Abstract
The bacterial type IV secretion systems (T4SSs) comprise a biologically diverse group of translocation systems functioning to deliver DNA or protein substrates from donor to target cells generally by a mechanism dependent on establishment of direct cell-to-cell contact. Members of one T4SS subfamily, the conjugation systems, mediate the widespread and rapid dissemination of antibiotic resistance and virulence traits among bacterial pathogens. Members of a second subfamily, the effector translocators, are used by often medically-important pathogens to deliver effector proteins to eukaryotic target cells during the course of infection. Here we summarize our current understanding of the structural and functional diversity of T4SSs and of the evolutionary processes shaping this diversity. We compare mechanistic and architectural features of T4SSs from Gram-negative and �positive species. Finally, we introduce the concept of the ‘minimized’ T4SSs; these are systems composed of a conserved set of 5–6 subunits that are distributed among many Gram-positive and some Gram-negative species.
Keywords: Type IV secretion, conjugation, pathogenesis, ATPase, translocation, DNA transfer, pilus, Gram-positive adhesins
1. Introduction
The bacterial type IV secretion systems (T4SSs) deliver protein and DNA substrates to bacterial or eukaryotic target cells generally by a contact-dependent mechanism (Alvarez-Martinez and Christie, 2009; Zechner et al., 2012). Studies exploring the structure and function of these nanomachines have focused mainly on a few paradigmatic systems of Gram-negative bacteria. However, T4SSs are broadly distributed phylogenetically and they have evolved exceptional diversity in their overall architectures, types of substrates secreted, and mechanisms employed for target cell attachment. This diversity can be understood fully only by characterizing various systems from different taxa. Broadly, these systems can be divided into three groups (Christie and Vogel, 2000; Guglielmini et al., 2013). The conjugation systems are the largest and most widely distributed subfamily (Alvarez-Martinez and Christie, 2009; Guglielmini et al., 2013). These systems operate to deliver single-stranded DNA substrates and one or more proteins across the donor cell envelope to bacterial or eukaryotic target cells by a mechanism requiring direct cell-to-cell contact (de la Cruz et al., 2010; Frost, 2009). Second are the effector translocators, these systems deliver protein effectors by a contact-dependent mechanism to the cytosols of eukaryotic target cells to aid in bacterial colonization and survival within host cells or tissues (Cascales and Christie, 2003). The third group comprises the release/uptake systems, which export substrates to or import them from the milieu (Cascales and Christie, 2003; Zechner et al., 2012). Members of each subfamily are distributed among Gram-negative bacterial species, but so far only the conjugation systems are known to exist in species of Gram-positive bacteria and archaea (Alvarez-Martinez and Christie, 2009; Grohmann, 2003).
In the first part of this review, we will update information on the structures and mechanisms of action of a few of the better-characterized Gram-negative bacterial T4SSs. Next, we will broaden the discussion to some increasingly studied Gram-positive T4SSs for structure-function comparisons with the Gram-negative systems. In recent years, an abundance of sequenced bacterial genomes has enabled detailed phylogenetic analyses of T4SSs; these studies have led to an understanding of the evolutionary processes shaping the T4SSs. We will summarize some of this information in order to introduce a new concept, the ‘minimized’ T4SSs. These systems, composed of only a subset of the building blocks employed by the well-characterized systems, recently were reported to exist in many streptococcal species (Zhang et al., 2012). We will present evidence that these ‘minimized’ T4SSs are widely spread among Gram-positive bacteria and some also exist in Gram-negative species. The ‘minimized’ T4SSs clearly evolved from ancestral conjugation systems, but some evidence exists for their alternative or additional function as effector translocators. Like their Gram-negative counterparts, therefore, medically-important Gram-positive species might have added T4SS-mediated effector translocation to their armament of pathogenic mechanisms for enhancing virulence potential.
2. General features of Gram-negative T4SSs
2a. The conjugation machines
Studies of T4SSs encoded by conjugative plasmids of Escherichia coli, e.g., F, R388, pKM101, RP4, and by the Agrobacterium tumefaciens T-DNA transfer system have shown that DNA transfer involves three biochemical reactions that are coordinated in space and time: i) the DNA transfer and replication (Dtr) proteins process the DNA substrate for transfer (de la Cruz et al., 2010; Wong et al., 2012) ii) the Dtr/DNA complex (the relaxosome) engages with the substrate receptor or type IV coupling protein (T4CP) (Zechner et al., 2012), and iii) substrate is translocated across the cell envelope via a transenvelope channel comprised of the mating pair formation (Mpf) proteins (Schroder and Lanka, 2005; Zechner et al., 2012). Conjugation also requires a fourth reaction that remains poorly understood at this time, establishment of the donor-target cell junction and propagation of accompanying signals to activate the transfer process (Lang et al., 2011)
The DNA processing reaction has been extensively characterized in vivo and in vitro; the reader is referred to excellent reviews for a more detailed discussion (de la Cruz et al., 2010; Zechner et al., 2012). Briefly, a nuclease termed the relaxase together with one or more accessory factors binds a cognate origin of transfer (oriT) sequence carried by an extrachromosomal plasmid or integrative conjugative element (ICE). Upon formation of the relaxosomal complex, the relaxase nicks the DNA strand destined for transfer (T-strand) and remains covalently bound via an active site tyrosine to the 5’ end of the T-strand. Formation of this transient bond stabilizes the 5’ end of the T-strand. The relaxase and accessory factors carry the translocation signals for recognition of the transfer intermediate by the cognate T4CP/Mpf complex. The relaxase also pilots the T-strand in a 5’ to 3’ direction through the channel, and in the recipient cell the relaxase participates in recircularization or integration of the T-strand into the chromosome (Cesar et al., 2006; Grandoso et al., 2000).
The relaxase and accessory factors bound at oriT mediate docking of the DNA transfer intermediate with the T4CP, also termed the substrate receptor. Dtr factors and other T4SS secretion substrates generally carry translocation signals at their C-termini, although internal signals also exist (Lang et al., 2010; Nagai et al., 2005; Vergunst et al., 2005). Various other factors, including the Dtr accessory proteins, membrane-associated proteins termed adaptors, or secretion chaperones can contribute to substrate -T4SS docking (for more details on substrate signals/recruitment reactions, see (Alvarez-Martinez and Christie, 2009; Sutherland et al., 2012; Zechner et al., 2012)). The T4CPs are ubiquitous receptors among the described conjugation systems and they are also associated with most effector translocator systems (Alvarez-Martinez and Christie, 2009). T4CPs typically possess an N-terminal transmembrane (TM) domain and a large C-terminal cytoplasmic domain with a nucleoside triphosphate binding domain (NBD). The characterized T4CPs bind ssDNA and dsDNA nonspecifically, and there is also evidence for DNA-binding-dependent ATP hydrolysis and oligomerization (Chen et al., 2008; Tato et al., 2007). An X-ray structure was solved for the soluble domain of TrwB, which presents as a homohexameric sphere with dimensions of 110 Å in diameter and 90 Å in height, and with a central channel of 20 Å in diameter (Gomis-Ruth et al., 2001). The N-terminal domain is predicted by electron microscopy image analysis to project across the cytoplasmic membrane (Hormaeche et al., 2002). The full-length protein is thus configured as a ball-stem F1-F0-like structure. Based on sequence and structural similarities with the FtsK and SpoIIIE DNA translocases (Gomis-Ruth et al., 2004), T4CPs might mediate DNA transfer through their central channels across the cytoplasmic membrane, although this remains to be shown.
In the third reaction, the docked DNA transfer intermediate is delivered to the translocation channel for delivery to the cell surface. The Mpf channels of Gram-negative bacterial conjugation systems are highly complex structures composed of one or two ATPases in addition to the T4CP, a cytoplasmic membrane translocon, and a translocation channel that is likely housed within a structural scaffold termed the core complex (Fronzes et al., 2009a; Wallden et al., 2010). The Mpf channel directs passage of the relaxase-T-strand intermediate across the cell envelope in one step, thereby protecting the substrate from periplasmic nucleases. In addition to the translocation channel, the Mpf proteins elaborate an attachment organelle, the conjugative pilus, which extends from the donor cell surface to establish productive contacts with recipient cells (Thanassi et al., 2012). Mutations in various Mpf subunits abolish pilus biogenesis without disrupting substrate transfer, indicating that the pilus functions exclusively to mediate target cell contact and mating junction formation and not as a conduit for substrate passage at a distance between donor and recipient cells (Alvarez-Martinez and Christie, 2009). Somewhat surprisingly, it is still unknown whether the translocation channel and conjugative pilus are physically linked or configured as distinct organelles at the cell surface. However, recent studies exploring the requirements for bacteriophage utilizing the F plasmid-encoded pilus as a receptor for entry into the host cell have supplied strong evidence for the physical conjunction of these two organelles (see below).
2b. Effector translocators
The effector translocators function in many medically-important pathogens, including Bordetella pertussis, Helicobacter pylori, Legionella pneumophila, Coxiella burnetii, Bartonella spp., Brucella spp., and Rickettsial spp. (Cascales and Christie, 2003; Llosa et al., 2009; Terradot and Waksman, 2011). A. tumefaciens also utilizes the VirB/VirD4 T4SS to induce Crown Gall disease on susceptible plant species, and several other phytopathogens employ effector translocators for infection or symbiosis (Alvarez-Martinez and Christie, 2009). The VirB/VirD4 T4SS actually functions both as a conjugation machine and an effector translocator, and at this time three such dual-function systems have been identified: i) the A. tumefaciens VirB/VirD4 system, which translocates oncogenic T-DNA and effector proteins to plants during infection but also a nonself-transmissible IncQ plasmid or other T-DNA-based substrates to bacterial, plant, yeast, and human target cells (Christie, 2004); ii) the L. pneumophila Dot/Icm system, which mobilizes many hundreds of effector proteins during infection as well as the IncQ plasmid substrate to recipient bacteria (Nagai and Kubori, 2011), and iii) the Bartonella spp. VirB/VirD4 system, which translocates the Bep (Bartonella effector proteins) substrates into mammalian cells, and plasmid DNA into human host cells (Schroder et al., 2011). Interestingly, the Bartonella spp. T4SS also can interface with the related E. coli plasmid R388 transfer system, such that chimeric machines composed of subunits from each system transfer R388 between bacteria or from Bartonella spp. into human host cells (Fernandez-Gonzalez et al., 2011).
For T4SSs employed by mammalian pathogens, studies have focused largely on defining the repertoire of effector proteins and their cellular actions in the eukaryotic host. This expanding field of study is beyond the scope of this review, but the specialized functions of these machines can be illustrated by the fact that systems are known to deliver from none to many hundreds of effector molecules into the host cell during infection. At one end of the spectrum, the Bartonella spp. Trw system is thought to function not as a translocation machine but as an attachment device for binding eukaryotic cell receptor (Eicher and Dehio, 2012). The cytotoxin-associated-gene (Cag) T4SS of H. pylori translocates one protein, CagA, which is responsible for inducing a pro-inflammatory response and a wide range of gene regulatory effects in gastric epithelial cells (Tegtmeyer et al. 2011). Brucella spp., Bartonella spp., and Rickettsial spp. T4SSs translocate a few to a few dozen effectors, but further work is needed to determine the complete substrate repertoires. At the opposite end of the spectrum, the Dot/Icm systems of L. pneumophila and C. burnetii are thought to deliver over 300 and 75 effector proteins, respectively, for intracellular infection of the human host (Nagai and Kubori, 2011).
A common feature of the effector translocator systems is the evolution of specialized functions through modification of their T4SS machine subunits. The H. pylori Cag system, for example, carries highly modified forms of subunits that are conserved among many T4SSs of Gram-negative species. Most T4SSs possess a small lipoprotein, VirB7, and a bitopic subunit, VirB10, that together with a periplasmic subunit VirB9 form a stable, transenvelope substructure required for biogenesis of the translocation channel (see below). The VirB7- and VirB10-like subunits of the H. pylori Cag system, CagT and CagY, respectively, actually bear little structural resemblance to their counterparts in other systems. They are large proteins with variable or multiple repeat domains, and they associate with a large, extracellular appendage (Rohde et al., 2003). Their surface variability probably allows H. pylori to attach to different types of host cell receptors or evade the immune system, reflecting an adaptive response to the host environment. Similarly, the Bartonella spp. Trw system codes for multiple pilus-associated proteins, and this surface diversity likely also evolved for binding to cell receptors of different hosts or immune evasion (Franz and Kempf, 2011). The intracellular Rickettsial spp. also have multiple copies of variable pilus genes that are presumed to facilitate long-term fitness or proliferation in the host cell environment (Gillespie et al., 2009).
Some effector translocators also lack subunits that are highly conserved among the conjugation systems. The Bartonella spp. Trw system, for example, lacks a VirD4-like T4CP and as noted above its role in pathogenesis is thought to be restricted to elaboration of different pilus structures for surface variability (Vayssier-Taussat et al., 2010). The VirB system of Brucella spp. similarly lacks a VirD4-like T4CP; however, recent studies determined that this system translocates several effector proteins. As discussed further below, a couple of T4SSs translocate substrates in two-steps, utilizing a dedicated translocase for transport across the cytoplasmic membrane and the T4SS for translocation across the outer membrane. In studies of the Brucella effectors, one study identified four candidate effectors and further established that the first 25 amino acids of these effectors are required for translocation, suggesting that these effectors are translocated across the cytoplasmic membrane via the general secretory pathway (GSP) (Marchesini et al., 2011). These substrates would then be recruited to the VirB machine for export across the outer membrane. Other groups alternatively have identified effectors by use of reporter translocation assays in which the reporter protein is fused to the N terminus of the effector (de Barsy et al., 2011; de Jong et al., 2008). Such fusions would block translocation through the GSP, and argue for VirB-mediated translocation in one step across both the cytoplasmic and outer membrane. It is intriguing to think that the Brucella spp. VirB system functions uniquely among the known T4SSs by recruiting secretion substrates from both intracellular and periplasmic locations.
2c. Substrate release/uptake systems
These systems have evolved to translocate substrates independently of target cell contact, likely by mutation of a conjugation or effector translocator T4SS. For example, many strains of Neisseria gonorrhoeae carry a ∼57-kb gonococcal genetic island (GGI) that codes for a T4SS (Ramsey et al., 2011). This T4SS is related in gene content and order to the F plasmid-encoded T4SS and like other conjugation machines this system translocates ssDNA. However, this T4SS secretes chromosomal DNA via relaxase-mediated nicking at oriT sequence located within the GGI. The GGI-encoded TraI relaxase is unusual in that it carries an HD phosphohydrolase domain, which might functionally substitute for His-rich motifs carried by other relaxases for metal ion coordination (Salgado-Pabon et al., 2007). Most strikingly, the GGI T4SS secretes its DNA substrate into the milieu by a mechanism independent of target cell contact (Ramsey et al., 2011). This chromosomal T4SS thus seems to have evolved through a combination of conjugative plasmid integration events followed by mutations that ultimately resulted in a loss of a requirement for target-cell activation of channel gating.
This T4SS subfamily also includes the B. pertussis Ptl system, which exports the multisubunit pertussis toxin (PT) across the outer membrane (Locht et al., 2011), and the H. pylori ComB system, which functions as a competence system to import exogenous DNA (Stingl et al., 2010). Both of these systems resemble the A. tumefaciens VirB/VirD4 system in subunit composition, except that both lack genes for the pilus-associated protein VirB5. Unlike other T4SSs, both of these systems translocate their respective substrates by two mechanistically and temporally disconnected processes. For PT translocation, the PT subunits are first secreted across the cytoplasmic membrane via the GSP (Locht et al., 2011). Once PT assembles in the periplasm, it is then recruited by the Ptl system for export across the outer membrane. The H. pylori ComB system translocates DNA across the outer membrane into the periplasm. Next, Hp-ComEC, a competence protein of unrelated ancestry to the T4SS, mediates DNA transport across the cytoplasmic membrane (Stingl et al., 2010). Conceivably, loss of pilus-associated VirB5 was an evolutionary adaptation the ultimately led to the capacity of these T4SSs to translocate substrates across the outer membrane by a contact-independent mechanism.
3. T4SS building blocks of Gram-negative T4SSs
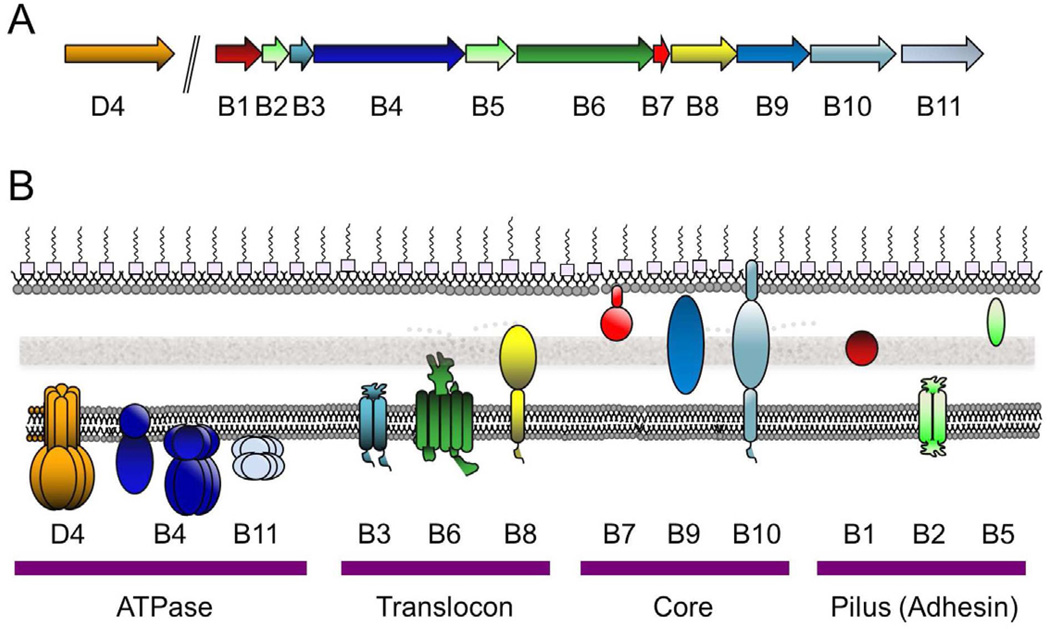
The T4SSs display considerable variation in their subunit composition, but most Gram-negative systems are built minimally from a conserved set of components that are related to those of the well-characterized A. tumefaciens T-DNA transfer system. Consequently, here, we will use the A. tumefaciens T4SS as a reference and the VirB/VirD4 nomenclature when referring to homologs or orthologs in the various systems. The A. tumefaciens T4SS is composed of 11 VirB Mpf subunits and the VirD4 T4CP. These subunits can be divided into four groups on the basis of subcellular location and function (Fig. 1). (Henceforth, when referring to a specific protein as opposed to a protein family, a subscript will denote the plasmid or bacterial species origin, e.g., A. tumefaciens VirD4 is VirD4At; plasmid R388-encoded TrwB is TrwBR388.)
Fig. 1.
The A. tumefaciens VirB/VirD4 T4SS gene organization and subunit subcellular locations. (A) virD4 is co-transcribed with the virD1 relaxase and virD2 accessory factor genes from the virD promoter and the eleven virB genes are transcribed from the virB promoter. (B) VirD4 and VirB proteins are associated with the cytoplasmic membrane (CM), cell-wall (CW) containing periplasm, or outer membrane (OM) as shown. General membrane topologies are denoted as are the oligomeric structures of the VirD4 (hexamer), VirB4 (monomer or hexamer, see text), VirB11 (hexamer) ATPases. VirB7 is an outer-membrane-associated lipoprotein and VirB10 spans both membrane with N- and C-proximal α-helical domains. The VirD4 and VirB subunits are clustered into four functional groups.
3a. VirD4, VirB4, VirB11 ATPases
Three ATPases, VirD4At, VirB4At, and VirB11At, are situated at the cytoplasmic entrance of the translocation channel (Figs. 1 & 2). VirD4At is related to TrwBR388 T4CP, the structural prototype for this protein family. While further studies are needed to determine whether DNA substrates pass through the lumen of the T4CP hexamer, several lines of evidence support the idea that T4CPs function as substrate receptors. Genetic, biochemical, and structural evidence exists for T4CP binding to DNA as well as cognate relaxases and other components of the relaxosome (Atmakuri et al., 2007; Lu et al., 2008). Purified TrwBR388 exhibits DNA-stimulated ATPase and oligomerization activities (Tato et al., 2005), and is also stabilized by insertion into membrane lipids (Vecino et al., 2011). TrwBR388 reconstitution in liposomes enhances nucleotide binding affinity as well as ATP-specific binding (Vecino et al., 2010). These findings support the notion that the TM domains and membrane insertion contribute to T4CP structure and activity, a point underscored by evidence that TrwB’s TM domains participate in interactions with other Mpf channel components, e.g., VirB10-like TrwER388 (Llosa et al., 2003; Alkorta, personal communication). In A. tumefaciens, the VirD4At T4CP - VirB10 interaction is likely important for coupling of ATP energy with channel gating, as it was previously shown that VirD4At together with the VirB11At ATPase induce a conformational change in VirB10At that is required for passage of DNA substrates through the translocation channel (Atmakuri et al., 2004; Cascales and Christie, 2004a).
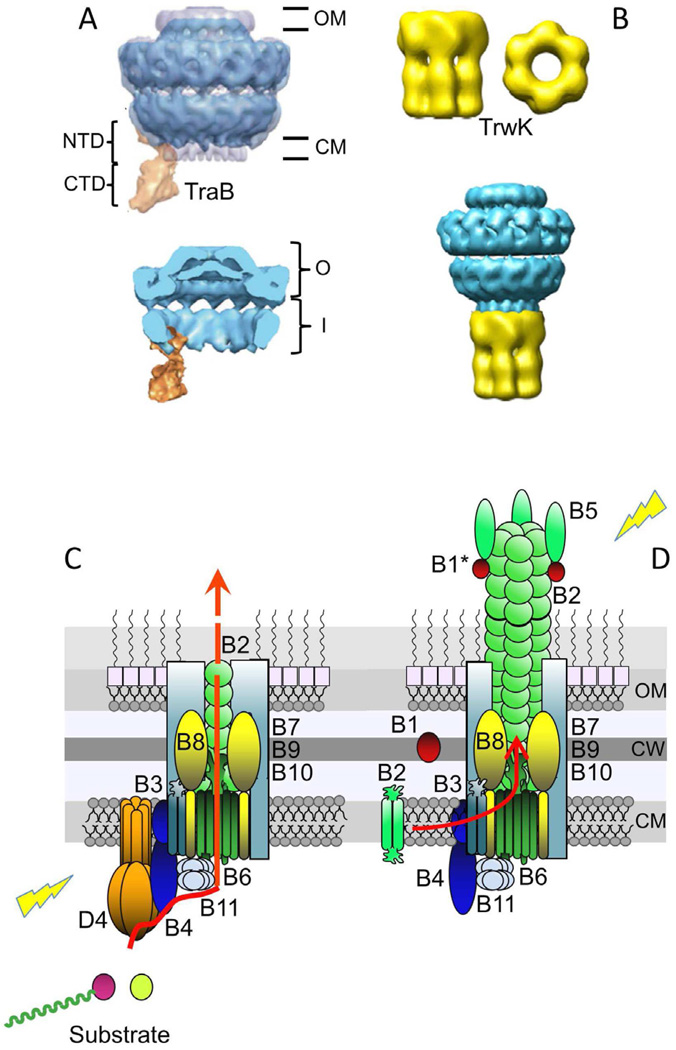
Fig. 2.
Gram-negative T4SS subassemblies and architectural models. (A) Upper: Structure of the pKM101-encoded core complex with the VirB4-like TraB homolog bound to the side. The core is shown in blue and TraB is in orange. The O and I layers are indicated as O and I, respectively. TraBN-terminal domain (NTD) and C-terminal domain (CTD) are shown. The negative stain core/TraB (in blue) is superimposed over the cryoelectron microscopy structure of the core complex alone (half-gray transparent). Lower: Section view of the core/TraB structure showing the core chamber, which is envisioned to house the translocation channel and pilus (Wallden et al., 2012). Images were published with permission. (B) Upper: Three dimensional reconstruction of VirB4-like TrwK of plasmid R388 from negative-stained images. Side and bottom views of the hexamer are shown. Lower: Model of interaction between the T4SS core complex and TrwKR388. The TrwKR388 hexameric structure was attached to the cytoplasmic side of the inner membrane region of the pKM101 core complex obtained from (Fronzes et al., 2009b). These images were reproduced with permission by (Pena et al., 2012). The figure is not to scale, dimensions of the core are ∼185 Å in width and length (Fronzes et al., 2009b), and those of the TrwKR388 hexamer are ∼132 Å and ∼165 Å in width and length, respectively (Pena et al., 2012). (C) Model depicting the architecture of the VirB/VirD4 translocation channel in the Gram-negative cell envelope, with the substrate translocation pathway depicted (red arrow). VirD4At and VirBAt channel subunits are shown (color code matches Fig. 1), with the shaded aqua rectangles corresponding to the VirB7/VirB9/VirB10 core complex. VirD4 T4CP is required for substrate transfer. Substrates include relaxase-T-strand (left) and effector protein (right). (D) Model of VirB/VirD4 conjugative pilus. Pilus biogenesis involves VirB4-mediated dislocation of membrane pilins and entry into the core chamber for polymerization. Pilus polymerization requires substantial conformational changes in the core complex. VirB1/VirB1*, but not VirD4 are required for pilus assembly. Lightning bolts: The T4SS apparatus is activated by substrate engagement (left) and target cell or bacteriophage binding to the extracellular pilus (right). Cytoplasmic membrane (CM), cell wall (CW), outer membrane (OM).
T4CP receptor activity was further shown by use of a ChIP-based, formaldehyde crosslinking assay termed transfer DNA immunoprecipitation (TrIP). In those studies, it was shown that the translocating T-DNA substrate forms a close contact with the A. tumefaciens VirD4At T4CP by a mechanism requiring T-DNA processing but not the VirBAt channel components (Cascales and Christie, 2004b). Analyses with mutant strains further showed that the T4CP-substrate contact does not require T4CP ATP hydrolysis activity (Atmakuri et al., 2004). Another important line of investigation has involved the construction of Mpf/T4CP chimeric systems for studies of substrate-T4CP and T4CP-Mpf engagement reactions (Cabezon et al., 1997). Several such hybrid systems have now been shown to mediate translocation of T4CP-specific substrates through heterologous Mpf channels. Recently, such an approach was applied to the Bartonella spp. VirB/VirD4 system, which as mentioned above has the intriguing ability to deliver substrate DNA into mammalian host cells (Schroder et al., 2011). Capitalizing on the observation that Bartonella spp. VirB/VirD4 system is highly similar in subunit composition to the E. coli R388 Trw system, Llosa and colleagues engineered a TrwBR388 T4CP/VirBBh Mpf chimeric machine in Bartonella henselae and showed that this T4SS translocates the R388 plasmid into mammalian cells (Fernandez-Gonzalez et al., 2011). This work establishes for the first time that chimeric T4SSs can function to deliver heterologous DNA into different human cell types, and it paves the way for design of chimeric T4SSs for targeted delivery of therapeutic DNA into specific human tissues (Llosa et al., 2012).
VirB4 is a signature subunit of all T4SSs described to date (Alvarez-Martinez and Christie, 2009). These subunits are ancestrally related to the T4CPs (Guglielmini et al., 2013); their C-terminal nucleotide binding domains (NBDs) are also probably structurally related to the corresponding NBDs of the T4CPs although there is still debate regarding the oligomeric structure of these ATPases. Fueling this debate, several VirB4 homologs have been purified as monomers (L. pneumophila VirB4Lvh, Thermoanaerobacter pseudethanolicus VirB4) (Durand et al., 2011; Wallden et al., 2012) or dimers (TraBpKM101, PrgJpCF10) (Chen et al., 2008; Durand et al., 2010; Durand et al., 2011). Low-resolution structures of VirB4Lvh and VirBpKM101 were solved by small-angle X-ray scattering and an X-ray structure was obtained for T. pseudethanolicus VirB4 C-terminal domain, the latter confirming the structural similarity of a VirB4 homolog with TrwBR388 T4CP (Durand et al., 2011). Waksman and his colleagues also recently solved a structure of a larger subassembly termed the core complex (see below) from the pKM101 T4SS by cryoelectron microscopy; in that structure the monomeric form of TraBpKM101 is observed bound to the side of the core complex (Wallden et al., 2012) (Fig. 2A). Evidence also was presented that the purified, dimeric form of VirB4-like PrgJ from the E. faecalis pCF10 transfer system displays both ATP hydrolysis and DNA-binding activities (Li et al., 2012). However, a very recent report has now provided evidence for hexameriation of VirB4-like TrwKR388 (Fig. 2B). The purified protein assembles as a double-ring barrel-shaped hexamer, as shown by negative-stain electron microscopy (Pena et al., 2012). In an alternative model to the structure solved for the pKM101 system, the TrwKR388 hexamer is proposed to sit at the base of the core complex (Fig. 2B).
The spatial relationship of VirB4 and the core complex is important to define given its possible role in substrate transfer or machine biogenesis. For example, with the demonstrated (Wallden et al. 2012) or predicted (Pena et al., 2012) configurations, it is easy to envision a role for VirB4 subunits in delivery of secretion substrates into the transfer channel. In the earlier TrIP studies, VirB4At was not observed to form a formaldehyde crosslink with the translocating T-DNA substrate (Cascales and Christie, 2004b). However, more recently, DNA-binding activities have been demonstrated for several VirB4 homologs, including TraBpKM101, TrwKR388, and PrgJpCF10 (Durand et al., 2010; Li et al., 2012; Pena et al., 2012). Furthermore, by TrIP, the pCF10 transfer intermediate did form a formaldehyde crosslink with PrgJpCF10 by a mechanism requiring synthesis of the Dtr processing factors and the PcfCpCF10 T4CP (Li et al., 2012). Thus, it seems likely that VirB4 subunits contribute more directly than previously envisioned to the delivery of substrate into the translocation channel (Fig. 2C).
The VirB4 ATPases fulfill other functions relating to machine biogenesis. Specifically, during biogenesis of conjugative pili, pilin monomers are initially processed and integrated into the cytoplasmic membrane. The integrated pilins serve as a pool for subsequent use in building the conjugative pilus (see below). In studies exploring the A. tumefaciens pilus assembly pathway, VirB4At was shown to induce changes in the membrane topological state of VirB2At pilin by an ATP-dependent mechanism; these observed changes were consistent with a role for the ATPase in catalyzing pilin dislocation (Kerr and Christie, 2010). The findings prompted a model in which VirB4 subunits catalyze membrane extraction of pilin monomers at an early step in the pilus polymerization pathway. The recent evidence for a VirB4-like TraBpKM101 - core complex interaction (Wallden et al., 2012) supports a model in which these ATPases catalyze pilin dislocation reiteratively and then shunt the pilin monomers into the core complex for pilus polymerization within the core chamber (Fig. 2D).
The VirB11 ATPases are double-ring-shaped hexamers related to a large family of AAA ATPases that also include the GspE traffic ATPases associated with type II secretion (T2S) and type IV pilus (T4P) assembly systems (Savvides et al., 2003). VirB11 ATPases are commonly associated with T4SSs of Gram-negative species but not Gram-positive species (Alvarez-Martinez and Christie, 2009). By TrIP, it was shown that VirB11At forms a formaldehyde crosslink with the translocating T-DNA substrate and that production of the VirD4At T4CP is required for this contact (Cascales and Christie, 2004b). VirB11 thus also mediates an early step of the substrate transfer and likely coordinates its activities with VirB4 to deliver the substrate into the transfer channel (Fig. 2C) (Atmakuri et al., 2004). Surprisingly little additional information is available describing how the widely-distributed T4SS ATPases coordinate their activities to energize machine biogenesis and substrate translocation. Finally, it is important to note that while VirD4 and VirB4 homologs are associated with conjugation machines and most effector translocator systems, VirB11 ATPases are missing from some conjugation systems in Gram-negative bacteria and all systems in Gram-positive bacteria. The VirB11 ATPases might endow the cognate systems with a specialized, yet unknown activity, or other machine adaptations might have compensated for the contributions of these ATPases to machine morphogenesis or function.
3b. Membrane translocase components VirB3, VirB6, VirB8
These subunits are predicted to assemble as the cytoplasmic membrane translocon together with the ATPases (Figs 1 and 2). VirB3-like subunits are small, hydrophobic proteins invariably found together with VirB4-like ATPases; both subunits are fused together as a single protein in some systems suggestive of a coordination of function (Alvarez-Martinez and Christie, 2009). VirB6 subunits have evolved as two distinct subtypes. Those resembling A. tumefaciens VirB6 are ∼300 residues with 5 or more membrane-spanning domains and a large central, periplasmic domain. The ‘extended-VirB6’ subunits are larger (>500 residues) with an N-terminal polytopic membrane domain and a large C-terminal hydrophilic domain (Alvarez-Martinez and Christie, 2009). The role of this C-terminal domain is unspecified, although there is evidence in the F plasmid (Audette et al., 2007) and Vibrio cholerae SXT ICE (Marrero and Waldor, 2007) transfer systems for protrusion or release of this domain to the cell surface and into target cells. Reminiscent of other polytopic membrane proteins associated with membrane transport, VirB6 subunits are thought to form part of the translocation channel at the cytoplasmic membrane (Jakubowski et al., 2004). Consistent with this proposed function, VirB6At formed fomaldehyde crosslinks with the translocating T-DNA substrate by TrIP, and complex formation required cosynthesis of the VirD4, VirB4, and VirB11 ATPases (Cascales and Christie, 2004b).
VirB8 subunits are bitopic proteins with a short cytoplasmic N-terminal domain, TM domain, and large C-terminal periplasmic domain (Baron, 2006). X-ray structures have been solved for the periplasmic fragments of Brucella suis and A. tumefaciens VirB8 subunits (Bailey et al., 2006; Terradot et al., 2005) and, recently, the VirB8-like subunit TcpC encoded by the Clostridium perfringens pCW3 conjugation system (Porter et al., 2012). All VirB8 domains present as large extended β-sheets with five α-helices, giving rise to an overall globular fold. These subunits packed as dimers or trimers in the crystal structures, and results of mutational analyses suggest that oligomerization is physiologically relevant (Paschos et al., 2006). VirB8At also formed a formaldehyde crosslink with the translocating T-DNA in the TrIP assay, suggesting that it also comprises part of the translocation channel (Cascales and Christie, 2004b). Interestingly, specific mutations in VirB6At enabled formation of the VirB6At-T-DNA crosslink but blocked the VirB8At-T-DNA formaldehyde crosslink (Jakubowski et al., 2003). These findings suggest that the translocating DNA substrate forms a close contact initially with polytopic VirB6At and then with VirB8At, reflecting a possible architectural arrangement for the translocation channel (Fig. 2C). VirB8 subunits are common constituents of Gram-negative T4SSs, and there is increasing evidence for association of proteins with VirB8-like folds among many Gram-positive T4SSs (see below).
3c. VirB7/VirB9/VirB10 core complex
A. tumefaciens VirB7 is a small outer membrane lipoproteins that forms a disulfide crosslinked dimer with VirB9. VirB9At is a periplasmic subunit that forms extensive contacts with VirB10 (Alvarez-Martinez and Christie, 2009). VirB10 has a novel architecture among known bacterial proteins. It is a bitopic protein with a short cytoplasmic domain and a TM domain, and its large periplasmic region is comprised of a proline-rich or coiled-coiled domain and a large globular C-terminal domain (Jakubowski et al., 2009). Most interestingly, an α-helical projection extending from the C-terminal domain spans the outer membrane, giving rise to a protein that extends across the entire cell envelope (Fig. 1). Strikingly, VirB7At, VirB9At, and VirB10At homologs from the pKM101 transfer system assemble as a 1 MDa barrel of ∼185 Å in diameter and height, as shown by cryoelectron microscopy (Fronzes et al., 2009b) (Fig. 2A). This structure, termed the core complex, is composed of 14 copies each of these subunits and presents as two layers (I- and O-layers) conforming to the portions of the barrel located near the cytoplasmic and outer membranes, respectively. The I-layer is composed of the N-terminal domains of the VirB9 and VirB10 homologs. The VirB10 TM domain anchors the core complex at the cytoplasmic membrane, such that the 14 TM domains form a ring of 55 Å in diameter. The O-layer is composed of VirB7 and the C-terminal domains of VirB9 and VirB10 (Fronzes et al., 2009b). This O-layer was purified and its structure was solved by X-ray crystallography (Chandran et al., 2009). The structure presents with a main body located in the periplasm comprised of the C terminus of VirB10 forming the interior of the structure and lining the core chamber and VirB9 on the periphery. VirB7 is also peripheral and positioned near a narrow cap composed of a hydrophobic ring of two-helix bundles contributed by VirB10. The cap has a 10Å diameter hole, too small to allow substrates to pass, but there is structural evidence for conformational flexibility of this domain (Fronzes et al., 2009a). As mentioned above, VirB10At undergoes a conformational change upon sensing of ATP utilization by the VirD4At and VirB11At ATPases that is required for substrate passage through the distal portion of the translocation channel (Cascales and Christie, 2004a). Energy coupling might regulate opening of the outer membrane pore comprised of the VirB10At α-helical cap. Consistent with such a proposal, mutation of a conserved Gly residue located near the cap locked A. tumefaciens VirB10 in the energized conformation, disrupting channel gating and leading to substrate leakage to the cell surface (Banta et al., 2011).
3d. Pilus-associated proteins VirB2, VirB5, VirB1
VirB2At and VirB5At are required for substrate transfer although their roles in the transfer process are largely unspecified. VirB2At pilin formed a formaldehyde crosslink with the translocating T-DNA substrate, suggesting that the pilin comprises part of the transfer channel (Cascales and Christie, 2004b). VirB1At is a cell wall hydrolase whose enzymatic activity is dispensable for elaboration of the translocation channel (Berger and Christie, 1994). These three subunits, however, are required for pilus biogenesis (Figs. 1 & 2D) (Fullner et al., 1996). VirB2At is the major pilin subunit comprising the pilus shaft. VirB5At is a pilus-tip adhesin likely responsible for initiating target cell contact (Aly and Baron, 2007). The role of VirB1At in pilus biogenesis is less clear although a degradation product, VirB1*, is released across the outer membrane and is thought to form part of the pilus fiber (Zupan et al., 2007).
Early steps in the pilus assembly pathway have been defined for the F, RP4, and A. tumefaciens VirB/VirD4 systems. For each of these systems, the propilin is delivered into the cytoplasmic membrane via the GSP followed by cleavage of an unusually long leader peptide. The N-termini are further modified through acetylation (TraAF) or covalent joining with processed or unprocessed C termini to generate a cyclic peptide (VirB2At, TrbBRP4) (Eisenbrandt et al., 1999; Lawley et al., 2003). As noted above, mature pilin forms a pool in the cytoplasmic membrane and VirB4 ATPase participates in some way to pilin dislocation for subsequent pilus polymerization. Pili extend by addition of pilin monomers at the cell-proximal end. Accordingly, given its role as a tip adhesin VirB5At might initiate the polymerization process, possibly aided by VirB1At or VirB1At*. Interaction networks have been described for VirB1At, VirB5At, and VirB8At, further suggesting a role for VirB8 subunits - or more specifically a VirB1/VirB5/VirB8 complex - for initiation of pilus assembly (Fig. 2D) (Yuan et al., 2005).
3e. Mosaicism of T4SSs
T4SSs have evolved structural and functional complexity through acquisition of proteins or domains whose ancestries are unrelated to the VirB/VirD4 subunits. In general, the acquisition of such features is thought to have evolved for specialized functions relating to niche adaptation. For example, the E. coli F plasmid transfer region consists of nearly fourty tra genes, eighteen of which are involved in assembly of the T4SS. Eight are VirB/VirD4 homologs, although notably this system lacks homologs for VirB8 and VirB11. The remaining components are specific to the F T4SS and function in mating pair stabilization and F pilus outgrowth and retraction, a dynamic process that seems to be unique to this T4SS (Lawley et al., 2003). The H. pylori Cag system is composed of a few of the highly-conserved VirB/VirD4-like subunits plus additional components or domains thought to be uniquely adapted for infection of the mammalian host (Tegtmeyer et al., 2011). The Dot/Icm systems of L. pneumophila and C. burnetii have some discernible VirB/VirD4-like subunits, but the bulk of the machine components are of an unrelated ancestry, again reflecting an evolutionary process of adaptation through acquisition of functions specific for colonization and infection (Nagai and Kubori, 2011). The mosaic nature of T4SSs appears to be the rule rather than the exception, especially for highly evolved effector translocator systems functioning in specific host tissues or environments during infection.
4. Architectures of Gram-negative T4SSs
With this mosaicism in mind, do T4SSs display any common architectural themes? Considerable structural information for various machines is needed to fully address this question, but recent significant advances have enabled structural definition of the pKM101 T4SS, which now serves as a structural prototype at least for T4SSs composed predominantly or exclusively of VirB/VirD4-like subunits.
4a. The translocation channel
The groundbreaking discovery of the VirB7/VirB9/VirB10 core complex yielded important insights into the overall T4SS architecture. The interior chamber of the core complex is sufficiently large to house the translocation channel, the composition of which can be inferred from results of genetic, two-hybrid interaction, and TrIP studies (Fig. 2). Accordingly, an ATP-energizing motor complex consisting of VirD4 T4CP and the VirB ATPases are positioned at the channel entrance; this complex coordinates machine biogenesis as well as energy-dependent steps of translocation (Fronzes et al., 2009a). The ATPase complex most probably forms specific contacts with the VirB7/VirB9/VirB10 core complex, but the ATPases are not necessary for assembly of the core complex. Thus, in an ordered assembly pathway, the core would first assemble as a structural scaffold. The ATPases would engage with the core complex, and a combination of ATP-dependent and -independent reactions would drive further steps of machine assembly.
The ATPase ternary complex likely interacts with the cytoplasmic membrane translocon. This translocon, composed of VirB3, VirB6, and the N-terminal region of VirB8, probably embeds within the 55 Å diameter ring formed by the 14 TM domains of VirB10 in the core complex. As discussed further below, the translocon but not the core complex, is conserved among T4SSs of both Gram-negative and -positive bacteria. This raises the interesting possibility that the membrane translocon in Gram-negative systems is a stable substructure capable of functioning independently of the core complex. Indeed, by TrIP it was shown that the ATPases, VirB6At, and VirB8At formed formaldehyde crosslinked complexes with the T-DNA substrate even in strains lacking core subunits or other components, e.g., VirB2At and VirB5At, implicated in outer membrane channel assembly (Cascales and Christie, 2004b). The cytoplasmic membrane translocon thus might engage and possibly even translocate substrates across the cytoplasmic membrane in the absence of core or the distal portion of the translocation channel.
The available data suggest that the portion of the translocation channel extending through the core complex and across the outer membrane consists of the VirB6 periplasmic domain, the C-terminal region of VirB8, VirB2 pilin, and VirB9 (Fig. 2C) (Christie et al., 2005; Fronzes et al., 2009a). At the outer membrane, the core’s cap formed by the helical bundles of VirB10 has a pore through which substrate might pass. However, given that the pore diameter is insufficient for substrate delivery, and also the lack of detection of a VirB10At -T-DNA substrate close contact by the TrIP assay (Cascales and Christie, 2004b), the outer membrane portion of the translocation channel probably has additional, yet unspecified structural complexity.
4b. Target Cell Attachment – Conjugative pili
In Gram-negative systems, conjugative pili are a major mechanism for T4SS-mediated target cell attachment. Assembly of pili and the translocation channel require the same T4SS genes with two exceptions: i) the VirB1-like cell wall hydrolase is required only for pilus biogenesis and ii) the VirD4 T4CP is required only for assembly of functional channels. Moreover, mutations have been isolated in several Mpf subunits that selectively disrupt assembly of one but not the second organelle. Such mutations rendering cells pilus-minus (Pil−) without affecting substrate transfer (Tra+) were mapped within the VirB2At pilin, VirB6At, VirB8At, VirB9At, VirB10At, and VirB11At. Conversely, mutations conferring the opposite Tra−, Pil+ phenotype have been identified mainly in pilin genes (Christie et al., 2005; Jakubowski et al., 2009; Kerr and Christie, 2010).
These findings prompted a model in which the Gram-negative T4SSs are configured not as a single channel/pilus structure but as two physically distinct organelles (Figs. 2C & D) (Christie et al., 2005). While this model is useful for formulation of mechanistic studies aimed at defining the requirements for biogenesis and function of each organelle, recent work on the F-like R1–16 transfer system strongly indicate that the two structures are physical linked (Lang et al., 2011). In this system, the filamentous phage R17 binds the R1–16 pilus as an initial step for entry of the ssRNA phage genome into the bacterial cell. Phage entry requires the Mpf proteins, as expected because these are involved in pilus assembly. Entry also requires the VirD4-like T4CP, which is surprising given that the T4CP is dispensable for pilus production. Even more remarkably, phage entry also required R1–16 processing by the Dtr factors and relaxosome engagement with the T4CP (Lang et al., 2011). These findings suggest that an activating signal, initiated within the cell by relaxosome-T4CP docking, is propagated across the cell envelope to the pilus where the phage is bound. Signal exchange across the T4SS/pilus in fact is not a new concept; early studies supplied evidence for ‘recipient-stimulated’ donor conjugal transfer whereby pilus binding to a recipient cell results in propagation of a signal to the donor cell interior to activate plasmid transfer. Thus, signal communication between the DNA processing/T4CP docking reactions within the cell and the extracellular pilus, best achieved if the translocation channel and pilus were physically joined, is required both for R17 phage entry and R1–16 plasmid transfer (Figs. 2C,D) (Zechner et al., 2012).
Conjugative pili can be long (2–20 mm) and flexible with a diameter of 8 nm and a central lumen of 2 nm, as represented by the F-type pili, or short (< 1 µm) and rigid with a diameter of 8–12 nm, as represented by P-type pili (Thanassi et al., 2012; Zechner et al., 2012). F-type pili enable E. coli donor cells to transfer DNA in liquid media, whereas P-type pili support efficient transfer only on solid surfaces. F-type pili are easily detected on the surfaces of F plasmid–carrying cells and undergo cycles of extension and retraction to the promote formation of conjugative junctions. F pilus extension and retraction has been visualized on living cells by laser-scanning confocal microscopy (Clarke et al., 2008). E. coli donor cells appear to extend and retract their pili to sample the immediate surroundings. Extension involves the addition of pilin subunits to the cell–proximal base of the pilus. If the extended pilus establishes contact with a recipient cell, retraction generates a force sufficient to bring the cells together. VirB4-like TraCF is the only ATPase involved in F pilus dynamics, but at this time there is little mechanistic understanding of how energy drives extension or retraction. In contrast to the F pili, P-type pili are rarely detected on donor cell surfaces; instead, they appear to be sloughed from the cell surface where they mediate aggregation of donor and recipient cells through hydrophobic interactions.
Pili of Gram-negative bacterial conjugation systems are the best characterized of the surface structures elaborated by T4SSs; however, other T4SS surface structures also have been visualized. These include a sheathed structure much larger than a pilus elaborated by the H. pylori Cag system (Rohde et al., 2003) and cell surface fibrous material elaborated by the L. pneumophila Dot/Icm system (Watarai et al., 2000). A few systems carry a VirB2 pilin-like subunit but lack a VirB5 homolog, and these systems are not thought to elaborate pili. Yet, these pilin proteins might still mediate target cell contacts through localization of pilin monomers or short oligomers at the cell surface.
5. Gram-positive T4SSs: comparisons with the Gram-negative machines
5a. Early substrate processing and translocation reactions
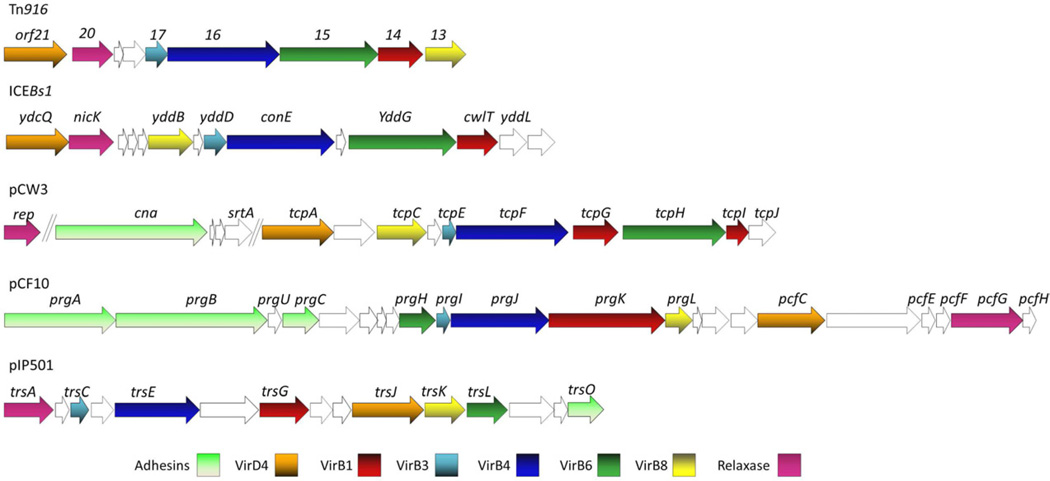
Studies of Gram-positive conjugation systems are important because these systems are widely responsible for rapid dissemination of multi-drug resistance and other virulence determinants among many medically important pathogens (Grohmann, 2003). Basic studies will also generate a mechanistic and structural understanding of macromolecular transport processes across the comparatively simple single-membrane (monoderm) cell envelope, allowing for comparisons with the T4SSs mediating transport across the double-membrane (diderm) envelopes. General properties of several conjugation systems, three plasmid-encoded (E. faecalis pCF10, Streptococcus agalactiae pIP501, Clostridium perfringens pCW3) and two ICE-encoded (Tn916, Bacillus subtilis ICEBs1), were previously reviewed (Alvarez-Martinez and Christie, 2009). We will first update the structure-function information of these systems and then present a working model depicting a possible architecture (Fig. 3).
Fig. 3.
The arrangement of T4SS genes carried by 5 representative Gram-positive ICEs and plasmids. The genes with similar predicted functions are depicted with the same color.
In spite of the comparatively limited study of Gram-positive systems, the available data suggest the early substrate processing and substrate-T4CP docking reactions mechanistically resemble those of the Gram-negative systems. Plasmid-encoded (PcfGpCF10, TraApIP501, MobMpMV158, MobApC221) and ICE-encoded (NicKICEBs1, Orf20Tn916) relaxases display strand-specific cleavage at cognate oriT sequences with or without a dependency on accessory factors (see (Alvarez-Martinez and Christie, 2009; Grohmann, 2003). The relaxases possess at least one active-site tyrosine thought to form a transient covalent adduct with T-strand. Although nothing is known about the substrate secretion signals, it is reasonable to propose that the relaxases and associated accessory factors carry such signals and mediate docking of the relaxase-T-strand transfer intermediate with the T4CP. Relaxase-T4CP interactions have been demonstrated with two-hybrid screens or pull-down assays (Abajy et al., 2007; Chen et al., 2008). In the case of the pCF10 transfer system, both the relaxase PcfG and its accessory factor PcfF interact with the PcfC T4CP (Chen et al., 2008). PcfC also binds DNA substrates in vitro, as well as pCF10 in vivo as shown with the TrIP assay. Reminiscent of findings in the A. tumefaciens system, formation of the pCF10-PcfC formaldehyde crosslink requires production of the Dtr factors (PcfF/PcfG) but not the Mpf channel components (Prg/Pcf) or ATP hydrolysis by PcfC (Chen et al., 2008).
Accumulating evidence further suggests that substrate transfer across the cytoplasmic membrane is probably also mechanistically and structurally conserved among Gram-negative and -positive bacteria. The pCF10, pCW3, pIP501, Tn916, and ICEBs1 transfer systems possess six conserved subunits bearing similarities to the A. tumefaciens VirD4 T4CP, VirB1, VirB3, VirB4, VirB6, and VirB8 channel subunits (Fig. 3). Indeed, as discussed below, these six subunits appear to be building blocks of most if not all of the Gram-positive T4SSs. Except for VirB1, these VirB/VirD4 subunits are required for substrate transfer across the cytoplasmic membrane of Gram-negative cells, and it is likely that the homologs or orthologs fulfill this same function in the Gram-positive systems. Presently, only limited information is available describing subunit - subunit interactions, but the networks identified thus far support this prediction (Abajy et al., 2007; Porter et al., 2012; Steen et al., 2009; Teng et al., 2008).
5b. The Gram-positive transocation channel
In contrast to the predicted mechanistic similarities in early DNA transfer reactions, fundamental differences exist in the latter reactions mediating i) substrate transfer across the Gram-positive cell wall ii) establishment of Gram-positive donor-target cell junctions. With respect to the cell-wall-spanning structure, Gram-positive systems lack genes for subunits comprising the core complex which serves both as a structural scaffold for the translocation channel and also to couple ATP energy with outer membrane channel gating. The channel architecture and dynamics of channel gating thus must be strikingly different in Gram-negative and -positive systems.
Only two of the conserved components of the Gram-positive systems are predicted to localize on the exterior face of the cytoplasmic membrane, the VirB1-like cell wall hydrolase and the C-terminal domain of the VirB8-like subunit. In fact, the hydrolases associated with Gram-positive T4SSs differ from prototypical VirB1At, which is secreted to the periplasm and has a single hydrolase domain. The Gram-positive subunits instead possess N-proximal TM domains and thus are likely anchored in the membrane (Alvarez-Martinez and Christie, 2009). Membrane anchoring might account for observed interactions between cell wall hydrolases and cognate T4CPs and the VirB6-like channel subunits (Abajy et al., 2007; Bantwal et al., 2012).
The hydrolases also can be classified into at least two subtypes, the first represented by pCW3-encoded TcpG and IceBs1-encoded YddH (renamed CwlT) (Bantwal et al., 2012; Fukushima et al., 2008). Both proteins carry two hydrolase domains; for CwlTIceBs1 these domains were shown to exhibit N-acetylmuramidase and DL-endopeptidase activities. In both proteins, mutations of catalytic residues abolished or diminished hydrolase activities of the respective domain and, correspondingly, a tcpG null mutation strongly diminished pCW3 transfer (Bantwal et al., 2012; Fukushima et al., 2008). The second hydrolase subtype is represented by E. faecalis PrgKpCF10 (Alvarez-Martinez and Christie, 2009). This is a large, ∼700 residue subunit with an N-proximal TM domain and three predicted hydrolase domains of the LytM, Phi29, and CHAP (NlpC/P60) families. Our recent studies have shown that the latter two domains, but not the LytM domain, are catalytically active and that one but not both of the catalytically active domains is necessary for assembly of a functional T4SS. Deletions or catalytic site mutations of both Phi29 and CHAP domains completely abolish pCF10 transfer. In contrast to the Gram-negative systems, the pCF10-encoded hydrolase therefore is absolutely required for T4SS function (Laverde-Gomez et al., submitted).
The second candidate channel-forming subunit is related to TcpCpCW3. Rood and colleagues recently solved the structure of the periplasmic region of TcpC, showing that it has two domains each with a VirB8-like structural fold. This fold consists of a highly curved antiparallel β-sheet folded around a central α-helix. Neither domain is related in primary sequence to VirB8At, and each exhibits only 16 % identity with the other. TcpC crystallized as a trimer, and interactions were identified with the TcpA T4CP, VirB6-like TcpH, and VirB1-like TcpC (Porter et al., 2012).
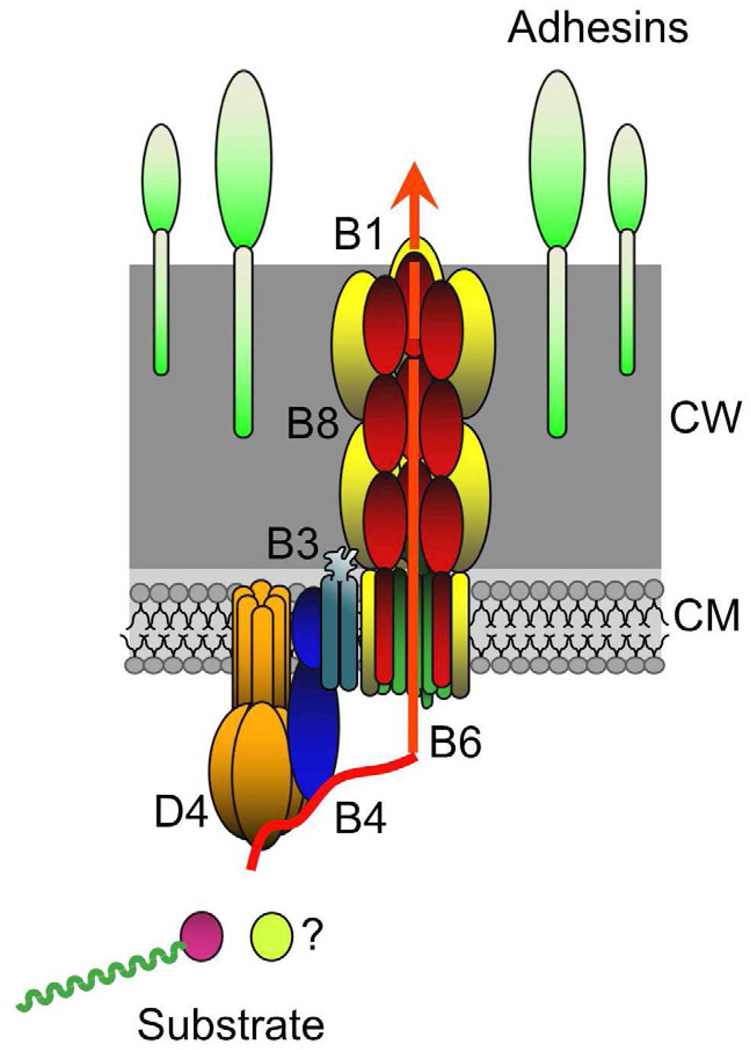
In view of these findings, we propose a working model in which the Gram-positive T4SSs are architecturally configured as depicted in Fig. 4. Accordingly, the ATPase/cytoplasmic membrane translocon complex is depicted as being structurally and mechanistically similar to the Gram-negative systems with the notable exceptions that the Gram-positive systems lack a VirB11 ATPase and the N-terminal TM domain of the VirB1 hydrolase forms part of the membrane translocon. The C-terminal hydrolase and VirB8-like domains extend across the cell wall and form a channel or fiber through or along which the secretion substrates pass. This structure would extend to the cell surface where, upon formation of the mating junction (see below), it would establish productive contacts with the recipient cell. While speculative at this time, this model depicts the configuration of a channel composed only of the six highly-conserved subunits of Gram-positive T4SSs. Notably, some T4SSs gene clusters such as those of pCW3, Tn916, ICEBs1 (see Fig. 3) and other systems described below, encode only these conserved subunits, placing significant constraints on the channel composition and architecture. Other Gram-positive systems, e.g., pCF10 and pIP501, code for other possible channel constituents that could add structural complexity to the translocation complex. Finally, it is also conceivable that assembly of the translocation channel is a dynamic process more akin to the pilus biogenesis systems of Gram-negative systems. The VirB4 subunit, for example, might catalyze dislocation of small membrane proteins that are typically also encoded within the T4SS gene clusters to build a wall-spanning fiber or pilus; this mechanism is reminiscent of the pseudopili elaborated by type II secretion or the B. subtilis competence systems.
Fig. 4.
Model for a T4SS channel spanning the Gram-positive cell envelope with the substrate translocation pathway depicted (red arrow). Channel subunits are identified according to the VirB/VirD4 nomenclature (color code matches Fig. 1). The cytoplasmic translocon/ATPase complex is depicted as resembling that of Gram-negative bacterial T4SSs. The cell-wall-spanning complex is composed of the multidomain VirB1-like hydrolase and VirB8-like subunits through or along which substrates pass. The Gram-positive systems cell-wall-anchored adhesins as opposed to pili for enhanced mating pair formation. Substrates include the relaxase-T-strand transfer intermediates and, possibly (?), protein effectors. Cytoplasmic membrane (CM), cell wall (CW).
5c. T4SS-mediated target cell attachment
The Gram-negative systems elaborate conjugative pili, but to date no Gram-positive T4SS has been shown to elaborate similar structures. Instead, these systems mediate target cell attachment at least in part through production of surface adhesins (Fig. 4). The best-characterized adhesin is aggregation substance produced by pheromone-dependent transfer systems such as E. faecalis pCF10 (Hirt et al., 2005; Olmsted et al., 1991). pCF10-encoded aggregation substance (PrgB, also called AS or Asc10), is a large ∼137-kDa protein with homologs identified primarily in related pheromone-inducible conjugation systems of enterococci. PrgBpCF10 possesses an N-terminal signal sequence, a C-terminal LPXTG cell wall anchor motif, RGD motifs, and a glucan binding or aggregation domain related to those found in the Streptococcus glucan-binding protein C (GbpC) and surface protein antigen (Spa)-family proteins (Waters and Dunny, 2001; Waters et al., 2003). In addition to its role in enhancement of interbacterial plasmid transfer, PrgBpCF10 binds bacterial lipoteichoic acid, mediates adherence to bacterial and eukaryotic cells, and promotes bacterial internalization by epithelial cells (Chuang-Smith et al., 2010).
A prgB mutant still transfers pCF10 at appreciable frequencies in solid-surface matings, suggesting that other surface adhesins or the translocation channel itself also can promote target cell contacts. Besides PrgBpCF10, the pCF10 tra region codes for two other cell-wall-anchored surface proteins, PrgApCF10 and PrgCpCF10 (Alvarez-Martinez and Christie, 2009). PrgApCF10 functions in surface exclusion, but it might also contribute to formation of productive mating junctions (Olmsted et al., 1993). PrgCpCF10 is related to Orf15pIP501 (now renamed TraO) and, interestingly, both proteins contain highly repetitive sequence motifs of a 3-residue periodicity comprised of Pro - uncharged -Glu/Asp (Alvarez-Martinez and Christie, 2009). Such repeat regions are features of other Gram-positive surface adhesins, including Streptococcus pyogenes Sfb, Iga-binding proteins of S. agalactiae, and S. aureus and streptococcal fibronectin binding proteins (Krishnan and Narayana, 2011). These features indicate that PrgCpCF10 and TraOpIP501 might also mediate specific contacts with bacterial recipients and, possibly, eukaryotic cells.
Among the 3 other representative conjugation systems under discussion, C. perfringens pCW3 codes for sortase and Cna, a surface-anchored protein with multiple repeats of a collagen-binding domain. Cna also might mediate contacts with bacterial and/or eukaryotic cell targets, the former possibly accounting for the observed high-frequency transfer rates of this plasmid. In contrast, neither ICEBs1 nor Tn916 code for obvious cell surface adhesins (Fig. 3). These systems do code for extended-VirB6 subtypes and the C-termini of these subunits might project to the cell surface to promote donor-target cell contacts.
6. Phylogenetic studies of T4SSs
6a. Evolution of T4SSs
With expansion of available genome sequences, phylogenetic studies have sought to describe evolutionary processes shaping the modern lineages. Most attention has focused on the mobile DNA elements, using relaxases, signature VirB4 and T4CP ATPases, or Mpf proteins for classification (Guglielmini et al., 2013; Guglielmini et al., 2011). The phylogenetic relationships were traced for relaxases encoded by more than 1700 plasmids among proteobacteria, ultimately resulting in classification into one of six MOB families (Garcillan-Barcia et al., 2011; Smillie et al., 2010). The phylogeny of T4CPs was found to closely match that of the relaxases, suggesting that the substrate (the relaxase) and the receptor (the T4CP) have evolved in parallel. This work has reinforced the notion that the T4CP is part of the conjugative DNA processing module, for which there is also increasing experimental evidence (Zechner et al., 2012). The VirB4 phylogeny shows a more scattered distribution and clades tend to follow prokaryotic phylogeny, implying that plasmids and their associated T4SSs adapt over evolutionary time to specific bacterial hosts. Distinct clades of VirB4 subunits exist among the Gram-negative, Gram-positive, and archaeal systems, reflecting limited movement of T4SSs between these groups. Thus, while the T4SSs have the potential for translocation across distant phylogenetic boundaries, mobility between the distant clades is sporadic and transient (Guglielmini et al., 2013; Guglielmini et al., 2011).
Phylogenetic studies of the VirB4 signatures suggest that T4SSs arose from an ancestral conjugation function, most probably among the diderms. The ancestral conjugation systems evolved either as ssDNA or dsDNA translocation systems, the former as the broadly distributed conjugation machines and the latter as the dsDNA translocases associated with chromosome segregation during division (FtsK), sporulation (SpoIIIE), and chromosomal transfer among Streptomyces spp. mycelia (TraB). The ssDNA conjugation systems diversified through genetic linkage of the T4CP with VirB4 and membrane proteins comprising the mating channel. The VirB4/Mpf systems were shaped further by effects of the cell envelope and ultimately resulted in membrane-specialized clades. As noted above, these clades tended to remain fixed among species bearing similar envelope structures/compositions (Guglielmini et al., 2013).
At much shorter evolutionary distances, conjugative systems diversified further among the ICEs and plasmids and, ultimately, as the effector translocator and substrate release/uptake subfamilies. Recent phylogenetic studies of Rickettsia spp., and Bartonella spp. have offered further insights into the expansion of the dedicated effector translocators. The Rickettsia spp. are intracellular pathogens that have coevolved with the host, resulting in reduction of their genomes and strict dependence on host resources. Interestingly, the Rickettsial T4SSs are characterized by gene duplication and distribution of component genes in islets around the genome. These genomes carry multiple copies of genes for VirB6 components, as well as duplications of genes encoding VirB4, VirB8, and VirB9 subunits. There is also evidence for lineage specific diversification of VirB2, VirB9 and VirB1, possibly reflecting differences in the cell envelope structure/composition (Gillespie et al., 2009; Gillespie et al., 2012).
As mentioned above, subunit composition can be mechanistically informative. For example, the absence of a VirB5 subunit suggests the Rickettsia spp. T4SSs do not elaborate pili, possibly because these intracellular pathogens would not need such a structure for substrate delivery into the host cell. Also of potential interest are the observations that: i) the B. pertussis Ptl and H. pylori ComB systems lack VirB5 homologs and these systems utilize a two-step translocation mechanism, ii) phylogenetic studies indicate that systems most closely related to the Rickettsia spp. T4SSs branch from the H. pylori ComB DNA uptake system, and iii) the Rickettsia spp. T4SSs are composed of multiple copies of extended-VirB6 subunits bearing resemblance to the ComEC channel proteins involved in DNA uptake. It is tempting to suggest that Rickettsia spp. elaborate dual- or alternative-function T4SSs with the capacity to export effector proteins and/or import DNA. The Rickettsia spp. systems also might translocate substrates by a two-step, contact-independent pathway(s).
Bartonella spp. carry at least two T4SSs, a VirB/VirD4-like system responsible for translocating effector proteins to the host cell and the Trw system which appears to function exclusively in mediating attachment to target cell receptors (Saenz et al., 2007). Corresponding phylogenomic analyses suggest the two T4SS gene clusters evolved through adaptive radiation in two sister clades of this a-proteobacterial pathogen (Engel et al., 2011). In lineage four, genes for the VirB, VirD4, and five to seven Bep effector substrates cluster on the chromosome, but in lineage three, the T4SS genes and genes for twelve to sixteen Bep effectors are scattered around the chromosome. The T4SSs likely evolved after divergence of the two lineages by independent events - and multiple events for lineage three - of plasmid integration. The bep effector genes also evolved separately by gene duplication, diversification, and domain reshuffling events resulting in modular gene architectures. Finally, the Trw system, which is confined to lineage four, has also undergone diversification, presumably reflecting the adaptation of this adhesin factor to the erythrocyte surface of different host species (Engel et al., 2011).
6b. T4SS classification and identification: Web tools for curation and identification
Recent studies have enabled formulation of phylogeny-based classification schemes. Early on, the Gram-negative T4SSs were classified as types IVA and IVB, reflecting common ancestries with the A. tumefaciens VirB/VirD4 and the Col1b–P9/R64 (IncI) plasmid transfer systems, respectively (Christie and Vogel, 2000). A subsequent scheme gave rise to four classes: (i) F-T4SS or MpfF, ii) P-T4SS or MpfT (for A. tumefaciens Ti plasmid), iii) I-T4SS or MpfI (for IncI plasmid R64), and GI-T4SS or MpfG (for genomic island-encoded based on ICEHIN1056) (Smillie et al., 2010). The most recent phylogenetic analyses of VirB4/VirD4 T4CPs identified four additional T4SS subgroups: v) MpfC (specific to Cyanobacteria) and vi) MpfB (Bacteriodes), and monoderm-specific subgroups vii) MpfFA (Gram-positives) and viii) MpfFATA (Gram-positives and archaea) (Guglielmini et al., 2013). These classification schemes are valuable for understanding evolutionary processes, defining mechanistic themes and variations, and predicting machine or machine subunit functions.
With expansion of sequenced genomes, it is also evident that only a fraction of the possible T4SSs have been identified. VirB4 signature subunits are typically well-annotated in the databases, but diversification of other T4SS components has hindered their detection and, hence, T4SS assignments. To curate the available information and for high-throughput prospecting for new T4SSs, various web-based algorithms have been described. We list some here for the interested reader: i) Named entity recognition for bacterial T4SSs (http://www.nactem.ac.uk/T4SS_NER/top.py, http://patricbrc.vbi.vt.edu/portal/partic/NACTEM) (Ananiadou et al., 2011), ii) AtlasT4SS (htt://www.t4ss.lncc.br), a curated database for T4SSs (Souza et al., 2012), iii) MobilomeFINDER (http://mml.sjtu.edu.cn/MobilomeFINDER), for identification of bacterial genomic islands (Ou et al., 2007), iv) A T4SS identification program (http://t4ss.bioinfoicdc.org/), (Zhang et al., 2012) v) (http://www.tau.ac.il/~tlap/LegionellaMachineLearning), a machine learning approach for genome-scale identification of L. pneumophila effectors (Burstein et al., 2009), and vi) identification of Anaplasma marginale T4SS effectors (Lockwood et al., 2011).
7. Expansion of the T4SS lexicon: The ‘minimized’ systems of Gram-positive species
7a. Streptococcal T4SSs
A question surfacing from the VirB4 phylogenetic studies is whether the identified genes are members of functional T4SSs or orphans from ancestral mobile elements. We addressed this question by examining flanking regions of virB4-like genes in Gram-positive genomes for the presence of other T4SS genes (see Fig. 5; Table 1). Zhang et al. carried out a similar analysis, and they reported the presence of T4SS gene clusters conserved both in composition and order in the genomes of many streptococcal species (Zhang et al., 2012). The conserved genes code for proteins similar to A. tumefaciens VirD4, VirB6, VirB4, and VirB1. These new T4SS gene clusters were classified as type IVC systems. Because the type IVA/B distinction is already inadequate for classification of known systems, for the present we suggest designating these gene clusters as ‘minimized’ or ‘minimal’ T4SS’s.
Fig. 5.
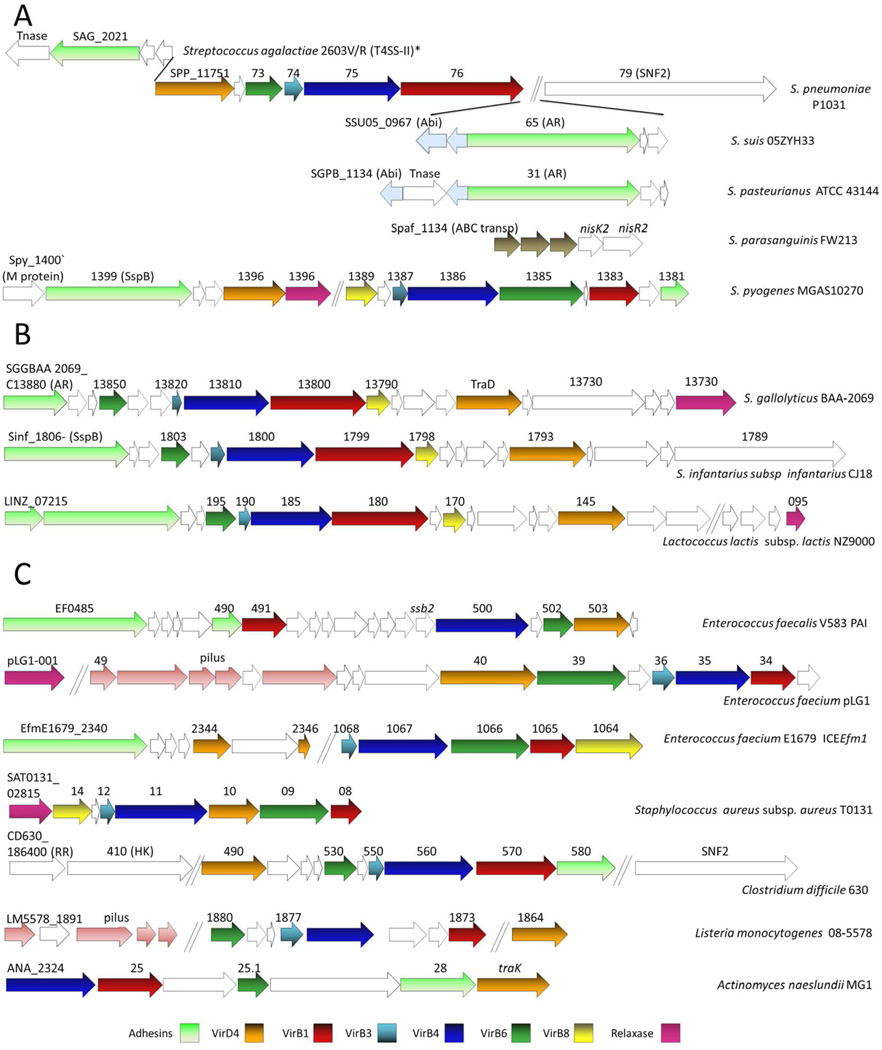
The arrangements of T4SS genes in different Gram-positive species. Most of the presented gene clusters were identified through an initial screen for virB4 signature genes in Gram-positive genomes. The function of each gene was predicted based on sequence relatedness as shown by pBLAST searches or, the case of virB6, a characteristic hydropathy profile of the encoded product. For each strain, the locus tag or the gene annotation of the first gene and the genes predicted to code for VirB4, VirB3, VirB6, VirB8 and VirD4 functions are denoted. The genes with similar predicted functions are depicted with the same color. (A) Gene arrangement of T4SSs likely part of an intact or vestigial Tn5253 element in streptococcal species. T4SS genes and associated SNF2 gene in S. pneumoniae P1031 are depicted to illustrate the signature gene arrangement found in most streptococcal species examined (for a complete list see Table 1). S. agalactiae 2603V/R T4SS II* represents the gene arrangement of one of two T4SSs present in this strain. (B) Gene arrangements of T4SSs bearing similarities to the Pcf/Prg system of E. faecalis pCF10 (C) Gene arrangements of T4SS clusters in other Gram positive strains. Abbreviations: Abi, abortive infection protein; Tnase, transposase; AR, agglutinin receptor; ABC transp, ABC transporter; PAI, pathogenicity island; RR, response regulator; HK, histidine kinase. Genes with similar functions (ABC transporter, pilus assembly) are similarly-colored.
Table 1.
Gram-positive strains or plasmids evaluated for the presence of ‘minimized’ type IV secretion systems.
| Strain/Plasmid name | Accession number |
|---|---|
| S. suis 05ZYH33 | CP000407.1 |
| S. suis SC84 | FM252031.1 |
| S. suis 98HAH33 | CP000408.1 |
| S. suis BM407 | FM252032.1 |
| S. suis D9 | CP002641.1 |
| S. suis SS12 | CP002640.1 |
| S. suis D12 | CP002644.1 |
| S. suis JS14 | CP002465.1 |
| S. equi subsp. zooepidemicus H70 | FM204884.1 |
| S. equi subsp. zooepidemicus ATCC 35246 | CP002904.1 |
| S. thermophilus JIM8232 | FR875178.1 |
| S. pneumoniae CGSP14 | CP001033.1 |
| S. pneumoniae ATCC 700669 | FM211187.1 |
| S. pneumoniae G54 | CP001015.1 |
| S. pneumoniae P1031 | CP000920.1 |
| S. pneumoniae 670-6B | CP002176.1 |
| S. pneumoniae Hungary 19A-6 | CP000936.1 |
| S. pneumoniae 70585 | CP000918.1 |
| S. agalactiae 2603V/R | AE009948.1 |
| S. agalactiae NEM316 | NC_004368.1 |
| S. dysagalactiae subsp. equisimilis ATCC 12394 | CP002215.1 |
| S. bovis ATCC 70338 contig 00014 | AEEL01000014.1 |
| S. pasteurianus ATCC 43144 | AP012054.1 |
| S. macedonicus ACA-DC198 | HE613569.1 |
| S. parasanguinus FW213 | CP003122.1 |
| S. infantarius subsp. infantarius CJ18 | NC_016826.1 |
| S. pyogenes MGAS10270 | NC_008022.1 |
| S. pyogenes MGAS2096 | NC_008023.1 |
| S. pyogenes MGAS10750 | NC_008024.1 |
| S. gallolyticus BA-2069 | NC_015215.1 |
| S. gallolyticus UCN34 | NC_013798.1 |
| S. aureus T0131 | CP002643.1 |
| S. aureus M809 | NZ_ACUS00000000.1 |
| S. aureus USA300_FPR3757 | NC_007793.1 |
| L. lactis subsp. cremoris NZ9000 | CP002094.1 |
| C. difficile 630 | NC_009089.1 |
| L. monocytogenes 08–5578 | NC_013766.1 |
| A. naeslandii MG1 | 12510 (NCBI project ID) |
| E. faecalis V583 PAI | AF454824.1 |
| E. faecalis pAM373 | NC_002630 |
| E. faecium 64/3×UW2774 pLG1 | HM565183.1 |
| E. faecium Aus0004 | CP003351.1 |
| E. faecium E1679 contig00369 | NZ_ABSC01000340.1 |
Our analyses suggest that most or all of the streptococcal T4SS gene clusters are intact Tn5253 ICEs or remnants of these elements (Fig. 5). Tn5253 elements are composed of two ICEs, Tn5252 into which Tn916 has inserted (Henderson-Begg et al., 2009; Mingoia et al., 2011). The ‘minimized’ T4SS gene clusters identified by Zhang et al. originated from Tn5252, and as these authors noted a Tn916-like element is often located nearby (Zhang et al., 2012). Additional Tn5253-like features often flank these T4SS genes; for example, downstream of these clusters are genes for single-stranded DNA-binding and Ca++-binding proteins, and an SNF2/DEAD helicase. Upstream of the virD4-like gene are typically those coding for cytosine methylase, replication initiation factors, and topoisomerase.
The streptococcal T4SS gene clusters themselves have several conserved features of interest (Fig. 5):
The “virB1-like” genes are homologs of E. faecalis pCF10-encoded PrgK and are much larger (>700 residues) than the cell wall hydrolases of Gram-negative bacterial T4SSs (∼250 residues).
The “virB6-like’ genes are invariably misannotated; they are typically multi-pass (5–7 TM domains) membrane proteins of which pCF10-encoded PrgH can be considered an archetype. Among the identified streptococcal clusters, none coded for the ‘extended-VirB6’ subtype.
Small genes are located upstream of the virD4-, virB6-, and virB4-like genes. These genes bear similarities to those at corresponding positions in the E. faecalis pCF10 prg/pcf transfer operon (Hirt, et al., 2005;Alvarez-Martinez and Christie, 2009). In the pCF10 system, these gene pairs are pcfB-pcfC (virD4-like), prgF-prgH (virB6-like), and prgI-prgJ (virB4-like). The gene pairs are conserved among most other examined Gram-positive T4SS gene clusters and, among these the prgI-prgJ (virB3-virB4) pairing is invariably found in the Gram-negative systems.
These streptococcal T4SS gene clusters possess another intriguing feature, the region between the virB1-like and SNF2 genes is variable in gene composition among the different species (Fig. 5). For example, although S. pneumoniae T4SS gene clusters typically lack additional genes in this region, most other species (S. suis, S. equi, S. agalactiae, S. dysagalactiae, S. macedonicus, S. thermophilus, and S. pasteurianus) carry genes for phage exclusion (Abi) and surface attachment proteins (e.g., agglutinin receptor). Abi proteins inhibit bacteriophage infection/propagation (Chopin et al., 2005). Such genes are often associated with mobile elements, likely reflecting a strategy to minimize competition between phage that use T4SSs for gaining entry into the cell and lateral gene transfer. The Abi/T4SS genetic linkage thus might have evolved to inhibit propagation of phages utilizing streptocococcal T4SSs for entry across the Gram-positive envelope. The adhesion genes are generally annotated as agglutinin receptor, surface antigen I/II, or glucan-binding proteins; these are sortase-anchored proteins that probably mediate attachment to bacterial or eukaryotic target cell receptors (Brady et al., 2010; Hendrickx et al., 2009; Vengadesan and Narayana, 2011). These functions resemble those described above for E. faecalis pCF10-encoded aggregation substance.
Further examples of plasticity within the virB1-SNF2 intervening region are found in a S. pasteurianus T4SS, which codes for a transposase in addition to the Abi and agglutinin receptor functions and a S. parasanguinis T4SS, which lacks genes for the Abi and surface proteins but codes for a sensor kinase/response regulator two-component system and an ATP-binding cassette (ABC) transporter system.
Genes for surface proteins associated with virulence can also be found upstream of the virD4-like gene in some streptococcal T4SS clusters (Fig. 5). For example, next to the T4SS cluster in S. agalactiae strain 2603V/R is a surface antigen I/II protein family (SspA) gene, and next to a T4SS gene cluster in S. pyogenes strain MGAS10270 are genes with functions annotated as SspB, a peptidase (annotated as an M protein), and Esp.
Our analyses of the streptococcal genomes also revealed T4SS gene clusters resembling the E. faecalis pCF10 transfer system (Fig. 5). These are carried by S. gallolyticus strain BAA-2069 and S. infantarius subsp. infantarius strain CJ18. These clusters code for most or all of the pCF10 transfer genes and, like pCF10, upstream of the transfer genes are genes coding for surface proteins although they are not homologs of pCF10-encoded PrgA, PrgB, and PrgC.
Are the streptococcal T4SS gene clusters functional? Several of the identified systems might be associated with mobile ICE’s, but several also lack associated antibiotic resistance genes, genes required for conjugative transposition, e.g., relaxase, xis, int, or att sites. That the T4SS genes in the latter systems are intact and linked with surface protein genes suggests the interesting possibility that they have evolved as effector translocators. Indeed, Zhang et al. have supplied experimental support for this proposal in their studies of S. suis strain 05ZYH33, a clinical isolate from a streptococcal toxic shock large outbreak (Zhang et al., 2012). This strain was shown to carry an 89 K pathogenicity island (PAI), which as noted above appears to be comprised in part of a Tn5253 (Tn916/Tn5252) element. Mutations in the virB4- or virD4-like genes display two phenotypes of interest. First, the mutant strains were avirulent and impaired in their ability to trigger a host immune response in a mouse infection model. Second, these mutations abolished transmission of the 89 K PAI to recipient streptococci. This T4SS thus might have retained its ancestral function as a mobile DNA element while also evolving the capacity to translocate effector protein(s) to the cell surface or into eukaryotic target cells.
7b. ‘Minimized’ T4SSs in other Gram-positive species
We asked if virB4-like genes in other Gram-positive species are flanked with other T4SS and surface protein genes. In general, we found this to be the case, although we acknowledge that our dataset is currently limited to a few genomes of prominent Gram-positive species. Some T4SS gene arrangements are depicted in Fig. 5. Notable features include:
Lactococcus lactis supbsp. cremoris strain NZ900: Carries most of the pCF10-like tra gene cluster. Two upstream genes code for predicted surface proteins.
E. faecalis V583 PAI: Genes coding for homologs of pCF10 surface proteins PrgA, PrgB, and PrgC are adjacent to the T4SS gene cluster. The T4SS gene cluster differs from that of pCF10, e.g., the virB1-like gene codes for a cell wall hydrolase resembling those encoded by Tn916 and pCW3, and the virB6-like gene codes for an ‘extended-VirB6 ‘subtype.
E. faecium: The virulence plasmid pLG1 carries a T4SS cluster coding for a pCW3-like VirB1 hydrolase and an extended-VirB6. Upstream of the virD4-like gene are genes coding for a probable surface protein annotated as a viral A-type inclusion protein and a pilus assembly cluster. Two other E. faecium genomes, Aus0004 (not shown) and the ICEEfm1 element in strain E1679, also carry T4SS gene clusters with flanking genes for surface proteins and ABC transporters.
Staphylococcus aureus: T4SS gene clusters were found in strains T0131 (Fig. 5) as well as in strains M809 and USA300_FPR3757 and plasmid pLW043 (not shown), but the gene orders and virB1 and virB6 subtypes are variable. Adjacent genes encode exported and surface proteins, β-lactamase, and vancomycin resistance, suggestive of possible associations with ICEs.
Clostridium perfringens strain 630: The T4SS is similar in content and arrangement to the Tn5253-like, streptocococal T4SS’s, e.g., downstream genes code for surface protein and SNF2. Upstream of the T4SS genes are genes for a two-component regulatory system.
Listeria monocytogenes strain 08–5578: Associated with the T4SS gene cluster are genes for a tetracopeptide repeat protein, pilus assembly proteins, and other virulence-associated proteins. vi) Actinomyces naeslundii strain MG1: Sandwiched within the T4SS gene cluster are large genes of which one codes for a predicted surface adhesin.
In sum, adjacent to the virB4 signature genes in Gram-positive genomes we typically detect intact T4SS genes encoding proteins similar to VirB1, VirB3, VirB6, and VirD4. Genes coding for a VirB8-like structural fold might also be found, but these are more difficult to identify due to low sequence similarity among the VirB8 family members. Additionally, the ‘minimized’ type IV secretion machines are commonly associated with surface attachment/virulence factors, e.g., surface adhesins, pili, as well as phage immunity systems. Other hallmarks of functional mobile elements may or may not be present. Finally, while many of the identified gene clusters might be associated with mobile elements, there is experimental evidence that a ‘minimized’ T4SS encoded by the S. suis 89K PAI evolved virulence functions reminiscent of the effector translocators of Gram-negative systems.
7c. Tn916-like elements: A promiscuous ‘minimized’ T4SS
As mentioned above, a driving force in the evolution of T4SS clades is the cell envelope itself. The VirB7/VirB9/VirB10 core complex represents one adaptation for T4SS function across the Gram-negative outer membrane, and is generally considered to be a ubiquitous feature of Gram-negative systems. By contrast, the ‘minimized’ T4SSs might have evolved from the ancestral Gram-negative systems through loss of nonessential functions to allow transport across the monoderm cell envelope. On the basis of the recent phylogenetic analyses, such adaptations to the cell envelope should have resulted in fixation of these T4SSs among the monoderms. In fact, however, at least one group of ‘minimized’ T4SSs - those encoded by the Tn916/Tn1545 ICE’s -also exist in Gram-negative species (Clewell et al., 1995; Roberts and Mullany, 2009, 2011). Recently, a Tn916-like element, designated Tn6009, was identified in Klebsiella pneumoniae, Pseudomonas spp., and Serratia liquefaciens, and this element was further shown to transfer from both Gram-negative and -positive species to E. faecalis recipients (Soge et al., 2008). As noted above, the Tn916-like elements carry the ‘minimized’ T4SS gene set coding for VirB1-, VirB3-, VirB4-, VirB6-, VirB8-, and VirD4-like functions (See Fig. 3). These elements lack genes for the Gram-negative VirB7/VirB9/VirB10 core complex, raising the interesting question of how these promiscuous systems mediate transport across the Gram-negative cell envelope. These elements also lack known attachment devices, raising a further question of how these T4SSs mediate target cell attachment and mating junction formation.
8. Summary and Future Directions
The recent structure-function and phylogenetic studies continue to shape our understanding of these fascinating and complex machines. The structural advances are accelerating and promise to generate a detailed view of how paradigmatic systems in Gram-negative species are architecturally configured. Continued progress on the structural front ultimately will allow comparisons of machine architectures across phylogenetic boundaries. At this time, however, many fundamental questions remain unanswered. In the pilus-based systems, what is the physical relationship of the pilus with the translocation channel and how does the core complex mediate assembly of both structures? For systems in both Gram-negative and –positive species, what is the structure of the energy-powering ATPase complex, how does the complex associate with the membrane translocon, and how does it drive machine morphogenesis? What are the architectures of the membrane translocons, the periplasm-spanning translocation channels, and the outer membrane pores? And finally, what are the architectures of mating junctions? Answers to some of these questions will come from structural analyses of machine subassemblies in vitro and to others will require high-resolution imaging of machines in their native environments within the cell envelope.
An important accompaniment to the structural studies are in vivo functional tests for assigning biological relevance of identified structures and defining dynamic processes, e.g., machine biogenesis, substrate transfer, and mating junction formation. Early steps of DNA substrate processing have been described in biochemical detail, but mechanisms regulating spatiotemporal control of substrate presentation and trafficking through the T4SS remain poorly understood. Other long-standing questions relate to activation of machine function at the cell interior by Dtr processing and ATPase activities and at the cell exterior by target cell or phage binding; answers to these questions will continue to require creative new genetic, functional, and cell biological assays. One of the most interesting dynamic processes associated with T4SS function, which can now be visualized in real time through use of fluorescent phage (Clarke et al., 2008), is F pilus extension and retraction. Mechanistically, how is this achieved?
Recent phylogenetic analyses have identified evolutionary processes shaping the modern T4SSs, and also suggest an exciting new concept in the field of type IV secretion, that of the so-called ‘minimized’ T4SSs found mainly in Gram-positive species but also in some Gram-negative species. At a fundamental level, these systems are excellent subjects for detailed structure-function studies of the membrane translocon stripped of other T4SS adaptations acquired over evolutionary time. Also of considerable medical importance, at least one such T4SS encoded by the S. suis 89K PAI functions as a mobile DNA element and, apparently, also as an effector translocator required for virulence and immune regulation. Do these systems in fact translocate proteins and, if so, what types of substrates are translocated and how do the effectors contribute to infection processes?
Finally, an overarching theme of this review is that a comprehensive understanding of T4SS structure, mechanism, and phylogeny is essential for future applied studies aimed at design of T4SS inhibitors or manipulations for therapeutic applications. One case in point is the recent discovery of small molecule inhibitors of the Brucella VirB T4SS, afforded through the early finding that VirB8 proteins function as dimers to promote assembly and function of T4SSs. Relying on X-ray crystallography data, molecular docking, a high-throughput two-hybrid screen, and site-directed mutagenesis, Baron and colleagues identified small molecule inhibitors of VirB8 dimerization (Villamil Giraldo et al., 2012), and they further showed that these inhibitors block intracellular proliferation of B. abortus in a macrophage infection model (Paschos et al., 2011). Interestingly, one of the more potent inhibitors is salicylidene acylhydrazide, a class of molecules known to also inhibit type III secretion systems. In view of the recent discovery that oligomeric VirB8-like subunits are conserved among Gram-negative and -positive T4SSs, it can now be asked whether these molecules exert broad-spectrum activity against distant T4SS clades.
Finally, the discovery that Bartonella spp. deliver DNA substrates into human cells has potentially broad ramifications (Fernandez-Gonzalez et al., 2011; Llosa et al., 2012; Schroder et al., 2011). Conceivably, T4SS-mediated DNA transfer to eukaryotic host cells is more prevalent than previously envisaged. If so, it is critical to understand how interkingdom DNA transfer contributes to infection processes. For therapeutic application, manipulation of T4SSs carried by attenuated pathogens or commensals might prove efficacious for targeted delivery of DNA vaccines or other therapeutic DNAs to specific mammalian cell types or tissues. In fact, T4SS-mediated genetic engineering is a decades-old concept arising from the discovery of A. tumefaciens VirB/VirD4-mediated transfer of oncogenic T-DNA to plant cells. The past is linked to the present and, clearly, the future of T4SS research remains exciting and challenging.
Acknowledgements
We thank members of the Christie laboratory for helpful discussions. This work was supported by NIH grant R01GM48746.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abajy MY, Kopec J, Schiwon K, Burzynski M, Doring M, Bohn C, Grohmann E. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria. J. Bacteriol. 2007;189:2487–2496. doi: 10.1128/JB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens . Microbiology. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- Ananiadou S, Sullivan D, Black W, Levow GA, Gillespie JJ, Mao C, Pyysalo S, Kolluru B, Tsujii J, Sobral B. Named entity recognition for bacterial Type IV secretion systems. PloS ONE. 2011;6:e14780. doi: 10.1371/journal.pone.0014780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26:2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153:442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- Bailey S, Ward D, Middleton R, Grossmann JG, Zambryski PC. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Kerr JE, Cascales E, Giuliano ME, Bailey ME, McKay C, Chandran V, Waksman G, Christie PJ. An Agrobacterium VirB10 mutation conferring a type IV secretion system gating defect. J. Bacteriol. 2011;193:2566–2574. doi: 10.1128/JB.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantwal R, Bannam TL, Porter CJ, Quinsey NS, Lyras D, Adams V, Rood JI. The peptidoglycan hydrolase TcpG is required for efficient conjugative transfer of pCW3 in Clostridium perfringens . Plasmid. 2012;67:139–147. doi: 10.1016/j.plasmid.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Baron C. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 2006;84:890–899. doi: 10.1139/o06-148. [DOI] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol. Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U. S. A. 2004a;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004b;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesar CE, Machon C, de la Cruz F, Llosa M. A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol. Microbiol. 2006;62:984–996. doi: 10.1111/j.1365-2958.2006.05437.x. [DOI] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang X, Manias D, Yeo HJ, Dunny GM, Christie PJ. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J. Bacteriol. 2008;190:3632–3645. doi: 10.1128/JB.01999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Current Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Christie PJ. Bacterial type IV secretion: The Agrobacterium VirB/D4 and related conjugation systems. Biochem. Biophys. Acta. 2004;1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS ONE. 2010;5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Maddera L, Harris RL, Silverman PM. F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17978–17981. doi: 10.1073/pnas.0806786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell DB, Flannagan SE, Jaworski DD. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, Muller A, Twizere JC, Nkengfac B, Vandenhaute J, et al. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell. Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- Durand E, Oomen C, Waksman G. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J. Bacteriol. 2010;192:2315–2323. doi: 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Waksman G, Receveur-Brechot V. Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system. BMC Struct. Biol. 2011;11:4. doi: 10.1186/1472-6807-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher SC, Dehio C. Bartonella entry mechanisms into mammalian host cells. Cell Microbiol. 2012;14:1166–1173. doi: 10.1111/j.1462-5822.2012.01806.x. [DOI] [PubMed] [Google Scholar]
- Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Conjugative pili of IncP plasmids, the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- Engel P, Salzburger W, Liesch M, Chang CC, Maruyama S, Lanz C, Calteau A, Lajus A, Medigue C, Schuster SC, et al. Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella . PLoS Genet. 2011;7:e1001296. doi: 10.1371/journal.pgen.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez E, de Paz HD, Alperi A, Agundez L, Faustmann M, Sangari FJ, Dehio C, Llosa M. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J. Bacteriol. 2011;193:6257–6265. doi: 10.1128/JB.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz B, Kempf VA. Adhesion and host cell modulation: critical pathogenicity determinants of Bartonella henselae . Parasit. Vectors. 2011;4:54. doi: 10.1186/1756-3305-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 2009a;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009b;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS. Conjugation, Bacterial. In: Schaechter M, editor. Desk Encyclopedia of Microbiology. San Diego, CA: 2009. pp. 294–308. [Google Scholar]
- Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, Shida T, Sekiguchi J. Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting n-acetylmuramidase and DL-endopeptidase activities. J. Biol. Chem. 2008;283:11117–11125. doi: 10.1074/jbc.M706626200. [DOI] [PubMed] [Google Scholar]
- Fullner KJ, Lara JC, Nester EW. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol. Rev. 2011;35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2012;4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Joardar V, Williams KP, Driscoll T, Hostetler JB, Nordberg E, Shukla M, Walenz B, Hill CA, Nene VM, et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 2012;194:376–394. doi: 10.1128/JB.06244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1- ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J. Mol. Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microb. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. The repertoire of ICE in prokaryotes underscores the unity, diversity, ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson-Begg SK, Roberts AP, Hall LM. Diversity of putative Tn5253-like elements in Streptococcus pneumoniae . Int. J. Antimicrob. Agents. 2009;33:364–367. doi: 10.1016/j.ijantimicag.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17:423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J. Bacteriol. 2005;187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaeche I, Alkorta I, Moro F, Valpuesta JM, Goni FM, De La Cruz F. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 2002;277:46456–46462. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 2004;341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JE, Christie PJ. Evidence for VirB4-mediated dislocation of membrane-integrated VirB2 pilin during biogenesis of the Agrobacterium VirB/VirD4 type IV secretion system. J. Bacteriol. 2010;192:4923–4934. doi: 10.1128/JB.00557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Narayana SV. Crystallography of gram-positive bacterial adhesins. Adv. Exp. Med. Biol. 2011;715:175–195. doi: 10.1007/978-94-007-0940-9_11. [DOI] [PubMed] [Google Scholar]
- Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Frohlich KU, Zechner EL. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol. 2010;78:1539–1555. doi: 10.1111/j.1365-2958.2010.07423.x. [DOI] [PubMed] [Google Scholar]
- Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol. Microbiol. 2011;82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Li F, Alvarez-Martinez C, Chen Y, Choi KJ, Yeo HJ, Christie PJ. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J. Bacteriol. 2012;194:4041–4051. doi: 10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Roy C, Dehio C. Bacterial type IV secretion systems in human disease. Mol. Microbiol. 2009;73:141–151. doi: 10.1111/j.1365-2958.2009.06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Schroder G, Dehio C. New perspectives into bacterial DNA transfer to human cells. Trends Microbiol. 2012;20:355–359. doi: 10.1016/j.tim.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278:4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS ONE. 2011;6:e27724. doi: 10.1371/journal.pone.0027724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 2008;70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell. Microbiol. 2011;13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J. Bacteriol. 2007;189:6469–6473. doi: 10.1128/JB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingoia M, Tili E, Manso E, Varaldo PE, Montanari MP. Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 2011;55:1453–1459. doi: 10.1128/AAC.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kubori T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front. Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted SB, Erlandsen SL, Dunny GM, Wells CL. High-resolution visualization by field emission scanning electron microscopy of Enterococcus faecalis surface proteins encoded by the pheromone-inducible conjugative plasmid pCF10. J. Bacteriol. 1993;175:6229–6237. doi: 10.1128/jb.175.19.6229-6237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted SB, Kao SM, van Putte LJ, Gallo JC, Dunny GM. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J. Bacteriol. 1991;173:7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HY, He X, Harrison EM, Kulasekara BR, Thani AB, Kadioglu A, Lory S, Hinton JC, Barer MR, Deng Z, et al. MobilomeFINDER: web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucl. Ac. Res. 2007;35:97–104. doi: 10.1093/nar/gkm380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos A, den Hartigh A, Smith MA, Atluri VL, Sivanesan D, Tsolis RM, Baron C. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun. 2011;79:1033–1043. doi: 10.1128/IAI.00993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos A, Patey G, Sivanesan D, Gao C, Bayliss R, Waksman G, O’Callaghan D, Baron C. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena A, Matilla I, Martin-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezon E, Arechaga I. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J. Biol. Chem. 2012;287:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CJ, Bantwal R, Bannam TL, Rosado CJ, Pearce MC, Adams V, Lyras D, Whisstock JC, Rood JI. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol. Microbiol. 2012;83:275–288. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]
- Ramsey ME, Woodhams KL, Dillard JP. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria . Front. Microbiol. 2011;2:61. doi: 10.3389/fmicb.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 2011;35:856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC, Dehio C. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat. Genet. 2007;39:1469–1476. doi: 10.1038/ng.2007.38. [DOI] [PubMed] [Google Scholar]
- Salgado-Pabon W, Jain S, Turner N, van der Does C, Dillard JP. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae . Mol. Microbiol. 2007;66:930–947. doi: 10.1111/j.1365-2958.2007.05966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder G, Lanka E. The mating pair formation system of conjugative plasmids - A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Schroder G, Schuelein R, Quebatte M, Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae . Proc. Natl. Acad. Sci. U. S. A. 2011;108:14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Mol. Microbiol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soge OO, Beck NK, White TM, No DB, Roberts MC. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J. Antimicrob. Chemother. 2008;62:674–680. doi: 10.1093/jac/dkn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza RC, Del Rosario Quispe Saji G, Costa MO, Netto DS, Lima NC, Klein CC, Vasconcelos AT, Nicolas MF. AtlasT4SS: A curated database for type IV secretion systems. BMC Microbiol. 2012;12:172. doi: 10.1186/1471-2180-12-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen JA, Bannam TL, Teng WL, Devenish RJ, Rood JI. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 2009;191:2926–2933. doi: 10.1128/JB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori . Proc. Natl. Acad. Sci. U. S. A. 2010;107:1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW Complex sirectly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 2007;282:25569–25576. doi: 10.1074/jbc.M703464200. [DOI] [PubMed] [Google Scholar]
- Tato I, Zunzunegui S, de la Cruz F, Cabezon E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA dependent ATPase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8156–8161. doi: 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278:1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng WL, Bannam TL, Parsons JA, Rood JI. Functional characterization and localization of the TcpH conjugation protein from Clostridium perfringens . J. Bacteriol. 2008;190:5075–5086. doi: 10.1128/JB.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Bayliss R, Oomen C, Leonard Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori . Proc. Natl. Acad. Sci. U. S. A. 2005;102:4956–4961. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Waksman G. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 2011;278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol. Rev. 2012;36:1046–82. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, Danchin A, Marignac G, Lenaour E, Boulouis HJ, Mavris M, et al. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 2010;6:e1000946. doi: 10.1371/journal.ppat.1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecino AJ, de la Arada I, Segura RL, Goni FM, de la Cruz F, Arrondo JL, Alkorta I. Membrane insertion stabilizes the structure of TrwB, the R388 conjugative plasmid coupling protein. Biochim. Biophys. Acta. 2011;1808:1032–1039. doi: 10.1016/j.bbamem.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Vecino AJ, Segura RL, Ugarte-Uribe B, Aguila S, Hormaeche I, de la Cruz F, Goni FM, Alkorta I. Reconstitution in liposome bilayers enhances nucleotide binding affinity and ATP-specificity of TrwB conjugative coupling protein. Biochim. Biophys. Acta. 2010;1798:2160–2169. doi: 10.1016/j.bbamem.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Vengadesan K, Narayana SV. Structural biology of Gram-positive bacterial adhesins. Protein Sci. 2011;20:759–772. doi: 10.1002/pro.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium . Proc. Natl. Acad. Sci. U. S. A. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamil Giraldo AM, Sivanesan D, Carle A, Paschos A, Smith MA, Plesa M, Coulton J, Baron C. Type IV secretion system core component VirB8 from Brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry. 2012;51:3881–3890. doi: 10.1021/bi300298v. [DOI] [PubMed] [Google Scholar]
- Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 2010;12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallden K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc. Nat. Acad. Sci. U. S. A. 2012;109:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai M, Andrews HL, Isberg R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 2000;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- Waters CM, Dunny GM. Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance. J. Bacteriol. 2001;183:5659–5667. doi: 10.1128/JB.183.19.5659-5667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Wells CL, Dunny GM. The aggregation domain of aggregation substance, not the RGD motifs, is critical for efficient internalization by HT-29 enterocytes. Infect. Immun. 2003;71:5682–5689. doi: 10.1128/IAI.71.10.5682-5689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F-like plasmids - a structural biology perspective. Mol. Microbiol. 2012;85:602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Carle A, Gao C, Sivanesan D, Aly KA, Hoppner C, Krall L, Domke N, Baron C. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 2005;280:26349–26359. doi: 10.1074/jbc.M502347200. [DOI] [PubMed] [Google Scholar]
- Zechner EL, Lang S, Schildbach JF. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Rong C, Chen C, Gao GF. Type-IVC secretion system: A novel subclass of type IV secretion system (T4SS) common existing in Gram-positive genus Streptococcus . PloS ONE. 2012;7:e46390. doi: 10.1371/journal.pone.0046390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J, Hackworth CA, Aguilar J, Ward D, Zambryski P. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens . J. Bacteriol. 2007;189:6551–6563. doi: 10.1128/JB.00480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]