in MRI alone, as indicated in Table 1. Further studies on a population with less advanced symptoms are warranted to test the full benefit of LLE for predictions.
In summary, we introduced LLE to improve classifications based on high-dimensional MRI data of regional brain atrophy. In combination with linear classifiers such as regularized logistic regression, support vector machine, and linear discriminant analysis, LLE significantly improved classification accuracy, especially for predictions of which MCI subjects would convert to AD and which one would remain stable based on baseline MRI scan. We conclude, LLE is a very effective tool for classification studies of AD using multivariate MRI data. The improvement in predicting conversion to AD in MCI could have important implications for health management and for powering therapeutic trials by targeting non-demented subjects who later convert to AD.
Supplementary Material
Highlights.
Locally linear embedding (LLE) is an unsupervised learning algorithm.
It was used to extract characteristic MR features of brain alternations.
It was used to classify normal aging subjects, MCI and AD patients from ADNI data.
The performance of predicting AD in MCIs was significantly improved by using LLE.
LLE benefitted various classifiers, such as SVM, LDA and regularized regressions.
Acknowledgment
We thank Mr. Raptentsetsang, Ms. Miriam Hartig and Ms. Diana Truran-Sacrey for their dedicated work in processing the images through FreeSurfer. This work was funded in part by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) [T32 EB001631-05], and by a research resource grant from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (grant: P41 RR 023953). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimers Association; Alzheimers Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is Rev March 26, 2012 coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. The work was possible by using resources of the Veterans Affairs Medical Center, San Francisco, California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitive normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see www.adni-info.org.
References
- Akhbardeh A, Jacobs M. Comparative analysis of nonlinear dimensionality reduction techniques for breast mri segmentation. Medical Physics. 2012;39:2275–2289. doi: 10.1118/1.3682173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin M, Niyogi P. Advances in Neural Information Processing Systems 14. MIT press; 2001. Laplacian eigenmaps and spectral techniques for embedding and clustering; pp. 585–591. [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi H. Forecasting the global burnden of alzheimer’s disease. Alzheimer Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Casanova R, Whitlow C, Wagner B, Williamson J, Shumaker S, Maldjian J, Espeland M. High dimensional classification of structural mri alzheimer’s disease data based on large scale regularization. Front Neuroinform. 2011:5. doi: 10.3389/fninf.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou L, Yetik IS, Wernick MN. New methods for fMRI data processing based on locally linear embeddings. Proceedings of the SPIE 6814, 681412-681412-9. 2008 [Google Scholar]
- Chang H, Yeung DY. Robust locally linear embedding. Pattern Recognition. 2006;39:1053–1065. [Google Scholar]
- Chen J, Wang R, Yan S, Shan S, Chen X, Gao W. Enhancing human face detection by resampling examples through manifolds. Trans. Sys. Man Cyber. 2007;Part A 37:1017–1028. [Google Scholar]
- Cho Y, Seong JK, Jeong Y, Shin SY. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. NeuroImage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Hsu AL, Chou KH, Bandettini P, Lin C. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. NeuroImage. 2012;60:59–70. doi: 10.1016/j.neuroimage.2011.11.066. [DOI] [PubMed] [Google Scholar]
- Chupin M, Gérardin E, Cuingnet R, Boutet C, Lemieux L, Lehéricy S, Benali H, Garnero L, Colliot O. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- Costafreda S, Dinov I, Tu Z, et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. NeuroImage. 2011;56:212–219. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. NeuroImage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Why voxel-based morphometric analysis should be used with great caution when characterizing group differences. NeuroImage. 2004;23:17–20. doi: 10.1016/j.neuroimage.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of aging. 2011;32 doi: 10.1016/j.neurobiolaging.2010.05.023. 2322.e19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen SF, Coup P, Garca-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Prediction of alzheimer’s disease in subjects with mild cognitive impairment from the adni cohort using patterns of cortical thinning. NeuroImage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini L, Frisoni GB, Pievani M, Reiber JH, Ganzola R, Milles J. Morphological hippocampal markers for automated detection of alzheimer’s disease and mild cognitive impairment converters in magnetic resonance images. Journal of Alzheimer’s Disease. 2009;17:643–659. doi: 10.3233/JAD-2009-1082. [DOI] [PubMed] [Google Scholar]
- Filipovych R, Davatzikos C. Semi-supervised pattern classification of medical images: Application to mild cognitive impairment (mci) NeuroImage. 2011;55:1109–1119. doi: 10.1016/j.neuroimage.2010.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. Automatically Parcellating the Human Cerebral Cortex. Cerebral Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. Edinburgh: Oliver and Boyd; 1925. [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Klöppel S, Gaser C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. NeuroImage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Geng X, Zhan DC, Zhou ZH. Supervised nonlinear dimensionality reduction for visualization and classification. Trans. Sys. Man Cyber. 2005;Part B 35:1098–1107. doi: 10.1109/tsmcb.2005.850151. [DOI] [PubMed] [Google Scholar]
- Goh A, Vidal R. Segmenting fiber bundles in diffusion tensor images; Berlin, Heidelberg. Springer-Verlag; 2008. pp. 238–250. Proceedings of the 10th European Conference on Computer Vision: Part III. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Second Edition. Springer: Springer Series in Statistics; 2009. 2nd ed. 2009. corr. 3rd printing 5th printing. edition. [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam T, Ting K, Kamruzzaman J. z-svm: An svm for improved classification of imbalanced data. In: Sattar A, Kang Bh, editors. Springer Berlin / Heidelberg: AI 2006: Advances in Artificial Intelligence; 2006. pp. 264–273. volume 4304 of Lecture Notes in Computer Science. [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L. Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The alzheimer’s disease neuroimaging initiative (adni): Mri methods. Journal of Magnetic Resonance Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayo O. Locally Linear Embedding Algorithm: Extensions and Applications. MS Thesis: University of Oulu; 2006. [Google Scholar]
- Koikkalainen J, Lötjönen J, Thurfjell L, Rueckert D, Waldemar G, Soininen H. Multi-template tensor-based morphometry: application to analysis of Alzheimer’s disease. NeuroImage. 2011;56:1134–1144. doi: 10.1016/j.neuroimage.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinats S, Verleysen M, Giron A, Fertil G. Dd-hds: A method for visualization and exploration of high-dimensional data. Trans. Neur. Netw. 2007;18:1265–1279. doi: 10.1109/tnn.2007.891682. [DOI] [PubMed] [Google Scholar]
- Ma L, Member S, Crawford MM, Tian J. Local manifold learning-based k -nearest-neighbor for hyperspectral image classification. IEEE trans. on Geoscience and Remote Sensing. 2010;48:4099–4109. [Google Scholar]
- Mannfolk P, Wirestam R, Nilsson M, Sthlberg F, Olsrud J. Dimensionality reduction of fmri time series data using locally linear embedding. Magnetic Resonance Materials in Physics, Biology and Medicine. 2010;23:327–338. doi: 10.1007/s10334-010-0204-0. [DOI] [PubMed] [Google Scholar]
- Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in mci patients, and their use in prediction of short-term conversion to ad: Results from adni. NeuroImage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohs RC, Knopman D, Petersen RC, Ferris SH, Ernesto C, Grundman M, Sano M, Bieliauskas L, Geldmacher D, Clark C, Thal LJ. Development of cognitive instruments for use in clinical trials of antidementia drugs: additions to the alzheimes disease assessment scale that broaden its scope. Alzheimer Dis Assoc Disord. 1997;11(Suppl 2):S13–S21. [PubMed] [Google Scholar]
- Morris JC. The clinical dementia rating (cdr): Current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 1979;9:62–66. [Google Scholar]
- Pelaez-Coca M, Bossa M, Olmos S. Discrimination of ad and normal subjects from mri: Anatomical versus statistical regions. Neuroscience Letters. 2011;487:113–117. doi: 10.1016/j.neulet.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Phan TG, Chen J, Donnan G, Srikanth V, Wood A, Reutens DC. Development of a new tool to correlate stroke outcome with infarct topography: A proof-of-concept study. NeuroImage. 2010;49:127–133. doi: 10.1016/j.neuroimage.2009.07.067. [DOI] [PubMed] [Google Scholar]
- Plant C, Teipel SJ, Oswald A, Bhm C, Meindl T, Mourao-Miranda J, Bokde AW, Hampel H, Ewers M. Automated detection of brain atrophy patterns based on mri for the prediction of alzheimer’s disease. NeuroImage. 2010;50:162–174. doi: 10.1016/j.neuroimage.2009.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querbes O, Aubry F, Pariente J, Lotterie JA, Dmonet JF, Duret V, Puel M, Berry I, Fort JC, Celsis P, Initiative TADN. Early diagnosis of alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132:2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. NeuroImage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder DD, Duin RPW. Technical Report PH-2002-01. Delft University of Technology; 2002. Locally linear embedding for classification. [Google Scholar]
- Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290:2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- Scholz M, Kaplan F, Guy CL, Kopka J, Selbig J. Non-linear pca: a missing data approach. Bioinformatics. 2005;21:3887–3895. doi: 10.1093/bioinformatics/bti634. [DOI] [PubMed] [Google Scholar]
- Stonnington CM, Chu C, Klppel S, Jr, CRJ, Ashburner J, Frackowiak RS. Predicting clinical scores from magnetic resonance scans in alzheimer’s disease. NeuroImage. 2010;51:1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BY, Zhang XM, Li J, Mao XM. Feature fusion using locally linear embedding for classification. Trans. Neur. Netw. 2010;21:163–168. doi: 10.1109/TNN.2009.2036363. [DOI] [PubMed] [Google Scholar]
- Suplber G, Simmons A, Muehlboeck J, et al. An mri-based index to measure the severity of alzheimer’s disease-like structural pattern in subjects with mild cognitive impairment. J Intern Med. 2010 doi: 10.1111/joim.12028. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel S, Ewers M, Reisig V, Schweikert B, Hampel H, Happich M. Long-term cost-effectiveness of donepezil for the treatment of alzheimers disease. European Archives of Psychiatry and Clinical Neuroscience. 2007;257:330–336. doi: 10.1007/s00406-007-0727-1. 10.1007/s00406-007-0727-1. [DOI] [PubMed] [Google Scholar]
- Tenenbaum JB, Silva Vd, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jr, CRJ Alzheimer’s disease diagnosis in individual subjects using structural mr images: Validation studies. NeuroImage. 2008;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zheng J, Yao Z, Li L. In: Improved locally linear embedding through new distance computing. Wang J, Yi Z, Zurada J, Lu BL, Yin H, editors. Springer Berlin Heidelberg: Advances in Neural Networks - ISNN 2006; 2006. pp. 1326–1333. volume 3971 of Lecture Notes in Computer Science. [Google Scholar]
- Wang J, Saligrama V, Castanon DA. Structural similarity and distance in learning. ArXiv e-prints; 2011. [Google Scholar]
- Wechsler D. Wechsler Adult Intelligence Scale. 4th Edition. TX: Pearson: San Antonio; 2008. [Google Scholar]
- Weinberger K, Saul L. Unsupervised learning of image manifolds by semidefinite programming. Computer Vision and Pattern Recognition, 2004; CVPR 2004. Proceedings of the 2004 IEEE Computer Society Conference on; 2004. pp. II–988–II–995. [Google Scholar]
- Westman E, Simmons A, Zhang Y, Muehlboeck JS, Tunnard C, Liu Y, Collins L, Evans A, Mecocci P, Vellas B, Tsolaki M, Koszewska I, Soininen H, Lovestone S, Spenger C, Wahlund LO. Multivariate analysis of mri data for alzheimer’s disease, mild cognitive impairment and healthy controls. NeuroImage. 2011;54:1178–1187. doi: 10.1016/j.neuroimage.2010.08.044. [DOI] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Ltjnen J the Alzheimer’s Disease Neuroimaging Initiative. Multi-method analysis of mri images in early diagnostics of alzheimer’s disease. PLoS ONE. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lui R, Gao JH, et al. Independent componenet analysis-based classification of alzheimer’s disease mri data. Journal of Alzheimer’s Disease. 2011;24:775–783. doi: 10.3233/JAD-2011-101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Hu D, Zhou Z. Growing locally linear embedding for manifold learning. Journal of Pattern Recognition Research. 2007:1–16. [Google Scholar]
- Zhang D, Shen D. Predicting future clinical changes of mci patients using longitudinal and multimodal biomarkers. PLoSone. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Jie Y. Non-negative matrix factorization with log gabor wavelets for image representation and classification. Systems Engineering and Electronics, Journal of 16. 2005:738–745. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.