Abstract
The Rrm3 DNA helicase of Saccharomyces cerevisiae interacts with proliferating cell nuclear antigen and is required for replication fork progression through ribosomal DNA repeats and subtelomeric and telomeric DNA. Here, we show that rrm3 srs2 and rrm3 sgs1 mutants, in which two different DNA helicases have been inactivated, exhibit a severe growth defect and undergo frequent cell death. Cells lacking Rrm3 and Srs2 arrest in the G2/M phase of the cell cycle with 2N DNA content and frequently contain only a single nucleus. The phenotypes of rrm3 srs2 and rrm3 sgs1 mutants were suppressed by disrupting early steps of homologous recombination. These observations identify Rrm3 as a new member of a network of pathways, involving Sgs1 and Srs2 helicases and Mus81 endonuclease, suggested to act during repair of stalled replication forks.
Some DNA helicases are known to play a central role in the preservation of genome stability (6, 9, 18, 28, 46, 47, 74), which is disregulated in many cancers. Bloom's syndrome (22), Werner syndrome (16), and Rothmund-Thomson syndrome (69), which arise from mutations in genes coding for DNA helicases (BLM, WRN, and RECQL4, respectively), are characterized by increased genome rearrangements, increased sister chromatid exchange, increased mutation rates and, consequently, a wide variety of cancers. Recent studies of closely related DNA helicases in lower eukaryotes have led to a more detailed understanding of the molecular basis of the genomic instability that is characteristic for these diseases. Saccharomyces cerevisiae cells lacking the BLM homologue, Sgs1, exhibit a hyperrecombination phenotype especially at repetitive sites in the genome and between modestly divergent DNA sequences, are hypersensitive to certain DNA damaging agents, and have an increased rate of chromosomal rearrangements (49, 70, 75). Like its human counterpart, Sgs1 is a 3′-to-5′ DNA helicase (4) that favors forked and branched DNA substrates compared to single- or double-stranded DNA. Hence, possible roles for these helicases in Holliday junction migration and resolution during homologous recombination have been suggested (4, 11, 35).
sgs1 mutants that also lack Srs2, another 3′-to-5′ DNA helicase of S. cerevisiae (52), exhibit a severe growth defect and show a high degree of inviability (21, 41), which can be suppressed by mutations that disrupt early steps of homologous recombination (17, 37). Previous studies demonstrated that srs2 mutations suppressed the UV sensitivity of rad6 and rad18 mutants when homologous recombination pathways were functional and suggested that Srs2 might act to channel damage away from homologous recombination into alternative repair pathways, such as the Rad6/Rad18 postreplication repair pathway (55). Genetic evidence has suggested that homologous recombination in sgs1 srs2 mutants is initiated at single-strand gaps (rather than at double-strand breaks), leading to the proposal that Sgs1 and Srs2 are required to prevent the accumulation of recombination intermediates that are generated during the repair of stalled replication forks (17). In addition to roles in DNA replication and recombination, both Srs2 and Sgs1 have been implicated in the activation of Rad53 in S-phase-specific checkpoints (18, 19, 42). Consequently, it has been proposed that the inability of srs2 mutants to slow down DNA replication in response to DNA damage could lead to the formation of recombinogenic (gapped) DNA molecules because of repriming downstream of the blocking lesion rather than lesion bypass and fork restart, which would involve more lengthy processes like template switching and reverse branch migration (42). The concept that Srs2 functions during DNA replication is further supported by the finding that Srs2 interacts with Pol32, a structural subunit of the polymerase δ complex (29).
Our laboratory has previously shown that the Rrm3 helicase physically interacts with another key component of DNA replication, proliferating cell nuclear antigen (PCNA), raising the possibility that the Rrm3 helicase, too, may be required for repairing DNA damage that arises from stalled replication forks (57). In contrast to Sgs1 and Srs2, Rrm3 unwinds DNA with a 5′-to-3′ polarity (32). Rrm3 was originally discovered as an inhibitor of recombination between ribosomal DNA (rDNA) repeats as well as between other naturally occurring direct repeats (36). The formation of extrachromosomal rDNA circles, a sign of rDNA instability, is increased in rrm3 mutants, and this increased recombination is suppressed by a rad52 mutation (33). This observation has suggested that Rrm3 might be an antirecombinase or may prevent some type of aberrant replication structure or damage that leads to increased rDNA recombination. Recently, roles in replication of telomeric and subtelomeric DNA (32) and in regulation of Ty1 transposition have also been assigned to Rrm3 (58). Interestingly, the Ty1 hypermobility phenotypes of rrm3 and sgs1 mutants are almost identical. The observation that the increased Ty1 mobility in sgs1 mutants was eliminated by mutations that inactivated homologous recombination has suggested that Sgs1, and possibly Rrm3, may regulate transposition by regulating homologous recombination (58) or possibly by preventing some type of DNA damage or aberrant replication structure that results in increased recombination.
Sgs1 and Srs2 helicases share homology with the RecQ and UvrD/PcrA/Rep DNA helicases of Escherichia coli, respectively (1, 20, 71). Interestingly, Rrm3 is most highly related to the RecD family of DNA helicases (COG0507; RPS-BLAST 2.2.6 at http://www.ncbi.nlm.nih.gov/blast). An alignment between Rrm3 and RecD spanning the entire length of both proteins includes 14% identical residues and 14% similar residues (Fig. 1). Biochemical studies indicating that Rrm3 and RecD both have single-strand DNA-dependent ATPase activity and 5′-to-3′ DNA helicase activity lend further support to an evolutionary link between RecD and Rrm3 (15, 63). RecD is a subunit of the RecBCD complex, which possesses 5′-to-3′ and 3′-to-5′ DNA helicase activities as well as nuclease activities that are essential for homologous recombination and DNA repair in E. coli (61). It is conceivable that the opposite polarities of Rrm3 and Srs2 (or Sgs1) helicase activities may cooperate in a similar fashion to regulate DNA repair or recombination at sites of replication damage.
FIG. 1.
Alignment of Rrm3 helicase of S. cerevisiae with RecD helicase of E. coli based on the location of seven conserved helicase motifs characteristic of superfamily I helicases. Helicase motifs are shaded light gray. Identical residues are indicated by letters, and similar residues are indicated by a + symbol.
In this study, we have investigated genetic interactions between rrm3 mutations and mutations in either SGS1 or SRS2. The results of these studies indicate that the Rrm3 helicase is essential for normal growth of cells that have a functional homologous recombination pathway when either Sgs1 or Srs2 is absent. Our analysis of the severe growth defect exhibited by rrm3 srs2 mutants with respect to morphology, DNA distribution, and cell cycle progression revealed that the double mutant frequently fails to undergo nuclear division and cytokinesis due to G2/M arrest. We propose a model for a role of Rrm3 in the recombinational repair of DNA lesions. Our observations on RRM3 are supported by evidence presented in an accompanying paper by Torres et al. (65).
MATERIALS AND METHODS
Yeast strains.
The S. cerevisiae strains used in this study are isogenic derivatives of S288C. The isogenic parental strains RDKY2664 (MATα ura3-52 trp1Δ63 his3Δ200), RDKY2666 (MATa ura3-52 trp1Δ63 his3Δ200), and RDKY2669 (MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8) were used to construct the strains used in the studies described here. Gene deletions in RDKY2664, RDKY2666, and RDKY2669 were generated by homologous-recombination-mediated integration of PCR fragments. Primer sequences to generate the knockout cassettes are available upon request. Derivatives of RDKY2664, RDKY2666, and RDKY2669 that were constructed for this study are listed in Table 1.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genetic background | Reference |
|---|---|---|
| RDKY4069a | MATα ura3-52 trp1Δ63 his3Δ200 rad51::HIS3 | Lab collection |
| RDKY4067a | MATα ura3-52 trp1Δ63 his3Δ200 rad52::HIS3 | Lab collection |
| RDKY4714b | MATaura3-52 trp1Δ63 his3Δ200 rrm3::URA3 | This study |
| RDKY5109b | MATaura3-52 trp1Δ63 his3Δ200 rrm3::URA3 rad52::TRP1 | This study |
| RDKY5111b | MATaura3-52 trp1Δ63 his3Δ200 rrm3::URA3 rad54::TRP1 | This study |
| RDKY5112b | MATaura3-52 trp1Δ63 his3Δ200 rrm3::URA3 rad55::TRP1 | This study |
| RDKY5114c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 sgs1::HIS3 | This study |
| RDKY5116c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 sgs1::HIS3 rad52::TRP1 | This study |
| RDKY5117c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 srs2::TRP1 | This study |
| RDKY5120c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 srs2::HIS3 | This study |
| RDKY5121c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 mre11::HIS3 | This study |
| RDKY5123c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 rad50::HIS3 | This study |
| RDKY5125c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 rad54::TRP1 | This study |
| RDKY5127c | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 mus81::HIS3 | This study |
| RDKY5289b | MATaura3-52 trp1Δ63 his3Δ200 sgs1::TRP1 | This study |
| RDKY5291b | MATaura3-52 trp1Δ63 his3Δ200 srs2::TRP1 | This study |
| RDKY5293a | MATα ura3-52 trp1Δ63 his3Δ200 mre11::TRP1 | This study |
Derivative of RDKY2664 (MATα ura3-52 trp1Δ63 his3Δ200).
Derivative of RDKY2666 (MATa ura3-52 trp1Δ63 his3Δ200).
Derivative of RDKY2669 (MATα ura3-52 leu2Δ1trp1Δ63his3Δ200lys2ΔBgl hom3-10 ade2Δ1 ade8).
Tetrad analysis.
Diploid strains for tetrad analysis were obtained by mixing haploids of opposite mating types that carry the desired mutations (derivatives of RDKY2666 and RDKY2669) followed by selection for the presence of auxotrophic markers. Diploids were grown overnight at 30°C in rich yeast extract-peptone-dextrose (YPD) medium, washed, transferred to 0.1% potassium acetate, and incubated for 5 days at 30°C with vigorous shaking. Asci were incubated in the presence of zymolase (500 μg/ml in 1 M sorbitol) for 3 min at 30°C and dissected on YPD agar plates using a micromanipulator mounted on a microscope (Carl Zeiss). The YPD plates were then incubated for 4 days at 30°C and photographed. Note that in some of the crosses performed here, ade2 and ade8 mutations segregated in the progeny, resulting in minor differences in growth rates on tetrad dissection plates. Therefore, for the determination of doubling times a set of strains without ade2, ade8, hom3, and lys2 mutations was constructed by mating RDKY2664- and RDKY2666-derived haploids which contained the mutations of interest. We observed no differences (other than those caused by the ade2 and ade8 mutations) in tetrad dissections obtained from diploids derived from the RDKY2666 and RDKY2664 strains compared to tetrad dissections obtained from diploids derived from the RDKY2666 and RDKY2669 strains.
Doubling time measurement.
After incubation of tetrad dissection plates for 4 days at 30°C, colonies were resuspended in water and spotted on YPD plates. After further incubation for 4 days at 30°C, cells from these spots were used to inoculate 5-ml YPD cultures at an optical density at 600 nm (OD600) of 0.2. To determine the doubling time, the OD of cultures was measured in 2-h intervals for 8 to 12 h. Doubling times reported are the average doubling times of three to nine cultures established from independent spores. Error bars shown with the graphs are standard deviations.
DNA distribution analysis.
Cells from cultures in mid-logarithmic phase were fixed by incubation in 3.7% formaldehyde and then in 70% ethanol, each for 1 h at room temperature. To visualize the DNA, cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and analyzed microscopically (Nikon Eclipse TE300). Digital images were acquired and analyzed using the MetaMorph imaging system (Universal Imaging Corporation).
DNA content analysis.
Cells were grown overnight at 30°C in YPD medium. Cultures were diluted in YPD to OD650 = 0.2 (∼3 × 106/ml), and incubation was continued until cultures reached OD650 = 0.6 to 0.8. Cells were then fixed in 70% ethanol for 1 h at room temperature and sonicated in 50 mM sodium citrate (pH 7). The cells were then washed once in 50 mM sodium citrate (pH 7), and RNase A was added to a final concentration of 250 μg/ml. After overnight incubation at 37°C, the cells were washed twice in 50 mM sodium citrate. To stain the DNA, Sytox Green (Molecular Probes, Eugene, Oreg.) was added to a final concentration of 1 μM and the cells were incubated in the dark at room temperature for 1 h immediately prior to fluorescence-activated cell sorting (FACS; FACSCalibur; Becton Dickinson). All of the strains for FACS analysis were freshly isolated meiotic products from a cross of strains RDKY5112 and RDKY5120.
Morphological analysis.
Cell morphology of mutants and a wild-type control was analyzed in aliquots taken from the same cultures that were analyzed by FACS (for growth conditions, see above). Cells were inspected microscopically, and the number of single cells, small-budded cells (bud smaller than one-third of the mother cell), large-budded cells (bud equal to or larger than one-third of the mother cell), and other cells (cells with protruded or multiple buds) was recorded. Differential interference contrast microscopy was carried out on the rrm3 srs2 mutant strain to record typical morphological abnormalities (Nikon Eclipse TE300). Digital images were acquired and analyzed using the MetaMorph imaging system (Universal Imaging Corporation).
RESULTS
Rrm3 acts in a network with Sgs1 and Srs2 to process recombination intermediates.
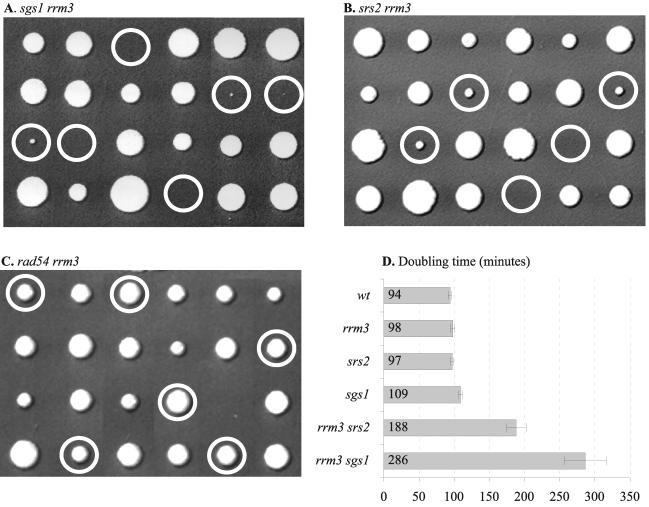
The Srs2 and Sgs1 helicases have been suggested to act in two alternative pathways that prevent the accumulation of recombination intermediates generated from single-stranded gaps thought to arise as a result of impaired DNA replication (17). The observed interaction between Rrm3 and PCNA in combination with the observation that rrm3 mutations do not cause an overt DNA replication defect raised the possibility that the Rrm3 helicase may function during DNA replication in pathways similar to those involving Sgs1 and Srs2. This prompted us to investigate functional relationships between Rrm3, Srs2, and Sgs1. Dissection of tetrads produced by sporulation of diploids that were heterozygous for sgs1 and rrm3 mutations revealed that rrm3 sgs1 haploids exhibited a severe growth defect (Fig. 2A). The majority of those haploids predicted by segregation analysis of genetic markers to contain rrm3 and sgs1 mutations were viable but formed very small colonies (Fig. 2A). A less severe negative genetic interaction was similarly observed between rrm3 and srs2 (Fig. 2B). Doubling times of rrm3 sgs1 and rrm3 srs2 mutants and appropriate controls were determined in liquid YPD medium, and the results are shown in Fig. 2D. The wild-type strain (94 ± 2 min), the rrm3 mutant (98 ± 1 min), and the srs2 mutant (97 ± 3 min) had similar doubling times, whereas an sgs1 mutation caused a slight growth delay (109 ± 3 min). When RRM3 was deleted in srs2 and sgs1 mutants, the doubling times increased significantly, to approximately 188 ± 20 and 286 ± 32 min, respectively, indicating that Rrm3 is required for normal growth in the absence of Srs2 and even more so in the absence of Sgs1.
FIG. 2.
Cells lacking Rrm3 and Srs2 or Rrm3 and Sgs1 exhibit a severe growth defect and low viability (A, B, and D). Unlike Srs2, Rrm3 is not required for the viability of rad54 mutants (C). Tetrads from diploids heterozygous for rrm3 and sgs1 (A), rrm3 and srs2 (B), and rrm3 and rad54 (C) were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3, sgs1::HIS3, srs2::TRP1, and rad54::TRP1). Circles indicate rrm3 sgs1, rrm3 srs2, and rrm3 rad54 mutants. Doubling times of the mutant strains and appropriate controls in rich medium (YPD) are shown in panel D.
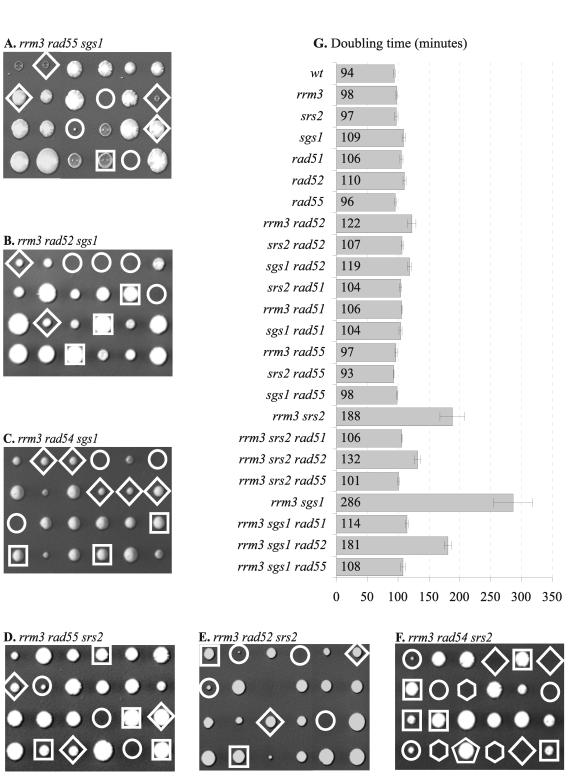
Since the severe growth defect of srs2 sgs1 mutants is suppressed by defects in homologous recombination (21), we tested if mutations in homologous recombination genes could suppress the growth defects of rrm3 sgs1 and rrm3 srs2 mutants. For this purpose, we analyzed the meiotic products obtained from diploids that were heterozygous for rrm3, sgs1 and rad51, rad52, rad54, or rad55 deletions as well as diploids that were heterozygous for rrm3, srs2 and rad51, rad52, rad54, or rad55 deletions. We observed that deletion of RAD51 or RAD55 fully suppressed the growth defects of both sgs1 rrm3 and srs2 rrm3 double mutants, resulting in increased recovery of double mutant colonies with normal growth rates (Fig. 3A, D, and G). In contrast, deletion of RAD52 failed to fully restore normal growth rates (Fig. 3B, E, and G), reducing the doubling time of the rrm3 srs2 mutant from ∼188 to ∼132 min and that of the rrm3 sgs1 mutant from ∼286 to ∼181 min (Fig. 3G). A similar observation has recently been made for the top3 mutant (∼262 min), whose severe growth defect is suppressed by a rad51 or rad55 deletion (∼155 min) more than by a rad52 deletion (191 min) (60). Differential suppression of rrm3 srs2 and rrm3 sgs1 growth defects by rad51 and rad52 deletions has also been reported by Torres et al. (65).
FIG. 3.
Effects of deletions of homologous recombination genes on the viability of rrm3 sgs1 and rrm3 srs2 mutants. (A, B, and C) Tetrads from diploids heterozygous for rrm3, sgs1, and either rad55, rad52, or rad54 were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3, sgs1::HIS3, and rad::TRP1). (D, E, and F) Tetrads from diploids heterozygous for rrm3, srs2, and either rad55, rad52, or rad54 were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3, srs2::HIS3, and rad::TRP1). Circles indicate rrm3 sgs1 and rrm3 srs2 mutants; diamonds indicate rrm3 sgs1 rad and rrm3 srs2 rad mutants; squares indicate rad52, rad54, and rad55 single mutants; pentagons indicate rrm3 rad54 mutants; hexagons indicate srs2 rad54 mutants. (F) Since srs2 rad54 spores and rrm3 srs2 rad54 spores are inviable, it was not possible to assign the genotypes with certainty when both spore types were present in the same tetrad; only one possible assignment is shown here. (G) Doubling times of the mutant strains and appropriate controls in rich medium (YPD) are shown with standard deviations. Deletion of RAD51 or RAD55 fully suppressed the growth defects of sgs1 rrm3 and srs2 rrm3 mutants, whereas deletion of RAD52 partially rescued these mutants.
Since synthetic lethality between srs2 and a rad54 deletion had previously been reported, we also analyzed the genetic interaction between rrm3 and rad54 and found that the rrm3 rad54 double mutant was viable (Fig. 2C) and that a rad54 deletion fully suppressed the rrm3 sgs1 growth defect (Fig. 3C). However, when we dissected 24 tetrads obtained from a diploid that was heterozygous for srs2, rrm3, and rad54 mutations, rad54 srs2 double mutants were not obtained (8 predicted), which was consistent with previous results (37, 56), and srs2 rrm3 rad54 triple mutants were also not obtained (14 predicted) (Fig. 3F). Microscopic inspection revealed that the predicted srs2 rad54 and rrm3 srs2 rad54 spores had divided only a few times to form clusters of fewer than 100 cells. These observations indicated that the severe growth defect of rrm3 sgs1 and rrm3 srs2 mutants is caused by homologous recombination. However, the incomplete rescue by a rad52 deletion suggested that a Rad52-dependent, Rad51-independent recombination pathway may be utilized for the repair of a subset of lesions in rrm3 srs2 and rrm3 sgs1 mutants.
Cells lacking Rrm3 and Srs2 frequently arrest with protruded or multiple buds.
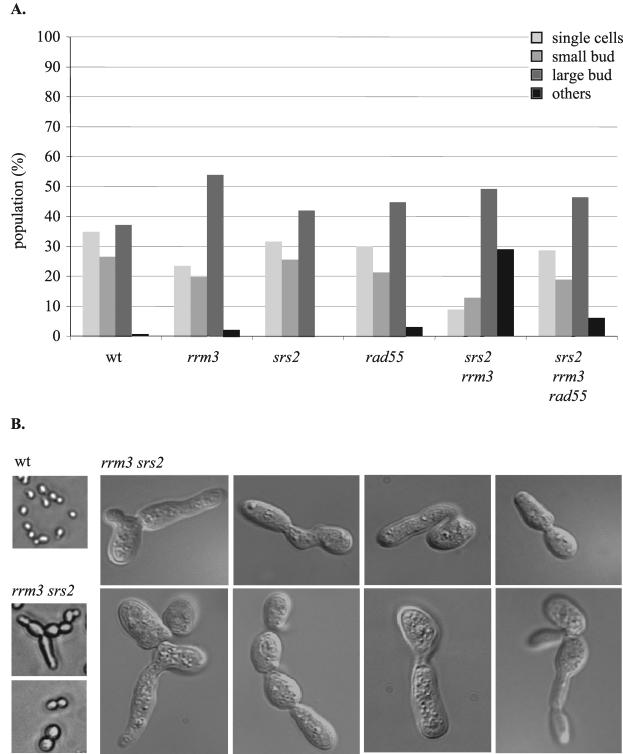
When the inferred rrm3 srs2 and rrm3 sgs1 double mutants were inspected microscopically, we found that spores that did not develop into colonies had arrested after a few cell divisions or as single cells with multiple and protruded buds. Because the viable rrm3 sgs1 double mutants exhibited a very severe growth defect, we did not consider it feasible to analyze them further. In contrast, the higher viability and less severe growth defect of the rrm3 srs2 double mutant permitted a more detailed analysis. Cultures of wild-type cells, srs2, rrm3, rrm3 srs2, and rrm3 srs2 rad55 mutants were grown to mid-logarithmic phase (OD650 of ∼0.7 to 1) and analyzed microscopically (Fig. 4). This analysis showed that 49% of rrm3 srs2 cells were present as large-budded cells typical of the G2 and M phases of the yeast cell cycle, while only 9% of cells were in G1 phase (single cells) and 13% had entered S phase (small-budded cells). The remaining 29% of the cells had an abnormal appearance and had protruded and/or multiple buds suggestive of a replication defect (27). Examples of these abnormal rrm3 srs2 cells are shown in Fig. 4B. In contrast, 37% of wild-type cells had large buds, 35% were single cells, 27% had small buds, and cells with multiple or protruded buds were extremely rare (1 of 179). Both rrm3 and srs2 mutants showed a slight increase in the percentage of large-budded cells (54 and 42%, respectively) compared to the wild-type culture (37%), but cells with multiple and protruded buds were still rare (13 of 560 and 0 of 298, respectively), and most of these were large-budded cells with a single protruded bud rather than the multiply budded cells that were typical of rrm3 srs2 double mutants. The decrease in G1- and S-phase cells in rrm3 srs2 double mutant cultures compared to that in wild-type cells and in rrm3 or srs2 single mutants was accounted for by the presence of the multiply budded rrm3 srs2 cells. The rrm3 srs2 rad55 mutant had a distribution of single, small-budded, and large-budded cells that was similar to that seen for wild-type cells and rrm3 or srs2 single mutants and had a significantly reduced proportion of cells with multiple or protruded buds (6%) compared to the rrm3 srs2 mutant. As in cultures established from single mutants, the abnormal cells in rrm3 srs2 rad55 cultures were mainly large-budded cells with a single protruded bud rather than chains of elongated cells with multiple deformed buds found in rrm3 srs2 cultures. These results are consistent with the idea that rrm3 srs2 cells undergo some type of arrest at G2/M as the result of a recombination-dependent process.
FIG. 4.
G2/M arrest and morphological abnormalities of rrm3 srs2 mutants are caused by homologous recombination. (A) Mutants were grown to mid-logarithmic phase and analyzed microscopically for the presence of single cells, small-budded cells (bud smaller than one-third of the mother cell), large-budded cells (bud equal to or larger than one-third of the mother cell), and other cells (malformed cells with protruded or multiple buds). The numbers of cells scored were as follows: wild-type (wt), 179; rrm3, 560; srs2, 298; rad55, 393; rrm3 srs2, 530; rrm3 srs2 rad55, 248. (B) rrm3 srs2 mutants are characterized by an increase in cell size (left panel) and the formation of protruded and multiple buds. Magnification: left panel, ×600; right panel (differential contrast interference microscopy), ×1,000.
Cells lacking Rrm3 and Srs2 frequently fail to undergo nuclear division.
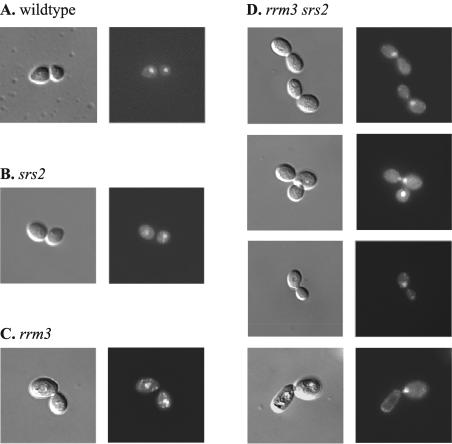
To determine the cause of the G2/M arrest of rrm3 srs2 mutants, the DNA distribution in large-budded and abnormal rrm3 srs2 cells was analyzed by staining with DAPI (Fig. 5). DAPI staining revealed that large-budded rrm3 srs2 mutants with apparently normal morphology frequently had only one nucleus that had migrated to the bud site but had failed to undergo nuclear division (Fig. 5D). In some cases, the anuclear bud showed signs of malformation. Moreover, many buds in the multiply budded cells were anuclear, suggesting that these cells were terminally arrested. The large number of cells with abnormal DNA distribution suggested that rrm3 srs2 cells frequently arrest in G2/M-phase either due to a failure to completely duplicate their genome or failure to segregate their chromosomes prior to cell division.
FIG. 5.
Abnormal DNA distribution in large-budded rrm3 srs2 mutants. Wild-type cells (A) and srs2 (B), rrm3 (C), and rrm3 srs2 (D) mutants were fixed and stained with DAPI to detect DNA. The number of large-budded cells with only a single nucleus at the bud site or a bowtie nucleus, indicative of mitotic arrest, was consistently increased among cells lacking both Rrm3 and Srs2 compared to wild-type cells or cells lacking either Rrm3 or Srs2. For example, in one experiment 73% (16 of 22) of large-budded rrm3 srs2 mutants had a single nucleus. Anuclear buds eventually deteriorate and become protruded.
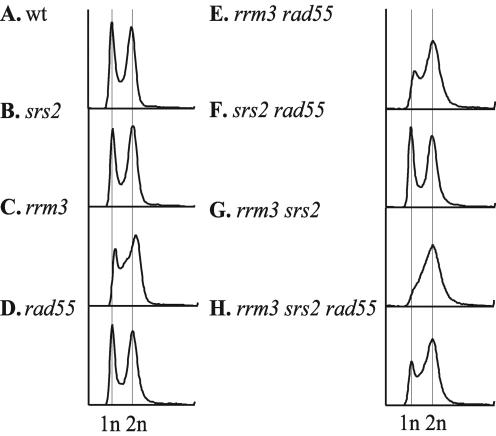
Cells lacking Rrm3 and Srs2 accumulate in G2/M phase with a 2N DNA content.
To ascertain the capability of rrm3 srs2 mutants to replicate their genome, the DNA content of logarithmically growing rrm3 srs2 cells was analyzed using FACS. The FACS profiles revealed that wild-type cells and the srs2 and rad55 mutants had a similar distribution of cells with 1N (G1 phase) and 2N (G2/M phase) DNA content, whereas the rrm3 mutant showed a modest increase in the proportion of cells with an intermediate DNA content (Fig. 6A to D), suggesting that in the absence of Rrm3 cells may take longer to complete S phase. The DNA content distribution of rrm3 and srs2 mutants was not affected by deletion of the RAD55 gene (Fig. 6E and F). In contrast, the FACS profile of the rrm3 srs2 mutant showed only a single peak consistent with a population of cells that have a 2N DNA content, while the 1N peak (cells in G1 phase) was virtually absent (Fig. 6G). This result suggests that the rrm3 srs2 cells completed genome duplication but failed to undergo nuclear and cellular divisions at normal rates, leading to an accumulation of G2/M cells and the absence of cells in G1 phase. Introduction of a rad55 mutation into the rrm3 srs2 strain caused the reappearance of a G1 population of cells (Fig. 6H) and resulted in a FACS profile that was indistinguishable from that obtained for either the rrm3 mutant or the rrm3 rad55 double mutant. This suggests that aberrant homologous recombination in cells lacking Rrm3 and Srs2 results in the accumulation of DNA structures that underlie the accumulation of G2/M cells.
FIG. 6.
rrm3 srs2 mutants accumulate in G2/M phase with a 2N or larger DNA content. Wild-type cells and mutants were grown to mid-logarithmic phase, fixed, and stained with Sytox Green to measure the DNA content by FACS analysis. Wild-type cells (A) and srs2 (B), rad55 (D), and srs2 rad55 (F) mutants showed similar FACS profiles. rrm3 (C) and rrm3 rad55 (E) mutants showed an increased percentage of cells in S phase. (G) rrm3 srs2 mutants accumulated in G2/M phase with a 2N or larger DNA content while being devoid of a G1 (1N DNA content) population. (H) Introduction of a rad55 mutation suppressed the G2/M arrest of rrm3 srs2 mutants, as signified by the presence of a G1 population.
Rrm3 has an additional role outside of homologous recombination.
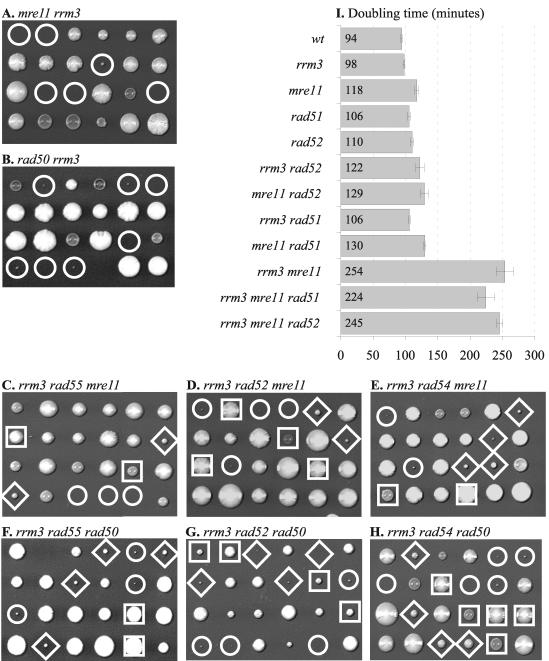
To further elucidate the role of Rrm3 in recombinational repair, we tested whether Rrm3 was required in cells lacking Mre11 or Rad50, both of which are subunits of the Mre11-Rad50-Xrs2 (MRX) complex. The MRX complex is required for multiple pathways of double-strand break repair, including homologous recombination and nonhomologous end joining (reviewed in references 10 and 13). In addition, the MRX complex has also recently been implicated in DNA replication and activation of S-phase checkpoints (14, 23, 45, 67), suggesting a central role for the MRX complex in DNA damage responses and the maintenance of genome integrity. When mre11 and rad50 haploids were crossed with an rrm3 haploid and the meiotic products were analyzed, we found that rrm3 mre11 and rrm3 rad50 double mutants exhibited a severe growth defect (Fig. 7A and B). Deletion of homologous recombination genes (RAD51, RAD52, RAD54, and RAD55) could not restore normal growth to the rrm3 mre11 and rrm3 rad50 mutants, as judged by colony size (Fig. 7C to H). When we determined doubling times in liquid YPD medium, we found that the rrm3 mre11 rad52 mutant (245 ± 4 min) grew as slowly as the rrm3 mre11 mutant (254 ± 13 min) and deletion of RAD51 caused a minor improvement (224 ± 13 min). We obtained similar results when we analyzed the meiotic products of a diploid strain that was derived from a different genetic background (YPH501; gift from V. Zakian, Princeton University) and was heterozygous for rrm3, mre11, and rad51 mutations. These findings demonstrate that a functional MRX complex is required for normal growth of rrm3 mutants and that, in contrast to rrm3 srs2 and rrm3 sgs1 mutants, aberrant homologous recombination is not the main cause of the growth defect of rrm3 mrx mutants.
FIG. 7.
The severe growth defect and decreased viability of rrm3 mre11 and rrm3 rad50 mutants were partially suppressed by rad52, rad54, and rad55 mutations. Tetrads from diploids heterozygous for rrm3 and mre11 mutations (A) or rrm3 and mre11 and either rad55 (C), rad52 (D), or rad54 (E) mutations were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3, mre11::HIS3, and rad::TRP1). Similarly, tetrads from diploids heterozygous for rrm3 and rad50 mutations (B) or rrm3 and rad50 and either rad55 (F), rad52 (G), or rad54 (H) mutations were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3, rad50::HIS3, and rad::TRP1). Circles indicate rrm3 mre11 and rrm3 rad50 mutants; diamonds indicate rrm3 mre11 rad and rrm3 rad50 rad mutants; squares indicate rad52, rad54, and rad55 single mutants. Doubling times of the mutant strains and appropriate controls in rich medium (YPD) are shown with standard deviations. Deletion of RAD52 did not rescue the rrm3 mre11 mutant, and deletion of RAD51 resulted in a very minor improvement of the growth rate.
Mus81 is not required in the absence of Rrm3.
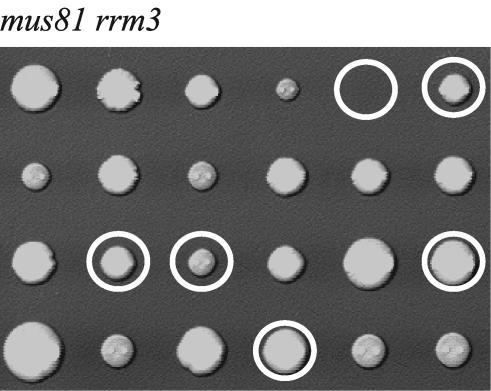
The Mus81/Mms4 complex has been implicated in DNA repair (30) and has recently been shown to possess structure-specific endonuclease activity (3, 8, 26, 34, 73). Mutations in MUS81 and MMS4 are lethal in strains lacking Sgs1 helicase (17, 48), and the suppression of the synthetic lethality by rad51, rad52, and rad54 mutations shows that this endonuclease acts during processing and/or resolution of recombination intermediates in a different pathway than Sgs1 (17). In contrast, the srs2 mus81 double mutant was found to be viable (17), suggesting that Srs2 and Mus81 may act in the same pathway. When the genetic interaction between rrm3 and mus81 deletions was analyzed, we found that rrm3 mus81 double mutants were viable (Fig. 8). Colonies formed by rrm3 mus81 mutants were of the same size as those formed by the single mutants, suggesting no negative genetic interaction between rrm3 and mus81. Therefore, Mus81 endonuclease may be needed to resolve DNA structures when Sgs1 is lacking, but not when Rrm3 or Srs2 are lacking. The normal growth of mus81 and rrm3 mus81 mutants, as judged by the size of colonies formed by mutant spores on dissection plates, as opposed to the severe growth defect of the rrm3 srs2 mutant, is consistent with the exclusive assignment of the Mus81 endonuclease complex to a pathway that is only required in the absence of Sgs1 (17).
FIG. 8.
Unlike Sgs1, Rrm3 is not required in cells lacking Mus81 endonuclease. Tetrads from diploids heterozygous for rrm3 and mus81 mutations were dissected and analyzed for the presence of auxotrophic markers (rrm3::URA3 and mus81::HIS3). Circles indicate rrm3 mus81 mutants.
DISCUSSION
Previous studies (17, 21) have led to the hypothesis that potentially toxic recombination intermediates arise during DNA replication and that two pathways, one involving a 3′-to-5′ DNA helicase, Sgs1, and the other involving a different 3′-to-5′ DNA helicase, Srs2, and the structure-specific endonuclease Mus81-Mms4, act in parallel to somehow process these intermediates. Thus, double mutants where both pathways are inactivated (e.g., sgs1 srs2 or sgs1 mus81) show a severe growth defect that is suppressed by a mutation that inactivates a gene encoding an early recombination function, like RAD51, RAD52, or RAD55. In this study, we found that a mutation in RRM3, which encodes a 5′-to-3′ DNA helicase that interacts with the replication factor PCNA, caused a severe growth defect when combined with either sgs1 or srs2 mutations and that in each case the growth defect was suppressed by mutations that inactivated genes encoding early recombination functions (summarized in Table 2). Synthetic lethality between sgs1 and rrm3 mutations was first suggested by Tong et al. (64) and by Ooi et al. (51), who performed a synthetic genetic array analysis and a synthetic lethality analysis by microarray, respectively, to identify mutations that are colethal with an sgs1 mutation. Recent work in the laboratory of V. Zakian (Princeton University) that is presented in an accompanying paper has also led to similar conclusions about Rrm3 (65). Morphological and FACS analysis of log-phase cultures of the rrm3 single mutant suggested that rrm3 mutants had a small delay in transiting S phase and a small increase in the proportion cells present in G2/M. In contrast, rrm3 srs2 mutant cells accumulated in G2/M as large-budded cells with a single nucleus and 2N DNA content, and this arrest phenotype was completely suppressed by rad51 and rad55 mutations, consistent with the view that the rrm3 srs2 growth defects are due to the accumulation of toxic recombination intermediates resulting from some type of aberrant replication structure. Unlike an sgs1 mutation, an rrm3 mutation did not result in a severe growth defect when combined with a mus81 mutation and, unlike an srs2 mutation, an rrm3 mutation did not result in lethality when combined with a rad54 mutation.
TABLE 2.
Summary of rrm3 interactions examined in this study
| Genotype | Phenotype | Extent of suppression when combined with indicated mutation
|
||
|---|---|---|---|---|
| Complete | Partiala | None | ||
| rrm3 srs2 | Very slow growth | rad51, rad55 | rad52 | rad54 |
| rrm3 sgs1 | Extremely slow growth | rad51, rad54, rad55 | rad52 | |
| rrm3 mre11 | Extremely slow growth | rad51 | rad52 | |
| rrm3 rad51 | Normal growthb | |||
| rrm3 rad52 | Normal growth | |||
| rrm3 rad54 | Normal growth | |||
| rrm3 rad55 | Normal growth | |||
| rrm3 mus81 | Normal growth | |||
Partial suppression was detected when doubling times were measured; colony size was minimally affected by these mutations.
For genotypes that resulted in a normal growth phenotype, the suppression tests were not applicable.
An rrm3 mutation caused a strong growth defect when combined with either an mre11 or rad50 mutation. But in contrast to the full rescue of sgs1 rrm3 and srs2 rrm3 mutants by rad51 and rad55 mutations, we found that rrm3 mre11 and rrm3 rad50 mutants were not rescued by disrupting homologous recombination. Similarly, it was previously shown that sgs1 mutants depend on the MRX complex in order to maintain normal growth and that the growth defect of sgs1 mrx mutants is only partially suppressed by a rad51 mutation (60). Our observations suggests that Rrm3 has a role in at least one other pathway outside of homologous recombination that involves the MRX complex. In addition to homologous recombination, the MRX complex is also required for nonhomologous end joining, telomere maintenance, and checkpoint functions (5, 10, 13, 14, 25, 66). Rrm3 has previously been shown to be required during replication of telomeres and subtelomeric Y′ elements (32). It is therefore plausible that the growth defect of rrm3 mre11/rad50 mutants may be caused by a deficiency in telomere maintenance. However, since both Sgs1 and Srs2 have important checkpoint functions, it is possible that Rrm3 may have a comparable, yet undiscovered, checkpoint function. If so, cells lacking the Mre11 complex, which is required to detect and modify DNA damage, may have to rely more heavily on the Rrm3, Srs2, and Sgs1 pathways that process DNA lesions that arise during DNA replication, thereby signaling the presence of DNA damage. Alternatively, some type of DNA damage might arise in an rrm3 mutant that requires the Mre11 checkpoint for complete repair. Genetic interactions between Rrm3 and the MRX complex in telomere maintenance or checkpoint function, which are likely to require the ability of Mre11 to recognize, bind, and anneal DNA ends rather than depend on its nuclease activity to process double-strand breaks, is supported by previous evidence that the nuclease activity of Mre11 is not required for the viability of srs2 mutants, whereas a deletion of the MRE11 gene is lethal in combination with an srs2 mutation (37).
The absence of Rrm3 in srs2 and sgs1 mutants caused frequent cell death or a severe growth defect that was characterized by the presence of cells with protruded and multiple buds. A detailed analysis of the growth defect of the rrm3 srs2 mutant using FACS and morphological analysis of log-phase cultures revealed the accumulation of morphologically abnormal cells in the G2/M phase of the cell cycle and a greatly reduced proportion of cells in the G1 and S phases. These cells had a 2N DNA content and a single nucleus that had migrated to the bud site; however, nuclear division and cytokinesis frequently failed, consistent with mitotic arrest or delay prior to anaphase. The observation of a bowtie nucleus in some large-budded rrm3 srs2 mutants (Fig. 5D) indicated that some cells had proceeded through the early phases of mitosis. The majority of cells lacking Rrm3 and Srs2 had 2N DNA content, and there was a virtual absence of cells with 1N DNA content. In addition, an increased number of cells with 4N or larger DNA content were detected by FACS analysis of rrm3 srs2 mutants compared to wild-type cells and the rrm3 and srs2 single mutants (data not shown). Taken together, these observations suggest that in the absence of Rrm3 and Srs2, cells either delay prior to mitosis due to arrest at the G2/M checkpoint or occasionally enter a new round of the cell cycle, including genome replication without completing cell division. The restoration of growth and the reappearance of the G1 cell population characteristic of normally cycling rrm3 mutant cells by disrupting early steps of homologous recombination in rrm3 srs2 double mutants suggest that this G2/M arrest is due to homologous recombination that occurs in the absence of Rrm3 and Srs2.
Holliday junctions are typically thought to be formed during recombination and most notably during double-strand break repair by the invasion of 3′ overhangs of DNA double-strand breaks into an intact double-strand DNA followed by appropriate processing steps (62, 72). However, Holliday junctions are also generated at arrested replication forks in E. coli by regression of the replication fork and subsequent annealing of the two displaced nascent strands (59, 76), and in S. cerevisiae cells Holliday junctions are more frequent in S phase than in any other phase of the vegetative cell cycle (76). These results are consistent with the idea that aberrant replication structures leading to stalled replication forks result in either regression of the stalled forks or processing of the fork to a double-strand break followed by recombination, each of which would result in the formation of a branched DNA structure (12, 24, 38, 43, 54). A link between Rrm3 and DNA replication was previously suggested by the demonstration of an interaction between Rrm3 and PCNA (57). Furthermore, an accumulation of branched DNA structures has been observed when genomic DNA from rrm3 mutants was analyzed by two-dimensional gel electrophoresis (32) and, consistent with this, rrm3 mutations cause increased recombination between some types of duplicated sequences (36). FACS analysis of log-phase rrm3 mutants revealed a higher proportion of cells with a DNA content intermediate between 1N and 2N as well as a higher proportion of cells with 2N DNA content than in wild-type cells; this could result from a weak replication defect or an increase in aberrant replication structures in rrm3 mutants, which could each result in the Holliday junctions observed in rrm3 mutants. The observation that the cell cycle progression defect of rrm3 mutants is not suppressed by a rad55 mutation is consistent with the idea that the Holliday junctions seen in rrm3 mutants are formed by replication rather than recombination, although additional experimentation will be required to establish this. If this idea were correct, then Rrm3 could play at least two different roles in processing stalled replication forks: Rrm3 could act to prevent stalled replication forks from being converted into Holliday junctions, and Rrm3 could act in the processing of Holliday junctions once they are formed.
Previous studies have proposed two different models for how Srs2, Mus81, and Sgs1 might function in dealing with the consequences of aberrant replication structures (17). In one type of model, it has been proposed that DNA structures formed during replication are acted on by recombination proteins, leading to intermediates that are potentially toxic unless further processed. In this model, Srs2 along with Mus81-Mms4 and Sgs1 define two different pathways capable of processing these intermediates, such that absence of both pathways leads to a severe growth defect that can be suppressed by recombination defects. Our observations that rrm3 mutations cause a severe growth defect when combined with either an sgs1 or srs2 mutation but not a mus81 mutation and that this growth defect is suppressed by mutations that eliminate early recombination functions seem inconsistent with this model. In a second type of model, Sgs1 acts on replication intermediates to prevent them from being processed to a form that is acted on by recombination functions, leading to an intermediate whose correct processing requires Srs2 and Mus81. This model also accounts for the genetic interactions between mutations in SGS1, SRS2, MUS81, and various RAD genes.
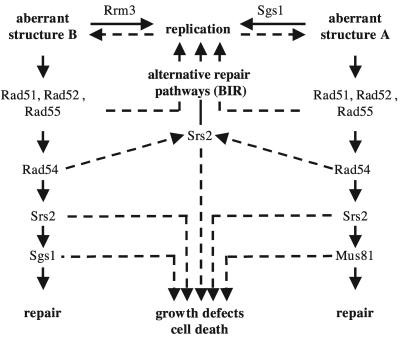
The data presented here on the genetics of RRM3 can be accounted for by a modification of this latter model (Fig. 9). The critical modification is the assumption that there are at least two possible aberrant replication structures that can form, one that is suppressed by the action of Rrm3 and one that is suppressed by the action of Sgs1. Consistent with the idea that Rrm3 prevents the formation of some type of aberrant replication structure, increased pausing of replication has been observed in rrm3 mutants (31). The intermediate suppressed by Rrm3 is acted on by early recombination functions in the absence of Rrm3 and is acted on by Sgs1 and Srs2. Similarly, the intermediate suppressed by Sgs1 is acted on by early recombination functions in the absence of Sgs1 and is acted on by Srs2 and Mus81. This model can account for all of the known genetic interactions between mutations in the genes analyzed and, in addition, is consistent with observations that Sgs1 assembles at replication forks in addition to its antirecombination function, while rrm3 mutations are known to cause some type of replication defect leading to increased stalled replication forks and Holliday junctions (19, 32, 33). The known antirecombination functions of Srs2 (39, 53, 68) and the ability of Mus81-Mms4 to cleave branched DNAs (3, 8, 26, 34, 73) are also consistent with this model. It is also possible to construct variations of this model with somewhat different roles for Srs2 and Mus81 relative to the two different intermediates postulated to arise from aberrant replication structures. However, critical to all variations on this type of model is the idea that there are at least two types of aberrant replication structures. Interestingly, the partial suppression of the rrm3 srs2 and rrm3 sgs1 growth defects by a rad52 deletion, as opposed to complete suppression by rad51 or rad55 deletions, suggests that Rad52-dependent mechanisms are required for the generation as well as for the repair of aberrant recombination intermediates in these helicase mutants. Based on their observation that an rrm3 sgs1 rad51 mutant that also lacks Rad59, a recombination protein required for certain types of break-induced replication (BIR), grows poorly, Torres et al. (65) propose that BIR, which can occur in the absence of Rad51 but not in the absence of Rad52, is the major pathway by which aberrant replication structures in rrm3 sgs1 and rrm3 srs2 mutants are repaired. As more members of these pathways are identified, it will be possible to put these models to more critical tests.
FIG. 9.
Model of the Rrm3/Sgs1/Srs2 network of DNA lesion avoidance and repair. Bold lines mark pathways normally utilized for DNA replication and repair, and dashed lines mark pathways used in the absence of the indicated protein function. For example, in the absence of Sgs1, aberrant replication structure A cannot be prevented and is converted by early homologous recombination functions (Rad51, Rad52, and Rad55/Rad57) into a potentially toxic recombination intermediate whose effective repair requires Srs2 and Mus81. Hence, sgs1 mutants lacking either Srs2 or Mus81 accumulate the toxic recombination intermediate, causing a severe growth defect. In the absence of Rad51 or Rad55, the toxic recombination intermediate is not formed and Srs2 and Mus81 are therefore not required, explaining, for example, the normal growth rate of the sgs1 srs2 rad51 strain. In strains lacking homologous recombination functions, the aberrant structure A is effectively processed by alternative DNA repair pathways and DNA replication can resume. If homologous recombination is disrupted at a later stage by the lack of Rad54, then Srs2 (but not Mus81) is required to process the recombination intermediate before alternative pathways can effectively repair it. Similarly, in the absence of Rrm3, formation of a different aberrant replication structure (B) cannot be prevented and is converted by early homologous recombination functions into a potentially toxic DNA structure that requires Srs2 and Sgs1 to be effectively repaired. Hence, rrm3 mutants lacking either Srs2 or Sgs1 accumulate this unresolved structure during DNA replication and exhibit a severe growth defect. As in sgs1 mutants, disruption of homologous recombination in rrm3 mutants prevents the formation of the toxic recombination intermediate and makes aberrant structure B accessible to alternative pathways. The partial suppression by a rad52 mutation, as opposed to full suppression by rad51 or rad55, indicates that Rad52 may be required for these alternative pathways, suggesting that some lesions may be repaired by BIR. The requirement of Sgs1 for both the reversal of aberrant replication structure A and the resolution of the potentially toxic recombination intermediate formed in the absence of Rrm3 is consistent with our observation that an rrm3 sgs1 mutant exhibits a more severe growth defect than an rrm3 srs2 mutant. Adapted from model 1 of Fabre et al. (17).
In conclusion, our results demonstrate that when homologous recombination is present, at least two of the three DNA helicases, Rrm3, Sgs1, and Srs2, are required for normal cell growth. Although the exact roles of these DNA helicases in these processes are unclear, Rrm3 and Sgs1 seem likely to function in an error avoidance mechanism at the replication fork, while Srs2 reverses potentially lethal recombination intermediates once they are initiated at DNA lesions. This function of Srs2 becomes essential if the incidence of DNA lesions is increased, for example, in the absence of Rrm3 or Sgs1 or when homologous recombination is defective due to the absence of Rad54. The demonstration of genomic instability in cells of more-complex eukaryotes, including Drosophila melanogaster (2, 40, 50), mouse (7, 44), and humans (22, 28), that carry mutations in Sgs1 homologues (DmBlm, mBlm, and hBlm, respectively) indicates that this repair network may be evolutionarily conserved and that yet-undiscovered Srs2 and Rrm3 homologues may play critical roles in multicellular organisms.
Acknowledgments
We thank Meng-Er Huang, Dan Mazur, and Christopher Putnam for helpful comments on the manuscript, Vincent Pennaneach for discussions, Karen Oegema and Ashad Desai for help with microscopy, Virginia Zakian and Jorge Torres (Princeton University) for communication of unpublished results, and Dennis Young for FACS analysis.
This work was supported by National Institutes of Health grant GM26017 to R.D.K.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, M. D., M. McVey, and J. J. Sekelsky. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299:265-267. [DOI] [PubMed] [Google Scholar]
- 3.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, R. J., J. A. Sharp, and J. C. Wang. 1998. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem. 273:9644-9650. [DOI] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosh, R. M., Jr., and V. A. Bohr. 2002. Roles of the Werner syndrome protein in pathways required for maintenance of genome stability. Exp. Gerontol 37:491-506. [DOI] [PubMed] [Google Scholar]
- 7.Chester, N., F. Kuo, C. Kozak, C. D. O'Hara, and P. Leder. 1998. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 12:3382-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccia, A., A. Constantinou, and S. C. West. 2003. Identification and characterization of the human Mus81/Eme1 endonuclease. J. Biol. Chem. 278:25172-25178. [DOI] [PubMed] [Google Scholar]
- 9.Cobb, J. A., L. Bjergbaek, and S. M. Gasser. 2002. RecQ helicases: at the heart of genetic stability. FEBS Lett. 529:43-48. [DOI] [PubMed] [Google Scholar]
- 10.Connelly, J. C., and D. R. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 11.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, M. M. 2002. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 510:107-120. [DOI] [PubMed] [Google Scholar]
- 13.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillingham, M. S., M. Spies, and S. C. Kowalczykowski. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423:893-897. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, C. J., and A. G. Motulsky. 1996. Werner syndrome: entering the helicase era. Bioessays 18:1025-1027. [DOI] [PubMed] [Google Scholar]
- 17.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frei, C., and S. M. Gasser. 2000. RecQ-like helicases: the DNA replication checkpoint connection. J. Cell Sci. 113:2641-2646. [DOI] [PubMed] [Google Scholar]
- 19.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 22.German, J. 1995. Bloom's syndrome. Dermatol. Clin. 13:7-18. [PubMed] [Google Scholar]
- 23.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 24.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 25.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 26.Haber, J. E., and W. D. Heyer. 2001. The fuss about Mus81. Cell 107:551-554. [DOI] [PubMed] [Google Scholar]
- 27.Heyer, W. D., M. R. Rao, L. F. Erdile, T. J. Kelly, and R. D. Kolodner. 1990. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 9:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hickson, I. D., S. L. Davies, J. L. Li, N. C. Levitt, P. Mohaghegh, P. S. North, and L. Wu. 2001. Role of the Bloom's syndrome helicase in maintenance of genome stability. Biochem. Soc. Trans. 29:201-204. [DOI] [PubMed] [Google Scholar]
- 29.Huang, M. E., A. de Calignon, A. Nicolas, and F. Galibert. 2000. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38:178-187. [DOI] [PubMed] [Google Scholar]
- 30.Interthal, H., and W. D. Heyer. 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:812-827. [DOI] [PubMed] [Google Scholar]
- 31.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication fork progression past non-histone protein-DNA complexes. Mol. Cell 12:1525-1536. [DOI] [PubMed] [Google Scholar]
- 32.Ivessa, A. S., J. Q. Zhou, V. P. Schulz, E. K. Monson, and V. A. Zakian. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 34.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keil, R. L., and A. D. McWilliams. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135:711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein, H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein, H. L., and K. N. Kreuzer. 2002. Replication, recombination, and repair: going for the gold. Mol. Cell 9:471-480. [DOI] [PubMed] [Google Scholar]
- 39.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 40.Kusano, K., M. E. Berres, and W. R. Engels. 1999. Evolution of the RECQ family of helicases: a Drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics 151:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, S. K., R. E. Johnson, S. L. Yu, L. Prakash, and S. Prakash. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339-2342. [DOI] [PubMed] [Google Scholar]
- 42.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Muzi-Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopes, M., C. Cotta-Ramusino, G. Liberi, and M. Foiani. 2003. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell 12:1499-1510. [DOI] [PubMed] [Google Scholar]
- 44.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 45.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyagawa, K. 1998. Genetic instability and cancer. Int. J. Hematol. 67:3-14. [DOI] [PubMed] [Google Scholar]
- 47.Modesti, M., and R. Kanaar. 2001. Homologous recombination: from model organisms to human disease. Genome Biol. 2:1014.1-1014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 50.Ohhata, T., R. Araki, R. Fukumura, A. Kuroiwa, Y. Matsuda, and M. Abe. 2001. Cloning, genomic structure and chromosomal localization of the gene encoding mouse DNA helicase RECQL5β. Gene 280:59-66. [DOI] [PubMed] [Google Scholar]
- 51.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 52.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 53.Rong, L., F. Palladino, A. Aguilera, and H. L. Klein. 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 55.Schiestl, R. H., S. Prakash, and L. Prakash. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, K. H., K. L. Derry, and R. D. Kolodner. 2002. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 277:45331-45337. [DOI] [PubMed] [Google Scholar]
- 58.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 60.Shor, E., S. Gangloff, M. Wagner, J. Weinstein, G. Price, and R. Rothstein. 2002. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162:647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35:243-274. [DOI] [PubMed] [Google Scholar]
- 62.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 63.Taylor, A. F., and G. R. Smith. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423:889-893. [DOI] [PubMed] [Google Scholar]
- 64.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 65.Torres, J. Z., S. L. Schnakenberg, and V. A. Zakian. 2004. The Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24:3198-3212. [DOI] [PMC free article] [PubMed]
- 66.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 67.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 68.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 69.Vennos, E. M., and W. D. James. 1995. Rothmund-Thomson syndrome. Dermatol. Clin. 13:143-150. [PubMed] [Google Scholar]
- 70.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 72.West, S. C. 1995. Formation, translocation and resolution of Holliday junctions during homologous genetic recombination. Philos. Trans. R. Soc. London B 347:21-25. [DOI] [PubMed] [Google Scholar]
- 73.Whitby, M. C., F. Osman, and J. Dixon. 2003. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 278:6928-6935. [DOI] [PubMed] [Google Scholar]
- 74.Wu, L., and I. D. Hickson. 2002. RecQ helicases and cellular responses to DNA damage. Mutat. Res. 509:35-47. [DOI] [PubMed] [Google Scholar]
- 75.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]