Abstract
The advent of somatic cell reprogramming technologies, which enables the generation of patient-specific, induced pluripotent stem cell (iPSC) and other trans-differentiated human neuronal cell models, provides new means of gaining insight into the molecular mechanisms and neural substrates of psychiatric disorders. By allowing a more precise understanding of genotype-phenotype relationship in disease-relevant human cell types, the use of reprogramming technologies in tandem with emerging genome engineering approaches provides a previously ‘missing link’ between basic research and translational efforts. In this review, we summarize advances in applying human pluripotent stem cell and reprogramming technologies to generate specific neural subtypes with a focus on the use of these in vitro systems for the discovery of small molecule-probes and novel therapeutics. Examples are given where human cell models of psychiatric disorders have begun to reveal new mechanistic insight into pathophysiology and simultaneously have provided the foundation for developing disease-relevant, phenotypic assays suitable for both functional genomic and chemical screens. A number of areas for future research are discussed, including the need to develop robust methodology for the reproducible, large-scale production of disease-relevant, neural cell types in formats compatible with high-throughput screening modalities, including high-content imaging, multidimensional, signature-based screening, and in vitro network using multielectrode arrays. Limitations, including the challenges in recapitulating neurocircuits and non-cell autonomous phenotypes are discussed. While these technologies are still in active development, we conclude that as our understanding of how to efficiently generate and probe the plasticity of patient-specific stem models improves, their utility is likely to advance rapidly.
Keywords: reprogramming, induced pluripotent stem cells, neural progenitors, disease-relevant cell type, phenotypic assays, neuroplasticity, neuropharmacology, high-throughput screening
1. Introduction
The development of effective treatments for neuropsychiatric disorders presents one of the greatest challenges and areas of unmet medical need in the 21st century. Affecting millions of individuals worldwide, they present a tremendous burden to individuals, their families, and society as whole (1, 2). As the causes of severe mental illnesses are likely to be complex and heterogeneous in nature, the application of new approaches and tools to gain insight into the underling etiology and pathophysiology is critically needed. Recent advances in human genetics have led to an explosion of our understanding of the role that genetic (3), and epigenetic (4), variation plays in determining the susceptibility to a wide range of psychiatric disorders. However, despite these important advances, our increased knowledge has yet to be translated to the discovery and validation of significantly improved, targeted therapeutics (5, 6). Moreover, the polypharmacology of many psychopharmacological agents, along with poor understanding of their precise mechanism of therapeutic action, have limited the development of improved therapeutics that offer either disease-modifying or prophylactic effects, in marked contrast to other disorders, such as rheumatoid arthritis (7).
One of the main challenges to developing next-generation therapeutics for psychiatric disorders is the fact that no common genetic variants of large effect have been identified. Instead, the genetic susceptibility for common disorders, such as bipolar disorder, major depression, schizophrenia, and autism is clearly polygenic in nature (3). This polygenicity renders it impossible to create truly genetically accurate models of human psychiatric disorders in animal models that additionally often lack neuroanatomical regions of the brain thought to contribute to such pathologies (8). Moreover, recent comparisons of mouse and human model systems provide a cautionary tale about the challenges in extrapolating from one to the other(9).
Given these challenges and limitations to solely using rodent model systems, the ability to use genetically accurate human models to investigate the molecular and cellular mechanisms of disease and to investigate the step-by-step development of pathophysiology would have a major impact on our understanding of psychiatric disease mechanisms. Moreover, by developing human cellular model systems capable of supporting screening for novel targets and lead structures for therapeutic development could help address current bottlenecks in the drug discovery process for psychiatric disorders leading to the next-generation of therapeutics (6). To date, an obstacle in developing such models has been the inaccessibility of the relevant human tissue in patients, making routine sample collection by biopsy infeasible. Even where tissue is available, terminally-differentiated cells such as neurons cannot be maintained in culture, so experiments cannot be repeated and these resources cannot be scaled up to the extent required to utilize them in chemical screens. Conversely, more accessible tissues such as lymphocytes do not necessarily recapitulate all of the signaling pathways and processes needed to explore pathophysiologic processes in neurons and other brain tissue; this is particularly the case for phenotypes which are not cell-autonomous, but rather require interaction between multiple cell types.
Fortunately, recent advances in the field of human stem cell biology, namely the ability to create patient-specific induced pluripotent stem cells (iPSC) that can be differentiated into a growing number of defined cell types using reprogramming technology (10–12), provides new avenues to investigate the pathophysiological mechanisms of psychiatric disorders. This approach allows more accessible cell types, such as dermal fibroblasts from a skin biopsy or lymphocytes in peripheral blood, to be reprogrammed to pluripotent cells. Using this technique, which relies on the expression of cocktails of transcription factors, such as OCT4, SOX2, KLF4, and c-MYC (10, 12), or OCT4, SOX2, NANOG, and LIN28 (11), there now exist a growing number of human iPSCs models of monogenic psychiatric disorders such as Fragile X syndrome (13–15), Rett syndrome (16–22), along with smaller number of examples of complex, polygenic psychiatric disorders, including schizophrenia (23–26), and bipolar disorder (27).
These new patient-derived cell lines effectively model a human disease genome in a form amenable to in vitro investigation. These models allow access to otherwise difficult or impossible to obtain living cells that comprise the human nervous system. As important, they enable repeated experiments and larger-scale investigations, in contrast to tissue obtained from neurosurgery or through post-mortem studies. Overall, disease-specific, human iPSC models provides an emerging, scalable platform from which to build a set of tools and an integrated approach for human chemical neurobiology that will allow: 1) genotype-phenotype correlations to be understood for complex genetic disorders; and 2) to develop phenotypic assays capable of supporting high-throughput screening for novel therapeutic agents that target molecular mechanisms not currently modulated by the existing pharmacopeia used to treat psychiatric disorders (Figure 1). As additional encouraging signs of the potential of this approach, outside of the field of psychiatry large-scale therapeutic screening using iPSC-derived disease models has already been successfully applied in a number of examples (28–30), pointing to the generality of the approach for studying human disease biology.
Figure 1.
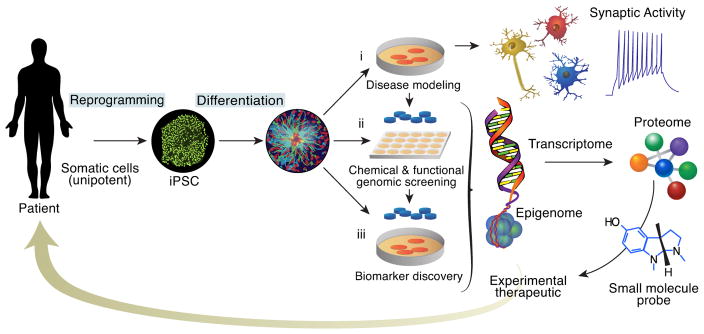
Overview of an Integrated Platform for Biological and Therapeutic Discovery Using Patient-Specific iPSC Models and Chemical Neurobiology.
2. Neurons Generated In Vitro from Multipotent, Self-Renewing, Neural Progenitor Cells
The isolation of multipotent, self-renewing, neural stem and progenitor cells from tissues derived from the rodent central nervous system (CNS) was first described over two decades ago by Reynolds and Weiss (31). Building on these studies, Carpenter and colleagues described the existence of similar multipotent progenitor cells in the human embryonic forebrain that could be expanded in vitro in the presence of basic neurons, astrocytes, and oligodendrocytes (32). However, these initial studies were limited at the time to working with post-mortem human brain tissues, which, for the reasons described below, limited the full potential of human disease modeling. Ultimately, the ability to expand multipotent neural stem and progenitor cells from human pluripotent stem cells as either non-adherent neurospheres, adherent monolayer cultures, or 3-dimensional structures that can form synaptically active, excitatory and inhibitory neuron subtypes by multiple groups over the past few years has brought this approach to modeling human CNS disorders to the forefront (33–44). Examples of these iPSC-derived neural progenitor cells (NPCs) and neurons that can now be generated and used for functional genomic studies and for chemical neurobiology studies are depicted in Figure 2.
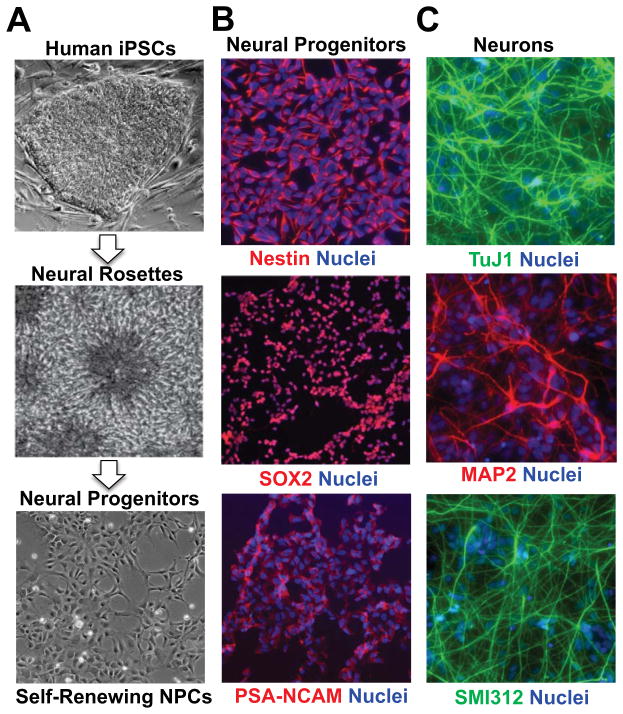
Figure 2. Generation of Long-Term, Self-Renewing Human iPSC-Derived Neural Progenitors for Use in Chemical Neurobiology Studies and Novel Therapeutic Screening.
(A) Human iPSC-derived NPCs can be derived from the manual isolation of neural rosette structures from iPSC colonies subject to neuroepithelial patterning by a variety of methods (33–44), including “dual SMAD” inhibition, growth factor removal, and from spontaneous formation. Isolated NPCs can be expanded in neural progenitor selective conditions on poly-ornithine/laminin coated surfaces in the presence of the mitogens EGF and FGF2 (bFGF). (B) Example of immunocytochemical analyses of the neural progenitor markers (Nestin, SOX2 and PSA-NCAM) and (C) neuronal markers (TuJ1+, MAP2+, SMI312+) after differentiation for 7 days that is initiated by removal of EGF and FGF2 (bFGF) mitogens. Images adapted from (63).
As a scalable platform for chemical neurobiology and novel therapeutic discovery, the use of these patient-specific, human iPSC-derived NPCs and differentiated neurons extends previous efforts with rodent stem cell-derived neurons (45–50), as well as efforts to use postmortem, human brain-derived immortalized (51, 52), or non-immortalized (53, 54) NPCs. First, while the rodent and human nervous systems share a number of evolutionary conserved properties there are also radical differences in terms of neurogenesis and neural patterning, most apparent grossly in the lissencephalic nature of rodent brain. Thus, in order to ultimately develop human disease-relevant neuronal cell models, it will be important to be able to routinely access neural cell types found within regions of the human brain, including the cerebral cortex (8, 39, 40, 55). Moreover, many aspects of the epigenetic regulation of the genome, including non-coding RNAs and regulator enhancers are unique to the human genome and cannot be adequately investigated in non-human systems (56–58). Third, since immortalization process itself interferes with the normal mechanisms of cell proliferation, the use of non-immortalized cells allows access to a more physiologically relevant state of self-renewing NPCs (59). Fourth, the use of iPSCs from living patients removes the dependency on post-mortem tissue-derived cells thereby allowing study of neurodevelopmental mechanisms by providing access to less-mature cell types in which terminal differentiation can be manipulated. It also allows application of human iPSC models as biomarkers, for diagnosis or treatment selection in a particular patient (60). Finally, the accessibility of somatic tissues allows efficient development of biobanks large enough to represent most common genetic variation, enabling investigation of particular genetic variants without needing to engineer each variant individually, particularly where they cannot readily be created with existing genome engineering technology, as with some large copy number variants, for example.
2. Phenotypic Screens Using Stem Cell-Derived Neural Progenitors and Neurons
As the example of Huntington’s disease illustrates, identifying a causal genetic variation is only the first step in understanding the process by which that gene contributes to the pathophysiology of disease (61). To complement the use of genetic approaches, the use of chemical biology provides a means to identify small-molecule probes that can help reveal pathophysiology and identify novel targets for therapeutic intervention. Recent reports in non-neuropsychiatric patient-derived disease models illustrate the potential for this approach (28–30). Moving beyond the testing of individual drugs described in a number of published studies to date, large-scale, high-throughput screens have only recently begun to be performed with stem-cell derived NPCs and differentiated neurons with potential for screens to be performed at multiple stages of human neurodevelopment (Figure 3).
Figure 3. Developing Disease-Relevant Cell-Based Assays Through Directed Differentiation of Human iPSCs to.
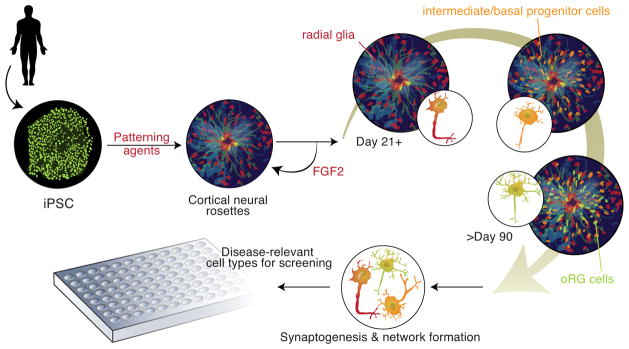
Example of human corticogenesis based upon recent studies of human pluripotent stem cells Directed differentiation of iPSCs to cortical neurons through rosette-derived neural progenitors cells. The use of morphogens and patterning agents, including inhibitors of SMAD (e.g. Noggin, dorsomorphin, and SB431542) that attenuate bone morphogenetic protein (BMP) and transforming growth factor beta (TGF-β) signal transduction pathways, and/or retinoic acid (vitamin A), leads to the formation within neural rosettes of self-renewing, cortical progenitor cells expressing the transcription factors PAX6+, FOXG1+, OTX1/2+, and TBR2+ (40, 55). Removal of the mitogen FGF2 initiates neurogenesis with the concomitant production of intermediate/basal progenitor cells and then outer radial glia (oRG) cells that produce a diverse range of upper and lower layer cortical, glutamatergic projection neuron subtypes that follows the temporal course of neuronal production observed in vivo in the human brain over a 21–90 day period (40, 55). By addition of other patterning agents the fate of the neural progenitors can be altered. For example, addition of the hedgehog signaling agonist puromorphine can ventralize neural rosettes to generate GABAergic (GAD67+) interneurons (40). Scaling these procedures and optimizing protocols for robustness and reproducibility will be critical milestones to achieve in order to support high-throughputscreening using patient-derived neuronal subtypes that are relevant to particular psychiatric disorders.
2.1 Identification of glutamate receptor potentiators as cognitive enhancers
One of the first, and still largest-scale phenotypic screens performed using stem cell-derived neurons that has been publicly disclosed is that of McNeish and colleagues that sought to identify novel structural classes of the ionotropic AMPA subtype of glutamate receptors using a combination of mouse embryonic stem cell (mESC) and hESC-derived neurons (62). To perform the primary screen, mESC-derived NPCs obtained through embryoid body differentiation were further differentiated in 384-well plates into a mixed population of neural and glial subtypes (62). Using a Fluorimetric Imaging Plate Reader (FLIPR) assay to measure calcium influx, the screen was performed in the presence of the AMPA receptor potentiator cyclothiazide to enable the identification of potentiators that were dependent upon partial AMPA receptor activation. In total, the high-throughput screen was performed on an impressively large 2.4 million compound library resulting in the identification of 5,911 initial hits, 17 of which were confirmed as positive upon retesting in dose responses, with secondary assays using human NPCs (ArunA Biomedical Inc.) derived from WA09 hESCs (62). These results provide an exciting example of screening a challenging CNS relevant target in stem cell-derived neurons to produce chemical matter that can be further optimized as leads for therapeutic development of cognitive enhancers.
2.2 Screens for neuroplasticity regulators using Human iPSC-derived neural progenitors
To date there have been two examples of large-scale screening using human iPSC-derived CNS NPCs, which are illustrative of what is likely to emerge with differentiated human iPSC-derived neurons.
2.2.1 Modulators of the Wnt/GSK3-mediated neuroplasticity pathways
In order to identify novel small-molecule probes of a pathway critical to neurogenesis and neuroplasticity, Zhao and colleagues recently reported on a large-scale screen of the Wnt/GSK3 signaling pathway using human iPSC-derived CNS NPCs (63). Dysregulation of Wnt/GSK pathway has been implicated in the pathophysiology of a number of neuropsychiatric disorders and in the response to a wide variety of clinically used drugs, including the mood stabilizers lithium and valproate, antidepressants, and antipsychotics (64–66). The putative psychosis risk gene DISC1 has been shown to be a direct inhibitor of GSK3 kinase activity, such that its absence leads to inhibition of Wnt signaling and alteration of adult neurogenesis as well as behavior (67). Similarly, loss of FMRP in Fragile X syndrome leads to the dysregulation of Wnt/GSK3 signaling (68–70). These findings are consistent with the observed adult neurogenesis phenotype seen in Fmr1-KO mice (71), as well as the neurodevelopmental defects observed in the Fragile X syndrome human iPSC model (14).
In light of these data, the identification of novel compounds that selectively modulate Wnt/GSK3 signaling in human NPCs and neurons may prove therapeutically useful for multiple psychiatric disorders. To advance testing of this notion, Zhao and colleagues introduced a TCF/LEF-responsive luciferase reporter into human iPSC-derived NPCs and validated the assay using Wnt3a conditioned media, lithium, and the GSK3 inhibitor, CHIR-99021 (63). To perform a pilot screen, a collection of ~1,500 compounds from a library of F.D.A.-approved drugs and known bioactive compounds was screened with cells subject to media alone, or the addition of an EC25 amount of Wnt3a-conditioned media or 10 mM lithium. With follow-up dose responses, this pilot screen identified a number of known bioactive molecules that could potentiate Wnt or lithium signaling (or both). This included potent mGluR1 antagonists and riluzole (Rilutek), an F.D.A.-approved drug, which was also identified in previous screens using immortalized mouse hippocampal progenitors (HT-22) (72), providing confidence in the validity of the iPSC-derived NPCs for characterizing Wnt pathway modulation.
2.2.2 Enhancers of human neural progenitor proliferation/viability
With the goal of identifying novel pro-neurogenic compounds, McLaren and colleagues recently reported on a screen of 1,000 compounds for their ability to enhance proliferation and/or viability of human iPSC-derived NPCs using a Cell Titer-Glo ATP bioluminescence-based, end-point assay (50). After two rounds of screening and testing, 5 compounds were validated as increasing the proliferation or viability of the cells, which may ultimately prove to be useful tools for enhancing the ability to expand NPCs on a large scale and potentially as therapeutic agents for modulating neurogenesis (50).
3. Future of Phenotype-Based Screens Using Neuropsychiatric Disorders iPSC Models
A number of new tools, technologies and screening approaches are likely to be applied in the near future to impact the discovery of new targets and leads for therapeutic development. These developments will enable screens to be performed at multiple stages of human neurodevelopment, including defined NPCs and neuron subtypes (Figure 3).
3.1 Prospective isolation of defined neural progenitors using cell surface markers
While a number of approaches exist for the patterning of iPSCs into early neuroectodermal cells that can subsequently be patterned and differentiated into neurons, the derivation of neural rosette-derived progenitors has emerged as a particularly useful method for generating cells expandable into telenchephalic NPCs that can subsequently be differentiated into a range of neuron subtypes (33–36, 73). Directly isolated neural rosette-derived progenitor cell cultures, however, contain a mixture of cell types with the capability to form cells of both the CNS and as well as neural crest-derived cells that form the peripheral nervous system. To enable purification of more homogenous CNS iPSC-derived NPCs from heterogenous populations of cells, recent studies performed by Yuan and colleagues sought to identity cell-surface markers on hESC and human iPSCs subject to neural induction that could be used for prospective isolation of defined subpopulations of progenitor cells (74). Using fluorescence activated cell sorting (FACS), these studies identified a cell-surface signature that enables the isolation of progenitors that could be expanded for multiple passages and retained the ability to differentiate into electrophysiologically active neurons (74). Importantly, since this method selects against CD271+ cells, its use enables the isolation of an enriched population of progenitors that yield CNS rather than peripheral nervous system subtypes of neurons. Given the importance of being able to work with distinct neuron subtypes for disease modeling and therapeutic screening, a critical need for the field moving forward will be to develop additional markers that allow, ideally prospectively, the isolation of different types of NPCs. These tools will also enable the standardization of methods and results between laboratories—a key issue that if not addressed will limit progress as investigators struggle to replicate each others’ work.
3.2 Multidimensional signature-based neuronal screening
As an alternative to selecting individual genes or molecular events such as phosphorylation for performing high-throughput screens, an alternative strategy is the selection of a multigene mRNA expression signature that can serve as a surrogate for complex cellular states (75). Once defined, a high-throughput, inexpensive, multiplexed mRNA profiling assay, such as that using Luminex-bead platform can be performed (76). Recent development of this technology have led to the ability to measure up to 1000 distinct transcripts that have been selected on their basis to allow through a computational inference model to allow assessment of over 80% of the variation that has been observed in the human transcriptome in the form the L1000 assay (T. Golub, A. Subramanian, manuscript in preparation). The L1000 assay is well-suited for the characterization of complex disease states were multiple gene expression changes often occur and where it may be desirable to screen for therapeutic agents that can reverse many of the changes observed. As part of the NIH-sponsored LINCS project (http://www.lincscloud.org), the Haggarty laboratory in collaboration with members of the Broad Institute’s Connectivity Map project (CMap) has recently completed a successful pilot study of this strategy for signature-based screening using human iPSC-derived NPCs and neurons treated with a collection of CNS-active drugs and probe compounds, at multiple time points, with multiple replicates, and in multiple states (e.g. +/− KCl-induced depolarization) (W. Zhao, A. Subramanian, T. Golub,, S. Haggarty, manuscript in preparation). Extension of this multidimensional signature-based screening approach to include specific disease models, a larger collection of compounds, alteration of cell states, and different cell types derived from iPSC models are in the works. Overall, the expansion of this “Neuro-CMap” to include diverse human iPSC-based disease models and perturbations could prove a valuable resource for the psychiatric disease research community.
3.3 Neuronal circuit-based and information processing assays
While not yet adapted to the point of being able to perform large-scale, high-throughput screens, yet another area of great potential utility for the generation of functional networks of pluripotent stem cell-derived neurons with the capacity for information processing applies multielectrode array technology (77–80). Using mESC-derived neurons, Ban and colleagues demonstrated the feasibility of generating functional networks in vitro that exhibit spontaneous bursting activity and evoked action potentials leading to a propagation of the electrical activity throughout the culture (77). In the case of hESC-derived neurons, Heikkila and colleagues have demonstrated that the spontaneous network electrical activity could be blocked by antagonizing sodium ion channels and multiple glutamate receptor subtypes (79). Importantly, these assays with pharmacological agents demonstrate the existence of functional synapses that corroborate immunocytochemical studies and receptor expression. Multiplexed versions of multielectrode arrays with neural circuits formed from defined neuronal subtypes rather than the heterogeneous cultures may reveal subtle phenotypes that would otherwise be missed by other functional assays.
4. Alternatives to Human iPSC Models for Chemical Neurobiology Studies
The ability to directly reprogram fibroblasts into so-called induced neurons (iNs) has been now demonstrated by multiple groups (81–85). Variations of this iN approach with different transcription factors have yielded defined neuronal subtypes, such as dopaminergic neurons through the expression of Nr4a1, Lmx1a, and Ascl1 (85). For further details of this approach, see other articles in this issue. By eliminating the need to generate iPSCs, iNs may substantially shorten the time between sample acquisition and establishment of a cellular model. A further advantage, while largely hypothetical, is the possibility that eliminating this intermediate step would reduce the disruption in epigenetic marks associated with iPSC generation. However, a key current limitation of this approach as it applies to high-throughput screening is the relatively small number of neurons produced: the methodology in its current form is highly inefficient, with only a small % of fibroblasts transdifferentiating to iNs, which would be limiting for large-scale screens. One potential solution to limited number of neurons is the creation of induced neural progenitor cells (iNPCs), rather than iNs, along with approaches using olfactory biopsies, which we discuss in more detail in the accompanying supplemental information.
5. Important Considerations and Remaining Challenges
A summary of the considerations for high-throughput screening with patient-derived stem cell models is provided in Table 1. Besides these consideration and the complementary approaches described above, more general technical issues emerge that require focused attention before the promise of human stem cell-based therapeutic screening can be fully realized. First, despite rapid progress in identifying the developmental trajectory of specific types of neurons and glia (86, 87) reducing these techniques to practice remains challenging. At present, while remarkable progress has been made (40, 55), it is still difficult to generate a specific cell type reliably and in adequate numbers, as might be required for a large-scale chemical screen, with the exception of a few cell types which have been the focus of intense study over the past decade (30, 88). On the other hand, while the mixed cell populations that result from standard differentiation protocols are generally framed as a limitation, they may also represent an opportunity: for diseases where the cell type of interest is unknown, the opportunity to interrogate multiple cell types simultaneously could be advantageous with emerging approaches for single cell analysis by imaging and transcriptomic approaches.
Table 1.
Considerations for High-throughput Screening with Patient-Derived Stem Cell Models.
| Requirement | Current State of Field | Challenges |
|---|---|---|
| Characterization of multiple iPSC clones from well clinically phenotyped patients and from multiple patients for each disorder |
|
|
| Delineation of disease-relevant cell types |
|
|
| Ability to efficiently generate large numbers of cells |
|
|
| Disease-relevant readouts |
|
|
| Well-defined and robust assay readout |
|
|
A second limitation is apparent in most published work to date using patient-derived cellular models, which assumes that cell-autonomous models of disease will suffice – in other words, that disease-relevant phenotypes can be observed in individual cultured cells regardless of their milieu. While this is a reasonable initial step, and preliminary work suggests it may be possible to identify such phenotypes, it neglects two key aspects of the developing brain. Specifically, at present it is difficult to study cell-cell interactions with these cultured cells, either in terms of differentiation or synapse formation and maintenance. Whether the synapses observed in vitro, which can show evidence of being intact and functional, adequately model those in vivo where multiple cell types act in concern, remains to be investigated. Understanding diseases that arise at the network, rather than the single cell, level may require other approaches to reconstruct complex connectivity amongst neurons, for example using brain slices from animal models or in some cases discarded human tissue (89, 90). On the other hand, just because a disease is most apparent at the network level, it does not necessarily follow that no deficit can be observed in single cells.
6. Summary and Future Directions
Assuming many of the current methodological challenges can be overcome, one can imagine having access to cohorts of iPSCs consisting of multiple clones per patient with defined genotypes (at the level of the whole genome) and rich phenotypic profiles, including knowledge of illness features and course and response to a range of therapeutics. Additionally there would be ready availability of engineered iPSC lines with specific gain- or loss-of-function genetic variants, on multiple genetic backgrounds, introduced using emerging technologies such as TALE nucleases (TALENs) (91–93) and the CRISPR/Cas tools (94, 95).
As part of efforts to identify phenotype in disease relevant cell types that can be used for high-throughput screening, it will be critical that studies rigorously address variation (both genetic and otherwise) between individual iPSC clones of the same patient and between patients with the same disorder. While substantial progress has been made in moving towards non-integrating factors for this purpose, it cannot be forgotten that reprogramming likely entails massive change in cellular states, in particular the epigenome and transcriptome. In certain cases, pooling of data between patients with the same disorder may be appropriate, but careful inspection of the individual iPSC clone performance and its reproducibility over time will be critical if one is to develop robust, reproducible, and scalable assays that can support high-throughput screening. Exactly the number of clones and patients required to rigorously assess genotype-phenotype will depend on the phenotype being investigated and the contribution of background genetic variation to its expressivity of the phenotype. Here, for single gene phenotypes, the use of genome-engineering technologies will be instrumental to establish causality (91–93), and in other cases (e.g. large deletions and duplications), where such engineering technologies are less developed, the consideration of other appropriate genetic controls will be required.
In closing, a general theme emerging from investigation of the human nervous system is the remarkable degree of plasticity it can exhibit (4). Nowhere is this plasticity more evident than the studies emerging from the field of human stem cell biology and the in vitro generation of patient-specific iPSC-derived NPCs and differentiated neuronal subtypes. Understanding how to control and direct this neuroplasticity for the purpose of developing disease-relevant cell types and phenotypic assays promises to shed light on fundamental questions of pathogenesis of psychiatric disorders and enable new approaches to discover novel treatment targets.
Supplementary Material
Acknowledgments
Members of the Haggarty and Perlis laboratories and the MGH Center for Experimental Drugs & Diagnostics are thanked for their continued dedication to advancing patient-specific iPSC models of psychiatric disorders and spirited discussions. Research in the Haggarty and Perlis laboratories has been supported in part by the FRAXA Research Foundation, Harvard Stem Cell Institute Seed Grant, Marigold Foundation, Stanley Medical Research Institute, Tau Consortium, Pitt-Hopkins Research Foundation, National Institute of Mental Health (R33MH087896; R01MH091115; R01MH095088; R21MH093958) and National Institute of Aging (RF1AG042978). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health. Artwork by Applied Art, LLC.
Footnotes
FINANCIAL DISCLOSURES
Dr. Haggarty and Perlis reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kessler RC, Demler O, Frank RG, Olfson M, Pincus HA, Walters EE, et al. Prevalence and treatment of mental disorders, 1990 to 2003. The New England journal of medicine. 2005;352:2515–2523. doi: 10.1056/NEJMsa043266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler RC, Aguilar-Gaxiola S, Alonso J, Chatterji S, Lee S, Ormel J, et al. The global burden of mental disorders: an update from the WHO World Mental Health (WMH) surveys. Epidemiol Psichiatr Soc. 2009;18:23–33. doi: 10.1017/s1121189x00001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. Epigenetic Mechanisms in Mood Disorders: Targeting Neuroplasticity. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman SE. Revolution stalled. Science translational medicine. 2012;4:155cm111. doi: 10.1126/scitranslmed.3003142. [DOI] [PubMed] [Google Scholar]
- 6.Insel TR. Next-generation treatments for mental disorders. Science translational medicine. 2012;4:155ps119. doi: 10.1126/scitranslmed.3004873. [DOI] [PubMed] [Google Scholar]
- 7.Fleischmann R. Novel small-molecular therapeutics for rheumatoid arthritis. Current opinion in rheumatology. 2012;24:335–341. doi: 10.1097/BOR.0b013e32835190ef. [DOI] [PubMed] [Google Scholar]
- 8.Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 13.Alisch RS, Wang T, Chopra P, Visootsak J, Conneely KN, Warren ST. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC medical genetics. 2013;14:18. doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farra N, Zhang WB, Pasceri P, Eubanks JH, Salter MW, Ellis J. Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry. 2012;17:1261–1271. doi: 10.1038/mp.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung AY, Horvath LM, Carrel L, Ellis J. X-chromosome inactivation in rett syndrome human induced pluripotent stem cells. Frontiers in psychiatry / Frontiers Research Foundation. 2012;3:24. doi: 10.3389/fpsyt.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Squillaro T, Alessio N, Cipollaro M, Melone MA, Hayek G, Renieri A, et al. Reduced expression of MECP2 affects cell commitment and maintenance in neurons by triggering senescence: new perspective for Rett syndrome. Mol Biol Cell. 2012;23:1435–1445. doi: 10.1091/mbc.E11-09-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedrosa E, Sandler V, Shah A, Carroll R, Chang C, Rockowitz S, et al. Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. Journal of neurogenetics. 2011;25:88–103. doi: 10.3109/01677063.2011.597908. [DOI] [PubMed] [Google Scholar]
- 25.da Silveira Paulsen B, Souza da Silveira M, Galina A, Kastrup Rehen S. Pluripotent stem cells as a model to study oxygen metabolism in neurogenesis and neurodevelopmental disorders. Archives of biochemistry and biophysics. 2012 doi: 10.1016/j.abb.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison JM, Zhou F, Nigam A, Hussain A, Barker D, Nehme R, et al. Using human iPSC-derived neural cells from genetically related individuals to understand bipolar disorder. International Society for Stem Cell Research, Annual Meeting 2013 [Google Scholar]
- 28.Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol. 2012;30:1244–1248. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013 doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Science translational medicine. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 32.Carpenter MK, Cui X, Hu ZY, Jackson J, Sherman S, Seiger A, et al. In vitro expansion of a multipotent population of human neural progenitor cells. Exp Neurol. 1999;158:265–278. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- 33.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XJ, Zhang SC. In vitro differentiation of neural precursors from human embryonic stem cells. Methods Mol Biol. 2006;331:169–177. doi: 10.1385/1-59745-046-4:168. [DOI] [PubMed] [Google Scholar]
- 36.Falk A, Koch P, Kesavan J, Takashima Y, Ladewig J, Alexander M, et al. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One. 2012;7:e29597. doi: 10.1371/journal.pone.0029597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS biology. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y, Pollard S, Conti L, Toselli M, Biella G, Parkin G, et al. Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Molecular and cellular neurosciences. 2008;38:245–258. doi: 10.1016/j.mcn.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. S471. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morizane A, Doi D, Kikuchi T, Nishimura K, Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. Journal of neuroscience research. 2011;89:117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Su P, Li D, Tsang S, Duan E, Wang F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, et al. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Lacson R, Cassaday J, Ross DA, Kreamer A, Hudak E, et al. Identification of small-molecule modulators of mouse SVZ progenitor cell proliferation and differentiation through high-throughput screening. Journal of biomolecular screening. 2009;14:319–329. doi: 10.1177/1087057109332596. [DOI] [PubMed] [Google Scholar]
- 46.Warashina M, Min KH, Kuwabara T, Huynh A, Gage FH, Schultz PG, et al. A synthetic small molecule that induces neuronal differentiation of adult hippocampal neural progenitor cells. Angew Chem Int Ed Engl. 2006;45:591–593. doi: 10.1002/anie.200503089. [DOI] [PubMed] [Google Scholar]
- 47.Wurdak H, Zhu S, Min KH, Aimone L, Lairson LL, Watson J, et al. A small molecule accelerates neuronal differentiation in the adult rat. Proc Natl Acad Sci U S A. 2010;107:16542–16547. doi: 10.1073/pnas.1010300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diamandis P, Wildenhain J, Clarke ID, Sacher AG, Graham J, Bellows DS, et al. Chemical genetics reveals a complex functional ground state of neural stem cells. Nature chemical biology. 2007;3:268–273. doi: 10.1038/nchembio873. [DOI] [PubMed] [Google Scholar]
- 49.Garavaglia A, Moiana A, Camnasio S, Bolognini D, Papait R, Rigamonti D, et al. Adaptation of NS cells growth and differentiation to high-throughput screening-compatible plates. BMC neuroscience. 2010;11:7. doi: 10.1186/1471-2202-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casalino L, Magnani D, De Falco S, Filosa S, Minchiotti G, Patriarca EJ, et al. An automated high throughput screening-compatible assay to identify regulators of stem cell neural differentiation. Molecular biotechnology. 2012;50:171–180. doi: 10.1007/s12033-011-9413-7. [DOI] [PubMed] [Google Scholar]
- 51.Breier JM, Radio NM, Mundy WR, Shafer TJ. Development of a high-throughput screening assay for chemical effects on proliferation and viability of immortalized human neural progenitor cells. Toxicological sciences : an official journal of the Society of Toxicology. 2008;105:119–133. doi: 10.1093/toxsci/kfn115. [DOI] [PubMed] [Google Scholar]
- 52.Danovi D, Folarin AA, Baranowski B, Pollard SM. High content screening of defined chemical libraries using normal and glioma-derived neural stem cell lines. Methods Enzymol. 2012;506:311–329. doi: 10.1016/B978-0-12-391856-7.00040-8. [DOI] [PubMed] [Google Scholar]
- 53.Hook L, Vives J, Fulton N, Leveridge M, Lingard S, Bootman MD, et al. Non-immortalized human neural stem (NS) cells as a scalable platform for cellular assays. Neurochemistry international. 2011;59:432–444. doi: 10.1016/j.neuint.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 54.Kanemura Y, Mori H, Kobayashi S, Islam O, Kodama E, Yamamoto A, et al. Evaluation of in vitro proliferative activity of human fetal neural stem/progenitor cells using indirect measurements of viable cells based on cellular metabolic activity. Journal of neuroscience research. 2002;69:869–879. doi: 10.1002/jnr.10377. [DOI] [PubMed] [Google Scholar]
- 55.Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 57.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLaren D, Gorba T, Marguerie de Rotrou A, Pillai G, Chappell C, Stacey A, et al. Automated Large-Scale Culture and Medium-Throughput Chemical Screen for Modulators of Proliferation and Viability of Human Induced Pluripotent Stem Cell-Derived Neuroepithelial-like Stem Cells. Journal of biomolecular screening. 2012 doi: 10.1177/1087057112461446. [DOI] [PubMed] [Google Scholar]
- 60.Perlis RH. Translating biomarkers to clinical practice. Mol Psychiatry. 2011;16:1076–1087. doi: 10.1038/mp.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gusella JF, MacDonald ME. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends in biochemical sciences. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 62.McNeish J, Roach M, Hambor J, Mather RJ, Weibley L, Lazzaro J, et al. High-throughput screening in embryonic stem cell-derived neurons identifies potentiators of alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate-type glutamate receptors. J Biol Chem. 2010;285:17209–17217. doi: 10.1074/jbc.M109.098814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao WN, Cheng C, Theriault KM, Sheridan SD, Tsai LH, Haggarty SJ. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. Journal of biomolecular screening. 2012;17:1252–1263. doi: 10.1177/1087057112456876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 65.Haggarty SJSK, Perlis RH, Karmacharya R. Wnt Signaling in Mood & Psychotic Disorders. In: Moon SHR, editor. Wnt signaling in Development and Disease: Molecular Mechanisms and Biological Functions. Wiley; 2013. [Google Scholar]
- 66.Pan JQ, Lewis MC, Ketterman JK, Clore EL, Riley M, Richards KR, et al. AKT kinase activity is required for lithium to modulate mood-related behaviors in mice. Neuropsychopharmacology. 2011;36:1397–1411. doi: 10.1038/npp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mines MA, Jope RS. Glycogen synthase kinase-3: a promising therapeutic target for fragile x syndrome. Frontiers in molecular neuroscience. 2011;4:35. doi: 10.3389/fnmol.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS genetics. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lazarov O, Demars MP, da Zhao KT, Ali HM, Grauzas V, Kney A, et al. Impaired survival of neural progenitor cells in dentate gyrus of adult mice lacking FMRP. Hippocampus. 2012;22:1220–1224. doi: 10.1002/hipo.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biechele TL, Camp ND, Fass DM, Kulikauskas RM, Robin NC, White BD, et al. Chemical-genetic screen identifies riluzole as an enhancer of Wnt/beta-catenin signaling in melanoma. Chem Biol. 2010;17:1177–1182. doi: 10.1016/j.chembiol.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 76.Peck D, Crawford ED, Ross KN, Stegmaier K, Golub TR, Lamb J. A method for high-throughput gene expression signature analysis. Genome biology. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ban J, Bonifazi P, Pinato G, Broccard FD, Studer L, Torre V, et al. Embryonic stem cell-derived neurons form functional networks in vitro. Stem Cells. 2007;25:738–749. doi: 10.1634/stemcells.2006-0246. [DOI] [PubMed] [Google Scholar]
- 78.Illes S, Fleischer W, Siebler M, Hartung HP, Dihne M. Development and pharmacological modulation of embryonic stem cell-derived neuronal network activity. Exp Neurol. 2007;207:171–176. doi: 10.1016/j.expneurol.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 79.Heikkila TJ, Yla-Outinen L, Tanskanen JM, Lappalainen RS, Skottman H, Suuronen R, et al. Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp Neurol. 2009;218:109–116. doi: 10.1016/j.expneurol.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 80.Lappalainen RS, Salomaki M, Yla-Outinen L, Heikkila TJ, Hyttinen JA, Pihlajamaki H, et al. Similarly derived and cultured hESC lines show variation in their developmental potential towards neuronal cells in long-term culture. Regenerative medicine. 2010;5:749–762. doi: 10.2217/rme.10.58. [DOI] [PubMed] [Google Scholar]
- 81.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 86.Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Science translational medicine. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 90.Arguello PA, Gogos JA. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci. 2012;35:3–13. doi: 10.1016/j.tins.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 91.Ramalingam S, London V, Kandavelou K, Cebotaru L, Guggino W, Civin C, et al. Generation and genetic engineering of human induced pluripotent stem cells using designed zinc finger nucleases. Stem cells and development. 2013;22:595–610. doi: 10.1089/scd.2012.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013 doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013 doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.