The significance of interactions involving aromatic side-chains in stabilizing protein structure is well accepted, while the geometry and specificity of these interactions are more elusive. Hydrophobic clustering plays a significant role; these interactions can be distinguished as aromatic/aliphatic and aromatic/aromatic interactions, with the aromatic/aromatic interactions displaying two dominant geometries–edge-to-face (EtF) and parallel displaced (PD) stacking [1].
Cross-strand aryl/aryl pairings occur predominantly at non-H-bonded sites in β-sheets [2]. In β-hairpin models these have been found to be stabilizing at turn flanking positions [3]. The excised N-terminal hairpin of the B1 domain of Protein G has a Tyr/Phe pair at such a position and is required for hairpin formation [4]. Trpzip4 and its analogs display two EtF Trp/Trp interactions at non-H-bonded sites, the turn flanking pair accounts for the majority of stabilization. HP6 and HP7, two peptide series more remotely related to GB1, have also shown remarkable stability attributed to a turn flanking EtF Trp/Trp pair. In all of its incarnations the interaction has been seen to maintain a specific geometry, with the edge of the N-terminal Trp abutting the face of the C-terminal Trp.
The stability of HP7 and its truncated version, as well as chignolin [5], suggest that an EtF aromatic/aromatic interaction immediately flanking a turn sequence is particularly stabilizing in small β-hairpins. HP6V has the same stabilizing EtF interaction but a β-turn that is less favorable in systems with short β-strands, provided an excellent system to test the limit of the W/W EtF interaction. The first truncated peptide of this design, AW-SNGK-WT, displayed the usual CD exciton couplet, in fact larger than expected, and an upfield Trp Hε3 with a melting curve suggesting a Tm of circa 25°C, apparently more stable than Ac-WNPATGKW-NH2, the 8-mer with the optimized reversing loop. NOESY sequencing, however, indicated that the upfield Hε3 signal was in the C-terminal, rather than N-terminal Trp. A battery of small Trp containing peptides was examined to ascertain the determinants of the EtF geometry between the Trp sidechains. The transition was not found to possess an absolute boundary; a “middle ground” with two folded states corresponding to the two EtF geometries was observed.
A 2.7 ppm upfield shift (CSD) seen for the G8-HN of Ac-WTNGKWTG-NH2 (peptide WP) suggested a local aryl-amide interaction at the N-terminus.
Aryl-X-Gly i → i+2 interactions can act as modest structuring elements even in denatured proteins [6] and peptides, evidenced by Gly HN CSDs up to −1.4. Only in proteins have Aryl-XG shifts as large as that in peptide WP been observed.
Further Results and Discussion
Peptide Ac-W-NPATGK-W-NH2 maintained the Trp side-chain geometry seen in all of its longer predecessors indicating that the Trp/Trp flip reversal is, to some degree, dependent on loop length. But this doesn’t explain why a four residue loop can adopt either conformation. In the short strand peptides with four residue turns, mutations at the S-1 position and T1 positions did not alter the “reversed EtF” geometry, which has been confirmed by comparing NMR structure ensembles (Fig. 2). We therefore turned to strand length as the potential culprit and a series of mutants demonstrated that indeed the interaction geometry is dependent on strand length (Table 1).
Figure 2.
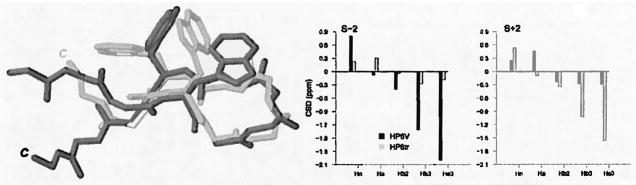
Figures 2a & b. Representative structures from the NMR ensembles of Ac-WINGKWTG-NH2 and KYVWINGKWTVE obtained exclusively from NOE distance constraints. Figure 2b displays the CSDs of the S+/−2 Trp residues at 280K.
Table 1.
CSDs that map the Trp flip transition.
| Sequence | S-2WHε3 | S+2WHε3 | S+4HN |
|---|---|---|---|
| Ac-W-NPATGK-W-NH2 | −0.87 | −0.17 | n.a. |
| KYVW- INGK -WTVE | −1.87 | −0.56 | 0.55 |
| ATW- INGK -WTG | −1.35 | −0.76 | 0.11 |
| Ac-TW- INGK -WTG-NH2 | −0.53 | −1.62 | −1.05 |
| TW- INGK -WTG | −0.41 | −1.92 | −0.48 |
| AW- SNGK -WT | −0.19 | −1.56 | n.a. |
| Ac-W- INGK -WT-NH2 | −0.39 | −1.68 | −2.70 |
| Ac-W- INGK -WTG-NH2 | −0.38 | −1.68 | −2.71 |
Synergy is observed between a W/W cross-strand interaction with flipped EtF geometry and an aryl-amide interaction at the C-terminus. The latter requires an indole ring that lies down on the C-terminal backbone. The aryl-amide interaction is disrupted by N-terminal extension of the hairpin (see Table 1), and is completely incompatible with the original EtF geometry.
Figure 1.
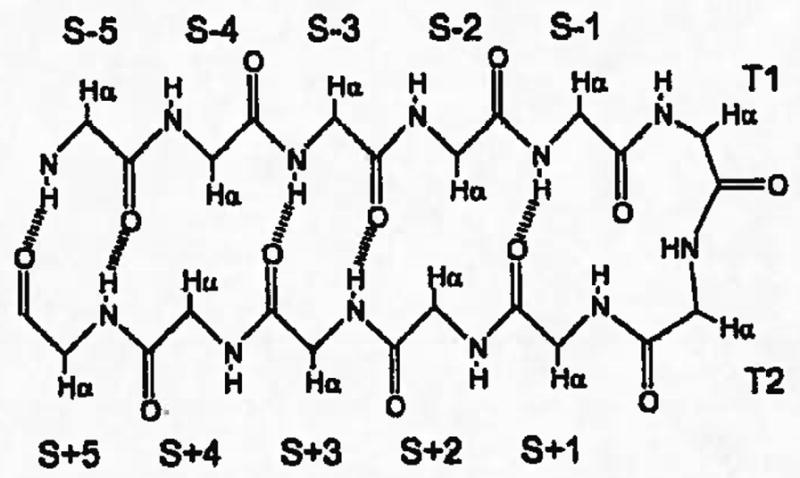
Nomenclature for residue postions in a β-hairpin with tight β turn.
Acknowledgments
The work was funded by NSF and NIH grants to Dr. Niels Andersen.
References
- 1.Guvench O, Brooks C. J of Am Chem Soc. 2005;127:4668–4674. doi: 10.1021/ja043492e. [DOI] [PubMed] [Google Scholar]
- 2.Jackups R, Jie Liang. J of Mol Biol. 2005;354:979–993. doi: 10.1016/j.jmb.2005.09.094. [DOI] [PubMed] [Google Scholar]
- 3.Andersen N, Olsen K, Fesinmeyer R, Tan X, Hudson F, Eidenschink L, Farazi S. J of Am Chem Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi N, Honda S, Yoshii H, Munekata E. Biochemistry. 2000;39:6564–6571. doi: 10.1021/bi000013p. [DOI] [PubMed] [Google Scholar]
- 5.Honda S, Yamasaki K, Sawada Y, Morii H. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Kemmink J, Mierlo C, Sheek R, Creighton T. J of Mol Biol. 1993;230:312–322. doi: 10.1006/jmbi.1993.1144. [DOI] [PubMed] [Google Scholar]


