Abstract
Basal layers of stratified epithelia express keratins K5, K14, and K15, which assemble into intermediate filament networks. Mutations in K5 or K14 genes cause epidermolysis bullosa simplex (EBS), a disorder with blistering in the basal layer due to cell fragility. Nonkeratinizing stratified epithelia, e.g., in the esophagus, produce more keratin K15 than epidermis, which alleviates the esophageal symptoms in patients with K14 mutations. Hypothesizing that increasing the cellular content of K15 could compensate for the mutant K14 and thus ease skin blistering in K14 EBS patients, we cloned the promoter of the K15 gene and examined its transcriptional regulation. Using cotransfection, gel mobility shifts, and DNase I footprinting, we have identified the regulators of K15 promoter activity and their binding sites. We focused on those that can be manipulated with extracellular agents, transcription factors C/EBP, AP-1, and NF-κB, nuclear receptors for thyroid hormone, retinoic acid, and glucocorticoids, and the cytokine gamma interferon (IFN-γ). We found that C/EBP-β and AP-1 induced, while retinoic acid, glucocorticoid receptors, and NF-κB suppressed, the K15 promoter, along with other keratin gene promoters. However, the thyroid hormone and IFN-γ uniquely and potently activated the K15 promoter. Using these agents, we could boost the amounts of K15 in human epidermis. Our findings suggest that treatments based on thyroid hormone and IFN-γ could become effective agents in therapy for patients with EBS.
Somatic gene therapy offers, ultimately, a promise to treat many inherited disorders; however, not all genetic diseases are amenable to this approach. Replacing a deficient enzyme or a signaling protein in a cell type that can easily repopulate the affected organ is conceptually straightforward (14, 29). It is much more difficult to treat a structural protein mutation in a tissue where the stem cells are concealed, such as the epidermis. While skin can be used to deliver a missing protein systemically, treating inherited skin diseases with replacement gene therapy, targeting the entire organ, is a much more difficult task and calls for alternative approaches (13, 15, 24, 32). At the same time, in gene families, one member can often substitute for another mutated or missing one, which commonly and surprisingly leads to a lack of a phenotype in knockouts of genes thought to be essential. Indeed, a whole section in this journal, “Mammalian genetic models with minimal or complex phenotypes,” is devoted largely to such phenomena. These considerations led us to hypothesize that increasing the expression of one structural protein, K15, could compensate for another, mutated protein of the same family, K14.
Keratins are a family of about 30 proteins that form intermediate filament in epithelial cells. Obligate heteropolymers of a type I and a type II subunit, they contribute to the structure and strength of the cytoskeleton (49). Mutations in keratin genes cause keratinopathies, inherited diseases of the skin and its appendages (39). For example, mutations in K5 or K14 genes give rise to epidermolysis bullosa simplex (EBS), those in K1, K2e, or K10 give rise to epidermolytic hyperkeratosis, those in K6, K16, or K17 give rise to pachyonychias, etc. Gene therapy approaches that replace the mutant gene are not feasible for treatment of keratinopathies because the entire integument needs to be treated. Therefore, searching for an alternative to gene therapy for EBS, we hypothesized that boosting the expression of K15 may compensate for the disrupted K14 and restore the mechanical strength of the epidermal basal layer. Increasing the expression of a related protein from the same gene family to compensate for the mutated one is a novel paradigm in gene therapy for inherited disorders and could offer real promise in the case of EBS.
The mitotically active keratinocytes of all stratified squamous epithelia are characterized by their contact with the basement membrane and expression of keratins K5 and K14 and, less abundantly, keratin K15 (8, 30, 36). Keratin K15 belongs to the “acidic” or type I keratin family and assembles into a keratin filament network paired with K5, its “basic,” type II keratin expression partner. In response to unknown stimuli, the basal cells are triggered to detach from the basement membrane, initiate their migration through the suprabasal layers, and terminally differentiate, ending transcription of K5, K14, and K15 while inducing a new sets of differentiation-specific keratins (18, 35). The differentiation of basal keratinocytes is also accompanied by changes in expression of several transcription factors, such as C/EBP, AP-1, and NF-κB (34, 44, 46).
Studies with mice with a deletion of the K14 gene have shown that K15 can be a major component of the basal keratin network of stratified, nonkeratinizing epithelia, protecting them from mechanical damage in the absence of K14 (30). Consequently, the blistering in patients with EBS due to a mutation in K14 is less severe in internal stratified epithelia, e.g., the esophagus, than in cornified epithelia, e.g., the epidermis. Such differences are not present in EBS patients with mutations in K5 (11, 17, 43). Elegant transgenic experiments have shown that decreased expression of the mutant keratin 14 relative to the healthy allele results in normal morphology and function of the skin (9, 10). The mechanisms of regulation of K15 expression thus become of major importance because increasing the content of K15 could significantly alleviate the symptoms in EBS patients with a mutation in K14.
The molecular mechanisms that control the expression of the K15 keratin gene are still unknown. Human K15 is encoded on chromosome 17 (3). The K14/K15 ratio can change dramatically during postnatal development (30). Under hyperproliferating conditions, in which keratinocytes are activated, the K15 protein and its mRNA are suppressed, suggesting that K15 expression may not be compatible with the activated phenotype (16, 58). Transcription of K15 appears to be suppressed by transforming growth factor beta, tumor necrosis factor alpha (TNF-α), epidermal growth factor, and keratinocyte growth factor in HaCaT cells (59).
With this in mind, we set out to clone the promoter of the K15 gene and to determine the molecular regulators of its expression. We were particularly interested in those regulators that can be affected by extracellular stimuli. These include nuclear receptors for retinoic acid, thyroid hormone, and glucocorticoids. We have shown previously that nuclear receptors for retinoic acid, thyroid hormone, and glucocorticoids and their ligands play an important role in regulating keratin synthesis (40, 41, 51, 52). Specifically, the receptors and their ligands suppress K5 and K14, the basal keratins, and K6, K16, and K17, the inflammation- and wound healing-associated keratins. Therefore, we hypothesized that retinoic acid, thyroid hormone, and glucocorticoids are involved in the regulation of K15 gene expression in the epidermis.
We also examined the regulation of K15 expression by C/EBP, AP-1, NF-κB, and STAT transcription factors because these also respond to extracellular stimuli (1, 5, 25, 47, 54). C/EBP-α and C/EBP-β are found in the suprabasal layers (34), the AP-1 proteins, JunB, JunD, and c-Fos, are found in the basal and granular layers, while Fra-1 and Fra-2 are in basal cells (44). NF-κB, an activation-associated transcription factor, has antiproliferative effects in the skin (12, 23, 46, 50), while STAT-1 is activated by gamma interferon (IFN-γ) and induces transcription of keratin K17 and other genes (21, 37, 48). We have found that the C/EBP, AP-1, NF-κB, and STAT proteins regulate expression of several epidermal keratin genes, including K5, K6, K14, and K17 (21, 22, 28, 31). Taken as a whole, these results led us to hypothesize that the C/EBP, AP-1, NF-κB, and STAT-1 proteins might be good candidates for regulation of K15 transcription.
To explore regulation of K15 transcription, we have used DNA-mediated cell transfection, electrophoretic mobility shift assays (EMSA), and DNase I footprinting of the K15 promoter. We have shown that the transcription of the K15 keratin gene is regulated by the receptors for thyroid hormone, retinoic acid, and glucocorticoids. C/EBPβ, AP-1, and IFN-γ are activators of K15 expression, while NF-κB is a suppressor. Thus, K15 promoter activity is, uniquely among keratin genes, increased by both IFN-γ and thyroid hormones. Furthermore, IFN-γ and thyroid hormones increase the amount of K15 protein produced in an ex vivo skin model. We expect that our findings regarding regulation of K15 expression may lead to a better understanding of the role of K15 keratin in normal and pathological conditions and perhaps to treatments of diseases such as EBS based on modulating keratin gene expression.
MATERIALS AND METHODS
DNA constructs.
The construct containing the keratin K15 gene promoter was obtained using the Human PromoterFinder DNA Walking kit (Clontech). For the PCRs, we used K15 GSP1 and K15 GSP2 primers, following the Clontech protocol (Table 1). K15 promoter deletion constructs were prepared by using PCR with the common reverse primer, K15R, in conjunction with a series of forward primers (Table 1). The reverse primer contains a HindIII restriction site, whereas the forward primers contain an XbaI site. The K14 promoter-containing construct has been described previously (31). It contains 2,300 bp of the upstream sequence. The constructs expressing the AP-1 proteins, c-Fos, c-Jun, and Fra1, were a gift from E. Ziff, those expressing NF-κB proteins p65, p50, and c/Rel were a gift from A. Beg, those expressing C/EBP-β were a gift from S. Chen-Kiang, and those expressing CHOP were a gift from D. Ron (5, 19, 56). The constructs expressing nuclear receptors hRAR-γ and cT3R-α were previously described (41, 52). Escherichia coli bacteria containing plasmids were grown to stationary phase in Luria broth, and the plasmid DNAs were purified using maxi-preps (Promega). Constructs GH-CAT and GRE-CAT, containing a well-characterized glucocorticoid receptor-responsive element and the consensus retinoic acid/thyroid hormone responsive elements, have been reported previously (41).
TABLE 1.
Primers used in this study
| Primera | Sequence |
|---|---|
| K15 GSP1 | 5′ TTT AAG CTT CCC CCA GCC AGG AGG GAA CCC 3′ |
| HindIII | |
| K15 GSP2 | 5′ TTT GGA TCC AAG TTT GCA GAA ATG TGG TGG TCA T 3′ |
| BamHI | |
| K15wt F | 5′ TTT CCC GGG CCT GAG GGA CTC CAG AGC 3′ |
| XmaI | |
| K15 921 F | 5′ TTT TCT AGA GCC AGC ACT TGA CAG GAC 3′ |
| XbaI | |
| K15 767 F | 5′ TTT TCT AGA CGT GCA GTT GGC AGG TGT 3′ |
| XbaI | |
| K15 440 F | 5′ TTT TCT AGA CAC AGC ATA ATG ATC GGC 3′ |
| XbaI | |
| K15 340 F | 5′ TTT TCT AGA CAC AGT TGG CCT GAG CTG 3′ |
| XbaI | |
| K15 R | 5′ TTT AAG CTT AAG TTT GCA GAA ATG TGG 3′ |
| HindIII | |
| K15 nuclear receptor −32/−102 F | 5′ GTC AGG AAG GCA GAA GGA GTT GGC TTT GCT TTA GGG GAG GAG ACG AGG TCC CAC AAC ACC CTC TGA AGG G 3′ |
| K15 nuclear receptor oligo F | 5′ CCC TTC AGA GGG TGT TGT GGG ACC 3′ |
| K15 C/EBP −266/−353 F | 5′ GTC CAG CTG AGG GCA CAG TTG GCC TGA GCT GCT CTC AGT ACA GGC AGA GGC CTT GGT AGC TGT GCT GTG ATG AGA GTT CGC TCC CTG 3′ |
| K15 C/EBP −266/−353 oligo F | 5′ AGG GAG CGA ACT CTC ATC ACA GCA 3′ |
| K15 C/EBP −186/−276 F | 5′ TCG CTC CCT GCT GCT CTC TTC TGG CAT GGA GAG ATG AAC CTG TAA TCC AAG TGT TAA AAC CGT GCC CTG GGG GAA AAC ACT ATT AAT TGT 3′ |
| K15 C/EBP −186/−276 oligo F | 5′ ACAATT AAT AGT GTT TTC CCC CAG 3′ |
F, forward; R, reverse. Numbers with shills are beginning and end positions upstream of the transcription start site.
Cells and transfection.
Normal human foreskin epidermal keratinocytes were obtained from M. Simon (Living Skin Bank, Burn Unit, State University of New York, Stony Brook, N.Y.). The cultures were initiated using 3T3 feeder layers as described previously (42) and then frozen in liquid nitrogen until used. Once thawed, the keratinocytes were grown without feeder cells in defined serum-free keratinocyte growth medium, supplemented with 5 ng of epidermal growth factor/ml and 0.05 mg of bovine pituitary extract (keratinocyte-SFM) (Gibco, San Diego, Calif.)/ml at 37°C in 5% CO2. The medium was replaced every 2 days, and cells were expanded through three passages for the experiments. They were trypsinized with 0.025% trypsin, which was neutralized with 0.5 mg of trypsin inhibitor/ml. For all experiments, third-passage keratinocytes were used at 50 to 70% confluence.
HeLa cells were grown in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% calf serum and incubated at 37°C in a 5% CO2 atmosphere. Subconfluent cultures were maintained in 100-mm-diameter dishes by periodically splitting cells with a solution of 0.25% trypsin in Hanks' balanced salt solution (Life Technologies). The day before transfection, cells were plated onto 60-mm-diameter dishes and grown to 15 to 30% confluency. Four hours before transfection, the medium was changed to DMEM supplemented with 10% calf serum. For studies of nuclear receptors, we used phenol-red-free DMEM supplemented with charcoal-pretreated 10% calf serum, depleted of RA, T3, and steroids (41). HeLa cells were cotransfected with K15-CAT and pRSVZ-β-GAL, as well as with constructs expressing c-Fos, c-Jun, Fra1, p65, p50, c/Rel, C/EBP-β, CHOP, retinoic acid receptor (RAR), and T3R using the transfection protocol described previously (20). Briefly, the DNA constructs were diluted in 100 μl of water. First, 25 μl of 2 M CaCl2 and then 125 μl of BES buffer, pH 6.95 (50 mM N-bis-92-hydrohyethyl-2-aminoethanesulfonic acid, 250 mM NaCl, 1.5 mM Na2HPO4), were added drop by drop. The transfection mixture was then incubated at room temperature for 10 to 15 min before being added to the cells. The final concentration of keratin promoter DNA was 1 μg/ml for K15-CAT and 1.5 μg/ml for deletion vectors (K15 921-CAT, K15 767-CAT, K15 440-CAT, and K15 340-CAT). The constructs expressing the transcription factors or nuclear receptors were always added in a 1:3 ratio relative to the keratin promoter constructs. In addition, 0.3 μg of pRSVZ/ml was added to each transfection. For each well, 250 μl of DNA solution was added to 2.25 ml of the growth medium. Cells were incubated with the transfection solution for 12 h, when fresh medium was added. Forty-eight hours following transfection, the cells were washed with phosphate-buffered saline and harvested by scraping.
Cells were disrupted in an extraction buffer (250 mM sucrose, 10 mM Tris [pH 7.8]) with four freeze-thaw cycles, and the transfection efficiency was measured using β-galactosidase assays (20). The chloramphenicol acetyltransferase (CAT) reaction mixture contained 69 μl of 1 M Tris-HCl (pH 7.8), 1 μl of [14C]chloramphenicol (40 to 50 mCi/mmol; New England Nuclear), 20 ml of 4 mM acetyl-coenzyme A solution, 30 to 60 μl of cell extract, and enough water to bring the total reaction volume to 150 μl. After incubation at 37°C for 1 h, the mixture was extracted into 1 ml of ethyl acetate, phases were separated by brief centrifugation, the organic layer was transferred to a new tube, and the solvent was evaporated. The residue was dissolved in 30 ml of ethyl acetate and separated by thin-layer chromatography on silica gel in chloroform-methanol (95:5). The plates were exposed to X-ray film for 12 to 24 h, and the intensity of radioactive spots was determined using a radioanalytic system (Ambis, Inc., San Diego, Calif.). The conversion of chloramphenicol to its monoacetylated derivative was kept below 50% by varying the amount of extract or the duration of the reaction. All CAT values were normalized for transfection efficiency by calculating the ratio of CAT activity to β-galactosidase in each transfected plate. Each transfection experiment was separately performed three or more times, with each data point resulting from a duplicate transfection.
Northern blot analyses.
Total RNA was extracted using the RNeasy Mini kit (Qiagen). Five micrograms of each RNA sample was loaded onto a 1.5% agarose-formaldehyde gel. The RNA was transferred to a nylon membrane (Amersham) and cross-linked in a UVC Stratalinker (Stratagene). DNA probes were labeled with [α-32P]dCTP by using the Random Primed DNA labeling kit (Roche Molecular Biochemicals). The cDNAs of K15, K14, and hypoxanthine phosphoribosyltransferase were previously described (31, 58). The probes were hybridized using ExpressHyb solution (Clontech) at 68°C for 1 h. The membrane was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.05% sodium dodecyl sulfate (SDS) solution, with continuous shaking three times for 30 min at room temperature and with 0.1× SSC-0.1% SDS at 50°C for 40 min. The membrane was exposed to BioMax film (Kodak) at −80°C.
Isolation of basal and differentiating keratinocytes from skin.
Skin, discarded after reduction mammoplasty, was obtained according to the Institutional Review Board (IRB)-approved protocol, generally 2 to 6 h after surgery. The tissue was cut into 3-mm strips and incubated with dispase (2.4 U/ml) and RNase inhibitor (4 U/ml; Roche) at 4°C overnight. Next, the epidermis was gently separated from the dermis and incubated in a solution containing 0.05% trypsin and 0.02% EDTA (Gibco-BRL) at 37°C. After 10 min, 2 volumes of 0.5-mg/ml trypsin inhibitor (Sigma) was added and the tissue was filtered through a cell strainer (Falcon). The trypsinization of the tissue is repeated twice more. The cells were collected by centrifugation, examined using trypan blue, and counted, and, if appropriate, the isolates were combined. Magnetic beads coated with M-450 rat anti-mouse-immunoglobulin G1 were incubated with the cells and the 3E1 clone β4 antibody from Gibco-BRL in 1× phosphate-buffered saline (PBS)-0.1% bovine serum albumin at 4o for 1 to 2 h. The beads were separated on a magnetic separator, and the suprabasal cell population was decanted. The beads with the basal cells were used in RNA isolation without removing the cells from the beads.
Purification of the C/EBP-β protein.
The plasmid expressing glutathione S-transferase (GST)-tagged C/EBP-β (19) was used to transform BL21(DE3) E. coli (US Biologicals), which was grown in Luria broth with ampicillin to an optical density at 600 nm of 0.8 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h. We used the GST-bulk purification kit that includes glutathione-Sepharose 4B and followed the procedures recommended by the manufacturer (Pharmacia). We prepared untagged protein using thrombin to remove the GST tag. The yield and purity of the proteins were assessed using standard SDS polyacrylamide gels.
EMSA.
E. coli-expressed wild-type cT3R and hRAR-α, the DNA-binding-domain portion of hGR, and GST-tagged C/EBP-β plasmid were previously described (19, 41, 52). One microgram of oligonucleotides was labeled using polynucleotide kinase (Promega) and [γ-32P]dATP (Amersham). Oligonucleotides (1.5 × 106 cpm each) were used in the primer extension reaction with Klenow DNA polymerase (Roche Molecular Biochemicals). Sequences of oligonucleotides used in EMSA and DNase I footprinting are listed in Table 1. The products were purified from a 2.5% agarose gel. The band corresponding to the size of the probe was cut out of the gel, eluted overnight into Tris-EDTA buffer (pH 8) at +4°C, and precipitated with ethanol. The resulting probe was mixed with 30 or 50 pg of purified T3R, RAR, and C/EBP-β and 10, 30, and 50 pg of GRE protein and incubated first for 30 min at room temperature and then for 10 min at +4°C. Higher concentrations of the protein contained 150 and 500 pg. The incubation was done in a solution containing 30 μl of 25 mM Tris (pH 7.8), 500 μM EDTA, 88 mM KCl, 10 mM 2-β-mercaptoethanol, 0.1 μg of aprotinin, 0.1 μg of poly(dI-dC), 0.05% Triton X-100 (vol/vol), and 10% glycerol (vol/vol). Samples were loaded onto a 4% polyacrylamide gel and separated by electrophoresis (20 to 25 mA) at +4°C for 2 h with a buffer containing 10 mM Tris (pH 7.8), 7.5 mM acetic acid, and 40 μM EDTA. Gels were dried and exposed to X-ray film for 4 h at −80°C.
DNase I footprinting.
The probes were labeled using the protocol described in the EMSA protocol above. Two different reactions were performed in parallel: A/G Maxam-Gilbert sequencing (using the reagents and protocols from the NEN Life Science Products sequencing kit) and DNase I footprinting (33). For the footprinting reaction, our gel shift protocol was used to allow binding of the protein to the DNA: 25 μl of the binding mix with 50 ng of purified receptor protein and 50,000 cpm of probe. In the experiments where increasing amounts of protein are used, the higher doses were 150 and 500 ng. When the ligand was added, e.g., thyroid hormone (see Fig. 4B), it was added to a final concentration of 10−7 M, which saturates the receptor. After 20 min of incubation at +4°C, 50 μl of solution containing 10 mM MgCl2 and 5 mM CaCl2 was added, and incubation continued for 1 min on ice. Next, 3 μl of the 1:25 dilution of DNase I (5 U/ml of stock), which we have found optimal for our conditions, was added, with incubation for exactly 1 min on ice. The reaction was stopped by adding 90 μl of stop solution containing 20 mM EDTA (pH 8.0), 1% SDS, 0.2 M NaCl, and 100 μg of yeast RNA/ml. DNA was purified by phenol extraction and ethanol precipitation. The pellet was resuspended in 1.4 μl of 9 M urea-1% NP-40, and after mixing, 4.6 μl of formamide loading buffer (US Biologicals) was added. All samples were heated at 90°C for 5 min, chilled on ice, and loaded on 12% sequencing polyacrylamide gels, together with samples from sequencing reactions of the same DNA. Gels were dried on the gel dryer and exposed to the X-ray film. The bands that are protected from cleavage by DNase I by the bound protein determined the footprint localization.
FIG. 4.
Specific binding of purified cT3R-α, hRAR-α, and GR-DBD to the K15 responsive element. (A) The gel shift analysis. Autoradiograms of the gel shift assays with K15 responsive element (bp −32 to −102) as a probe with purified receptor proteins show that the DNA-protein complexes were resolved on a nondenaturing polyacrylamide gel. The triangles above the lanes represent increasing amounts of protein, 30 and 150 pg. The arrows on the left point to the T3R binding complex, those in the middle point to the four GR binding complexes, and that on the right points to the RAR band. (B) Footprinting analysis. The DNA sequence on the left represents the K15 RE. The triangles represent 50, 150, and 500 ng. T3 was added to a concentration of 10−7 M. The vertical bars on the left represent the footprint sequences protected by the three nuclear receptors. The “unbound” lanes contain unprotected DNA.
Treatment of skin explants and immunohistology.
Human skin specimens were obtained from reduction mammoplasty following a protocol approved by the local IRB. The fat layer was removed, and circular biopsies were obtained using a 3-mm biopsy punch and submerged in keratinocyte-SFM medium (Gibco-BRL) containing penicillin and streptomycin (Gibco). Biopsies were maintained submerged for 48 h under the following conditions: control (no additives), human recombinant IFN-γ (100 ng/ml; Sigma), Thyroid hormone (1 μM final concentration, from 1 mM stock in 0.1 N NaOH; Sigma), or a combination of the two. Following incubation for 24 h, skin biopsies were embedded in OCT compound (Tissue Tek) and frozen in liquid nitrogen. Five-micrometer-thick skin sections were cut with a cryostat (Jung Frigocut 28006; Leica) and stored at −80°C. Slides containing frozen sections were washed in PBS and fixed in acetone for 1 min at room temperature. Following another washing with PBS, mouse monoclonal antibody detecting human K15 (58) was applied diluted 1:125 in PBS with 5% bovine serum albumin. After overnight incubation with the primary antibody at +4°C, slides were rinsed in PBS and incubated with secondary fluorescence-conjugated anti-mouse immunoglobulin G antibody diluted 1:60 (Sigma-Aldrich) for 3 h at room temperature. After the final wash in PBS, they were mounted with mounting media (Fluoromount-G; Southern Biotechnology Associates, Inc.) and covered with coverslips. As a negative control, untreated skin was stained, omitting the incubation with primary antibody. Stained sections were examined under a Carl Zeiss microscope, and digital images were collected using the Adobe Photoshop TWAIN_32 program.
RESULTS
Sequence features in the promoter of the human K15 gene.
K15 is expressed in vivo in the basal keratinocytes of stratified epithelia and in the hair bulge (30, 58, 60). However, the expression of K15 in cultured epidermal cells has not been reported, and because we intended to analyze the regulation of the K15 promoter by transfecting keratinocytes in culture, first we needed to ascertain that K15 is expressed under our experimental conditions. Therefore, we used Northern blot analyses to compare the expression of K15 and K14 in cultured primary human keratinocytes with their expression in the basal and differentiated layers of the epidermis. K15 is coexpressed with K14 in the cultured as well as in the basal epidermal keratinocytes, although it appears that the abundance of K15 mRNA is relatively reduced in the cultured cells. Neither K14 nor K15 is expressed in the differentiated cells (Fig. 1).
FIG. 1.
Expression of K15 keratin in human epidermal basal keratinocytes. Northern blot analysis of RNA from cultured primary human keratinocytes (1), total epidermis (2), separated epidermal differentiated keratinocytes (3), or basal keratinocytes (4) are shown. Five-microgram aliquots of RNA were resolved by electrophoresis through a formaldehyde agarose gel. After brief staining with ethidium bromide to visualize the rRNA bands, the gel was processed for Northern blot analysis. The blot was successively hybridized with [32P]dCTP-labeled DNA probes derived from cDNAs encoding K15, K14, and hypoxanthine phosphoribosyltransferase (internal control).
To investigate regulation of K15 expression in keratinocytes, we cloned the DNA 1,251 bp upstream of the human K15 gene and determined its nucleotide sequence (Fig. 2A). We matched the sequence with the previously published murine sequence (38). Using the TransFac program to screen the K15 promoter region (http://bimas.dcrt.nih.gov/molbio/signal/index.html) for the presence of binding sites for different transcription factors, we identified several potential binding sites, including a TATA box and sites for AP-1, AP-2, C/EBP, c-Myc, GATA-1, GATA-3, H4TF-2, IRF-2, and NF-1 (Fig. 2A and data not shown). A prominent feature of the promoter is a repetitive sequence consisting of 11 TCAC repeats at the position −459 to −503 bp upstream of the transcription start site. The murine K15 promoter does not contain the repeats (38).
FIG.2.
Promoter sequence of the K15 gene and comparison of the basal activities of the K15 promoter deletion constructs. (A) Sequence of the human K15 keratin gene promoter. Numbers indicate the positions of bases upstream from the transcription start site. The translated sequence is shown in boldface. The repetitive sequence, STAT-1, C/EBP-β, and AP-1 binding sites, the T3R, RAR, and GR recognition half-sites, and the TATA box sequence are underlined. (B) Basal activity of the deletion constructs. The constructs containing the K15 promoter DNA were transfected into HeLa cells. The basic activity of the K15 wt-CAT construct was considered to be 1, and the activities of all the deletion constructs are shown relative to this. The schematics of the promoter region correspond to the features described in Fig. 2A.
To examine the roles of the identified sites in regulating K15 expression, we constructed several promoter deletions using PCR (Fig. 2B). The activities of K15 921-CAT and K15 767-CAT constructs, which contain the TCAC repeats, were almost zero, while the activity of the K15 440-CAT construct, which does not, was the same as that of K15 wt-CAT. These data suggest the presence of a transcriptional silencer in the region containing the repeats, between bp −440 and −767, and also suggest that the sequences further upstream, at bp −921 to −1251, counteracts this silencing. To test the possibility that the sequence at bp −440 to −767 contains a context-independent silencer, we cloned it into a different context, in front of a K14-CAT vector. However, the basal activities of the constructs with and without the silencer were the same (data not shown). Therefore, if there is a silencer located in this 327-bp region, it seems to be specific for the context of the K15 promoter.
Regulation of the K15 gene promoter by thyroid hormone, retinoic acid, and glucocorticoids.
We have previously described a unique mechanism of transcriptional regulation of keratin genes by nuclear receptors. Most keratin genes are inhibited by ligand-occupied T3R and RAR but are activated by unliganded T3R (51, 53). Regulation occurs through a direct binding of the receptors to the responsive elements in the keratin gene promoters (40, 52). Glucocorticoids directly suppress the transcription of a subset of keratin genes, the K5, K14, K6, K16, and K17 genes, by a mechanism that involves binding of the receptor to the glucocorticoid-responsive elements in the keratin gene promoters (41). Because all three nuclear receptors, T3R, RAR, and GR, regulate expression of basal keratins, K5 and K14, we hypothesized that they also affect the K15 gene expression.
To test this hypothesis, we cotransfected HeLa cells with constructs expressing either K15 wt-CAT or K14 wt-CAT with cT3R-α in the absence or presence of thyroid hormone and with hRAR-γ in the absence or presence of RA (Fig. 3A and B). The GH-CAT construct, representing a native responsive element induced by the cT3R-α and hRAR-γ receptors in the presence of their ligands, served as the positive control (Fig. 3C). The activity of the K14 promoter was suppressed by the thyroid hormone receptor in the presence of thyroid hormone and induced by the unliganded receptor, as expected (Fig. 3B). Surprisingly, K15 promoter activity was significantly induced by the presence of thyroid hormone, while it was suppressed by the unliganded receptor (Fig. 3A). Thus, the transcriptional regulation of K15 gene expression by T3R is unique among those for keratin genes, because the thyroid hormone has been shown to suppress, not induce, the expression of all other keratin genes tested (40, 41, 51, 53). As expected, and in contrast to the case with T3R, in the presence of its ligand RAR suppresses K15 activity approximately threefold, as it does for other keratin genes including K14 (Fig. 3A and B).
FIG. 3.
Comparison of regulation of K15 and K14 keratin gene expression by T3, RA, and GR receptors in HeLa cells. The K15 construct (A) and K14 construct (B) were cotransfected along with plasmids expressing cT3R-α and hRAR-γ receptors, and the transfected cells were incubated in the presence or absence of T3, RA, or dexametasone (DEX). The basal activities of the promoters were considered to be 1, and the relative activities in the presence of ligands and cotransfected receptors were determined. The β-galactosidase activities from cotransfected LacZ-expressing vector, which do not change in response to nuclear receptors, were used to normalize the CAT activities. (C and D) Regulation of the positive control plasmids, GH-CAT and GRE-CAT. While GH-CAT and GRE-CAT are induced by the corresponding ligands, K14 is suppressed by all three. Importantly, the K15 and K14 promoters are affected in opposite ways from each other in response to T3 and T3R.
We compared the effect of GR on the expression of the K15 and K14 genes by transfecting HeLa cells with K15 wt-CAT or K14 wt-CAT in the absence and presence of dexametasone (Fig. 3B). Glucocorticoid receptor is present at sufficiently high levels in HeLa cells. Both K15 and K14 were suppressed by GR and its ligand (Fig. 3A and B), while the GRE-CAT construct used as a positive control was induced (Fig. 3D), as expected.
Identification of the responsive elements for T3R, RAR, and GR in the K15 gene promoter.
We have previously identified the DNA regions containing multiple binding sites for T3R, RAR, and GR in the keratin promoters K5, K14, K6, K16, and K17 (40, 41, 51, 53). The promoter deletion analyses showed that the K15 340-CAT vector was still regulated by T3R, RAR, and GR, indicating that the responsive elements of the K15 gene are located within the first 340 bp (data not shown). Detailed analyses of the sequence revealed a potential responsive element comprising the region located 44 to 65 bp upstream of the transcription start site, similar to the elements in other keratin genes. Therefore, we performed gel shift assays with the DNA fragment comprising bp −32 to −102 as a radioactively labeled probe for binding purified receptors cT3R-α, hRAR-α, and a recombinant GR protein segment containing the DNA binding domain (Fig. 4A). By analogy with other keratin gene promoters (41, 53), we find that the −32 to −102 DNA fragment binds cT3R-α, forming two complexes (Fig. 4A). The addition of thyroid hormone (T3) significantly changed the binding of cT3R to the DNA, increasing the amount of the faster-running complex and decreasing the slower one. A similar result was found on a positively regulated TRE (41).
hRAR-α also binds to the K15 probe, which is similar to what we found with other keratin genes (Fig. 4A). Using the same DNA fragment, we found that GR binds to the fragment as well (Fig. 4A). Again, similar to the case with other keratin gene promoters (41, 53), as the concentration of the receptor increases, up to four GR proteins bind this DNA. From these results, we concluded that all three receptors, cT3R-α, hRAR-α, and GR, bind to the same responsive element of the K15 DNA promoter.
To map the exact interaction sites between the K15 DNA-responsive region and the nuclear receptors, we used the −32 to −102 K15 DNA fragment in DNase I footprinting experiments (Fig. 4B). Each of these three nuclear receptors contains at least two binding sites within this region. The purified cT3R-α protected the K15 DNA sequence spanning bp −49 to −65 from cleavage by DNase I. We propose that AGGAGA (−60 to −65) and AGGTCC (−52 to −57) are the binding sites for cT3R-α. The addition of the ligand, T3, does not change the binding of T3R to the site comprising bp −49 to −56 but significantly reduces the binding to the other site, −57 to −65. The hRAR-α and GR receptors protected the K15 DNA sequences from bp −43 to −65 and −46to −65, respectively, from cleavage. We propose that AGGTCC (−52 to −57) and ACAACA (−45 to −50) constitute the RARE in the K15 promoter. This experiment also suggested that the binding sites for GR (−42 to −65) overlap with both the T3R and the RAR binding sites (Fig. 4B).
Transcriptional regulation of the K15 gene by C/EBP-β, a transcription factor associated with the basal layer.
C/EBPs and CHOP are expressed in the skin in a spatially organized manner that corresponds to the timing of epidermal differentiation (34). To test whether they are involved in transcriptional regulation of the K15 gene, we cotransfected into HeLa cells the K15 wt-CAT construct together with plasmids expressing the C/EBP-β and CHOP proteins (Fig. 5A). C/EBP-β increased the K15 promoter activity sixfold. CHOP, an inhibitor of C/EBPβ, decreased K15 activity twofold. When CHOP was cotransfected with C/EBP-β, CHOP suppressed induction by C/EBP-β, reducing K15 activity to the baseline level.
FIG. 5.
Transcriptional regulation of K15 keratin gene expression by the C/EBP transcription factors. (A) Cotransfection experiments. K15 wt-CAT was cotransfected into HeLa cells with constructs expressing C/EBP-β or CHOP. The basal activity of the K15 wt-CAT construct was considered to be 1, and the relative activities of the promoter in the presence of cotransfected constructs are shown. β-Galactosidase activity was used to normalize the CAT activities. (B) Binding of C/EBP-β to the K15 promoter region by gel shift analysis with the purified CEB/Pβ protein and the −266/−353 probe. Arrows point to the bound C/EBP-β protein on the DNA. (C) The footprinting analysis. The DNA sequence of the K15 promoter is given on the left alongside the gel. The leftmost lanes are sequencing lanes for T/C, A/G, and G. Lanes marked U contain the unbound, free DNA. The triangle on top represents 50 and 150 ng of the purified C/EBP-β protein. The rectangle marks the protected C/EBP-β binding site in the K15 promoter region.
Detailed analysis of the promoter sequence pointed to two sites similar to the C/EBP consensus, located at bp −282 to −291 and bp −309 to −318. To determine whether the sequences are bound by C/EBP-β, we used the −266/−353probe, which contains both sites, in a gel shift experiment and found that the probe binds CEBP-β protein with significant affinity (Fig. 5B). We used the K15 DNA fragment comprising bp −266 to −353 with purified C/EBP-β protein in DNase I footprint experiments and showed that the C/EBP-β transcription factor directly interacts with the K15 DNA region, protecting the sequence TGCTGTGATGA (bp −282 to −291) (Fig. 5C). This DNA region closely resembles the consensus-binding motif for C/EBPs, TGCTGTAATG, and is similar to the site shown by point mutations to be the primary C/EBP-β binding site in the K6 promoter, matching 7 of 11 nucleotides (28). The significance of the other near-consensus sequence in the K15 promoter at −309 to −318 is, at present, unexplored.
Transcriptional regulation of the K15 gene by AP-1 and NF-κB, the activation-associated transcription factors.
Proinflammatory cytokines, such as TNF-α and interleukin 1 (IL-1), and growth factors, such as epidermal growth factor, regulate gene expression through activation of transcription factors that belong to the AP-1 and NF-κB families. To investigate the transcriptional regulation of K15 keratin gene expression by these transcription factors, we cotransfected the K15 wt-CAT construct into HeLa cells together with constructs expressing the AP-1 proteins c-Fos, c-Jun, and Fra1 and the NF-κB proteins p50, p65, and c/Rel (Fig. 6). Although cotransfecting c-Fos or c-Jun separately increased the K15 promoter activity less than twofold, the simultaneous cotransfection of c-Fos and c-Jun produced a synergistic response of approximately sixfold (Fig. 6A). On the other hand, Fra1, another member of the AP-1 family, did not significantly affect the K15 promoter activity. Promoter deletion analyses showed that the K15 340-CAT vector was still regulated by the AP-1 transcription factor (data not shown). We detected one perfect AP-1 consensus sequence (bp −181 to −187) and a few AP-1-like sequences (−1144 to −1150, −1031 to −1037, −834 to −842, and −129 to −135). When we mutated 6 bp of the −181 to −187 AP-1 consensus sequence in K15 340-CAT into a SmaI site, the construct was still fully regulated by AP-1 (data not shown). Either this AP-1 site is not functional or in its absence other sites have a compensatory effect.
FIG. 6.
Transcriptional regulation of K15 keratin gene expression by the AP-1 and NF-κB transcription factors. Construct containing the K15 promoter was cotransfected into HeLa cells with constructs expressing AP-1 proteins c-Fos, c-Jun, and Fra-1, individually or in combination (A), or NF-κB proteins p50, p65, and c/Rel, individually or in combination (B). The basal activity of the K15 wt-CAT construct was considered to be 1.
The NF-κB proteins had a significant suppressive effect on K15 promoter activity (Fig. 6B). Cotransfection of p65 individually and in combination with p50 suppressed K15 promoter activity fourfold. c/Rel or p50 individually did not have any effect on the K15 promoter activity. Promoter deletion analyses showed that the K15 340-CAT construct was still regulated by the NF-κB transcription factor (data not shown). There is no NF-κB consensus site in the K15 340-CAT vector. It is possible that regulation of K15 promoter activity by NF-κB proteins occurs through interaction with some other transcription factor and that NF-κB only indirectly binds to the promoter region. We note that NF-κB regulates, e.g., K6 keratin expression through interaction with C/EBP-β (28). From the above data, we conclude that the expression of the K15 gene is induced by the AP-1 proteins c-Fos and c-Jun and suppressed by the p65 NF-κB transcription factor.
Transcriptional regulation of the K15 gene by IFN-γ.
Some inflammatory epidermal diseases, such as psoriasis, are associated with production of IFN-γ. Keratinocytes are an important target of IFN-γ, which activates transcription factor STAT-1 and induces the expression of keratin K17, among other proteins (4, 21, 26). Therefore, we examined the regulation of the K15 gene by this cytokine. We transfected K15 wt-CAT, K15 340-CAT, and K14 wt-CAT into HeLa cells and cultured the cells in the presence or absence of IFN-γ (Fig. 7). IFN-γ increased promoter activity of K15 wt-CAT sevenfold and of K15 340-CAT twofold. In contrast to the expression of K15, the expression of the major basal type I keratin, K14, was not regulated by IFN-γ. Sequence analysis of K15 wt-CAT detected a STAT-1 consensus binding sequence (TTCN2-4GAA) at position −834 to −842; this sequence is missing from the K15 340-CAT construct. It is not clear at present which sequences within the 340-bp construct mediate the twofold induction. The identification of the IFN-γ-responsive sequences is one of our future goals.
FIG. 7.
Transcriptional regulation of the K15 keratin gene expression by IFN-γ. Constructs K15 wt-CAT, K15 340-CAT, and K14 wt-CAT were transfected into HeLa cells, which were then incubated in the presence or absence of IFN-γ. Fold increase due to IFN-γ is shown. Note that the negative control plasmid, K14 wt-CAT, is not regulated by IFN-γ.
Overall, the results suggest that K15 expression, but not K14 expression, may be specifically inducible by thyroid hormones and IFN-γ in the epidermal basal cells. We hypothesize that such induction may alleviate the symptoms of K14 deficiency.
Thyroid hormones and IFN-γ increase the expression of K15 in human skin.
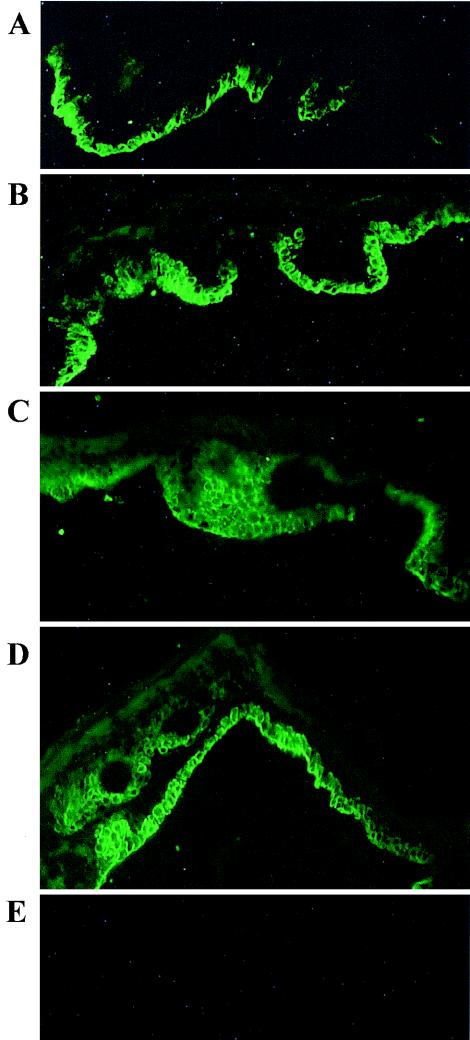
Having established the possibility of selectively inducing K15 by using thyroid hormones and IFN-γ, we decided to determine whether similar effects can be achieved in human skin. We used an ex vivo system, first developed by Varani et al. (55), which responds to extracellular signals, such as those of TNF-α and IL-1 (27, 28). Biopsies of skin obtained after breast reduction surgery were placed in a culture medium and either left untreated, as controls, or treated with thyroid hormones, IFN-γ, or both (Fig. 8). While in the control samples the basal layer shows relatively weak, patchy staining with the K15-specific antibody, in the thyroid hormone-treated samples much stronger staining overall was observed, with some regions showing very high expression of K15 but still some patches remaining unstained. On the other hand, the IFN-γ-treated samples show very strong staining throughout the basal layer. Moreover, we observed the presence of K15 keratin in suprabasal cells. Similar distribution of K15 is seen in the samples treated with both agents simultaneously. These ex vivo results confirm the results of transfection assays, namely, thyroid hormones and IFN-γ can increase the expression of K15 protein in human epidermal keratinocytes.
FIG. 8.
Induction of K15 protein synthesis in human skin in vivo. Samples of human skin were placed in culture and either left untreated as the control (A) or treated with thyroid hormone (B), IFN-γ (C), or both (D). Note the increased but still patchy staining in the thyroid hormone-treated sample and the contiguous staining in the basal layer of the IFN-γ-treated sample. (E) Control without the primary antibody.
DISCUSSION
The results presented here suggest that it is possible to enhance exclusively the expression of keratin K15 in epidermal keratinocytes. Specifically, thyroid hormone and IFN-γ activate the K15 promoter but do not affect the expression of other basal cell keratins. Skin samples treated with thyroid hormone and IFN-γ increase the production of K15 in the basal layer. Treatments based on these two extracellular signaling molecules may improve the mechanical strength of basal cells in EBS.
The effect of the thyroid hormone is particularly striking because this agent suppresses the expression of all other epidermal keratin genes tested (40, 41, 51, 53). Therefore, it may boost K15, while at the same time suppressing the expression of mutant K14. Thyroid hormone suppresses both K14 and K5 expression (41, 53). We believe that K5, which is expressed at much higher levels than K15, will be in excess in basal keratinocytes even in the presence of thyroid hormone and will therefore pair with K15, while the expression of mutant K14 is suppressed. The ratio of healthy to mutant keratin protein strongly affects the integrity of the intermediate filament network (2, 9, 10). Therefore, we expect that an increase of K15 expression, coupled with even a partial reduction of K14 (and K5), will be beneficial in EBS.
The data show that three nuclear receptors bind in the same DNA region. Interestingly, according to gel shift and footprinting results, the binding pattern of T3R for the K15 responsive element is more similar to the binding pattern for the positive, consensus responsive element than that for the negative elements found in other keratin genes (52). This correlates with the outcomes of transcriptional regulation, i.e., the K15 responsive element mediates positive regulation. In contrast, the patterns of binding of RAR and GR to the K15 responsive element are similar to the binding patterns of their receptors for other keratin responsive elements, which also correlates well with their effects on transcriptional regulation. The precise molecular mechanisms of the regulatory processes at present remain unexplained.
The induction of K15 by IFN-γ parallels the induction of K17 (21, 26). However, while K15 is expressed strictly in the basal layer, K17 is found only in the suprabasal layers. Therefore, replacement of K14 in EBS patients by K17 is not feasible. These results open the possibility, however, that the inflammatory processes that accompany the blistering, if associated with the production of IFN-γ, may ameliorate the condition by inducing K15.
Similar to the case with other keratin genes, K15 is regulated by the AP-1 and NF-κB proteins. AP-1 seems to enhance the expression of all keratin genes. It is therefore surprising that K15 is suppressed in activated keratinocytes (57, 59), associated with active AP-1 proteins (6, 7, 44, 45). On the other hand, NF-κB suppresses the basal cell-specific keratins, K5 and K14, in addition to K15, while strongly inducing the activated keratinocyte-specific keratins, K6 and K16 (23, 46). The NF-κB proteins may be instrumental in suppressing the expression of K15 in those processes that are associated with the production of TNF-α or IL-1, strong activators of NF-κB (5).
Two interesting features of the promoter sequence bear emphasis. The first is the presence of the silencer in the region comprising −440 to −767. The role of the silencer in the regulation of K15 expression is at present unknown. The second feature is the repetitive segment in the region comprising bp −459 to −503. While the functional significance of the segment is not clear, such simple repeats tend to be highly polymorphic. The variable number of these tandem repeats could potentially be used as a marker for studies of inheritance in the acidic type keratin gene locus.
In conclusion, this study shows that K15 keratin can be specifically induced at the transcriptional level by thyroid hormone and IFN-γ, and it suggests the possibility of alleviating the symptoms of EBS by inducing K15 production. This model, induction of expression of another protein of the same family to compensate for the loss of function of a mutated protein, could be a useful approach in treatment of some genetic disorders.
Acknowledgments
Our experiments have been supported by grants AR30682, AR41850, and AR40522 (M. Blumenberg) and grant AR45974 (M. Tomic-Canic) from the National Institutes of Health and by Cancer Research UK and the RAB Charitable Foundation (A. Waseem).
REFERENCES
- 1.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 2.Arin, M., M. Longley, X. Wang, and D. Roop. 2001. Focal activation of a mutant allele defines the role of stem cells in mosaic skin disorders. J. Cell Biol. 152:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader, B. L., L. Jahn, and W. W. Franke. 1988. Low level expression of cytokeratins 8, 18 and 19 in vascular smooth muscle cells of human umbilical cord and in cultured cells derived therefrom, with an analysis of the chromosomal locus containing the cytokeratin 19 gene Eur. J. Cell Biol. 47:300-319. [PubMed] [Google Scholar]
- 4.Banno, T., M. Adachi, L. Mukkamala, and M. Blumenberg. 2003. Unique keratinocyte-specific effects of interferon-g that protect skin from viruses, identified using transcriptional profiling. Antivir. Ther. 8:119-132. [PubMed] [Google Scholar]
- 5.Beg, A. A., T. S. Finco, P. V. Nantermet, and A. Baldwin, Jr. 1993. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of I kappa B alpha: a mechanism for NF-kappa B activation. Mol. Cell. Biol. 13:3301-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernerd, F., T. Magnaldo, and M. Darmon. 1992. Delayed onset of epidermal differentiation in psoriasis. J. Investig. Dermatol. 98:902-910. [DOI] [PubMed] [Google Scholar]
- 7.Bernerd, F., T. Magnaldo, I. M. Freedberg, and M. Blumenberg. 1993. Expression of the carcinoma-associated keratin K6 and the role of AP-1 proto-oncoproteins. Gene Expr. 3:187-199. [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenberg, M., and M. Tomic-Canic. 1997. Human epidermal keratinocyte: keratinization processes, p. 1-29. In P. Jolles et al. (ed.), Formation and structure of human hair, vol. 78. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 9.Cao, T., M. J. Arin, and D. R. Roop. 2001. Gene therapy for inherited skin diseases. Curr. Probl. Dermatol. 13:173-182. [Google Scholar]
- 10.Cao, T., M. A. Longley, X. J. Wang, and D. R. Roop. 2001. An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy J. Cell Biol. 152:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, Y., I. A. Lamprecht, Y. Qian-Chun, A. Jackel, B. Zabel, J. P. Ernst, and E. Fuchs. 1994. A human keratin 14 “knockout”: the absence of K14 leads to severe epidermolysis bullosa simplex and a function for an intermediate filament protein. Genes Dev. 8:2574-2587. [DOI] [PubMed] [Google Scholar]
- 12.Dajee, M., M. Lazarov, J. Y. Zhang, T. Cai, C. L. Green, A. J. Russell, M. P. Marinkovich, S. Tao, Q. Lin, Y. Kubo, and P. A. Khavari. 2003. NF-κB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature 421:639-643. [DOI] [PubMed] [Google Scholar]
- 13.Dellambra, E., G. Pellegrini, L. Guerra, G. Ferrari, G. Zambruno, F. Mavilio, and M. De Luca. 2000. Toward epidermal stem cell-mediated ex vivo gene therapy of junctional epidermolysis bullosa. Hum. Gene Ther. 11:2283-2287. [DOI] [PubMed] [Google Scholar]
- 14.Desnick, R. J. 2001. Enzyme replacement and beyond. J. Inherit. Metab. Dis. 24:251-265. [DOI] [PubMed] [Google Scholar]
- 15.Eady, R. A. 2001. Epidermolysis bullosa: scientific advances and therapeutic challenges. J. Dermatol. 28:638-640. [DOI] [PubMed] [Google Scholar]
- 16.Freedberg, I. M., M. Tomic-Canic, M. Komine, and M. Blumenberg. 2001. Keratins and the keratinocyte activation cycle. J. Investig. Dermatol. 116:633-640. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, E. 1994. Intermediate filaments and disease: mutations that cripple cell strength. J. Cell Biol. 125:511-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, E., and H. Green. 1980. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell 19:1033-1042. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, W., and S. Chen-Kiang. 1993. Convergent regulation of NF-IL6 and Oct-1 synthesis by interleukin-6 and retinoic acid signaling in embryonal carcinoma cells. Mol. Cell. Biol. 13:2515-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, C. K., D. Connolly, and M. Blumenberg. 1991. Comparison of methods for transfection of human epidermal keratinocytes. J. Investig. Dermatol. 97:969-973. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, C. K., S. Flanagan, M. Ohtsuki, K. Shuai, I. M. Freedberg, and M. Blumenberg. 1994. Disease-activated transcription factor: allergic reactions in human skin cause nuclear transcription of STAT-91 and induce synthesis of keratin K17. Mol. Cell. Biol. 14:4759-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, C. K., T. Magnaldo, M. Ohtsuki, I. M. Freedberg, F. Bernerd, and M. Blumenberg. 1993. Epidermal growth factor and transforming growth factor alpha specifically induce the activation- and hyperproliferation-associated keratins 6 and 16. Proc. Natl. Acad. Sci. USA 90:6786-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman, C. K., and E. Fuchs. 2000. It's got you covered. NF-kappaB in the epidermis. J. Cell Biol. 149:999-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khavari, P. A., O. Rollman, and A. Vahlquist. 2002. Cutaneous gene transfer for skin and systemic diseases. J. Intern. Med. 252:1-10. [DOI] [PubMed] [Google Scholar]
- 25.Klampfer, L., T. H. Lee, W. Hsu, J. Vilcek, and S. Chen-Kiang. 1994. NF-IL6 and AP-1 cooperatively modulate the activation of the TSG-6 gene by tumor necrosis factor alpha and interleukin-1. Mol. Cell. Biol. 14:6561-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komine, M., I. M. Freedberg, and M. Blumenberg. 1996. Regulation of epidermal expression of keratin K17 in inflammatory skin diseases. J. Investig. Dermatol. 107:569-575. [DOI] [PubMed] [Google Scholar]
- 27.Komine, M., L. S. Rao, I. M. Freedberg, M. Simon, V. Milisavljevic, and M. Blumenberg. 2001. Interleukin-1 induces transcription of keratin K6 in human epidermal keratinocytes J. Investig. Dermatol. 116:330-338. [DOI] [PubMed] [Google Scholar]
- 28.Komine, M., L. S. Rao, T. Kaneko, M. Tomic-Canic, K. Tamaki, I. M. Freedberg, and M. Blumenberg. 2000. Inflammatory versus proliferative processes in epidermis. Tumor necrosis factor alpha induces K6b keratin synthesis through a transcriptional complex containing NFkappa B and C/EBPbeta. J. Biol. Chem. 275:32077-32088. [DOI] [PubMed] [Google Scholar]
- 29.Kren, B. T., N. R. Chowdhury, J. R. Chowdhury, and C. J. Steer. 2002. Gene therapy as an alternative to liver transplantation. Liver Transpl. 8:1089-1108. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd, C., Q. C. Yu, J. Cheng, K. Turksen, L. Degenstein, E. Hutton, and E. Fuchs. 1995. The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J. Cell Biol. 129:1329-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, S., L. Rao, I. M. Freedberg, and M. Blumenberg. 1997. Transcriptional control of K5, K6, K14, and K17 keratin genes by AP-1 and NF-kappaB family members. Gene Expr. 6:361-370. [PMC free article] [PubMed] [Google Scholar]
- 32.Magnaldo, T., and A. Sarasin. 2002. Genetic reversion of inherited skin disorders. Mutat. Res. 509:211-220. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Maytin, E. V., and J. F. Habener. 1998. Transcription factors C/EBP alpha, C/EBP beta, and CHOP (Gadd153) expressed during the differentiation program of keratinocytes in vitro and in vivo. J. Investig. Dermatol. 110:238-246. [DOI] [PubMed] [Google Scholar]
- 35.Moll, R., W. W. Franke, D. L. Schiller, B. Geiger, and R. Krepler. 1982. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 31:11-24. [DOI] [PubMed] [Google Scholar]
- 36.Nelson, W. G., and T.-T. Sun. 1983. The 50- and 58-kdalton keratin classes as molecular markers for stratified squamous epithelia: cell culture studies. J. Cell Biol. 97:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickoloff, B. J., C. E. Griffiths, and J. N. Barker. 1990. The role of adhesion molecules, chemotactic factors, and cytokines in inflammatory and neoplastic skin disease—1990 update. J. Investig. Dermatol. 94:151S-157S. [DOI] [PubMed] [Google Scholar]
- 38.Nozaki, M., M. Mori, and A. Matsushiro. 1994. The complete sequence of the gene encoding mouse cytokeratin 15. Gene 138:197-200. [DOI] [PubMed] [Google Scholar]
- 39.Porter, R. M., and E. B. Lane. 2003. Phenotypes, genotypes and their contribution to understanding keratin function. Trends Genet. 19:278-285. [DOI] [PubMed] [Google Scholar]
- 40.Radoja, N., D. V. Diaz, T. J. Minars, I. M. Freedberg, M. Blumenberg, and M. Tomic-Canic. 1997. Specific organization of the negative response elements for retinoic acid and thyroid hormone receptors in keratin gene family. J. Investig. Dermatol. 109:566-572. [DOI] [PubMed] [Google Scholar]
- 41.Radoja, N., M. Komine, S. H. Jho, M. Blumenberg, and M. Tomic-Canic. 2000. Novel mechanism of steroid action in skin through glucocorticoid receptor monomers. Mol. Cell. Biol. 20:4328-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rheinwald, J. G., and H. Green. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6:331-344. [DOI] [PubMed] [Google Scholar]
- 43.Rugg, E. L., W. H. I. McLean, E. B. Lane, R. Pitera, J. R. McMillan, P. J. C. Dopping-Hepenstal, H. A. Navsaria, I. M. Leigh, and R. A. J. Eady. 1994. A functional “knockout” of human keratin 14. Genes Dev. 8:2563-2573. [DOI] [PubMed] [Google Scholar]
- 44.Rutberg, S. E., E. Saez, A. Glick, A. A. Dlugosz, B. M. Spiegelman, and S. H. Yuspa. 1996. Differentiation of mouse keratinocytes is accompanied by PKC-dependent changes in AP-1 proteins. Oncogene 13:167-176. [PubMed] [Google Scholar]
- 45.Saez, E., S. E. Rutberg, E. Mueller, H. Oppenheim, J. Smoluk, S. H. Yuspa, and B. M. Spiegelman. 1995. c-fos is required for malignant progression of skin tumors. Cell 82:721-732. [DOI] [PubMed] [Google Scholar]
- 46.Seitz, C. S., Q. Lin, H. Deng, and P. A. Khavari. 1998. Alterations in NF-kappaB function in transgenic epithelial tissue demonstrate a growth inhibitory role for NF-kappaB. Proc. Natl. Acad. Sci. USA 95:2307-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuai, K. 1994. Interferon-activated signal transduction to the nucleus. Curr. Opin. Cell Biol. 6:253-259. [DOI] [PubMed] [Google Scholar]
- 48.Shuai, K., C. M. Horvath, L. H. Huang, S. A. Qureshi, D. Cowburn, and J. E. Darnell. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 49.Steinert, P. M., and D. R. Roop. 1988. Molecular and cellular biology of intermediate filaments. Annu. Rev. Biochem. 57:593-625. [DOI] [PubMed] [Google Scholar]
- 50.Takeda, K., O. Takeuchi, T. Tsujimura, S. Itami, O. Adachi, T. Kawai, H. Sanjo, K. Yoshikawa, N. Terada, and S. Akira. 1999. Limb and skin abnormalities in mice lacking IKKα. Science 284:313-316. [DOI] [PubMed] [Google Scholar]
- 51.Tomic, M., C.-K. Jiang, H. S. Epstein, I. M. Freedberg, H. H. Samuels, and M. Blumenberg. 1990. Nuclear receptors for retinoic acid and thyroid hormone regulate transcription of keratin genes. Cell Regul. 1:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomic-Canic, M., I. M. Freedberg, and M. Blumenberg. 1996. Codominant regulation of keratin gene expression by cell surface receptors and nuclear receptors. Exp. Cell Res. 224:96-102. [DOI] [PubMed] [Google Scholar]
- 53.Tomic-Canic, M., I. Sunjevaric, I. M. Freedberg, and M. Blumenberg. 1992. Identification of the retinoic acid and thyroid hormone receptor-responsive element in the human K14 keratin gene. J. Investig. Dermatol. 99:842-847. [DOI] [PubMed] [Google Scholar]
- 54.Trautwein, C., C. Caelles, P. van der Geer, T. Hunter, M. Karin, and M. Chojkier. 1993. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature 364:544-547. [DOI] [PubMed] [Google Scholar]
- 55.Varani, J., S. E. Fligiel, L. Schuger, P. Perone, D. Inman, C. E. Griffiths, and J. J. Voorhees. 1993. Effects of all-trans retinoic acid and Ca++ on human skin in organ culture. Am. J. Pathol. 142:189-198. [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, X. Z., and D. Ron. 1996. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science 272:1347-1349. [DOI] [PubMed] [Google Scholar]
- 57.Waseem, A., C. M. Alexander, J. B. Steel, and E. B. Lane. 1990. Embryonic simple epithelial keratins 8 and 18: chromosomal location emphasizes difference from other keratin pairs. New Biol. 2:464-478. [PubMed] [Google Scholar]
- 58.Waseem, A., B. Dogan, N. Tidman, Y. Alam, P. Purkis, S. Jackson, A. Lalli, M. Machesney, and I. M. Leigh. 1999. Keratin 15 expression in stratified epithelia: downregulation in activated keratinocytes. J. Investig. Dermatol. 112:362-369. [DOI] [PubMed] [Google Scholar]
- 59.Werner, S., and B. Munz. 2000. Suppression of keratin 15 expression by transforming growth factor beta in vitro and by cutaneous injury in vivo. Exp. Cell Res. 254:80-90. [DOI] [PubMed] [Google Scholar]
- 60.Whitbread, L. A., and B. C. Powell. 1998. Expression of the intermediate filament keratin gene, K15, in the basal cell layers of epithelia and the hair follicle Exp. Cell Res. 244:448-459. [DOI] [PubMed] [Google Scholar]