Abstract
CHARGE syndrome is a sporadic autosomal-dominant genetic disorder characterized by a complex array of birth defects so named for its cardinal features of ocular coloboma, heart defects, choanal atresia, growth retardation, genital abnormalities, and ear abnormalities. Approximately two-thirds of individuals clinically diagnosed with CHARGE syndrome have heterozygous loss-of-function mutations in the gene encoding chromodomain helicase DNA-binding protein 7 (CHD7), an ATP-dependent chromatin remodeler. To examine the role of Chd7 in development, a zebrafish model was generated through morpholino (MO)-mediated targeting of the zebrafish chd7 transcript. High doses of chd7 MO induce lethality early in embryonic development. However, low dose-injected embryos are viable, and by 4 days post-fertilization, morphant fish display multiple defects in organ systems analogous to those affected in humans with CHARGE syndrome. The chd7 morphants show elevated expression of several potent cell-cycle inhibitors including ink4ab (p16/p15), p21 and p27, accompanied by reduced cell proliferation. We also show that Chd7 is required for proper organization of neural crest-derived craniofacial cartilage structures. Strikingly, MO-mediated knockdown of the jumonji domain-containing histone demethylase fbxl10/kdm2bb, a repressor of ribosomal RNA (rRNA) genes, rescues cell proliferation and cartilage defects in chd7 morphant embryos and can lead to complete rescue of the CHARGE syndrome phenotype. These results indicate that CHARGE-like phenotypes in zebrafish can be mitigated through modulation of fbxl10 levels and implicate FBXL10 as a possible therapeutic target in CHARGE syndrome.
Keywords: CHARGE syndrome, chd7, fbxl10, zebrafish, cell proliferation, rRNA
Introduction
CHARGE syndrome is an autosomal dominant genetic disorder that affects 1 in 10,000–18,000 newborns worldwide (1, 2). CHARGE is an acronym for ocular coloboma, heart defects, atresia of the choanae, retardation of growth and development, genital anomalies, and ear malformations/deafness. The clinical presentation of CHARGE syndrome is highly variable and may include additional features such as cleft lip/palate, cranial nerve dysfunction, kidney anomalies, and rare limb anomalies (3). Two-thirds of cases of CHARGE syndrome are caused by spontaneous mutation of the gene encoding chromodomain helicase DNA binding protein 7 (CHD7), an ATP-dependent chromatin remodeler (4, 5). Most CHD7 mutations are nonsense, frameshift, or splice-site, predicted to lead to loss of protein function, and thus CHARGE syndrome is likely due to reduced dosage of CHD7 (2, 6). Consistent with haploinsufficiency as the genetic mechanism underlying CHARGE syndrome, mice that are homozygous for Chd7 null mutations die around embryonic day 10.5, but heterozygous Chd7 mutants are viable and recapitulate many features of CHARGE syndrome, including heart defects, choanal atresia, postnatal growth retardation, genital abnormalities, abnormal semicircular canals, and cleft palate (7, 8).
CHD7 is a member of the CHD family of proteins. Nine proteins comprise this family in vertebrates, and all nine contain tandem N-terminal chromodomains and a central conserved SNF2-like ATPase domain presumed to mediate chromatin remodeling. In addition to the chromodomains and ATPase domain, CHD7 contains two BRK domains of unknown function and a SANT-like domain that may mediate DNA and/or histone binding (9). CHD7 is a nuclear protein and binds to gene enhancer elements and promoters, functioning as a transcriptional co-regulator (10–12). CHD7 cooperates with PBAF (polybromo- and BRG1-associated factor-containing complex) to regulate genes important for formation and migration of neural crest, including TWIST and SOX9 (13). In mouse neural stem cells, CHD7 collaborates with SOX2 to regulate a common set of target genes including Jag1, Gli3, and Mycn. Mutations in these and other genes co-regulated by CHD7 and SOX2 cause clinical malformation syndromes that show some clinical overlap with CHARGE syndrome (12). Thus, it is hypothesized that dysregulated expression of genes normally regulated by CHD7 during development gives rise to the developmental defects observed in CHARGE syndrome.
In addition to its role as a transcriptional regulator in the nucleoplasm, CHD7 localizes to the nucleolus (14, 15), where it associates with rDNA and functions as a positive regulator of rRNA transcription. Cell proliferation is tightly coupled to protein synthesis, ribosome biogenesis, and rRNA production (16–19). Accordingly, siRNA-mediated knockdown of CHD7 in cultured cells suppresses protein synthesis and cell proliferation (14). Affected tissues from Chd7 mutant mouse embryos also show deficiencies in rRNA levels as well as cell proliferation (14, 20, 21). These findings raise the possibility that the pathogenesis of CHARGE syndrome is related to that of other human disorders caused by deficiencies in ribosomal biogenesis. Collectively known as the “ribosomopathies”, these disorders include Schwachman-Diamond syndrome, dyskeratosis congenita, cartilage hair hypoplasia, Treacher Collins syndrome, and myelodysplastic syndrome (22). Despite these discoveries, it remains unclear if the multiple anomalies in CHARGE syndrome are due to dysregulated expression of nucleoplasmic gene targets, rRNA, or the combination of both deficits.
Here, we developed a zebrafish model of CHARGE syndrome through morpholinomediated targeting of the zebrafish chd7 homolog. At 4 days post fertilization (dpf) chd7 morphant fish show multiple defects in organ systems analogous to those affected in humans with CHARGE syndrome. The defects in the chd7 morphant fish are accompanied by a general deficiency in cell proliferation at the early stages of development, associated with elevated expression of potent cell cycle inhibitors including ink4ab, p21, and p27. Remarkably, reduction of the Fbxl10/Kdm2bb histone demethylase, a negative regulator of rRNA transcription, restores cell proliferation in the chd7 morphants, with concomitant rescue of CHARGE-like phenotypes. Our findings implicate cell proliferation deficiencies in the pathogenesis of CHARGE syndrome, and suggest that elevation of rRNA levels maybe a viable strategy for therapeutic intervention in CHARGE syndrome.
Results
Organization of the zebrafish chd7 gene
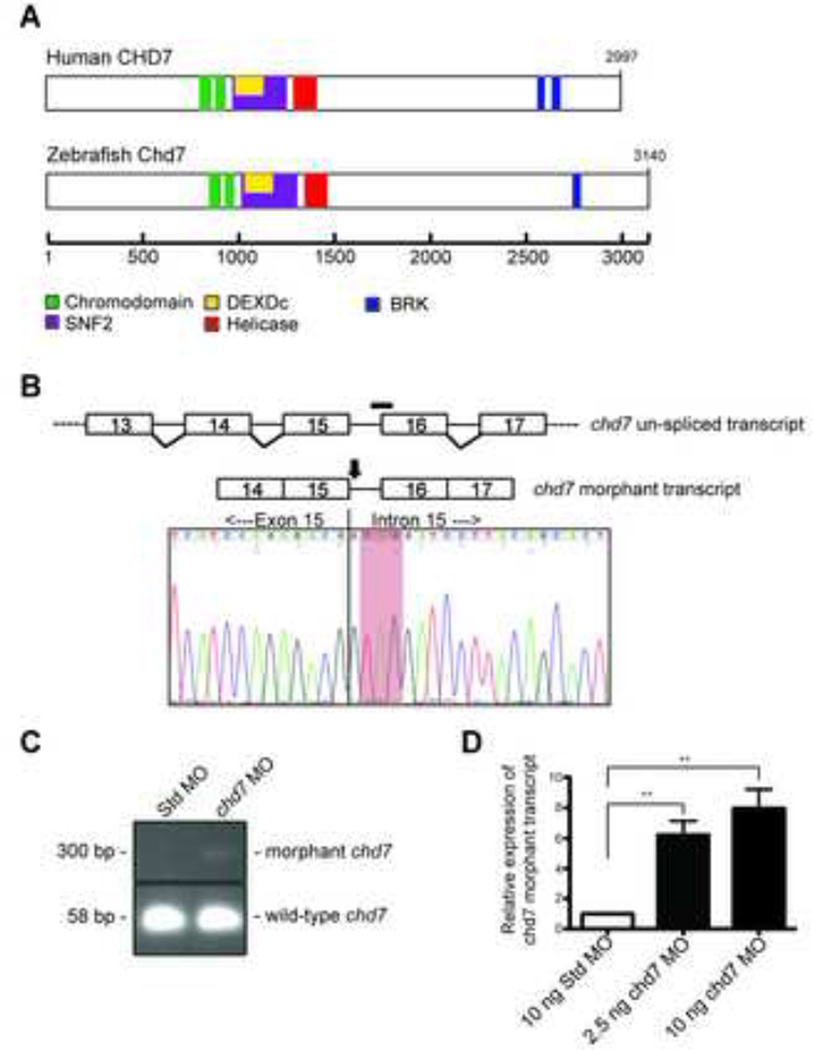
The sole zebrafish chd7 gene is located on chromosome 2 (Zv9 Ensembl). Of the five annotated transcripts, one is non-protein coding, two contain only the first two or three exons, and the remaining two are full-length. Both of the full-length transcripts have identical predicted protein coding sequences and have an exon-intron structure similar to that of human CHD7. These full-length transcripts code for a zebrafish Chd7 protein of 3140 amino acids, which is slightly longer than the human homolog of 2997 amino acids. Additionally, the zebrafish Chd7 protein contains a similar complement of protein domains including tandem N-terminal chromodomains, a central SNF2-like ATPase/helicase domain, and a C-terminal BRK domain (Figure 1A). The N-termini of zChd7 and hCHD7 are less conserved; however, the remainder of Chd7, including all of the functional domains, is highly similar. Overall, the aligning portion of the zChd7 sequence demonstrates 69% identity to hCHD7 at the amino acid level.
Figure 1. Morpholino targeting of the zebrafish chd7 RNA induces an aberrant transcript.
(A) Schematic of the human CHD7 and zebrafish Chd7 proteins with the location of the predicted protein domains. (B) Schematic of the zebrafish un-spliced chd7 transcript (upper) and the exon/intron splice site targeted by the chd7 morpholino (black bar). Schematic of the chd7 morphant transcript (lower) with the location of the predicted induced nonsense mutation (black arrow). Chromatogram of the sequenced morphant transcript PCR product reveals a nonsense mutation (red highlight). (C) Agarose gel with the amplified PCR products of both wild-type and morphant chd7 transcripts from Std control morphants and chd7 morphants. (D) Graph of qRT-PCR data quantifying the expression of the chd7 morphant transcript. Error bars represent standard error of the mean (SEM) (n = 4). Significance was determined by a Student’s two-tailed t-test and significant p-values are noted p < 0.01 (**).
chd7-morpholino gene targeting recapitulates major features of CHARGE syndrome
To model CHD7 haploinsufficiency in zebrafish, an antisense morpholino (MO) was designed to produce a mis-spliced chd7 transcript containing a premature stop codon. Specifically, the MO was targeted to the junction of chd7 intron 15 and exon 16 to induce production of a transcript either missing exon 16 or one including intron 15 (Figure 1B, upper). We performed RT-PCR analysis using multiple combinations of primers designed to amplify transcripts containing exon 15 mis-spliced to exon 17. However, such transcripts were not detected in chd7 MO-injected embryo, raising the possibility that the chd7 MO blocks splicing of exons 15 and 16, leading to intron inclusion. We therefore performed RT-PCR analysis using multiple primer sets designed to amplify transcript containing an intron 15 inclusion. The expected wild-type transcript was amplified with cDNA from control embryos. The wild-type transcript was also present in chd7 morphant embryos, even upon treatment with high doses of chd7 MO. However, chd7 morphant embryos also expressed a larger transcript, consistent with intron inclusion (Figure 1C). The levels of the chd7 morphant transcript increased in an MO dose-dependent manner (Figure 1D). Direct sequence analysis of the morphant transcript cDNA revealed inclusion of intron 15. In silico translation of the morphant transcript predicts mis-incorporation of a premature stop codon at 1383 amino acids past the first codon (Figure 1B, lower), presumably leading to degradation of the morphant chd7 message via nonsense-mediated decay.
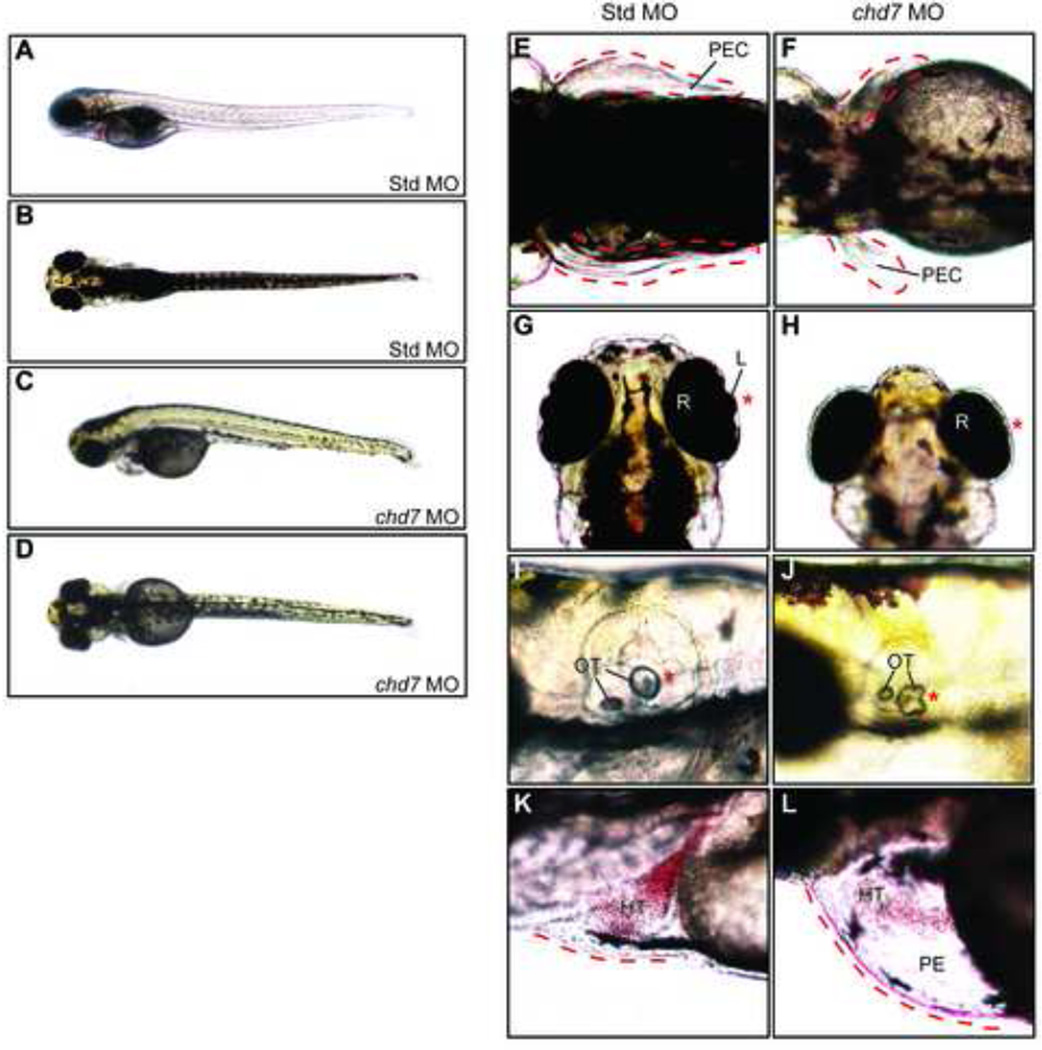
The chd7 morphants exhibited CHARGE-like defects in an MO dose-dependent manner. Specifically, high doses of chd7-MO (5–10 ng) induced lethality within 24 hours post-fertilization (hpf), reminiscent of embryonic lethality observed in homozygous null Chd7 mice (7, 8). Low-doses of chd7-MO (2.5 ng) yielded viable fish that were indistinguishable from wild-type and control-injected embryos at 3 days post-fertilization (dpf). However, multiple defects were apparent at 4 dpf (Figure 2A–D). Approximately 95% of low-dose chd7-MO injected embryos showed an abnormal phenotype (Figure 2E–L). chd7 morphant fish developed defects in organs analogous to those affected in humans and mice with heterozygous CHD7 mutations (Supplemental Table 1). Defects included pectoral fin hypoplasia (~60%), eye abnormalities (70%), and abnormalities in both otolith morphology and number (approximately 25% and 10% respectively) (Figure 2E–L). Moreover, ~60% of chd7 morphants presented with heart defects including pericardial edema, weak heartbeat with reduced circulatory flow, and occasionally a lack of proper heart tube folding. Importantly, 64% of chd7 morphants also exhibited craniofacial defects involving the nasal region and jaw (Figure 3A–B). None of these defects were apparent upon injection of a chd7 MO containing a 5-bp mismatch to the endogenous chd7 gene, indicating that the observed defects are specific to chd7 targeting. To further verify specificity, we tested a second splice-blocker chd7 MO, and a third chd7-MO designed to block translation. Both the second splice-blocker and chd7-translation blocker yielded similar phenotypes to that seen with the original splice blocker MO, although the translation blocker yielded more severe heart defects, and ear defects were observed at a higher frequency. No aspects of chd7 morphant phenotype were rescued upon co-injection of chd7 and p53 morpholinos, indicating that the observed defects in the chd7 morphants are unlikely to be related to non-specific MO-mediated cell death. Lastly, we note that the observed phenotype resembles that reported in a separate study in which the chd7 gene was targeted using a different splice-blocker MO (23, 24). Overall, the chd7 morphants displayed defects in many of the same organs affected in humans and mice with CHD7 mutations, including the ear, eye, heart, craniofacial region, and limbs. Thus, the requirement for CHD7 in the development of these organs appears to be conserved between mice, humans, and zebrafish, and suggests that MO-mediated targeting of chd7 creates a suitable system in which to examine Chd7 function in development.
Figure 2. Zebrafish chd7 targeting results in CHARGE-like phenotypes.
Whole-embryo lateral (A,C) and dorsal (B,D) views of representative Std control and chd7 morphant embryos at 4 dpf. The chd7 morphants display pectoral fin defects (E–F), eye abnormalities including under-developed or missing anterior eye structures (G–H), changes in otolith morphology (I–J), and pericardial edema (K–L). Missing or abnormal structures are highlighted with an asterisk (*) and/or dashed line. Structures are also highlighted in Std morphants for comparison. HT = heart, L = lens, OT = otolith, PE = pericardial edema, PEC = pectoral fin, R = retina.
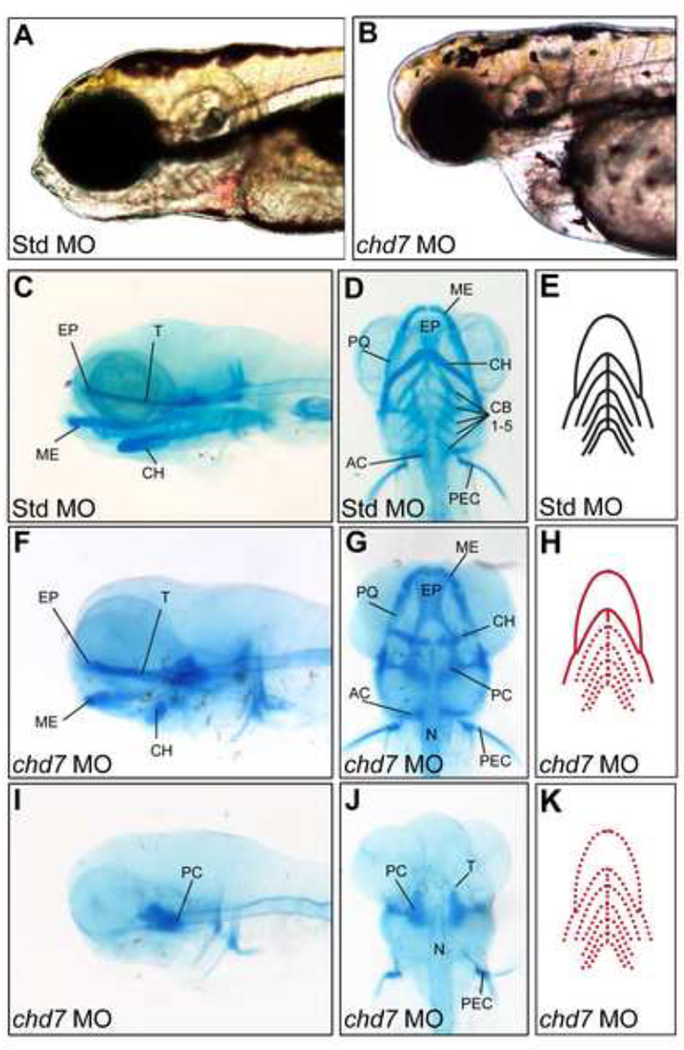
Figure 3. chd7 morphants display variable defects in craniofacial cartilage development.
(A–B) Bright-field lateral views of representative Std control morphants and chd7 morphants at 4 dpf. (C–D) Lateral and ventral views of Std morphants with wild-type craniofacial cartilage structures at 4 dpf. (F–G) Representative lateral and ventral views of the average chd7 morphant phenotype and is categorized as underdeveloped. The ceratohyal cartilages of the chd7 morphant are malformed and form a more linear shape. The five ceratobranchial cartilages were also undetectable with Alcian blue staining. (I–J) Lateral and ventral views of a severe chd7 morphant phenotype detected a highly underdeveloped neurocranium with the anterior and branchial arches absent. (E,H,K) Schematic views of the zebrafish craniofacial cartilage excluding the neurocranium. A solid red line indicates that the structure is present but malformed; while, a dashed red line indicates that the structure is absent. AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, N = notochord, PC = parachordal, PEC = pectoral fin, PQ, palatoquadrate, T = trabecula cranii.
chd7 morphants develop defects in neural crest-derived craniofacial cartilage
Early zebrafish craniofacial structure is dependent on proper migration of the multipotent cranial neural cells from the neural tube into the pharyngeal arches. These cells differentiate to form several tissues of the developing cranium such as the cranial nerves, bones, and cartilage (25). Based on these findings and published studies implicating a deficiency in neural crest cell migration in CHARGE syndrome (13, 24), we performed a detailed morphological analysis of craniofacial cartilage in low-dose chd7 morphant embryos. Compared to Standard MO-injected controls, chd7 morphants showed wide range of craniofacial cartilage abnormalities (Figure 3F–G). In approximately 50% of chd7 morphants, the first pharyngeal arch comprising Meckel’s cartilage and the palatoquadrate were morphologically normal. However, Meckel’s cartilage was located posteriorly to that observed in control morphants. In addition, the ceratobranchial cartilages were absent, and the ceratohyal was malformed (less V-shaped) compared to controls. In the majority of chd7 morphants, the ceratobranchial arches were undetectable at this resolution. However, microscopic analysis of sagittal sections revealed that the first ceratobranchial arch was occasionally present (Supplemental Figure 1). In more severely affected chd7 morphants, the only craniofacial cartilage structure observed was a severely underdeveloped neurocranium with malformed parachordal and trabecula cranii cartilages. Interestingly, parachordal and trabecula cranii cartilages are partially derived from mesoderm, while the more anterior cartilage structures that were typically absent in the severe chd7 morphants are mostly derived from neural crest (26). These results suggest that chd7 plays a critical role in the formation of cranial neural crest-derived cartilage tissues.
Chd7 is required for normal cellular proliferation during zebrafish development
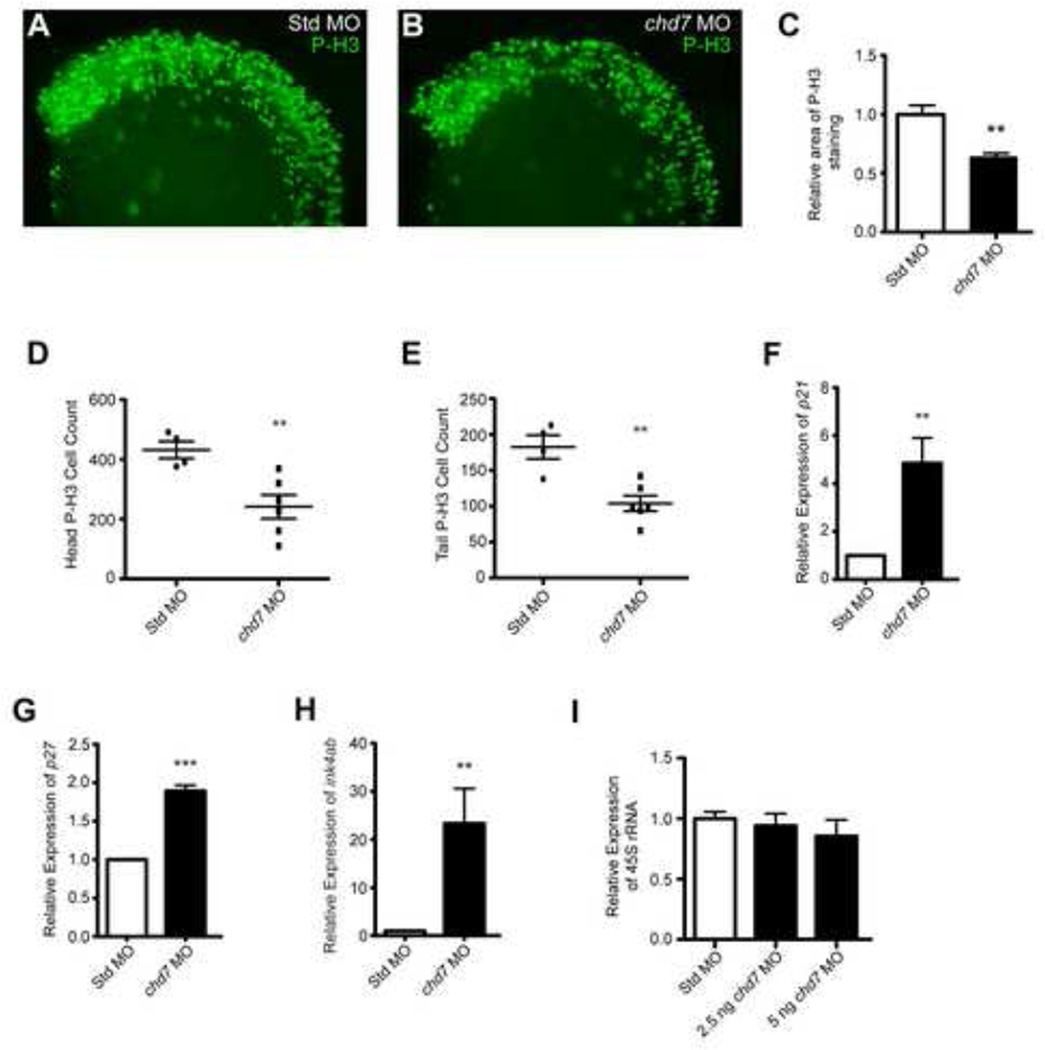
In E9.5–E10.5 mouse embryos, haploinsufficiency of CHD7 is associated with reduced proliferation of olfactory neural stem cells and cells of the otic epithelium and ganglion (20, 21). Moreover, we previously showed that siRNA-mediated knockdown of CHD7 in cultured cells attenuates their proliferation (14). These proliferative deficiencies occur without concomitant increases in cell death, indicating that CHD7 regulates cell proliferation and not apoptosis. We tested for proliferative defects in the chd7 morphants through immunofluorescence analysis of phosphorylated histone H3 at serine 10 (P-H3), a marker of mitosis (Figure 4A–B). Compared to control embryos, the total area positive for cellular P-H3 expression in chd7 morphants (25 hpf) was reduced by 37% in the anterior region of the embryo (Figure 4C). In addition, counts of P-H3 immuno-positive cells showed a 47% reduction in the total number of proliferating cells in chd7 morphants compared to controls (Figure 4D–E). A reduction in the number of proliferating cells was detected not only in the anterior region of the embryo (Figure 4D), where chd7 levels are high at this stage of development, but also the tail region (Figure 4E), where chd7 levels are low. Zebrafish chd7 expression is ubiquitous in early development and becomes more anteriorized around 3 dpf (27). Thus, the observed proliferative deficiencies are probably related to Chd7’s role during early development, prior to the time at which chd7 expression is restricted to the anterior region of the zebrafish embryo.
Figure 4. chd7 targeting impairs cell proliferation.
(A–B) Lateral views of representative P-H3 stained zebrafish morphants at 25 hpf. (C) Quantification of the area occupied by P-H3 positive cells in chd7 morphants relative to P-H3 positive cells in Std morphants (n = 5–6). (D–E) Graphs of P-H3 positive cell counts taken from confocal images of both the head and tail regions of Std and chd7 morphants (n = 4–6). (F–H) Graphs of qRT-PCR data measuring gene expression of several cell cycle regulator genes at 25 hpf in chd7 morphants relative to Std morphants (n = 5–6). (I) Expression of pre-rRNA in chd7 morphants at two separate morpholino dosages relative to Std morphants at 8 hpf (n = 3). All error bars represent SEM. Significance for all graphs was determined with a Student’s two-tailed t-test and significant values are noted p < 0.01 (**) and p < 0.001 (***).
Similar to the findings in mice and cultured cells, the proliferative defects in the chd7 morphants occurred in the absence of apoptosis, as determined through TUNEL-assays (data not shown). The proliferative deficiencies were accompanied by elevated expression of potent cell-cycle inhibitors, including p21, p27, and ink4ab (the zebrafish homolog of both p15 and p16 (28)) (Figure 4 F–H). Because we have previously shown that decreases in rRNA correlate with reduced proliferation, we also quantified the levels of 45S pre-rRNA, the product of Pol I transcription of rDNA that is ultimately processed into the mature 18S, 5S, and 28S ribosomal subunits (14, 29). No significant differences in pre-rRNA levels were detected between chd7 morphants and controls, even in embryos injected with high doses of chd7 MO (Figure 4I). Thus, either chd7 does not regulate rRNA in the zebrafish, or the effect is restricted to specific cell types or other stages of development. Overall, these findings indicate that Chd7 is required for normal cellular proliferation in the developing zebrafish embryo, although it is currently not clear from these data that the effect is mediated through rDNA regulation.
Fbxl10 regulates rRNA levels during zebrafish embryogenesis
Cell proliferation rates are tightly coupled to ribosomal RNA levels. Though our results were inconclusive as to whether zebrafish Chd7 regulates rRNA, we set out to modulate rRNA levels in chd7 morphants, reasoning that this might restore the proliferative deficiencies. To achieve this, we chose to knockdown the homolog of a known repressor of rRNA genes in mammals: FBXL10 (also known as NDY1, JHDM1B, and KDM2B). FBXL10 is a jumonji domain-containing histone demethylase that represses rRNA genes in the mammalian nucleolus, suppressing cell proliferation (30). Other studies contradict these findings, suggesting that FBXL10 increases proliferation by directly suppressing cell-cycle inhibitors in the nucleoplasm, including p15Ink4b, p16Ink4a, and p19Arf (31–33). Similarly to chd7, fbxl10 (kdm2bb) is expressed ubiquitously throughout early zebrafish embryogenesis. By 4 dpf, fbxl10 expression is restricted to the anterior embryo including the retina and central nervous system (34). Using a splice-blocker morpholino, we knocked down fbxl10 transcript levels by 35 to 50% (Figure 5A–B). When 5 ng of MO were injected, the fbxl10 knockdown yielded viable fish without an obvious gross morphological abnormalities or defects in craniofacial cartilage structures (Figure 5C–D, Supplemental Figure 2). Additionally, fbxl10 knockdown did not impact cell proliferation (Figure 5E–G). However, higher doses of fbxl10 morpholino induced early embryonic lethality by 24 hpf. Also, chd7 expression was also found to be normal in fbxl10 morphants when measured by qRT-PCR (not shown). We next tested if Fbxl10 regulates rRNA in the nucleolus or cell cycle genes in the nucleoplasm. 45S pre-rRNA levels were 30% higher in embryos injected with fbxl10-MO than control embryos (Figure 5H). ink4ab levels were not significantly different between controls and fbxl10 morphants (Figure 5I). These results support the reported mammalian function of Fbxl10 as a repressor of rRNA genes in the nucleolus.
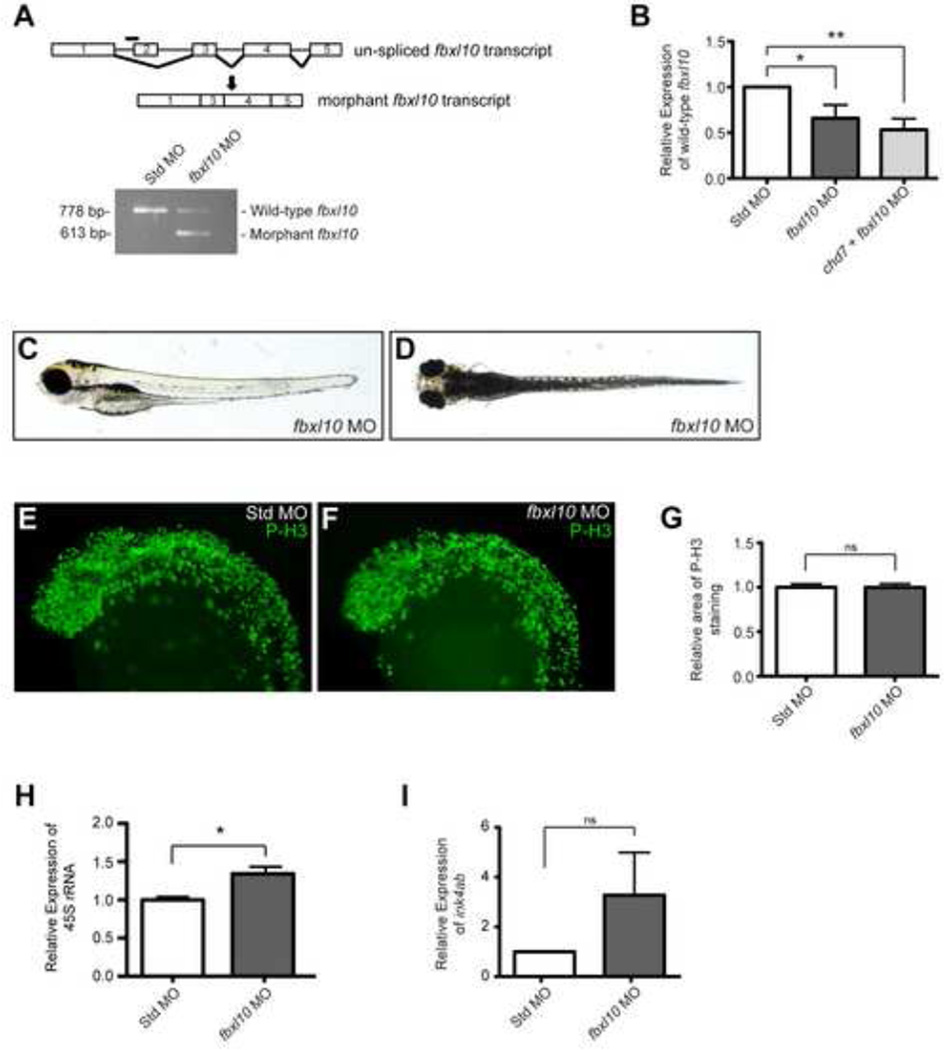
Figure 5. Targeting of zebrafish fbxl10 transcript modulates pre-rRNA expression.
(A) Schematic of the un-spliced fbxl10 transcript and the location of the fbxl10 morpholino (black bar). Injection of the morpholino results in an exclusion of exon 2 in the mature fbxl10 transcript and is predicted to induce a nonsense mutation (black arrow). Exclusion of exon 2 in the fbxl10 morphant transcript was confirmed by PCR. (B) Graph of qRT-PCR data measuring the expression of wild-type fbxl10 relative to the expression in Std control morphants at 8 hpf (n =4). (C–D) Representative lateral (C) and dorsal (D) bright-field images of fbxl10 morphants. (E–F) Lateral views of representative P-H3 stained zebrafish morphants at 25 hpf. (G) Quantification of the area occupied by P-H3 positive cells in fbxl10 morphants relative to P-H3 positive cells in Std morphants (n = 12–13). (H) Graph of qRT-PCR expression data for 45S pre-rRNA relative in fbxl10 morphants relative to Std morphants (n = 4). (I) Graph of qRT-PCR data measuring ink4ab expression relative to Std morphants (n = 6). Error bars in all graphs represent SEM. Significance determined by a Student’s two-tailed t-test and significant values are noted p < 0.05 (*) and p < 0.01 (**).
Rescue of chd7 morphant phenotype upon knockdown of fbxl10
Having established that Fbxl10 represses rRNA levels in the fish, we tested the effects of knocking down both fbxl10 and chd7, through co-injection of chd7 and fbxl10 morpholinos. Co-injection of fbxl10-MO and the chd7-translation blocker MO induced embryonic lethality within 24 hpf. However, co-injection of fbxl10-MO with the chd7 splice blocker MO suppressed the morphological defects induced by chd7 morpholino alone. Specifically, co-injection of chd7 and fbxl10 morpholinos reduced the penetrance of pericardial edema (13% from 58%), eye abnormalities (11% from 73%), pectoral fin defects (8% from 55%), craniofacial defects (27% from 64%), and otolith abnormalities (0% from 25%) (Figure 6A). In addition to the improvement of gross morphology discussed above, we also observed a significant restoration in overall head size in the chd7/fbxl10 double morphants compared to the chd7 morphant (Supplemental Figure 3).
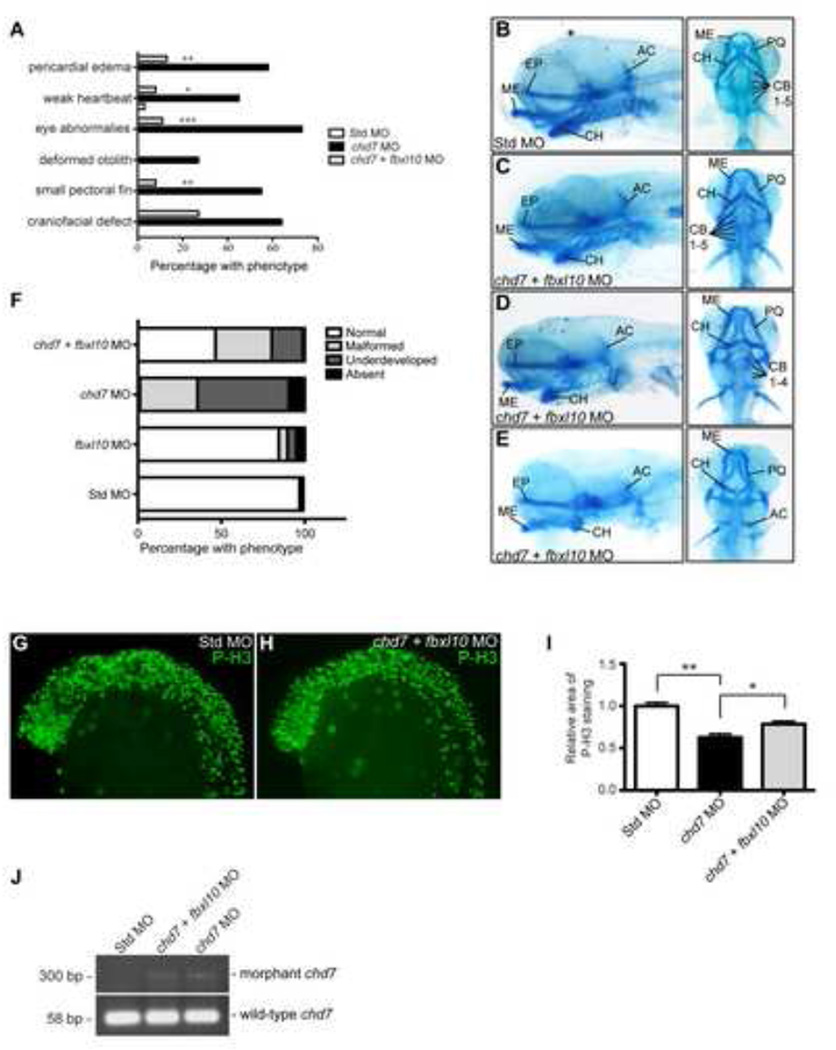
Figure 6. Modulation of fbxl10 expression rescues CHARGE-like phenotypes and improves cellular proliferation defects.
(A) Graph of the percentage of observed CHARGE-like phenotypes across the different zebrafish morphants. Significance was determined by chi-square tests. (B–I) Lateral and ventral views of representative cartilage staining in the chd7/fbxl10 double morphants compared to controls (B) at 4 dpf including morphants with normal phenotypes (C), mild malformations of the ceratohyal (D), and chd7/fbxl10 double morphants with severe ceratohyal malformations and ceratobranchial cartilage were undetectable (E). (F) Graph of the frequency of the observed craniofacial cartilage phenotypes in the developing zebrafish. Zebrafish were categorized on the severity of the craniofacial cartilage defect. “Malformed” morphants had all cartilages present but displayed morphological changes in the ceratohyal. “Underdeveloped” morphants had no detectable ceratobranchial arches in addition to a malformed ceratohyal. A morphant was categorized “Absent” with no detectable anterior and branchial arches. A highly underdeveloped neurocranium was present in these morphants. (G–H) Lateral views of representative P-H3 stained zebrafish Std and chd7/fbxl10 morphants at 25 hpf. (I) Quantification of the area occupied by P-H3 positive cells in chd7 morphants (re-plotted from Figure 4C) and chd7/fbxl10 morphants relative to P-H3 positive cells in Std morphants (n = 7). (J) Representative image of the amplified PCR products of both wild-type and morphant chd7 transcripts from 8 hpf across the panel of morphants. Significant p-values in all graphs are noted p < 0.05 (*), p < 0.01 (**), p < 0.001 (***). AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, PQ = palatoquadrate.
Forty-six percent of co-injected morphants showed restoration of the craniofacial cartilage to wild-type or near wild-type morphology with only minor malformations in the second pharyngeal arch (Figure 6B–C, Supplemental Figure 4A). Compared to the single chd7 morphants (Figure 3F–G), the chd7/fbxl10 morphants had more anteriorly developed Meckel’s cartilage, a wild-type or near wild-type ceratohyal morphology, and all ceratobranchial arches were detectable upon Alcian blue staining. The remaining 54% showed craniofacial cartilage abnormalities similar to those observed in chd7 single morphants, but in general, these abnormalities were less severe than those seen in the single chd7 morphants (Figure 6D–F). For example, the chd7/fbxl10 double morphants had a similar proportion of morphants categorized to be malformed compared to the chd7 single morphants due to more linear ceratohyal morphologies (Figure 6D, Supplemental Figure 4B). However, only a small proportion of the chd7/fbxl10 double morphants were scored as underdeveloped with both an inverted ceratohyal cartilage and undetectable ceratobranchial arches (Figure 6E, Supplemental Figure 4C). This indicates a significant restoration of the ceratobranchial arches in the double morphants. The craniofacial cartilage structures in the majority of single fbxl10 morphants injected in parallel were again indistinguishable from the standard-MO controls (Fig. 6F).
We also analyzed cell proliferation though quantitative analysis of P-H3 immuno-positive cells in morphant embryos. Relative to controls, the percentage of mitotic cells within measured areas increased from 63% in chd7 single morphants to 78% in co-injected embryos (Figure 6G–I). Importantly, these percentages reflect average counts from multiple co-injected embryos, from which the degree of rescue was variable. In fact, analysis of individual P-H3 immunostained embryos showed complete to near-complete restoration of cell proliferation in approximately two-thirds of all embryos analyzed (n = 29). Overall, these data indicate that knockdown of fbxl10 can mitigate chd7-morphant phenotypes in zebrafish. Importantly, co-injected embryos retained the chd7 morphant transcript, indicating that the effects of Fbxl10 depletion genuinely trump those of Chd7 depletion, and that rescue is not simply due to loss of the chd7 morphant transcript (Figure 6J).
Analysis of rRNA and cell cycle genes in fbxl10/chd7 double morphants
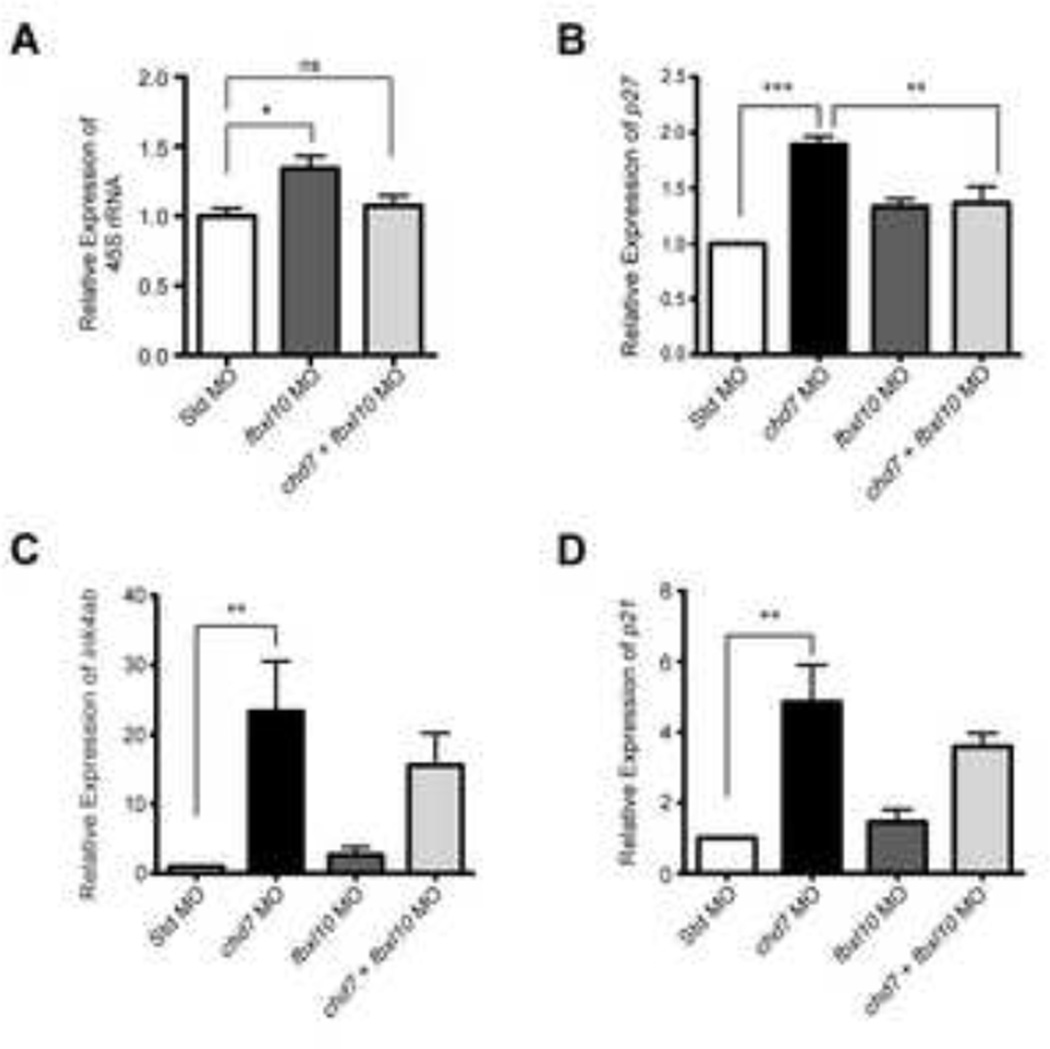
To gain insights into the mechanism underlying the fbxl10-mediated rescue, we compared the levels of rRNA, p21, p27, and ink4ab in fbxl10/chd7 co-injected morphant embryos to those in chd7 single morphants and standard-injected embryos. These experiments are inherently difficult to analyze for several reasons. First, 46% of co-injected embryos show rescue by 4 dpf, and, at the 25 hpf time point at which embryos are collected to perform the assay, one cannot distinguish rescued embryos from those that will fail to show rescue. In addition, embryos must be pooled to obtain sufficient quantities of RNA for the analysis. Nonetheless, we found that pre-rRNA levels were similar between co-injected and control-injected embryos, indicating that the fbxl10-mediated increases in rRNA are attenuated through targeted knockdown of chd7 (Figure 7A). Thus, even though reduced rRNA levels were not detected in the chd7 single morphants, elevated rRNA levels in the fbxl10 morphants are clearly attenuated upon knockdown of chd7, suggesting that Chd7’s role as a positive regulator of rRNA may be context specific. p27 levels, elevated in the chd7 single morphants, were also attenuated upon co-injection of fbxl10 and chd7 morpholinos (Figure 7B). p21 and ink4ab levels in chd7 single morphants were not significantly different from co-injected embryos (Figure 7C–D). However, these results reflect transcript levels measured from pooled co-injected embryos, of which 46% were rescued and 54% remained morphant. Thus, the levels of p21 and ink4ab shown in the plots, which are clearly trending downward, may be much lower in the 46% of embryos that were rescued. Based on these findings, we propose that elevated rRNA levels, induced upon knockdown of fbxl10, leads to suppression of cell cycle inhibitors activated upon chd7 targeting, thereby alleviating the chd7-associated cell proliferation defect.
Figure 7. Gene expression changes in cell-cycle regulators in chd7/fbxl10 double morphants.
(A) Expression of pre-rRNA in chd7/fbxl10 double morphants and fbxl10 single morphants (re-plotted for comparison from Figure 5F) relative to Std morphants at 8 hpf (n = 3). (B–D) Graphs of qRT-PCR data measuring gene expression of several cell cycle regulator genes at 25 hpf across the panel of morphant embryos relative to Std morphants (n = 5–6). Expression data for chd7 morphants re-plotted here from Figure 4D–F for comparison. Error bars represent SEM. Significance for all graphs was determined with a Student’s two-tailed t-test and significant values are noted p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
Discussion
To study the role of Chd7 during development, we generated a zebrafish model of CHARGE syndrome through MO-mediated gene targeting. The chd7 morphant phenotype is highly MO dose-dependent, with high doses leading to early embryonic lethality and lower doses yielding viable fish with craniofacial defects as well as malformations of the eye, heart, otoliths, and pectoral fins. The chd7 morphants also show cranial cartilage abnormalities, consistent with a defect in developing neural crest, the proposed cell type of origin for many of the anomalies observed in CHARGE syndrome patients (13, 24, 35–37). The chd7 morphant embryos also display widespread deficiencies in cellular proliferation, accompanied by elevated levels of potent cell-cycle inhibitors. Remarkably, the gross morphological alterations, cartilage abnormalities, and cell proliferation deficiencies that define the chd7 morphant phenotype are all restorable with knockdown of Fbxl10, a repressor of rRNA. We propose that the mechanism of rescue is due to suppression of cell cycle inhibitors that are activated upon chd7 targeting, possibly through global modulation of rRNA levels.
To date, zebrafish, frogs, and mice have been used to study the role of CHD7 in development (7, 8, 13, 23, 24, 38, 39). The overall phenotype and the organ systems affected in the chd7 morphants reported here are consistent with those previously observed in the other models (Supplemental Table 1). For example, defects in neural crest development and malformed craniofacial cartilage structures were previously reported in both zebrafish and frogs. Similarly to our zebrafish model, the frog model also shows facial width compression (13). As in the previously described zebrafish, frog, and mouse models, chd7 morphants described here show eye, heart, and otolith abnormalities. However the specific anomalies described vary somewhat between the models. With respect to the eyes for example, our zebrafish models show an underdeveloped lens, the previously described zebrafish models show retinal disorganization, the frog models develop colobomas, and the mouse models present with keratoconjunctivitis sicca (7, 13, 24, 39). The underlying basis for these differences is not known, but interestingly, this phenotypic variability is highly reminiscent of that seen among human patients with CHARGE syndrome and warrants further study.
To better understand the contribution of the proliferative deficiencies to the overall phenotype of the chd7 morphants, we knocked down the levels of fbxl10/kdm2bb, a histone demethylase that normally suppresses cell proliferation and has a similar spatiotemporal expression pattern to chd7 (27, 30–32, 34). To our knowledge, this is the first report of a zebrafish fbxl10 morphant model, and the first report that fbxl10 modulates rRNA levels in the zebrafish. Fifty percent reduction of fbxl10 did not yield any gross morphological abnormalities. However, more substantial decreases of fbxl10 induced early embryonic lethality by the end of the segmentation period (24 hpf). This phenotype is somewhat reminiscent of that observed in Fbxl10 mutant mice. Specifically, fbxl10 heterozygotes develop normally, while fbxl10-null homozygotes die during late embryogenesis or shortly after birth (40).
It has been demonstrated in several model systems that decreases in CHD7 levels impair cellular proliferation in multiple cell types (14, 20). Our studies not only corroborate these findings, but also implicate proliferative deficiencies as the basis for the developmental anomalies observed in the zebrafish model of CHARGE syndrome. The rescue of a multiple congenital anomaly syndrome caused by a deficiency in rRNA biogenesis is not unprecedented. Treacher-Collins Syndrome (TCS) is congenital disorder of craniofacial development caused by mutations in TCOF1 and POLR1D (41–43). These mutations lead to reduced rRNA biogenesis, and similarly to CHARGE syndrome, neural crest is implicated as the cell type of origin for most of the associated anomalies in TCS. In mouse models of TCS, impaired ribosome biogenesis triggers the nucleolar stress response, activating p53 and leading to proliferative deficiencies and apoptosis of neural crest cells. Moreover, inhibition of p53 blocks neural crest apoptosis and rescues the craniofacial defects (44). In our study, several genes associated with the nucleolar stress response, including p21, p27 and rRNA itself, responded upon MO-targeting of chd7 and fbxl10. These findings lead us to hypothesize that the cell proliferation defects observed in the chd7 morphants are due to activation of the nucleolar stress response, like in TCS. However, MO-mediated knockdown of p53 failed to rescue the chd7 morphant phenotype. Additionally, we previously showed that proliferative deficiencies induced upon knockdown of CHD7 in cultured cells were accompanied by changes in the levels of rRNA and p21, but not p53 (14). Thus, if the nucleolar stress response is the basis of the proliferative defect seen in the chd7 morphants, and the mechanism of rescue is due to suppression of the nucleolar stress response, it is likely to be p53-independent.
Together with the proposed role for CHD7 as a regulator of rRNA transcription in the nucleolus, the multiple anomalies in CHARGE syndrome are thought to due to insufficiencies in cell specification and proliferation during development. Our findings in the zebrafish suggest that restoring the cell proliferative deficiencies at the early stages of embryogenesis, even in the context of the other cellular deficits, could be sufficient to attenuate or altogether bypass the developmental defects associated with CHD7 mutation. Our studies lay the foundation to test this hypothesis in higher vertebrates, either through targeted modulation of FBXL10 or other genes that regulate rRNA levels, or genes that directly regulate the cell cycle. It is also noteworthy that histone demethylases are particularly amenable to targeting with small-molecule inhibitors (45, 46). If the findings here on Fbxl10 are validated in higher vertebrates, we might be able to tap into this growing area of therapeutic research for application to CHARGE syndrome.
Methods
Zebrafish Maintenance
Wild-type Tuebingen (TÜ) zebrafish (Danio rerio) embryos were raised at 28°C. The zebrafish were raised on 14-hour light cycle and 10-hour dark cycle.
Protein Sequence Comparison
Human CHD7 (NP_060250.2) and zebrafish Chd7 (ENSDART00000016208) protein sequences were aligned using the NCBI BLAST blastp suite to determine the degree of amino acid overlap between the two sequences (47). To retrieve the predicted domain composition of each protein sequence, we used the NCBI Conserved Domain Database (48).
Morpholino Design and Injection
A splice-blocking morpholino, 5’- ACCTACAATGAAGGAAATAGGCCGT-3’, a 5-bp mismatch control morpholino, 5’-ACGTAGAATCAAGCAAATACGCCGT-3’, a second confirmation splice-blocking morpholino, 5’-TGTGCCTGGAGGCAACAGCACAAAC-3’, and a translation-blocking morpholino, 5’-GGCTCATCATGCCTGGGTCAGCCAT-3’ were designed against the zebrafish chd7 transcript (ENSDART00000016208). To reduce fbxl10 expression, we used a splice-blocking morpholino designed against the zebrafish fbxl10 transcript (ENSDART00000102530), 5’-ACAACACCTGAGAACAGAAGCAGGA-3. To target p53 expression, we used a previously characterized morpholino (49). To control for phenotypes resulting from the injection procedure alone, a standard control morpholino that has no target or biological activity within the zebrafish was used in all experiments (Gene Tools). Phenotypes between standard morphants and those injected with the 5-bp mismatch chd7 morpholino were comparable. Zebrafish embryos received 2.5 ng of chd7 morpholino, 5 ng of standard control morpholino, 4 ng p53 morpholino, or 5 ng fbxl10 morpholino unless concentration is indicated otherwise. The chd7/fbxl10 double morphants received a simultaneous injection of 2.5 ng chd7 morpholino and 5 ng fbxl10 morpholino. The chd7/p53 double morphants received a simultaneous injection of 2.5 ng chd7 morpholino and 4 ng p53 morpholino. This p53 morpholino dosage was slightly higher than that recommended for testing for non-specific morpholino related cell death (50). For all co-injection experiments, chd7 and fbxl10 or chd7 and p53 morpholino single injections were performed in parallel as an additional control. All morpholinos were designed and manufactured by Gene Tools (Philomath, Oregon). Morpholinos were dissolved in sterile water to stock solution of 65 mg/ml. 5 mg/ml or 10 mg/ml working solutions were made by diluting the morpholino in water and 2% phenyl red. Zebrafish embryos were then injected at the 1 or 2 cell stage with using a microinjector.
Scoring of zebrafish morphant phenotypes
Zebrafish embryos were incubated at 28°C in fish water until the desired developmental stage was reached. The chd7 morphant phenotypes were monitored and scored using a Leica S6E stereomicroscope between 4–6 dpf. The bright-field images of chd7 morphant phenotypes were taken at 6 dpf and were images on a Leica DM6000. Significance was assessed by chi-square contingency test.
RNA extraction, cDNA synthesis, and qRT-PCR
Total RNA was extracted by homogenizing 20–30 pooled embryos at 8 hpf or 25 hpf in Trizol reagent (Invitrogen). Embryos were homogenized by drawing them through a 21G 1 ½ needle and 1 ml syringe (BD) approximately 20 times. RNA was further purified via the Trizol Reagent protocol (Invitrogen) and was re-dissolved in RNase-free water. From the total purified RNA, cDNA was synthesized using a High-capacity cDNA Archive Kit (Applied Biosystems). Using quantitative polymerase-chain reaction (PCR), gene expression was measured in triplicates across a set of three biological replicates. PCR reactions were performed using SybrGreen or Taqman chemistry on an ABI 7300 real-time thermocycler. Designed primer sequences for PCR reactions are listed in Table 2. Expression of the 45S rRNA transcript measured using a previously published primer set (51). Expression of several genes was amplified using several Taqman assays (Invitrogen) including p27 (ID: Dr03101119_ml) and β-actin1 (ID: Dr0332610_m1). Permission for the ink4ab Taqman probe (ID: AJ1RUB6) was provided graciously by Dr. Hatem Sabaawy.
Cartilage staining of whole mounts
At 4 dpf, zebrafish were anesthetized with tricaine and fixed with cold 4% paraformaldehyde solution (PFA). Zebrafish cartilage was stained with Alcian blue and Alzarian Red as previously described (52). Zebrafish were imaged in glycerol and bright-field images were taken using a Leica MZ10F fluorescent microscope.
Cartilage sectioning
Zebrafish morphants were anesthetized with tricaine and fixed with cold 4% PFA at 4 dpf. The sample preparation, sectioning, and Alcian blue staining of all zebrafish embryos was performed by Histoserv, Inc. (Germantown, MD). All embryos were sectioned at 10µm.
Whole-mount antibody labeling
At 25 hpf, zebrafish embryos were anesthetized with tricaine and fixed with cold 4% PFA overnight at 4°C. Fixed embryos were rinsed several times with PBS and permeablilized overnight at room temperature with 3% Triton in PBS. Embryos were rinsed in PBS again and blocked in 5% normal goat serum in PBS (blocking solution) for 3 hours at room temperature. Embryos were then incubated with rabbit anti-phosphorylated Histone H3 (phospho-ser10) antibody (1:200, Cell Signal #9701) diluted in blocking solution overnight at 4°C. Embryos were rinsed several times over 6 hours in blocking solution and incubated overnight at 4°C with goat anti-rabbit AlexaFluor 488 (1:200, Invitrogen A-11008). Embryos were rinsed in blocking solution for 2 hours. Zebrafish were stored in VECTASHIELD mounting medium (VECTOR Laboratories). Fluorescent images were taken on a Leica MZ10F microscope after mounting zebrafish in 0.1% agarose.
Quantification of proliferation
Quantification of the cellular area expressing fluorescently labeled P-H3 was performed using Adobe Photoshop as previously described (53). Mitotic cells were quantified by calculating the average fluorescent area (pixel2). Five cells within each fluorescent image were selected using the Photoshop Magic Wand Tool. On selection of the five mitotic cells, cells of similar fluorescent intensity were selected using the “Select Similar” command with a stringency factor of 20. Cells were not selected for based on intensity; however, selected cells in the plane of focus tended to be slightly higher in fluorescent intensity. The average pixel area was calculated for each image and the process was repeated across three separate z-planes for each zebrafish embryo. The three averaged pixel areas from the z-planes were again averaged to give a total average pixel area for each individual embryo. The total average pixel areas from multiple zebrafish embryos were plotted and statistical significance was determined using a Student’s t-test.
To count the number of P-H3-positive cells, zebrafish embryos were imaged on a Leica SP2 confocal microscope. Z-plane images were taken throughout the embryo at 40µm steps to avoid capturing cells in multiple z-planes. The cells were then manually counted by an individual whom was blinded to the identity of the morpholino used in the experiment, to avoid bias. The data were then stratified by the morpholino used (Standard versus chd7), and the results were plotted and tested for statistical significance using a Student’s two-tailed t-test.
Supplementary Material
(A–C) Alcian blue cartilage staining of 10µm sectioned zebrafish embryos at 4 dpf. Staining revealed wild-type structures of Std morphants (A), while chd7 morphants (B–C) had many of the cartilaginous structures absent. Sectioning confirmed the common chd7 morphant phenotype does not develop all of the ceratobranchial cartilages (B). Staining of severe chd7 morphants only detected some development of the neurocranium (C). AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
(A–B) Lateral and dorsal bright-field images of 4 dpf fbxl10 morphants. (C–D) Whole-mount lateral and ventral representative images of fbxl10 morphant craniofacial structure. (E) Sagittal sectioning of Alcian blue stained craniofacial cartilage in a representative fbxl10 morphant. AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
(A) A diagram indicating the locations of head measurements taken across the morphant zebrafish head region. Width measurements were taken across the forebrain (i) and midbrain (ii). The length of the zebrafish head was also measured from the anterior tip to the pectoral fins (iii). (i–iii) Graphs of the individual measurements for each morphant (n=7) correspond with the location on the diagram. All error bars represent SEM. Significance for all graphs were determined with a Student’s two-tailed t-test and significant values are noted p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
Bright-field images of 10µm sagittal sections of representative chd7/fbxl10 double morphants. The chd7/fbxl10 double morphants were classified into three categories based on the degree of restoration in craniofacial development. These images are sections from representative double morphants from each category including normal (A), malformed (B), and underdeveloped (C). AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
Supplemental Table 1: CHD7 mutant phenotypes comparison across multiple species.
Supplemental Table 2: PCR primers designed to measure gene expression.
Highlights.
Loss of chd7 in zebrafish causes extensive craniofacial defects
Alterations in chd7impairs cellular proliferation in the zebrafish embryo
Modulating rRNA regulator Fbxl10reverses features of CHARGE syndromein zebrafish
Acknowledgments
We thank Ronald Conlon, Radhika Atit, and Olivia Corradin for helpful comments and discussion. We also thank Jason Heaney, Stephanie Doerner, and Carol Fernando for technical assistance. This work was supported by RO1HD056369 (PCS), R01CA160356 (PCS), The CHARGE Syndrome Foundation (PCS), and 5T32GM008613 (SB). Imaging in this work was supported by the Office of Research Infrastructure Program of the National Institutes of Health under the award numbers S10RR021228 (PAC) and S10RR017980 (PAC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jongmans MC, Admiraal RJ, van der Donk KP, Vissers LE, Baas AF, Kapusta L, van Hagen JM, Donnai D, de Ravel TJ, Veltman JA, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM, Hoefsloot LH. Mutation update on the CHD7 gene involved in CHARGE syndrome. Human mutation. 2012;33:1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- 3.Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 5.Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KG, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartels CF, Scacheri C, White L, Scacheri PC, Bale S. Mutations in the CHD7 gene: the experience of a commercial laboratory. Genet Test Mol Biomarkers. 2010;14:881–891. doi: 10.1089/gtmb.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 8.Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK, Martin DM. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mammalian genome : official journal of the International Mammalian Genome Society. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 9.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 10.Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, Dekkers DH, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 13.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, et al. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 2010;19:3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kita Y, Nishiyama M, Nakayama KI. Identification of CHD7S as a novel splicing variant of CHD7 with functions similar and antagonistic to those of the full-length CHD7L. Genes to cells : devoted to molecular & cellular mechanisms. 2012;17:536–547. doi: 10.1111/j.1365-2443.2012.01606.x. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Santoro R, Koberna K, Grummt I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 2005;24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 18.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, et al. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet. 2011;20:3138–3150. doi: 10.1093/hmg/ddr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs-McDaniels NL, Albertson RC. Chd7 plays a critical role in controlling left-right symmetry during zebrafish somitogenesis. Dev Dyn. 2011;240:2272–2280. doi: 10.1002/dvdy.22722. [DOI] [PubMed] [Google Scholar]
- 24.Patten SA, Jacobs-McDaniels NL, Zaouter C, Drapeau P, Albertson RC, Moldovan F. Role of Chd7 in zebrafish: a model for CHARGE syndrome. PLoS One. 2012;7:e31650. doi: 10.1371/journal.pone.0031650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight RD, Schilling TF. Cranial neural crest and development of the head skeleton. Advances in experimental medicine and biology. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- 26.Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T, Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS One. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauch GJ, Lyons DA, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot WS. Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission. 2003 [Google Scholar]
- 28.Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:15166–15171. doi: 10.1073/pnas.0603349103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 30.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 31.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci U S A. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, Wong KK, Bardeesy N. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 [Google Scholar]
- 35.Siebert JR, Graham JM, Jr, MacDonald C. Pathologic features of the CHARGE association: support for involvement of the neural crest. Teratology. 1985;31:331–336. doi: 10.1002/tera.1420310303. [DOI] [PubMed] [Google Scholar]
- 36.Sanlaville D, Etchevers HC, Gonzales M, Martinovic J, Clement-Ziza M, Delezoide AL, Aubry MC, Pelet A, Chemouny S, Cruaud C, et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J Med Genet. 2006;43:211–217. doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall V, McCue K, Roberts C, Kyriakopoulou V, Beddow S, Barrett AN, Vitelli F, Prescott K, Shaw-Smith C, Devriendt K, et al. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest. 2009;119:3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Layman WS, McEwen DP, Beyer LA, Lalani SR, Fernbach SD, Oh E, Swaroop A, Hegg CC, Raphael Y, Martens JR, et al. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet. 2009;18:1909–1923. doi: 10.1093/hmg/ddp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian C, Yu H, Yang B, Han F, Zheng Y, Bartels CF, Schelling D, Arnold JE, Scacheri PC, Zheng QY. Otitis media in a new mouse model for CHARGE syndrome with a deletion in the Chd7 gene. PLoS One. 2012;7:e34944. doi: 10.1371/journal.pone.0034944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda T, Tokunaga A, Sakamoto R, Yoshida N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Molecular and cellular neurosciences. 2011;46:614–624. doi: 10.1016/j.mcn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Edwards SJ, Gladwin AJ, Dixon MJ. The mutational spectrum in Treacher Collins syndrome reveals a predominance of mutations that create a premature-termination codon. Am J Hum Genet. 1997;60:515–524. [PMC free article] [PubMed] [Google Scholar]
- 42.Gladwin AJ, Dixon J, Loftus SK, Edwards S, Wasmuth JJ, Hennekam RC, Dixon MJ. Treacher Collins syndrome may result from insertions, deletions or splicing mutations, which introduce a termination codon into the gene. Hum Mol Genet. 1996;5:1533–1538. doi: 10.1093/hmg/5.10.1533. [DOI] [PubMed] [Google Scholar]
- 43.Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, Hoefsloot LH, Peters DJ, Boers AC, Daumer-Haas C, Maiwald R, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 44.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole PA. Chemical probes for histone-modifying enzymes. Nature chemical biology. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohse B, Kristensen JL, Kristensen LH, Agger K, Helin K, Gajhede M, Clausen RP. Inhibitors of histone demethylases. Bioorganic & medicinal chemistry. 2011;19:3625–3636. doi: 10.1016/j.bmc.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langheinrich U, Hennen E, Stott G, Vacun G. Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol. 2002;12:2023–2028. doi: 10.1016/s0960-9822(02)01319-2. [DOI] [PubMed] [Google Scholar]
- 50.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azuma M, Toyama R, Laver E, Dawid IB. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J Biol Chem. 2006;281:13309–13316. doi: 10.1074/jbc.M601892200. [DOI] [PubMed] [Google Scholar]
- 52.Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotechnic & histochemistry : official publication of the Biological Stain Commission. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- 53.Lehr HA, van der Loos CM, Teeling P, Gown AM. Complete chromogen separation and analysis in double immunohistochemical stains using Photoshop-based image analysis. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1999;47:119–126. doi: 10.1177/002215549904700113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–C) Alcian blue cartilage staining of 10µm sectioned zebrafish embryos at 4 dpf. Staining revealed wild-type structures of Std morphants (A), while chd7 morphants (B–C) had many of the cartilaginous structures absent. Sectioning confirmed the common chd7 morphant phenotype does not develop all of the ceratobranchial cartilages (B). Staining of severe chd7 morphants only detected some development of the neurocranium (C). AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
(A–B) Lateral and dorsal bright-field images of 4 dpf fbxl10 morphants. (C–D) Whole-mount lateral and ventral representative images of fbxl10 morphant craniofacial structure. (E) Sagittal sectioning of Alcian blue stained craniofacial cartilage in a representative fbxl10 morphant. AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
(A) A diagram indicating the locations of head measurements taken across the morphant zebrafish head region. Width measurements were taken across the forebrain (i) and midbrain (ii). The length of the zebrafish head was also measured from the anterior tip to the pectoral fins (iii). (i–iii) Graphs of the individual measurements for each morphant (n=7) correspond with the location on the diagram. All error bars represent SEM. Significance for all graphs were determined with a Student’s two-tailed t-test and significant values are noted p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
Bright-field images of 10µm sagittal sections of representative chd7/fbxl10 double morphants. The chd7/fbxl10 double morphants were classified into three categories based on the degree of restoration in craniofacial development. These images are sections from representative double morphants from each category including normal (A), malformed (B), and underdeveloped (C). AC = auditory capsule, CB = ceratobranchial, CH = ceratohyal, EP = ethmoid plate, ME = Meckel’s cartilage, PC = parachordal, PQ = palatoquadrate, T = trabecula cranii.
Supplemental Table 1: CHD7 mutant phenotypes comparison across multiple species.
Supplemental Table 2: PCR primers designed to measure gene expression.