Summary
Agrobacterium tumefaciens VirB10 couples inner membrane (IM) ATP energy consumption to substrate transfer through the VirB/D4 type IV secretion (T4S) channel and also mediates biogenesis of the virB-encoded T pilus. Here, we determined the functional importance of VirB10 domains denoted as the: (i) N-terminal cytoplasmic region, (ii) transmembrane (TM) α-helix, (iii) proline-rich region (PRR) and (iv) C-terminal β-barrel domain. Mutations conferring a transfer- and pilus-minus (Tra−, Pil−) phenotype included PRR deletion and β-barrel substitution mutations that prevented VirB10 interaction with the outer membrane (OM) VirB7–VirB9 channel complex. Mutations permissive for substrate transfer but blocking pilus production (Tra+, Pil−) included a cytoplasmic domain deletion and TM domain insertion mutations. Another class of Tra+ mutations also selectively disrupted pilus biogenesis but caused release of pilin monomers to the milieu; these mutations included deletions of α-helical projections extending from the β-barrel domain. Our findings, together with results of Cys accessibility studies, indicate that VirB10 stably integrates into the IM, extends via its PRR across the periplasm, and interacts via its β-barrel domain with the VirB7–VirB9 channel complex. The data further support a model that distinct domains of VirB10 regulate formation of the secretion channel or the T pilus.
Introduction
Bacteria use type IV secretion (T4S) systems to deliver DNA and protein substrates to a diverse range of prokaryotic and eukaryotic target cells (Cascales and Christie, 2003; Backert and Meyer, 2006). The T4S systems of Gram-negative bacteria translocate substrates and also elaborate surface pili by mechanisms catalysed by two or three ATPases. In the Agrobacterium tumefaciens VirB/ VirD4 T4S system, biochemical and genetic findings support a working model that the three ATPases (VirD4, VirB4 and VirB11) located predominantly or exclusively at the cytoplasmic face of the inner membrane (IM) co-ordinate with polytopic VirB6 and the bitopic subunits VirB8 and VirB10 to deliver DNA and protein substrates across the IM. In the periplasm, C-terminal domains of VirB8 and VirB10, VirB2 pilin, and the OM-associated subunit VirB9 form the portion of the secretion channel that bridges IM and outer membrane (OM) machine subassemblies. At the OM, a channel presumptively comprised of the lipoprotein, VirB7, and its interaction partner, VirB9, conveys substrates to target cells by a mechanism that requires direct cell-to-cell contact (see Christie and Cascales, 2005; Christie et al., 2005; Backert and Meyer, 2006).
Many T4S systems of Gram-negative bacteria are comprised of VirB-like subunits and, in particular, the VirB7– VirB10 subunits are phylogenetically conserved (Cao and Saier, 2001; Christie et al., 2005). These subunits have been postulated to comprise a transenvelope ‘core’ structure for assembly of the secretion channel (Das and Xie, 2000). Supporting this model, we previously showed that VirB10 functions dynamically by coupling IM energy to stable and productive assembly of this ‘core’ complex (Cascales and Christie, 2004a). Upon sensing ATP energy consumption by the IM VirD4 and VirB11 ATPases, VirB10 undergoes a structural transition that is necessary for formation of a stable VirB7–VirB9–VirB10 complex. This ATP-mediated sequence of events in turn enables passage of DNA substrates through the distal portion of the translocation channel, as monitored by a formaldehyde-cross-linking assay which identifies close contacts between a translocating DNA substrate and secretion channel subunits (Cascales and Christie, 2004b). A proposal that VirB10 physically bridges IM and OM T4S machine subassemblies is supported by a variety of protein–protein interaction and ultrastructural data (see Christie and Cascales, 2005; Bayliss et al., 2007). For example, at the IM, A. tumefaciens VirB10 forms a precipitable ternary complex with the three ATPases, VirD4, VirB4 and VirB11 (Atmakuri et al., 2004). Homologues of VirB10 in the plasmids R27 and R388 conjugation systems also have been shown to interact with VirD4-like coupling proteins in two-hybrid screens (Gilmour et al., 2003; Llosa et al., 2003). In the periplasm, VirB10 interacts with a disulphide cross-linked dimer of the VirB7 lipoprotein and OM channel subunit VirB9 (Fernandez et al., 1996; Spudich et al., 1996; Baron et al., 1997; Beaupre et al., 1997; Das et al., 1997; Das and Xie, 2000). Finally, very recently, homologues of VirB7, VirB9 and VirB10 encoded by the pKM101 T4S system were shown to form a multimeric channel by negative staining and cryoelectron microscopy (CryoEM). The channel is composed of 14 of each subunit and presents as a ring of 185 Å in diameter with a central channel of 55 Å at the cytoplasmic membrane and 10 Å at the OM (Fronzes et al., 2008). VirB10-like TraF was found to be required for assembly of a ring structure composed of VirB7-like TraN and VirB9-like TraO, consistent with the notion that the VirB10 family of T4S subunits transduce IM energy to mediate assembly and possibly also gating of the OM channel complex.
VirB10 thus appears to function both as a structural scaffold and as an energy sensor/transducer to mediate substrate transfer across the OM (Christie and Cascales, 2005). This latter activity in particular is reminiscent of the Escherichia coli TonB protein (Postle and Larsen, 2007). Although VirB10 senses ATP energy consumption and TonB the proton motive force, both proteins are energized through interactions with an IM energy-harvesting complex, VirB10 with the VirD4/VirB11/VirB4 ATPase complex (Atmakuri et al., 2004), and TonB with the ExbB/ ExbD complex (Postle and Larsen, 2007). Both proteins also possess a similar overall architecture characterized by a: (i) short N-terminal cytoplasmic region, (ii) putative transmembrane (TM) α-helix, (iii) proline-rich region (PRR) and (iv) C-terminal periplasmic domain (Cao and Saier, 2001; Terradot et al., 2005; Chu et al., 2007). The molecular details of energy sensing/transduction are not yet described for either protein family, but there is experimental evidence that TonB activates transport by ‘shuttling’ from the IM energy-harvesting complex to a cognate OM transporter in a cyclical reaction (Larsen et al., 2003).
The structure of the C-terminal domain of a VirB10 homologue, Helicobacter pylori ComB10, was solved by X-ray crystallography (Terradot et al., 2005). This domain presents as an extensively modified β-barrel with regions of predicted structural flexibility. A large groove extends along the side of the β-barrel and two α-helical subdomains project away from the barrel. One of these projections, designated α1, sits at the side of the β-barrel and the second, designated α2,α3 or the antennae projection (AP), extends from the top of the barrel. The β-barrel of ComB10 crystallized as a head-to-tail dimer, such that the AP aligns along the length of the barrel groove. A flexible flap domain and a conserved XDLDF motif are situated at the base of the β-barrel. The C-termini of ComB10 and other VirB10-like proteins share extensive primary sequence and predicted secondary structural motifs, and thus the ComB10 β-barrel is considered a structural paradigm for the entire VirB10 protein family (Terradot et al., 2005).
In this study, we undertook a mutational analysis to define contributions of VirB10 domains and novel structural features to T-DNA translocation and T pilus biogenesis. Additionally, by use of the scanning cysteine accessibility method (SCAM) (Bogdanov et al., 2005), we investigated whether VirB10 activates translocation through a TonB-like ‘shuttling’ mechanism or remains tethered at the IM. Our findings indicate that VirB10 is stably anchored at the IM, extends via its PRR across the periplasm, and interacts via its β-barrel with the VirB7–VirB9 channel complex. We further isolated ‘uncoupling’ mutations that selectively block T pilus biogenesis but not substrate transfer. The isolation of such mutants supports a model that VirB10 can alternatively regulate assembly of the T4S secretion channel or the extracellular pilus.
Results
N-terminal mutations selectively disrupt T pilus biogenesis
We first sought to test the functional importance of the cytoplasmic and putative TM domains for VirB10 energy sensing. We introduced two-residue insertions (Ala–Cys; i2) every five residues in the N-terminal 50 residues (Fig. 1) with the aim of disrupting potential interactions with IM channel subunits, e.g. VirD4 (Gilmour et al., 2003; Llosa et al., 2003; Atmakuri et al., 2004). Interestingly, all 10 mutant proteins supported transfer of oncogenic T-DNA to plant cells as well as a mobilizable IncQ plasmid to agrobacterial recipient cells (Fig. 2 and data not shown). The i2 mutant proteins also formed precipitable complexes with VirD4 (Fig. 2A) as well as the disulphide cross-linked VirB7–VirB9 dimer associated with the OM (Fig. 2A; Jakubowski et al., 2003). These wild-type (WT) properties strongly indicate that the N-terminal i2 mutations did not alter the capacity of VirB10 to sense IM ATP energy or undergo the structural transition necessary for a stable and productive interaction with the OM channel subunits (Cascales and Christie, 2004a).
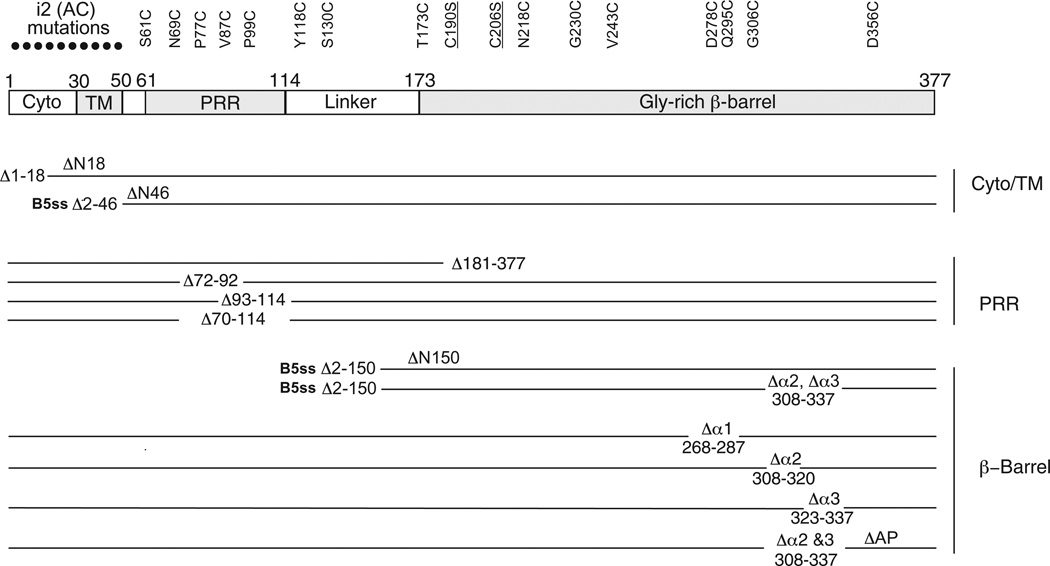
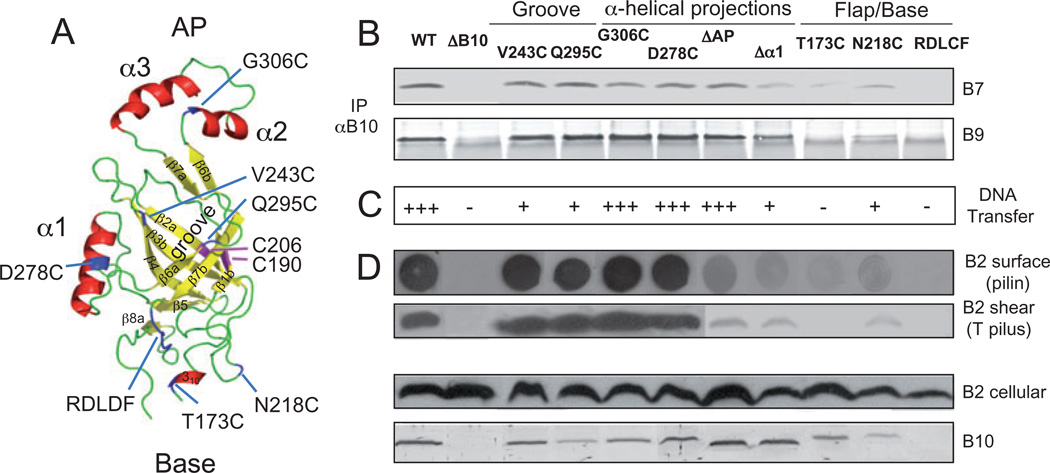
Fig. 1. Schematic of VirB10 mutations characterized in these studies.
Top: The various domains of VirB10 and their locations with residue numbers above are depicted. Domain abbreviations: Cyto, cytoplasmic domain; TM, transmembrane helix, PRR, proline-rich region. Ala, Cys (i2, AC; black dots) insertion mutations were engineered at five-residue intervals in the N-terminal 50 residues. Cys substitution mutations in the remainder of the protein are listed at top; Ser substitutions for the two endogenous Cys residues, Cys190 and Cys206 (underlined), also were characterized. Bottom: Deletion mutations of various domains with deleted residues indicated; alternative designations used in the text for some deletion mutations are listed above the corresponding line. The ΔN46 and ΔN150 fragments were exported to the periplasm by fusion to the signal sequence of VirB5 (B5ss). ΔAP, deletion of the antennae projection (see Terradot et al., 2005).
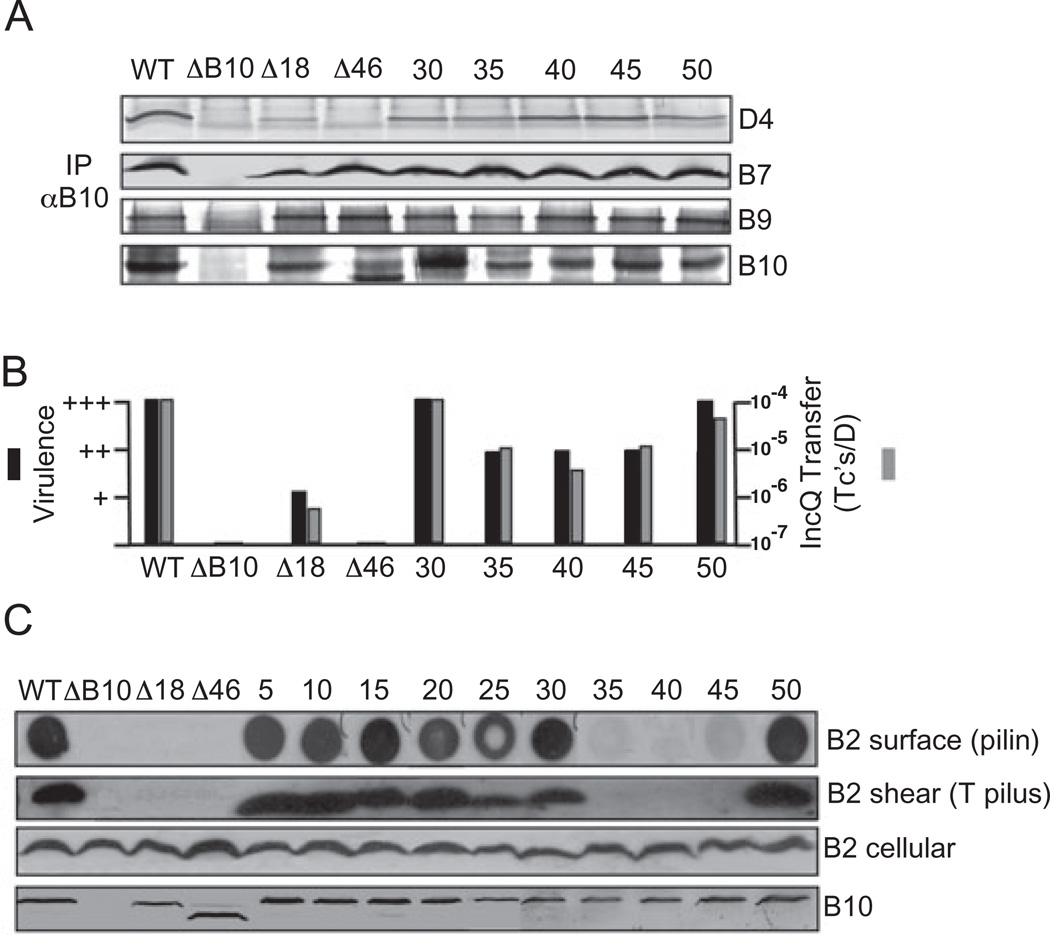
Fig. 2. Phenotypes of N-terminal insertion and deletion mutations.
A. Effects of mutations on VirB10 partner interactions. Material immunoprecipitated from detergent-solubilized cell extracts with anti-VirB10 antibodies (IP αB10) was analysed for the presence of VirD4, VirB7, VirB9 and VirB10 by immunoblot development with corresponding antibodies. Experiments were carried out under non-reducing conditions for co-precipitation of the VirB7–VirB9 dimer complex.
B. Effects of mutations on T-DNA transfer as monitored by virulence on wounded Kalanchoe leaves (black bars; - avirulent, +++ WT virulence) and transfer of the mobilizable IncQ plasmid pML122 to A. tumefaciens recipients (Grey bars; Tc’s/D, the number of transconjugants per donor cell).
C. Effects of mutations on T pilus production. Top-to-bottom: colony immunoblots developed with anti-VirB2 antibodies showing presence of surface-exposed pilin protein (B2 surface); extracellular shear fraction subjected to ultracentrifugation, gel electrophoresis and immunoblot development with anti-VirB2 antibodies revealing presence of high-molecular-weight T pilus (B2 shear); total cellular material subjected to gel electrophoresis and blot development with anti-VirB2 or anti-VirB10 antibodies revealing total cellular levels of the respective proteins. Strains: WT, wild-type Δ348; ΔB10, non-polar vIrB10 strain PC1010; Δ18, PC1010 producing VirB10Δ1–18; A46, PC1010 producing VirB10Δ1–46 fused to the VirB5 signal sequence for export of the C-terminal fragment to the periplasm; 5–50, PC1010 producing i2 (Ala-Cys) insertion mutations at the residue indicated. Strains producing mutant proteins with i2 insertions at residues 5, 10, 15, 20 and 25 displayed WT phenotypes with respect to co-immunoprecipitation (A) and substrate transfer (B) (data not shown).
Next, we introduced potentially more disruptive mutations into the N-terminal region. VirB10DN18, a mutant lacking the first 18 residues (Fig. 1), supported T-DNA and IncQ plasmid transfer albeit at reduced levels compared with the WT strain (Fig. 2B), and also formed precipitable complexes with VirD4 and the VirB7–VirB9 dimer (Fig. 2A). The extreme N-terminus thus is dispensable for energy sensing and assembly of a functional OM channel complex. Larger N-terminal deletions of the cytoplasmic and TM domain conferred protein instability as evidenced by low or undetectable protein accumulation, presumably because of improper membrane insertion or mislocalization in the cytoplasm. However, substitution of the VirB5 signal sequence for the N-terminal 46 residues of VirB10 (Fig. 1) resulted in abundant accumulation of the C-terminal fragment (residues 46–377) in the periplasm (Fig. 2C), as indicated by its release into the milieu upon osmotic shock (data not shown). Interestingly, VirB10ΔN46 formed a precipitable complex with the VirB7–VirB9 dimer although it failed to interact detectably with VirD4 (Fig. 2A). The IM-untethered, periplasmic region of VirB10 can therefore interact with the OM channel complex in the absence of ATP energy sensing. VirB10ΔN46 did not support substrate transfer, however, indicating that the TM domain and/or N-terminal cytoplasmic residues are nevertheless important for establishing protein–protein contacts required for a functional secretion channel (Fig. 2B).
The VirB proteins also mediate assembly of T pili (Christie et al., 2005). To monitor T pilus production, we developed a colony blot assay to detect surface-localized pilin (see Experimental procedures) and we also harvested T pili by shearing of cells and ultracentrifugation (Sagulenko et al., 2001a). In initial studies, we found the colony blot assay to reproducibly detect VirB2 pilin and the pilus-associated protein VirB5 on the surfaces of known Pil+ cells but not Pil− mutants. As expected, the assay did not detect IM-associated (VirD4) or periplasmic (VirB8, VirB10) subunits (data not shown). Interestingly, although several of the VirB10.i2-producing strains produced T pili, the ΔN18- and 40.i2-producing strains failed to elaborate detectable levels of surface pilin or T pili (Fig. 2C). Strains producing the 35.i2 and 45.i2 mutant proteins also did not possess detectable T pili although these cells possessed low levels of surface pilin as shown by colony immunoblotting (Fig. 2C). The ΔN18, 35.i2, 40.i2 and 45.i2 mutations did not affect accumulation of cellular VirB2 (Fig. 2C), and thus appear to specifically block polymerization of the T pilus in the absence of detectable effects on substrate transfer.
The TM domain anchors VirB10 in the IM
In view of the above findings, we sought to distinguish whether the energized form of VirB10 remains stably anchored in the IM or disengages from the membrane and physically traverses the periplasm for assembly of the VirB7–VirB9–VirB10 channel complex. The latter ‘shuttling’ mechanism was proposed for TonB on the basis of the finding that a Cys residue engineered in the N-terminal putative cytoplasmic region was accessible to a membrane-impermeable thiol-reactive reagent (Larsen et al., 2003). Similarly, we assayed for Cys accessibility to the biotinylated sulphydryl-specific maleimide probe, MPB, which passes readily through the pores of the OM but very inefficiently across the IM (Bogdanov et al., 2005). We analysed accessibility of the 10 Cys residues resulting from the Ala–Cys insertion mutations, as well as 15 Cys substitution mutations introduced into the PRR, linker and β-barrel domains (Fig. 1). As controls, we pretreated cells with the IM-impermeant, non-biotinylated maleimide derivative AMS to block periplasmic Cys residues from MPB labelling. We also assayed for accessibility of cytoplasmic and TM Cys residues by MPB treatment of sonicated cell extracts.
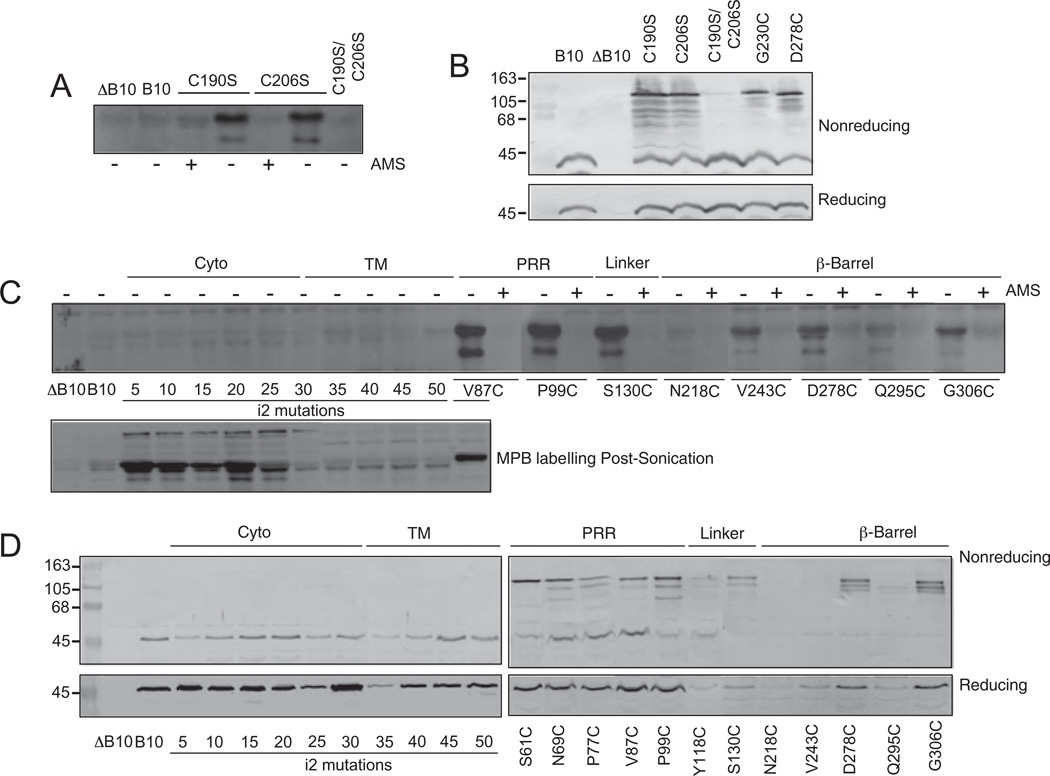
Agrobacterium tumefaciens VirB10 possesses two Cys residues at positions 190 and 206 within the β-barrel domain. In the VirB10 model derived from the H. pylori ComB10 X-ray structure, these Cys residues are in close proximity and might form an intramolecular disulphide bond (see below). MPB did not label native VirB10 or a double substitution mutant, C190S/C206S, but strongly labelled two single substitution mutants, C190S and C206S (Fig. 3A). The two native Cys residues thus likely form a disulphide bridge, although it is formally possible they could instead form an intermolecular cross-link. To examine this possibility, we assayed for the presence of higher-order cross-linked species in cell extracts maintained under non-reducing conditions. Reduced and non-reduced forms of VirB10 and C190S/C206S exhibited similar mobilities in protein gels, whereas non-reduced forms of the C190S and C206S mutant proteins formed higher-order cross-linked species of a size (∼100 kDa) roughly corresponding to a VirB10 homodimer (Fig. 3B). These findings strongly indicate that C190 and C206 form an intramolecular disulphide bridge. The C190S/C206S, C190S and C206S mutant proteins did not support substrate transfer or pilus biogenesis (data not shown), indicating that this C190–C206 cross-link is required for VirB10 function. In view of these results, for the Cys accessibility studies described below we introduced Cys mutations into native VirB10.
Fig. 3. Accessibility of introduced Cys residues to thiol-reactive reagents and disulphide cross-linking.
A and C top. Whole cells exposed to MPB without (−) or with (+) AMS pre-treatment.
B and D top. Immunoblots developed with anti-VirB10 antibodies showing migration of native VirB10 and mutant proteins in non-reducing SDS-polyacrylamide gels. For the MPB-labelling studies, results are shown only for Cys mutants that supported DNA transfer.
C bottom. Sonicated cell extracts exposed to MPB in the absence of AMS pre-treatment.
B and D bottom. Immunoblots showing migration of Cys mutants in reducing SDS-polyacrylamide gels.
Strains: ΔB10, non-polar virB10 strain PC1010; B10, PC1010 producing native VirB10 from an IncP replicon; C190S, C206S, C190S/C206S, PC1010 producing single or double Ser substitutions of native Cys residues; 5–50, PC1010 producing i2 (Ala-Cys) insertion mutations at the residue indicated in the cytoplasmic (Cyto) and putative transmembrane (TM) domains; PC1010 producing additional Cys substitution mutants listed in the proline-rich region (PRR), Linker and β-Barrel domains.
MPB did not label any of the N-terminal Cys residues predicted to reside in the cytoplasm and TM domain, and efficiently labelled most Cys residues in the PRR, linker and β-barrel domains (Fig. 3C). Additionally, AMS pretreatment completely blocked MPB labelling as expected of periplasmically localized Cys residues (Fig. 3C). In sonicated cell extracts, MPB strongly labelled Cys residues at positions 5, 10, 15 and 20, weakly labelled Cys-25, and failed to label Cys residues at positions 30, 35, 40, 45 and 50 (Fig. 3C).
Cys residues in the PRR, linker and β-barrel – but not those in the N-terminal cytoplasmic and TM domains – also formed presumptive intermolecular disulphide crosslinks when electrophoresed under non-reducing conditions (Fig. 3D). The higher-order species migrated at positions expected of VirB10 homodimers (∼100 kDa) in strains producing (Fig. 3D) and lacking (data not shown) other VirB proteins, suggesting that each of these periplasmic domains participates in assembly of VirB10 homodimers. The specific nature of the intermolecular contacts awaits further study; however, the absence of cross-linking by N-terminal cytoplasmic and TM domain Cys residues suggests that the N-terminal cytoplasmic domain does not gain access to the oxidizing environment of the periplasm. Thus, results of the SCAM and disulphide cross-linking studies together strongly indicate that VirB10 adopts a stable, bitopic membrane topology, such that the N-terminal 30 residues are cytoplasmic, residues ∼30–50 form the TM helix, and the remainder of the protein is in the periplasm. We conclude that VirB10 does not energize substrate translocation by ‘shuttling’ to the VirB7–VirB9 channel complex.
The PRR is required for VirB10 function and promotes VirB7–VirB9–VirB10 multimer formation
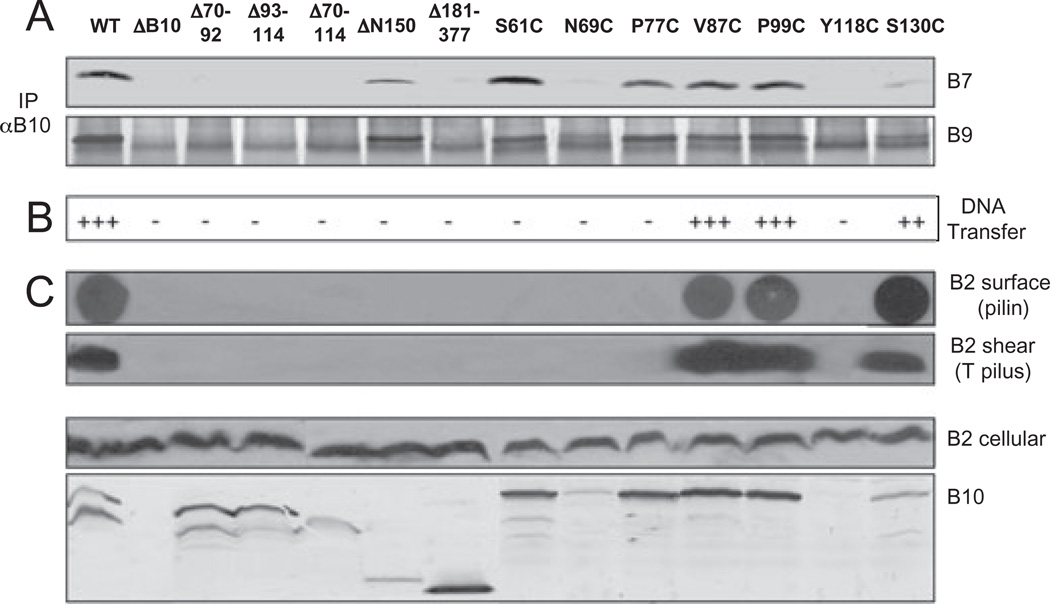
Structural studies of PRRs such as that of the TonB energy transducer have shown that these domains adopt extended structures (Evans et al., 1986). To explore whether the PRR of VirB10 might serve to bridge the IM energetic components with the OM VirB7–VirB9 channel complex, we assessed the effects of various deletion mutations on protein function. Mutant proteins deleted of some (Δ70–92, Δ93–114) or all (Δ70–114) of the PRR accumulated at detectable levels but failed to support substrate transfer or pilus biogenesis, or interact with the VirB7–VirB9 dimer as shown by immunoprecipitation (Fig. 4). These results are compatible with a model that the PRR adopts an extended structure required for protein function, but do not exclude a possibility that the PRR exerts its function by affecting the overall conformation of the periplasmic fragment or by directly binding the OM channel subunits. To examine the latter possibility, we constructed Δ181–377, a C-terminal truncation mutant lacking the C-terminal β-barrel domain (Fig. 1). This mutant did not interact detectably with the OM subunits or support substrate transfer or pilus production (Fig. 4). Conversely, the β-barrel domain alone (ΔN150; Fig. 1) accumulated at detectable levels and co-precipitated with the VirB7–VirB9 multimer when exported to the periplasm via the VirB5 signal sequence (Fig. 4). We attempted to export an epitope-tagged PRR fragment (residues 50–120) to assay directly for an interaction with the VirB7–VirB9 dimer, but this fragment was unstable in the A. tumefaciens periplasm. The β-barrel thus appears to interact with the VirB7–VirB9 multimer, but our data prevent firm conclusions regarding a possible PRR interaction with the OM channel complex.
Fig. 4. Phenotypes of PRR deletion and substitution mutations.
A. Effects of mutations on VirB10 interaction with the outer membrane channel subunits. Material immunoprecipitated from detergent-solubilized cell extracts with anti-VirB10 antibodies (IP αB10) was analysed for co-precipitation of VirB7 and VirB9 by immunoblot development with corresponding antibodies. Experiments were carried out under non-reducing conditions for co-precipitation of the VirB7–VirB9 dimer complex.
B. Effects of mutations on T-DNA transfer as monitored by virulence on wounded Kalanchoe leaves (− avirulent, +++ WT virulence); results correlated with frequencies of IncQ plasmid transfer to agrobacterial recipients (data not shown).
C. Effects of mutations on T pilus production. Extracellular pilin and T pilus, and total cellular pilin and VirB10 derivatives were detected as described in Fig. 2 legend. A 40 kDa proteolytic product of full-length VirB10 (48 kDa) is often detected in total-cell extracts.
Strains: WT, wild-type A348; ΔB10, non-polar virB10 strain PC1010; isogenic PC1010 producing the following PRR mutants − Δ70–92, Δ93–114, Δ70–114, PRR internal deletion mutants; ΔN150, residues 150–377 containing the β-barrel domain exported to the periplasm by fusion to the VirB5 signal sequence; Δ181–377, residues 1–180 containing the cytoplasmic, TM, PRR and linker domains; Cys substitution mutants as listed.
We further characterized the effects of Cys substitution mutations in and near the PRR. Two mutant proteins (V87C, P99C) displayed WT function, and two others (N69C, Y118C) accumulated at low levels and failed to support substrate transfer or pilus biogenesis (Fig. 4B and C). Two other mutant proteins (S61C, P77C) accumulated at abundant levels, but also conferred a Tra−, Pil− phenotype despite the fact that they formed precipitable complexes with the VirB7–VirB9 multimer (Fig. 4). Although the underlying mechanism(s) for these mutant phenotypes awaits further study, the findings highlight the importance of the PRR in VirB10 function and support a working model that the PRR functions to extend VirB10 across the periplasm and also to promote formation of stabilizing homo- and/or heteromultimers.
β-Barrel domain analysis: the helical projections are dispensable for substrate transfer
We introduced mutations into novel structural features in the β-barrel domain, including (i) a pronounced groove that extends along the β-barrel, (ii) two α-helical projections designated α1 and α2,α3 (termed the antennae projection or AP) that extend to the side and from the top of the β-barrel, respectively, and (iii) a flexible flap and a conserved RDLDF motif positioned near the base of the β-barrel (Fig. 5A; Terradot et al., 2005).
Fig. 5. Modelled β-barrel structure and phenotypes of substitution mutations.
A. A. tumefaciens VirB10 β-barrel ribbon structure modelled with co-ordinates from the H. pylori ComB10 X-ray structure using the Modeller package (Marti-Renom et al., 2000) and PyMOL (http://pymol.org/) to construct the figure. This C-terminal domain (residues 173–377) presents as a non-canonical β-barrel with a groove marked by the V243C and Q295C substitution mutations, a flexible base marked by the S173C and N218C mutations and a conserved RDLDF motif, and two helical projections, α1 marked by D278C and the α2,α3 antennae projection (AP) marked by G306C. Native Cys-190 and Cys-206 residues forming an intramolecular disulphide cross-link are shown in pink; Cys substitution mutations are in blue. β-Strands β1-β7 are identified.
B. Effects of mutations on VirB10 interaction with the outer membrane channel subunits. Material immunoprecipitated from detergent-solubilized cell extracts with anti-VirB10 antibodies (IP αB10) was analysed for co-precipitation of VirB7 and VirB9 by immunoblot development with corresponding antibodies. Experiments were carried out under non-reducing conditions for co-precipitation of the VirB7–VirB9 dimer complex.
C. Effects of mutations on T-DNA transfer as monitored by virulence on wounded Kalanchoe leaves (− avirulent, +++ WT virulence); results correlated with frequencies of IncQ plasmid transfer to agrobacterial recipients (data not shown).
D. Effects of mutations on T pilus production. Extracellular pilin and T pilus, and total cellular pilin and VirB10 derivatives were detected as described in Fig. 2 legend.
Strains: WT, wild-type A348; ΔB10, non-polar virB10 strain PC1010; isogenic PC1010 producing Cys substitution mutants as listed.
The groove of the β-barrel is lined with β-strands β6a and β6b on one side and β4 on the other (Fig. 5A). In the ComB10 X-ray structure, the groove was a site of a large crystal-packing interface, with residues in the AP interacting with residues in the β6a and β6b strands and with the β4 strand to create a head-to-tail dimer (Terradot et al., 2005). We reasoned that if this head-to-tail dimer forms stably in vivo, the AP might block accessibility of Cys residues located in the groove. To examine this possibility, we introduced Cys residues into the groove, one located deep in the pocket (V243C; β4) and a second more exposed at the edge (Q295C; β6a-β6b). Both mutations conferred attenuated DNA transfer efficiencies without detectably affecting T pilus production (Fig. 5C and D) or the ability of VirB10 to co-precipitate with the VirB7–VirB9 dimer (Fig. 5B). As shown above (Fig. 3C), the V243C and Q295C mutations were inefficiently labelled by MPB when compared with labelling of the PRR Cys mutations and, moreover, non-reduced forms of these mutant proteins did not efficiently assemble higher-order cross-linked species (Fig. 3D). These Cys mutations thus are comparatively inaccessible, possibly because the groove corresponds to a binding partner interface.
Given that the AP was implicated as fitting into this groove (Terradot et al., 2005), we sought to assess the functional importance of this projection. Three deletion mutations composed of the entire AP (Δ308–337), helix α2 only (Δ308–320) and helix α3 only (Δ323–337) displayed identical phenotypes and results are presented for the Δ308–337 (ΔAP) mutation. Interestingly, the ΔAP mutant accumulated at abundant levels, supported efficient DNA transfer and formed precipitable complexes with the VirB7–VirB9 multimer (Fig. 5). Therefore, the AP and by extension the head-to-tail dimer detected in the crystal structure are not required for assembly of a functional T4S channel. We also characterized a G306C substitution in the AP (Fig. 5A). This mutation was phenotypically silent (Fig. 5), accessible to MPB labelling (Fig. 3C), and formed higher-order disulphide cross-linked species in non-reducing gels (Fig. 3B). The AP is therefore probably solvent exposed and not buried in a hydrophobic pocket of the VirB10 groove, another binding partner interface, or the membrane.
We deleted helix α1 to assess the contribution of this side projection to VirB10 function (Fig. 5A). The Δα1 mutant accumulated at abundant levels (Fig. 5D) but displayed attenuated function as evidenced by inefficient complex formation with the VirB7–VirB9 multimer and diminished DNA transfer by the host strain (Fig. 5B and C). These findings indicate that helix α1 contributes to but is not required for assembly of a functional T4S channel. A D278C mutation in helix α1 did not affect VirB10 function (Fig. 5), was MPB labelled (Fig. 3C) and formed higher-order disulphide cross-linked species (Fig. 3D). These data support structural predictions that this α-helical projection also is solvent accessible (Terradot et al., 2005).
The base of the β-barrel was proposed to be a region of structural flexibility of potential importance for establishment of VirB10 partner interactions (Terradot et al., 2005). We introduced Cys substitutions in (i) 310 α-helix just N-proximal to the β-barrel base (S173C), (ii) a flap structure formed by the β3a-β2b sheet that seals the bottom of the β-barrel (N218C), and (iii) β7c also near the β-barrel base and characterized by the conserved xDLDF sequence (RDLDF for VirB10 and homologues in other α-proteobacteria) (Fig. 5A). In the RDLDF motif, three mutations were constructed – QNALM, RDLCF, RKLDF – and all displayed identical phenotypes, as presented for the RDLCF mutation (Fig. 5). Interestingly, mutant proteins with substitutions in the base of the β-barrel accumulated at low or undetectable levels (Fig. 5D). The mutations in this region thus were destabilizing, and as expected they also abrogated (N218C, S173) or abolished VirB10 (RDLDF mutations) function, as monitored by VirB7–VirB9 complex formation (Fig. 5B), and DNA transfer and pilus production (Fig. 5C and D). Also consistent with the intrinsic instabilities of these mutant proteins, Cys substitutions in the barrel base labelled poorly with MPB and did not form higher-order disulphide cross-linked species (Fig. 3 and data not shown). The base of the β-barrel – most notably the conserved RDLDF motif – thus is important for VirB10 stability and function.
The AP selectively contributes to T pilus biogenesis
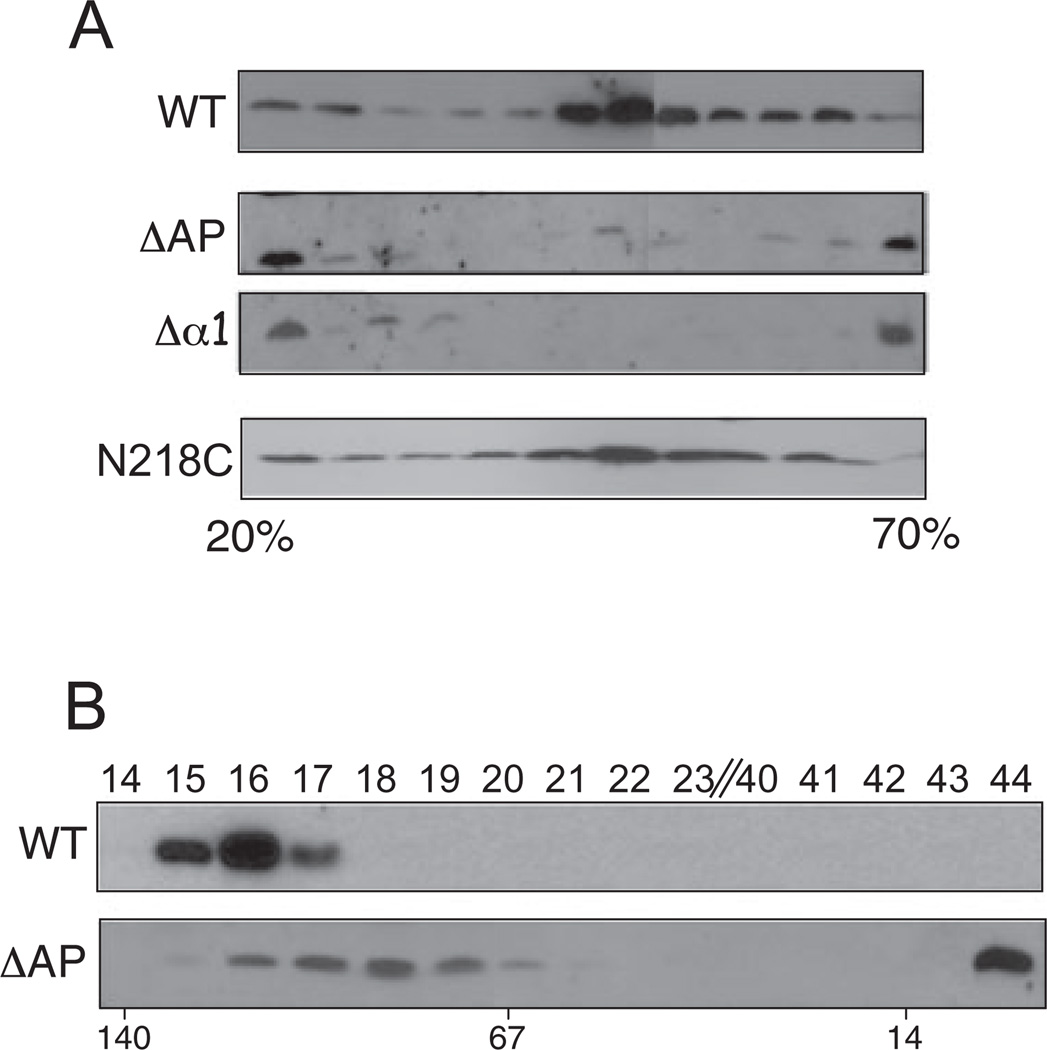
Although both helical projections were dispensable for assembly of a functional secretion channel, on further examination we discovered that both projections play critical roles in biogenesis of the T pilus. Strains producing the Δα1 or ΔAP mutant proteins exported low levels of VirB2 pilin to the cell surface as assessed by colony immunoblotting, as well as low levels of T pilus recoverable by ultracentrifugation of the shear fraction (Fig. 5D). Additionally, when extracellular material from WT and mutant cell cultures was fractionated through 20–70% linear sucrose gradients, T pili from WT cells distributed broadly with peak amounts in the centre of the gradient, as also shown previously (Fig. 6A; Sagulenko et al., 2001b). In striking contrast, T pili or pilin present in extracellular fractions of the Δα1 and ΔAP mutant cultures partitioned mainly in regions of low or high sucrose density (presumptively corresponding to VirB2 monomers or oligomers and aggregates respectively) and only a small amount of pilin was detected in the middle of the gradient. This pilus/pilin distribution profile was unique for the Δα1 and ΔAP mutant strains, as similar analyses of other mutant strains with aberrant levels of extracellular pilin or T pilus, e.g. N218C and T173C (Fig. 5D), 35.i2 and 45.i2 (Fig. 2C), yielded overall distribution profiles reminiscent of WT T pili; representative data are presented for the N218C mutant (Fig. 6A).
Fig. 6. Effects of VirB10 helix projection mutations on T pilus assembly.
A. Sucrose density gradient distribution profiles of extracellular VirB2 pilin/T pili produced by WT cells and the ΔAP and Δα1 helix mutants. For comparison, extracellular material from the N218C mutant strain, which also produces low amounts of surface pilin (see Fig. 5), was similarly analysed. Immunoblots developed with anti-VirB2 antibodies show the distribution profiles of extracellular pilin in identically prepared 20–70% sucrose density gradients.
B. Extracellular material from WT cells and the ΔAP mutant was fractionated through a Superdex 75 10/300 GL gel filtration column and fractions (listed at top) were analysed for the presence of VirB2 pilin by immunostaining. Molecular masses of reference proteins are shown below (in kDa).
We further fractionated extracellular material from the WT strain and the ΔAP mutant through a Superdex 75 10/300 GL gel filtration column. For these experiments, to detect pilin protein in the column fractions, the extracellular fraction was harvested from 10 times the number of ΔAP mutant cells relative to the WT strain. VirB2 from WT cells partitioned in a size range exceeding 100 kDa, consistent with the assembly of high-molecular-weight T pili (Fig. 6B). Some VirB2 from ΔAP mutant cells partitioned broadly in fractions corresponding to 80–100 kDa whereas the bulk of the protein partitioned in fraction 44 corresponding to a molecular size of < 10 kDa (Fig. 6B). The AP is therefore important for T pilus polymerization, but completely dispensable for assembly of a functional secretion channel. This projection also appears to contribute directly or indirectly to OM channel gating as suggested by the accumulation of VirB2 monomers or short oligomers in ΔAP mutant culture supernatants.
Discussion
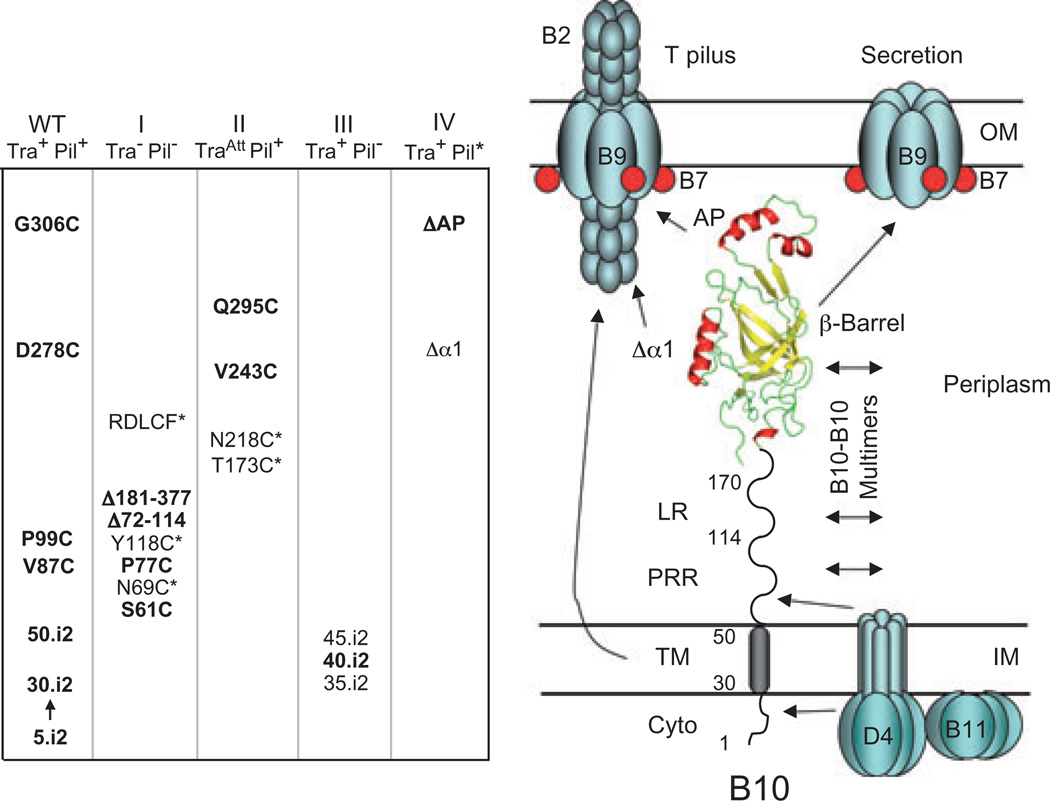
Mechanistic studies of T4S machines have been advanced by the availability of structural information for individual subunits and machine subcomplexes (Yeo et al., 2000; 2003; Gomis-Ruth et al., 2001; Savvides et al., 2003; Terradot et al., 2005; Bayliss et al., 2007; Fronzes et al., 2008). In this study, we carried out a structure–function analysis of domains and novel features of the A. tumefaciens VirB10 energy sensor (Cascales and Christie, 2004a). On the basis of our findings, we propose that VirB10, whose structure is modelled from the H. pylori ComB10 β-barrel X-ray structure (Terradot et al., 2005) and other sequence-based predictions, spans the cell envelope establishing critical contacts with IM and OM T4S machine subassemblies (Fig. 7). In its capacity as a ‘bridging’ subunit, VirB10 functions as a structural scaffold for the T4S system. However, VirB10 also functions dynamically, at least in part through energy-mediated conformational changes, to regulate T4S channel assembly or gating and morphogenesis of the T pilus. For purposes of discussion below, we have grouped mutations into four classes (Fig. 7).
Fig. 7.
VirB10 mutants characterized in these studies were phenotypically silent (WT) or grouped into four classes on the basis of Tra and Pil phenotypes. Class I (Tra−, Pil−) mutations abolished both activities and Class II (TraAtt, Pil+) mutations conferred attenuated substrate transfer without affecting pilus production; asterisked, unbolded mutations accumulated at low levels and might disrupt protein function through destabilizing effects on protein folding. Classes III and IV ‘uncoupling’ mutations permitted substrate transfer but either completely blocked pilus production (III) or resulted in defective pili and release of pilin monomers (IV); unbolded mutations displayed some variation in substrate transfer efficiencies and were classified on the basis of effects on pilus biogenesis (see text for details). Model on the right depicts the domains of VirB10 and regions proposed to be involved in intersubunit interactions; only a subset of potential contacts are shown. The TM helix and β-barrel helical projections contribute to OM channel gating and T pilus biogenesis through unspecified intersubunit contacts. The VirB/D4 subunits might assemble the secretion channel and T pilus as a single surface organelle or as two physically distinct structures.
VirB10, a structural scaffold bridging IM and OM machine subassemblies
Our phenotypic studies of various VirB10 mutants have supplied important information regarding the disposition of VirB10 at the cell envelope and requirements for stability and multimerization. With respect to functionality of the N-terminal domains, Cys accessibility and disulphide cross-linking studies supplied strong evidence that VirB10 stably integrates into the IM (Fig. 3). This finding prompted studies exploring the contribution(s) of the cytoplasmic and TM domains to VirB10 energy sensing. Interestingly, i2 mutations intended to disrupt potentially important protein–protein interactions, e.g. VirD4–VirB10, were phenotypically silent with respect to substrate transfer (Fig. 2). The i2 mutations in the cytoplasmic domain might be too small to disrupt intersubunit contacts, yet the ΔN18 mutation also supported low levels of DNA transfer (Fig. 2), suggesting that the extreme N-terminal cytoplasmic residues are dispensable for protein function. In an earlier study, we also showed that N-terminally GST-tagged VirB10 is fully functional (Jakubowski et al., 2003). Consequently, VirB10 can tolerate both a sizeable N-terminal deletion and the addition of a heterologous bulky moiety without loss of function.
By analogy to TonB, which forms TM–TM helix interactions with the ExbB energy-harvesting protein (Larsen and Postle, 2001), we envisioned that VirB10 might sense IM ATP energy use through TM–TM helix interactions. Of the two ATPases required for energization of VirB10, VirD4 possesses two N-terminal TM helices (Das and Xie, 1998), whereas VirB11 associates peripherally at the cytoplasmic face of the IM (Rashkova et al., 1997). The i2 mutations were introduced into the TM helix to shift the helix register and disrupt possible TM–TM contacts. Somewhat unexpectedly, the VirB10.i2 mutants retained the capacity to functionally interact with VirD4 (Fig. 2), arguing against a model that energy sensing is mediated exclusively through a TM–TM helix interaction (see below). Previously, it was reported that N-terminal fragments of VirB10 homologues in the R27 and R388 plasmid transfer systems interact with coupling proteins (Gilmour et al., 2003; Llosa et al., 2003) and two-hybrid evidence also was presented for possible binding of the PRR of VirB10-like TrhB to VirD4-like TraG (Gilmour et al, 2003). Here, we did not detect complex formation between the ΔN46 mutant protein (composed of the PRR and β-barrel domain) and VirD4 (Fig. 2), suggesting that the PRR cannot mediate this interaction independently of the TM domain. We suspect that the TM domain contributes indirectly to the VirB10–VirD4 interaction, possibly by spatially positioning VirB10 at specific sites at the membrane, e.g. the cell pole where the VirB/D4 machine assembles (Kumar et al., 2000). Such a function would be reminiscent of the role recently described for the TM domain of bitopic FtsQ in localization to the E. coli division site (Scheffers et al., 2007).
We also explored the contributions of the PRR and β-barrel domains to VirB10 function. The VirB10 PRR (61–114) is composed of 28% Pro residues. Structural studies have established that PRRs adopt extended structures that can be rigid or more flexible depending on Pro content and periodicity (Williamson, 1994). For TonB, one model envisages that an extended PRR is important for ‘bridging’ the IM energy-harvesting complex with TonB-dependent OM transporters. However, the PRR can be deleted with little impact on function (Larsen et al., 1993), a finding more consistent with the alternative proposal that energized TonB dynamically disengages from the IM and ‘shuttles’ to the OM transporters (Postle and Larsen, 2007). Our results showing that VirB10 is anchored at the IM (Fig. 3) and that the PRR is essential for substrate transfer, pilus production and complex formation with the VirB7– VirB9 multimer (Fig. 4) strongly favour a ‘bridging’ model. We thus propose that the VirB10 PRR extends the protein across the periplasm to allow the β-barrel to form critical contacts with the OM channel subunits. At this time, we cannot exclude the possibility that the PRR influences an overall structural fold of the periplasmic C-terminus required for productive engagement with the VirB7–VirB9 OM complex. However, a ‘bridging’ model also is compatible with the recently solved CryoEM structure, which shows that VirB10-like TraF adopts a structure extending from the IM to the OM (Fronzes et al, 2008). In the channel complex, the X-ray structure of the C-terminal domain of ComB10 (Terradot et al, 2005) could be fitted into a portion of the channel wall located near the OM (Fronzes et al, 2008). If this modelling is correct, positioning of the C-terminal β-barrel near the OM would require that the PRR and/or linker domain adopt an extended structure.
Studies also have shown that PRRs often participate in assembly of multiprotein complexes (Williamson, 1994). Consistent with a role for the VirB10 PRR in mediating assembly of higher-order T4S complexes, we found that PRR Cys mutants readily formed higher-order cross-linked species probably corresponding to VirB10 dimers (Fig. 3). Moreover, in a previous study we showed that the periplasmic region – not just the TM domain – is required for VirB10 self-association, as monitored with a cytology-based two-hybrid screen (Ding et al., 2002). Other groups also have supplied evidence for PRR-mediated heteromultimer formation. The list of candidate binding partners includes VirD4 (see above) and VirB8, another bitopic subunit shown to be important for channel assembly (Das and Xie, 2000; Kumar et al., 2000). Additionally, the PRR was implicated in binding to VirB9 (Das and Xie, 2000), and in this regard it is noteworthy that the CryoEM structure now shows that the N-terminal domain of VirB9-like TraO is in close contact with a region corresponding to the PRR and linker domains of VirB10-like TraF (Fronzes et al., 2008).
The β-barrel domain mutations abolished (Class I), abrogated (Class II) or ‘uncoupled’ (Class IV) VirB10 function with respect to substrate transfer and pilus biogenesis (Fig. 7). Mutations in the β-barrel groove were designed to disrupt possible protein–protein interactions, specifically, a head-to-tail dimer detected in the ComB10 crystal structure (Terradot et al., 2005). The phenotypes of these mutations are consistent with a proposal that the groove is a site of subunit–subunit interaction (Fig. 5); however, analyses of the AP deletion mutants strongly indicated that the head-to-tail dimer is a crystal-packing artefact. Mutations in the barrel base, specifically in a flexible flap domain that seals the bottom of the barrel (β3a,β2b; N218C) and a conserved RDLDF motif (β7C; RDLCF, QNALM, RKLDF), were highly destabilizing (Fig. 5). These findings highlight the importance of this region for stable folding or for mediating stabilizing intermolecular contacts. Finally, we had envisioned that the α-helical projections might mediate dynamic contacts with the VirB7–VirB9 channel complex, but phenotypes of the Δα1 and ΔAP mutants suggest instead that the β-barrel itself binds these OM channel subunits (Fig. 5).
VirB10, a dynamic modulator of secretion channel and T pilus biogenesis
Our long-term goal in introducing Cys mutations along the length of VirB10 is to use SCAM (Bogdanov et al., 2005) to refine our understanding of the conformational changes VirB10 undergoes during biogenesis of the secretion channel and T pilus. Our previous finding that ATP-energized VirB10 undergoes a structural transition necessary for formation of a productive contact with VirB9 and, in turn, DNA transfer across the distal portion of the secretion channel (Cascales and Christie, 2004a) points to a role for VirB10 as a regulator of OM secretion channel assembly or activity. Here, we further isolated ‘uncoupling’ mutations that are completely permissive for substrate transfer but disrupt polymerization of the T pilus (Fig. 7). It is important to note that the contribution of VirB10 to T pilus biogenesis, as detected through isolation of these ‘uncoupling’ mutations, must be mechanistically distinct from its role in mediating substrate transfer through sensing of ATP energy consumption by the VirD4 and VirB11 ATPases. This is because this T4S system elaborates T pili completely independently of VirD4 (Lai et al., 2000) and, hence, VirD4-mediated ATP energization of VirB10 (Cascales and Christie, 2004a). Conceivably, VirB10 dynamically regulates T pilus biogenesis through an alternative energy-sensing mechanism involving VirB11 and/or the VirB4 ATPase, both of which are required for assembly of T pili (Christie et al., 2005).
The isolation of ‘uncoupling’ mutations selectively disrupting T pilus biogenesis comprises genetic evidence that VirB10 differentially regulates secretion channel assembly or activity and morphogenesis of the T pilus. We isolated two subclasses of Tra+, Pil− mutations: (i) mutations that completely abolish detectable T pilus production (Class III), and (ii) mutations that disrupt T pilus assembly and release pilin monomers to the milieu (Class IV) (Fig. 7). The Class III mutations included the i2 insertions at positions 35, 40 and 45 of the TM domain. Intriguingly, these mutations were located in a sequence (33-LIV*GGVVL*ALSLS*L-46) that closely resembles a right-handed seven-residue motif shown through saturation mutational analyses to represent a consensus TM–TM dimer interface (LIxxGVxxGVxxT) (Arkin, 2002; Curran and Engelman, 2003). This motif highlights the importance of a GG4 dimer interface, but a GA4 sequence as occurs in VirB10 (underlined in the sequence) also is a common TM dimerization motif (Arkin, 2002). These mutations likely disrupt this motif, and thus a functionally important TM–TM interaction. Thus, while the TM appears to contribute indirectly to the VirD4–VirB10 interaction, a direct homo- or heterotypic TM–TM interaction is required specifically for T pilus assembly.
The Class IV mutations included the ΔAP and Δα1 mutations (Figs 5 and 7). In silico sequence analyses show that the AP is variable in primary sequence among the VirB10-like proteins (http://mmg.uth.tmc.edu/Assets/pdf/VirB10%20PD%20Allignment.pdf), although APs of all family members analysed were found to possess at least one α-helix. It is intriguing to speculate that the AP interacts directly with VirB2 pilin to mediate sequential addition of pilin monomers to a growing pilus. Alternatively or in addition, the AP could modulate the activity of an OM channel through which the T pilus extends (Fig. 7). The Δα1 mutation phenotypically resembles the ΔAP mutation in disrupting T pilus biogenesis and mediating pilin release, hence, its Class IV designation (Fig. 7). However, the Δα1 mutation also disrupted substrate transfer, indicating that this motif is more generally important for elaboration of the secretion channel and the T pilus. Helix α1 is composed of a Gly-rich motif (GxxGxxG/D) that is highly conserved among the VirB10 family members; this sequence might mediate formation of T4S subunit contacts important for T4S machine assembly.
If we assume that VirB/D4 proteins elaborate a single supramolecular structure composed of the secretion channel and T pilus, the uncoupling mutations might differentially affect functionality or assembly of one versus the other structure. However, the isolation of uncoupling mutations raises the intriguing alternative possibility that the secretion channel and T pilus are physically distinct structures requiring different VirB/D4 subunit activities for assembly (Fig. 7; Christie et al., 2005). In fact, at this time mutations in several T4S subunits have been shown to differentially affect channel versus T pilus assembly. Other mutations of the Tra+, Pil− subclass have been isolated in the VirB11 ATPase (Zhou and Christie, 1997; Sagulenko et al., 2001a), polytopic VirB6 (Jakubowski et al., 2004), OM channel subunit VirB9 (Jakubowski et al., 2005) and VirB2-like pilin proteins (Eisenbrandt et al., 2000). Deletion of virB1 (codes for a transglycosylase) also selectively disrupts T pilus assembly without abolishing substrate transfer (Berger and Christie, 1994; Lai et al., 2000). Conversely, deletion of virD4 (Lai et al., 2000) and a recently identified VirB2 mutant (J.E. Kerr and P.J. Christie, unpubl. data) fall into the Tra−, Pil+ subclass of uncoupling mutations. Presently, there is no supporting biochemical evidence for a proposal that the VirB/D4 proteins assemble alternatively as a secretion channel or T pilus. However, such evidence is difficult to obtain because biochemical extraction of intact channel or pilus basal structures from the bacterial cell envelope have proven challenging. In the future, ultrastructural information obtained by CryoEM might prove useful for visualizing distinct T4S organelles on intact cells.
In summary, our findings support a model that VirB10 functions as a structural scaffold by coupling IM and OM subassemblies of the T4S system. Additionally, the identification of ‘uncoupling’ mutations further suggests VirB10 differentially regulates biogenesis of the secretion channel or the T pilus, either in the context of one supramolecular structure or as two physically distinct organelles. Our ongoing studies are exploring whether the TM helix and the AP form molecular contacts exclusively required for T pilus biogenesis.
Experimental procedures
Bacterial strains and induction conditions
A. tumefaciens A348 was the WT strain and PC1010 a non-polar ΔvirB10 mutant respectively (Berger and Christie, 1994). Conditions for growth of A. tumefaciens and E. coli and for Vir gene induction with 200 µM acetosyringone (AS) in induction media (ABIM) have been described previously (Fernandez et al., 1996). Plasmids were maintained in A. tumefaciens and E. coli by addition of carbenicillin (100 µg ml−1), kanamycin (100 µg ml−1), tetracycline (5 µg ml−1), gentamicin sulphate (100 µg ml−1) and spectinomycin (600 µg ml−1).
Construction of virB10 insertion, cysteine substitution and deletion mutations
Plasmid pKVD10 expressing PIac-virB10 served as template for construction of Ala–Cys insertion and Cys substitution mutations by oligonucleotide-directed mutagenesis (see Berger and Christie, 1994). Oligonucleotides used in this study were designed with at least 15 bp of 5′ and 3′ sequence complementarity to regions of virB10 flanking the insertion or substitution sites (Table S1). In-frame insertions of Ala–Cys codons were constructed by introducing an Sphl (GCATGC) site at 15 bp intervals near the 5′ end of virB10. Additionally, Cys codons were substituted for codons along the length of VirB10 by insertion of Sphl or Pstl sites (each carries a Cys codon) such that adjacent codons were not altered. Mutations were identified by restriction enzyme digestion and confirmed by sequencing. The resulting plasmids were designated pIGxx, where xx denotes the codon immediately preceding the Ala–Cys codon insertion or Cys codon substitution.
Plasmids pKVD116 and pKVD136 synthesize VirB10 derivatives that lack the first 18 and 46 residues respectively. Both deletion derivatives were generated by PCR amplification of the respective gene fragments using pKVD10 as template and deoxyoligonucleotides listed in Table S1. virB10Δ1–18 (named virB10AN18) and virB10Δ188–377 were cloned as Ndel–Xhol fragments into pBSIISK+.Ndel (Berger and Christie, 1994). virB10Δ1–46 and virB10Δ1–150 generating VirB10 C-terminal fragments in the periplasm were constructed by PCR amplification of the respective gene fragments using pKVD10 as template and the oligonucleotides described (Table S1). The fragments were introduced as Ndel–Kpnl fragments into pZD74, resulting in fusion to the 5′ end of virB5 encoding a signal sequence (VirB5ss); in the text, the genes are named virB10ΔN46ss and virB10ΔN150ss, and mature proteins VirB10ΔN46 and VirB10ΔN150. Plasmid pZD74 was constructed by introduction of the virB5 signal sequence as a ∼100 bp PCR fragment flanked by Ncol and Ndel/Sall sites into pCR2.1-TOPO. The following VirB10 derivatives were constructed by PCR QuikChange® with pKVD10 as a template and oligonucleotides listed in Table S1. Plasmids pKVD117, pKVD145 and pKVD146 synthesize PRR deletion mutations VirB10Δ70–114, VirB10Δ72–92 and VirB10Δ93–114 respectively. Plasmids pKVD126, pKVD129, pKVD127 and pKVD128 synthesize helix projection deletion mutants, Δα1, Δα2,α3 (ΔAP), Δα2 and Δα3, respectively, by deletion of codons 268–287, 308–337, 308–320 and 323–337. Plasmids pKVD140, plG022 and pKVD112 produce VirB10 variants with the conserved RDLDF motif (residues 353–357) mutated to RKLDF, RDLCF and QNALM. Plasmids KVD109, KVD110 and KVD111 synthesize VirB10 variants in which serine has been substituted for the native cysteines at position C190S, C206S, or both C190S and C206S respectively. All of the above ColE1 plasmids were introduced into A. tumefaciens by ligation to the broad-host-range Tetr plasmid pSW172 (IncP) or a Tetr, Kanr derivative pXZ151, or Gentr or Kanr derivatives of pBBR1MCS (see Berger and Christie, 1994).
Protein analysis and immunoblotting
Total-cell extracts were suspended in Laemmli’s buffer with (reducing) or without (non-reducing) β-mercaptoethanol and proteins were electrophoresed through sodium dodecyl sulphate (SDS)-polyacrylamide gels using glycine or tricine buffer systems (Jakubowski et al., 2003). For Western blotting and immunostaining, proteins were transferred to nitrocellulose membranes and immunoblots were developed with anti-VirB or -VirD4 antibodies (Atmakuri et al., 2004; Cascales and Christie, 2004a; Jakubowski et al., 2005). Proteins were loaded on SDS-polyacrylamide gels on per cell equivalent basis for comparisons of steady-state levels of T4SS subunits between strains.
Co-immunoprecipitation
A 10 ml cell culture at an OD600 equivalent of 1.0 was centrifuged, and pelleted cells were washed twice in 50 mM HEPES, pH 8.0 and suspended in 500 µl of 50 mM HEPES, pH 8.0. Cells were incubated on ice for 15 min with EDTA (5 mM final concentration) and then for 45 min with lysozyme (100 µg ml−1 final concentration). Membranes were solubilized by incubation with dodecyl maltoside (DM; 2% final concentration) and protease inhibitors (1× final concentration; Boehringer Mannheim) on ice with shaking for 4 h. Solubilized material was collected by centrifugation for 15 min at 14 000 g. Protein A-Sepharose CL-4B (Pharmacia, 20 µl bed volume) was pre-incubated with the soluble fraction for 60 min at room temperature (RT) and the beads were removed by centrifugation at 500 g. The supernatant was incubated overnight at 4°C with anti-VirB10 antibodies and Protein A-Sepharose CL-4B (40 µl bed volume). The beads were washed once with 50 mM HEPES, pH 8.0, supplemented with 1% DM, and twice with 50 mM HEPES supplemented with 0.1% DM. The beads were then re-suspended in 75 µl of 50 mM HEPES, pH 8.0 and 25 µl of 5× Laemmli’s buffer, boiled for 10 min, and analysed by Western blotting and immunostaining.
Cellular fractionation and T pilus isolation
For T pilus isolation, A. tumefaciens strains were grown to an OD600 of 0.5 in MG/L media at 28°C. Cells were pelleted, diluted fivefold in ABIM and incubated for 6 h at 22°C. Next, 500 µl of an AS-induced culture was spread on ABIM agar plates, and the plates were incubated for 4 days at 18°C. Cells were then gently scraped off the plates in 1 ml of 50 mM KPO4 buffer (pH 5.5) and pelleted by centrifugation at 14 000 g for 15 min at RT. The supernatant representing the extracellular fraction was saved, and the cell pellet was again re-suspended in 500 µl of 50 mM KP04 pH 5.5. This suspension was passed through a 25-gauge needle to collect flagella, pili and surface proteins. The extracellular fraction and material collected by shearing were pooled, centrifuged at 14 000 g for 30 min at 4°C, and filtered through a 0.22-µm-pore-size cellulose acetate membrane to remove unpelleted cells. The filtered fraction was then centrifuged at 100 000 g for 1 h at 4°C to recover T pili, or layered on a 20–70% sucrose density gradient (5 ml) and ultracentrifuged in an SW55 Beckman rotor at 80 000 g for 20 h at 4°C. Alternatively, the filtered fraction was subjected to analytical gel filtration using a Tricorn high-performance column (Superdex 75 10/300 GL) pre-equilibrated and run with buffer A [50 mM HEPES (pH 7.0), 100 mM NaCI, 2 mM MgCI2, 0.1 mM EDTA, 5% glycerol]. The gel filtration column was calibrated by using lactate dehydrogenase (140 kDa), albumin (67 kDa), chymot-rypsinogen A (25 kDa) and RNase A (14 kDa) (Amersham Biosciences) as standards. Sucrose and gel filtration fractions and other cellular and extracellular material were analysed by tricine-SDS-PAGE and immunostaining (Sagulenko et al., 2001 a,b).
Extracellular VirB2 blot assay
Agrobacterium tumefaciens strains were grown to an OD600 of 0.5 in MG/L medium at 28°C. Cells were pelleted, diluted fivefold in ABIM and incubated for 6 h at 22°C. Cultures normalized to OD600 of 1.0 in ABIM were centrifuged at 14 000 g for 5 min at RT. The pellet was re-suspended in 100 µl of ABIM and 25 µl of the AS-induced culture was spotted on ABIM agar plates followed by incubation for 3 days at 18°C. A nitrocellulose membrane was placed on top of the colonies and allowed to sit for 5 min at RT. The membrane was washed twice with a blocking solution (10% milk, 0.01% Tween 20, 1× Tris buffered saline; TTBS), incubated overnight at RT with the specified antibody, and developed by immunostaining.
Conjugation assays
The RSF1010 derivative pML122 was introduced into various A. tumefaciens donor strains by diparental mating with E. coli strain S17-1(pML122). A. tumefaciens strains carrying pML122 were mated with A348Spcr recipient cells. Cells from a mid-logarithmic phase (OD600 = 0.5) culture were harvested and incubated in IM for 6 h at 22°C to induce vir gene expression. Five microlitres of pre-induced donor and recipient cells were mixed on a nitrocellulose filter on an IM agar plate, and the plate was incubated for 4 days at 18°C. Mating mixtures were recovered from filters and plated on media selective for transconjugants or serially diluted for determination of donor cell numbers. Transfer frequencies are reported as transconjugants recovered per donor cell. Experiments were repeated in triplicate, and results are reported for a representative experiment (Jakubowski et al., 2005).
Virulence assays
Agrobacterium tumefaciens strains were tested for virulence by inoculating wound sites of Kalanchoe daigremontiana leaves. As controls, all leaves were co-inoculated with WT A348 and isogenic null mutants as positive and negative controls (Berger and Christie, 1994). Relative virulence was assessed by inoculation of serially diluted cultures ranging from 102 to 108 colony-forming units (cfu) on plant wound sites. Tumour formation was monitored over a period of 6 weeks and scored on a scale of 0–3 pluses, with 0 (–, avirulent), 1 (formation of small tumours by inoculation of 108 cfu of a mutant strain that appeared several weeks after tumours being incited by the WT strain), 2 (formation of slightly smaller tumours with ∼1-week delay compared with the WT response), 3 (WT virulence).
Chemical labelling of cysteine residues
Cysteine accessibility experiments were carried out as described previously (Jakubowski et al., 2004) with minor revisions. Briefly, a 25 ml culture of A. tumefaciens strain PC1010 producing cysteine-substituted VirB10 derivatives was induced for vir gene expression by growth in ABIM to an A600 of 0.5. Cells were harvested, re-suspended in buffer B [100 mM HEPES (pH 7.5), 250 mM sucrose, 25 mM MgCI2, 0.1 mM KCI], and distributed into two centrifuge tubes to a final A600 of 12 in 500 µl of buffer A. To one cell suspension, 4-acetamido-40-maleimidylstilbene−2,20-disulphonic acid (AMS; Molecular Probes) was added to a final concentration of 5 mM and cells were incubated for 30 min at 25°C. AMS was removed by suspending cells in 10 ml of buffer A and centrifuging. The washed cell pellet was suspended in 500 µI of buffer A. To the AMS pre-treated cells and the second cell suspension, 3-(Nmaleimidylpropionyl) biocytin (MPB; Molecular Probes) was added to a final concentration of 100 µM (from a 20 mM stock freshly dissolved in DMSO) and the cells were incubated for 5 min at 25°C. The final concentration of DMSO in the reaction mixture did not exceed 0.5% (v/v). β-Mercaptoethanol (20 mM final concentration) was added to quench biotinylation, and cells were washed twice in buffer B containing 20 mM β-mercaptoethanol. As controls, sonicated extracts of induced cells served as the starting material for MPB labelling to assay for accessibility of Cys residues introduced in the presumptive cytoplasmic and TM domains.
Detection of labelled Cys residues
Labelled cells were suspended in TES [10 mM Tris (pH 7.5), 5 mM EDTA, 2% (w/v) SDS] and then vortex mixed for 30 min at 37°C. Samples were then diluted with 250 µl of buffer C [150 mM Tris (pH 8.0), 0.5 M sucrose, 10 mM EDTA]. Lysozyme (1 mg ml−1 final concentration) was added and the samples were incubated on ice for 1 h, then vortex mixed for 15 min at 37°C. Triton X-100 (20 µl), EDTA-free protease inhibitor cocktail (30 µl; Pierce Biochemicals) and 1 M MgCI2 stock solution (13 µl) were added and the samples were vortexed for 10 min at 25°C, then incubated with gentle rocking for 3 h at 4°C. Samples were diluted with 900 µl of buffer C, and cell debris was removed by centrifugation at 14 000 g for 15 min. Protein A-Sepharose CL4B (Pharmacia) (30 µl bed volume) was incubated with the supernatant for 60 min at RT and centrifuged at 5000 g to remove protein A-Sepharose and non-specifically bound proteins. Anti-VirB10 antibodies coupled to protein A-Sepharose were added to the supernatant and the mixture was incubated overnight at 4°C. The beads were washed twice in buffer C with 1% Triton X-100 for 10 min, once in buffer C with 0.1% Triton X-100 for 10 min, and once in buffer C for 5 min, and then re-suspended in 30 µl of 5× Laemmli’s buffer. Samples were boiled for 5 min, centrifuged at 400 g for 5 min, and the solubilized proteins were subjected to SDS-PAGE and transferred to nitrocellulose membranes (0.45 µm pore size). Membranes were incubated overnight in blocking buffer [1× PBS, 0.1%Tween 20, 5% (w/v) bovine serum albumin (BSA)] and then for 4 h in the presence of avidin HRP (Pierce; 1:10 000 dilution of 2 mg ml−1 stock solution). Blots were washed three times in buffer C (1× PBS, 0.1% Tween 20, 0.5% BSA) and biotinylated proteins were analysed by chemiluminescence according to the manufacturer’s (Amersham Pharmacia) instructions.
Supplementary Material
Acknowledgements
We thank Dr Krishnamohan Atmakuri for helpful comments and contributions during early phases of this study, and Dr Loic Coutte for development of the pilin colony blot assay. We thank Dr Hye-Jeong Yeo for help with the fractionation studies and Dr Mikhail Bogdanov for technical advice on the Cys accessibility studies. This work was supported by NIH Grant GM48746 to P.J.C.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Arkin IT. Structural aspects of oligomerization taking place between the transmembrane alpha-helices of bitopic membrane proteins. Biochim Biophys Acta. 2002;1565:347–363. doi: 10.1016/s0005-2736(02)00580-1. [DOI] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Baron C, Thorstenson YR, Zambryski PC. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Harris R, Coutte L, Monier A, Fronzes R, Christie PJ, et al. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc Natl Acad Sci USA. 2007;104:1673–1678. doi: 10.1073/pnas.0609535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupre CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TB, Saier MH., Jr Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology. 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004a;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004b;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion systems (review) Mol Membr Biol. 2005;22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu BCH, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20:467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- Das A, Xie YH. Construction of transposon Jn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Anderson LB, Xie YH. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Zhao Z, Jakubowski SJ, Atmakuri K, Margolin W, Christie PJ. A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbrandt R, Kalkum M, Lurz R, Lanka E. Maturation of IncP pilin precursors resembles the catalytic Dyad-like mechanism of leader peptidases. J Bacteriol. 2000;182:6751–6761. doi: 10.1128/jb.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JS, Levine BA, Trayer IP, Dorman CJ, Higgins CF. Sequence-imposed structural constraints in the TonB protein of E. coli. FEBS Lett. 1986;208:211–216. doi: 10.1016/0014-5793(86)81020-1. [DOI] [PubMed] [Google Scholar]
- Fernandez D, Spudich GM, Zhou XR, Christie PJ. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil H, Orlova E, Waksman G. Structure of a type IV secretion system core complex. Science. 2008 doi: 10.1126/science.1166101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Interaction between the IncHH plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol Microbiol. 2003;49:105–116. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187:3486–3495. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RB, Xie YH, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- Lai EM, Chesnokova O, Banta LM, Kado CI. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen RA, Postle K. Conserved residues Ser(16) and His(20) and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB, and the ability of TonB to respond to proton motive force. J Biol Chem. 2001;276:8111–8117. doi: 10.1074/jbc.M007479200. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Wood GE, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- Larsen RA, Letain TE, Postle K. In vivo evidence of TonB shuttling between the cytoplasmic and outer membrane in Escherichia coli. Mol Microbiol. 2003;49:211–218. doi: 10.1046/j.1365-2958.2003.03579.x. [DOI] [PubMed] [Google Scholar]
- Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci USA. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modelling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Postle K, Larsen RA. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. 2007;20:453–465. doi: 10.1007/s10534-006-9071-6. [DOI] [PubMed] [Google Scholar]
- Rashkova S, Spudich GM, Christie PJ. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol. 1997;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko Y, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001a;183:p5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J Bacteriol. 2001b;183:3642–3651. doi: 10.1128/JB.183.12.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Robichon C, Haan GJ, den Blaauwen T, Koningstein G, van Bloois E, et al. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J Bacteriol. 2007;189:7273–7280. doi: 10.1128/JB.00723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich GM, Fernandez D, Zhou XR, Christie PJ. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:4956–4961. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. The structure and function of prolinerich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- Yeo H-J, Yuan Q, Beck MR, Baron C, Waksman G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci USA. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-R, Christie PJ. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.