Abstract
The reciprocal translocation t(12;21)(p13;q22), the most common structural genomic alteration in B-cell precursor acute lymphoblastic leukaemia in children, results in a chimeric transcription factor TEL-AML1 (ETV6-RUNX1). We identified directly and indirectly regulated target genes utilizing an inducible TEL-AML1 system derived from the murine pro B-cell line BA/F3 and a monoclonal antibody directed against TEL-AML1. By integration of promoter binding identified with chromatin immunoprecipitation (ChIP)-on-chip, gene expression and protein output through microarray technology and stable labelling of amino acids in cell culture, we identified 217 directly and 118 indirectly regulated targets of the TEL-AML1 fusion protein. Directly, but not indirectly, regulated promoters were enriched in AML1-binding sites. The majority of promoter regions were specific for the fusion protein and not bound by native AML1 or TEL. Comparison with gene expression profiles from TEL-AML1-positive patients identified 56 concordantly misregulated genes with negative effects on proliferation and cellular transport mechanisms and positive effects on cellular migration, and stress responses including immunological responses. In summary, this work for the first time gives a comprehensive insight into how TEL-AML1 expression may directly and indirectly contribute to alter cells to become prone for leukemic transformation.
Keywords: TEL-AML1, ETV6-RUNX1, translocation, B-cell leukaemia, precursor B-cell, SILAC
Introduction
The reciprocal translocation t(12;21)(p13;q22) is the most frequent chromosomal rearrangement in childhood B-cell precursor acute lymphoblastic leukaemia (ALL) with an incidence of ∼25%.1, 2 The resulting TEL-AML1 (syn.: ETV6-RUNX1) expression leads to expansion of B-cell precursors with enhanced self-renewal capacity and impaired differentiation to more mature B-cell stages.3, 4, 5, 6, 7, 8 The translocation already occurs in utero in an early B-cell progenitor cell and leads to establishment of a pre-leukemic clone persisting in the bone marrow for several years, insufficient to generate an overt leukaemia.3, 9, 10 It is unclear if the translocation inevitably leads to the disease or if only a small portion progress, as conflicting reports about the incidence of this translocation in healthy newborns exist.3, 11
The global binding pattern of the TEL-AML1 fusion protein on promoter regions in precursor B-cells is not known and several mechanisms of action have been proposed so far. The runt-homology DNA-binding domain of AML1 retained in the TEL-AML1 fusion protein has been shown to be essential for DNA binding.12 Transiently transfected TEL-AML1 blocks AML1-dependent transcription of several promoters with requirement of both, the TEL and AML1 part of the fusion protein.13, 14, 15 These studies proposed that the TEL moiety of the chimeric protein converts AML1 from an activator to a transcriptional repressor. However, alternative mechanisms of TEL-AML1 activity have also been suggested like sequestration of transcriptional cofactors to the cytoplasm16 or dimerization with wild-type protein.17, 18
Studies comparing either patients with and without the TEL-AML1 fusion19 or TEL-AML1-positive cell lines with small hairpin RNA-mediated knock down of the fusion protein indicated expression differences for genes involved in differentiation, apoptosis and immune responses.20, 21 The two latter studies did not find any enrichment for the canonical AML1-binding motif in regulated genes in the acquired mRNA data.
In this study, we sought to globally identify promoter regions targeted and regulated by the TEL-AML1 fusion protein. We wanted to differentiate between direct and indirect regulatory effects of the TEL-AML1 fusion protein in a cell system void of secondary aberrations as seen in patients and patient-derived cell lines by using chromatin immunoprecipitation (ChIP)-on-chip to identify promoter-binding sites in combination with mRNA microarray analysis to assess the gene regulatory effect of TEL-AML1. Furthermore, we analyzed the effect of TEL-AML1 expression on the protein output using stable isotope labelling by amino acids in cell culture (SILAC) to deduce indirect regulatory effects of TEL-AML1 independent of promoter binding.
Materials and methods
Patient samples, control samples and cell lines
Four bone marrow samples of patients with TEL-AML1-positive precursor B-cell leukaemia at the time of diagnosis and CD19+ MACS (Miltenyi Biotech, Bergisch Gladbach, Germany)-sorted cells of two healthy donors were obtained after informed consent. No cytogenetic aberration other than t(12;21) was detected in those patient samples. The BA/F3-derived inducible murine cell system was kindly provided by Anthony Ford and has been described elsewhere.22 BA/F3TA+ bearing the inducible TEL-AML1 plasmid as well as the empty vector control cells (BA/F3TA−) were induced by 32.5 pM mifepristone treatment for 16 h. NALM-6 (DSMZ ACC 128) and REH (DSMZ ACC 22) cells were cultured according to the provider. TEL-AML1 cDNA was generated with overlap extension PCR from TEL (primers: 5′-GGCGCTCGCGAATGTCTGAGACTCCTGCTCAG-3′, 5′-GGATTCATTCCAAGTATGCATTCTGCTATTCTCCCAATGGGCATGG-3′) and AML1 (primers: 5′-CCATGCCCATTGGGAGAATAGCAGAATGCATAC TTGGAATGAATCC-3′, 5′-CCGCGACTAGTTCAGTAGGGCCTCCACACGGCCTC-3′),23 cloned into the expression vector pMC324 and transfected into NALM-6 cells using DMRIE-C (Invitrogen, Darmstadt, Germany). Stable cell clones were selected using 400 μg/ml hygromycin.
Generation and testing of monoclonal antibody against TEL-AML1 (ETV6-RUNX1) fusion protein
A peptide comprising amino acids PIGRIAECILGM (Figure 1a) of TEL- AML1 fusion protein was synthesized and coupled to keyhole limpet hemocyanin and ovalbumin. Lou/C rats were immunized subcutaneously and intraperitoneally with a mixture of 50 μg peptide-keyhole limpet hemocyanin, 5 nmol CpG oligonucleotide (Tib Molbiol, Berlin, Germany), 500 μl phosphate-buffered saline and 500 μl incomplete Freund's adjuvant. A boost without adjuvant was given 6 weeks after the primary injection and fusion of the rat spleen cells with the murine cell line P3 × 63-Ag8.653 (ATCC CRL-1580) was performed after 3 days. Monoclonal antibodies that reacted specifically in ELISA with the immunized peptide were further analyzed in western blot analysis. Clone ‘TEL 6F2' of rat IgG2a subclass was purified with protein G affinity chromatography with Vivapure maxiprep spin columns (Sartorius, Goettingen, Germany) and concentrated by ultracentrifugation (MWco=10 kDa).
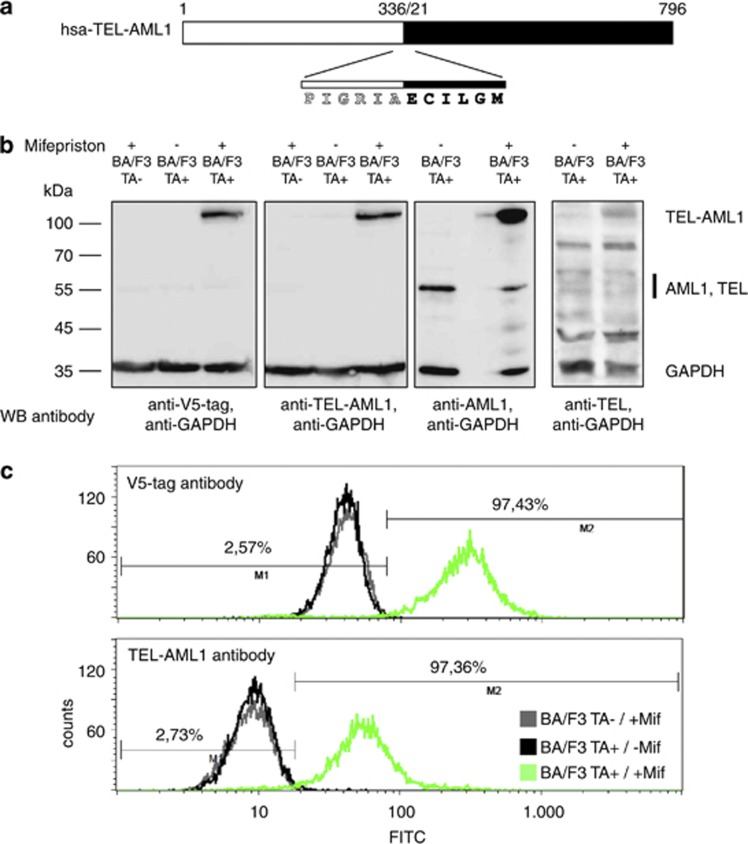
Figure 1.
TEL-AML1 antibody design and specificity testing. (a) Design strategy for the TEL-AML1 antibody. The immunization peptide spanning the fusion site between the TEL (white) and AML1 (black) fusion partners is indicated. (b) Specificity of the TEL-AML1 antibody. Western blots (WB) of the parental BA/F3 cell line (TA−) and stable cell lines carrying the inducible TEL-AML1 fusion construct (TA+) are treated with mifepristone as indicated. TEL-AML1 was specifically detected only in the induced cell lines, whereas the AML and TEL antibodies (right panels) detected both, the fusion protein and the native protein. Please note that the TEL antibody also detects numerous unspecific bands in the whole-cell lysates. (c) Detection of TEL-AML1 fusion protein by fluorescence-activated cell sorting (FACS) analysis. Induction with mifepriston resulted in on average 93.5±0.7% (n=15;±1 s.d.) cells carrying the TEL-AML1 fusion protein in FACS analysis using the TEL-AML1 antibody. A representative example is shown. Tightness of the induction system is shown in comparison to parental BA/F3 cells treated with mifepriston. Abbreviations: FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blotting and fluorescence-activated cell sorting analysis
Procedures were performed as directed by the manufacturer. For western blotting, antibodies directed against glyceraldehyde 3-phosphate dehydrogenase (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used. Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies against rat (R&D Systems, Minneapolis, MN, USA) and mouse (Santa Cruz Biotechnology) and detection was performed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA, USA). For fluorescence-activated cell sorting analysis, 500 000 cells were fixed and permeabilized using Cytofix/Cytoperm (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Cells were stained either with TEL-AML1 antibody and secondary anti-rat FITC-conjugated antibody (Santa Cruz Biotechnology) or with V5-tag antibody already FITC-conjugated (Abcam, Cambridge, UK).
ChIP and DNA amplification
ChIPs were performed as described elsewhere.25 Five microgram of TEL-AML1 antibody or rat isotype control IgG (Abcam) were incubated with protein G magnetic beads (Dynal, Invitrogen) pre-blocked with pre-immunization serum for 6 h at 4 °C. Beads were washed and incubated with DNA of 1 × 107 cells sheared to ∼500 bp length at 4 °C over night. The mouse endogenous TEL and AML1 proteins were immunoprecipitated with antibodies anti-TEL (N-19; Santa Cruz Biotechnology) and anti-AML1 (Ab-1; Calbiochem/Merck, Darmstadt, Germany). Enrichment of the previously described target promoter region of Gzmb26 in the samples was analyzed by quantitative real-time PCR using Sybr Green (Applied Biosystems, Foster City, CA, USA) and primers following standard procedure. Primer sequences were: Gzmb#1 forward 5′-GGACTCTGATACCATAGGCTA-3′, reverse 5′-GACTCAGAGAACCACCACTTA-3′; Gzmb#2 forward 5′-GTAAGTGGTGGTTCTCTGAGT-3′, reverse 5′-CCTGATGACGTCTTCTGAGTA-3′. Chromatin-immunoprecipitated DNA was amplified using the GenomePlex Complete Whole Genome Amplification Kit (Sigma-Aldrich Chemie GmbH, Munich, Germany). Labelling and microarray hybridization on 385K RefSeq mouse promoter arrays were performed by Source BioScience imaGenes (Berlin, Germany). Genes corresponding to a peak area with a false discovery rate ⩽0.05 as calculated from the log2 ratios between the TEL-AML1 ChIP experiment and input material were identified with NimbleScan software (Nimblegen Systems Inc., Madison, WI, USA). Promoter regions with at least 1.5-fold enrichment over the respective empty vector control were retained for further analysis. Data were submitted to Gene Expression Omnibus (GEO Accession number: GSE50736).
Gene expression analysis
Total RNA, isolated from 5 × 106 cells with standard TRIZOL procedure (Invitrogen), was labelled using a GeneChip 3′ in vitro transcription Express Kit (Affymetrix, Santa Clara, CA, USA) and hybridized to a GeneChip Mouse Genome 430 2.0 Array (Affymetrix) following standard procedure. Bioinformatic data analysis was performed using R 2.15 software,27 Bioconductor 2.1628 and biomaRt 2.12.29 Background correction and normalization of the probe signals were performed using the GCRMA package (v.2.28.0).30 mRNAs with a fold change of at least 1.5 between TEL-AML1 expressing and induced control cells and a P-value ⩽0.05 were considered significant and were retained for further analysis. Data were submitted to Gene Expression Omnibus (GEO accession number GSE50736). For human samples, 10 ng of RNA was labelled using Agilent Low Input Quick Amp Labelling Kit following the manufacturer's protocol (Agilent Technologies, Boeblingen, Germany). Hybridization was performed on the Agilent Whole Human Genome Oligo Microarrays 8 × 60K using Agilent Gene Expression Hybridization Kit (Agilent Technologies).
SILAC and mass spectrometric analysis
Cells were grown in cell culture medium deficient for L-arginine and L-lysine (SILAC RPMI, Thermo Fisher Scientific), supplemented with 2 mM L-glutamine (Gibco, Invitrogen), 10% dialyzed fetal bovine serum (Invitrogen) and amino acids of different molecular weight (0.389 mM L-arginine and 0.789 mM L-lysine; Sigma Aldrich) for at least five doublings. The ‘middle' medium was supplemented with L-arginine-13C6 and L-lysine-D4, whereas the ‘heavy' medium contained L-arginine-13C615N4 and L-lysine-13C615N2. Induction of TEL-AML1 expression was performed as described above. Cells were harvested 16 h after induction and pellets corresponding to 7 × 106 cells were flash frozen in liquid nitrogen. Mass spectrometric analysis of trypsin-digested size-fractionated proteins was performed as described elsewhere.31, 32 Peptides and proteins were identified with Mascot (Matrix Science, London, UK) and quantified with MSQuant (http://msquant.sourceforge.net) as described previously.32
Data and functional annotation analysis and correlation with gene ontology (GO)
For defined gene groups, we assigned GO terms, KEGG and Biocarta pathways, functional categories (COG_ontology, SP-PIR_keywords, UP_Seq_features), protein domains from interpro, PIR-superfamily and SMART databases using the DAVID bioinformatics resources 6.7, NIAID/NIH.33 Enrichment over the whole set of probed genes and heuristic multiple linkage clustering analysis was performed using the same tools and Benjamini-Hochberg correction was applied to correct for multiple testing on the data set.
Results
Identification of TEL-AML1 target genes
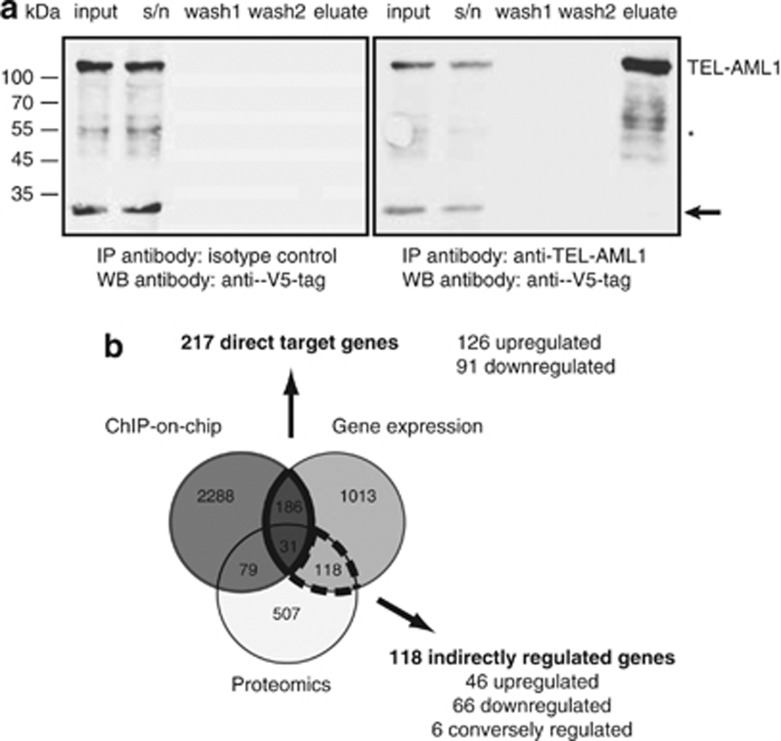
In an attempt to generate a monoclonal antibody detecting the TEL-AML1 fusion protein, we used a peptide immunization approach as outlined in Figure 1a. We confirmed a rapid, strong and stable TEL-AML1 expression after induction of the previously described murine BA/F3-related pro-B-cell line.22 In western blotting analysis, the antibody showed a strong detection of the fusion protein in BA/F3 cells (Figure 1b), similar to a V5-tag-specific antibody. On average, 93.5% of cells (±0.74; ±1 s.e.m.) exhibited expression of TEL-AML1 16 h after induction in fluorescence-activated cell sorting analysis, whereas non-induced cells did not show expression of the fusion protein (Figure 1c). However, in REH cells expressing the fusion protein at lower levels, we could also detect a band running at the height of either AML or TEL in western blotting (Supplementary Figure 1). We conclude that the antibody not only detects endogenous fusion protein at low levels, but also cross-reacts with AML1 in REH cells, but not in the inducible BA/F3 cell line system. Chromatin immunoprecipitation experiments of TEL-AML1-expressing BA/F3 cells were integrated with gene expression arrays and differential protein expression data (SILAC) to investigate and distinguish early direct target genes from indirect target genes of the chimeric transcription factor (outlined in Figure 2). The induction of TEL-AML1 results in binding to promoter regions of 2585 genes identified in ChIP experiments and also probed on the mRNA microarray platform, including granzyme B, a previously classified TEL-AML1 target.26 Of those 217 exhibited differentially expressed mRNA levels and were thus identified as direct targets of TEL-AML1 in our cell line model (Figure 3, Supplementary Table 1). As DNA binding of the fusion protein has been thought to be conferred through the domain of the AML1 part, we performed AML1 immunoprecipitation experiments in the same cell system. Unexpectedly, only 5 of the directly regulated promoters were also identified by AML1 ChIP, whereas 10 were identified by TEL ChIP in this study. Twelve promoter regions identified in AML1 ChIP-seq from another mouse pro-B-cell line BMiFLT3(15–3)34 overlapped with the here identified TEL-AML1 directly regulated promoters. Therefore, the majority (88%) of the identified gene promoter regions were specific for the TEL-AML1 fusion protein. The effect of TEL-AML1 promoter binding on protein output was assessed by SILAC. Surprisingly, only approximately one-third of the direct TEL-AML1 targets (31genes) showed a significant differential expression on protein level (Table 1). Even when considering only minor protein changes between induced and not induced cells, the overlap only increases by eight additional proteins. Indirect effects of TEL-AML1 overexpression were identified as 118 regulated proteins were also differentially regulated on the mRNA level but not immunoprecipitated by TEL-AML1 ChIP (Figure 3, Supplementary Table 2). The majority of regulated mRNAs and proteins (86.6%) were regulated in the same direction on protein and mRNA level. Rab8b was the only gene, whose mRNA was upregulated with concomitant downregulation of the protein upon TEL-AML1 stimulation. Interestingly, the 3′- untranslated region of this mRNA harbours a predicted binding site for microRNA miR-19a/b, both upregulated upon TEL-AML1 induction in our cell line model. We compared the TEL-AML1 occupancy between our mouse cell line model and the human precursor B-cell line NALM-6 stably transfected with TEL-AML1. Although the human cell line underwent 3 weeks of selection, many regions precipitated by ChIP of TEL-AML1-associated DNA overlapped (Figure 4).
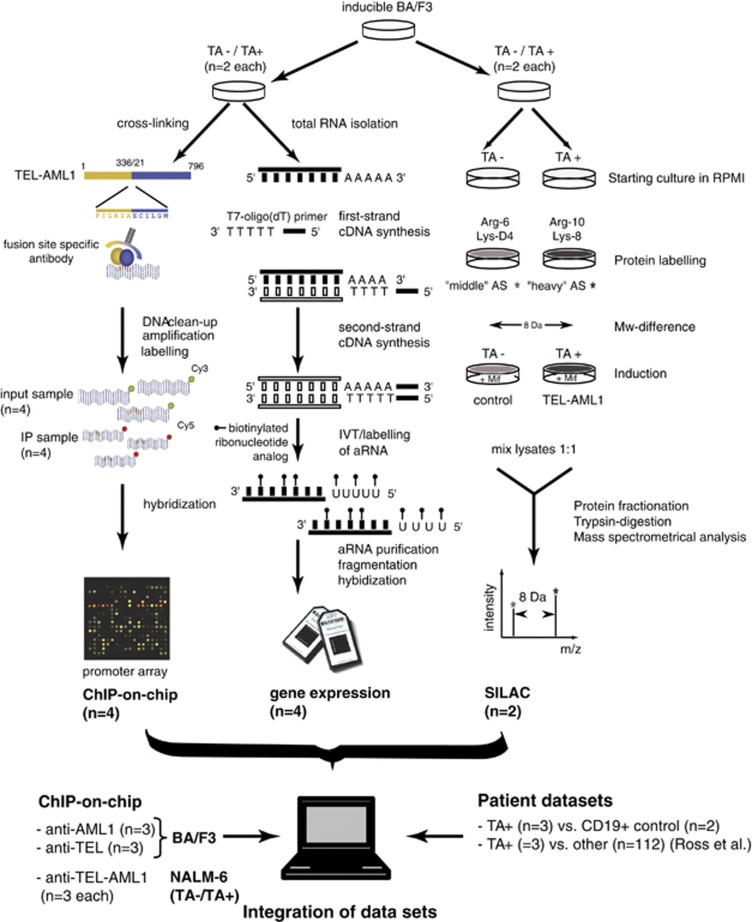
Figure 2.
Experimental design of the study. TEL-AML1 inducible BA/F3 cells were characterized by chromatin immunoprecipitation (ChIP-on-chip, left panel), gene expression microarrays (middle panel) or stable isotope labelling of amino acids in cell culture (SILAC, right panel). Empty vector controls (TA−) were compared with TEL-AML1-induced cell lines (TA+). In case of SILAC, both conditions were labelled with different molecular weight amino acids as indicated and analyzed by mass spectrometry. For ChIP-on-chip and gene expression microarrays results were obtained separately for induced and non-induced cells. The respective results were then compared with each other to identify differential regulated and expressed genes. Please note that both, ChIP and gene expression arrays, were performed on the very same cell culture populations in order to achieve high comparability of these two methods. For TEL-AML1, experiments were performed in biological duplicates, whereas ChIP-on-chip for TEL and AML1 were performed in triplicates. Abbreviation: IVT, in vitro transcription.
Figure 3.
TEL-AML1 chromatin immunoprecipitation in induced BA/F3 cells. (a) Western blot (WB) analysis of TEL-AML1 chromatin immunoprecipitation (right) show precipitation of TEL-AML1 compared with an unspecific isotype control (left). Light (arrow) and heavy (*) band of the IP antibodies are marked. (b) Intersections of the identified promoter regions bound by TEL-AML1 (ChIP-on-chip, dark grey circle), differentially expressed genes (light grey circle) and differentially expressed proteins (white circle). Directly regulated TEL-AML1 target genes were found in the intersection of ChIP-on-chip and gene expression analysis, whereas the intersection of gene expression and proteomics contained the indirectly affected genes. Abbreviations: IP, immunoprecipitation; WB, western blot.
Table 1. Directly regulated TEL-AML1 target genes with concomitant change in protein level.
| Mouse ensemble | Symbol | mRNA | Protein |
|---|---|---|---|
| gene ID | Log2-FC T/A+ vs T/A− | Ratio T/A+ vs. T/A− | |
| ensmusg21185 | 9030617O03Rik | 1.85 | 1.41 |
| ensmusg37405 | Icam1 | 1.26 | 2.43 |
| ensmusg24589 | Nedd4l | 1.16 | 2.12 |
| ensmusg15733 | Capza2 | 1.03 | 1.44 |
| ensmusg28559 | Osbpl9 | 0.95 | 2.10 |
| ensmusg29440 | Psmd9 | 0.75 | 1.18 |
| ensmusg03546 | Klc4 | 0.74 | 1.22 |
| ensmusg36112 | Metap2 | −0.61 | 0.80 |
| ensmusg04535 | Tax1bp1 | −0.66 | 0.61 |
| ensmusg56201 | Cfl1 | −0.7 | 1.18 |
| ensmusg18068 | Ints2 | −0.87 | 0.84 |
| ensmusg60093 | Hist1h4a | −0.89 | 1.21 |
| ensmusg69266 | Hist1h4b | −0.89 | 1.21 |
| ensmusg60678 | Hist1h4c | −0.89 | 1.21 |
| ensmusg61482 | Hist1h4d | −0.89 | 1.21 |
| ensmusg69274 | Hist1h4f | −0.89 | 1.21 |
| ensmusg60981 | Hist1h4h | −0.89 | 1.21 |
| ensmusg60639 | Hist1h4i | −0.89 | 1.21 |
| ensmusg67455 | Hist1h4j | −0.89 | 1.21 |
| ensmusg64288 | Hist1h4k | −0.89 | 1.21 |
| ensmusg69306 | Hist1h4m | −0.89 | 1.21 |
| ensmusg69305 | Hist1h4n | −0.89 | 1.21 |
| ensmusg91405 | Hist2h4 | −0.89 | 1.21 |
| ensmusg68851 | Hist4h4 | −0.89 | 1.21 |
| ensmusg59796 | Eif4a1 | −0.97 | 0.76 |
| ensmusg20706 | Ftsj3 | −1.08 | 0.78 |
| ensmusg32185 | Carm1 | −1.11 | 0.79 |
| ensmusg21692 | Dimt1 | −1.12 | 0.74 |
| ensmusg28273 | Pdlim5 | −1.21 | 0.60 |
| ensmusg05846 | Rsl1d1 | −1.29 | 0.79 |
| ensmusg27533 | Fabp5 | −1.42 | 0.75 |
Genes were identified in our BA/F3 cell line model using chromatin immunoprecipitation (ChIP)-on-chip and gene expression. Genes with changes in protein output as identified by stable isotope labelling by amino acids in cell culture (SILAC) are given. Please note that Cfl1 is downregulated on mRNA level, but exhibits higher protein levels because of enhanced protein stability.
Figure 4.
Comparison of immunoprecipitated chromosomal regions. Regions precipitated with the TEL-AML1 antibody from NALM-6 cells stably expressing the fusion transcript were mapped to human chromosomal regions (black bars) and compared with regions immunoprecipitated from the induced mouse Ba/F3 cell line (grey bars). The densities of regions are blotted horizontally.
Analysis of AML1-binding site enrichment
As most TEL-AML1-direct targets were not detected in AML1 ChIP assays, we tested whether the binding of the fusion protein to DNA was indeed conferred through the canonical AML1-binding motif ‘TGYGGTY' as expected from a previous study.12 To correct for different lengths of identified promoter regions in the ChIP experiments, we normalized the number of binding sites to 1 kb of promoter region. Promoter regions of the microarray used for the ChIP experiment carried on average 0.9 AML1-binding sites/1 kb (±0.4, ±1s.d.) and were used as background for statistical testing using a Student's t-test. Direct targets were significantly enriched over background in AML1-binding sites with 2.6 AML1-binding sites/1 kb (±1.7; ±1 s.d.; P-value <0.001), whereas promoters of genes whose mRNAs and proteins were regulated indirectly upon TEL-AML1 expression were not (0.9±0.5 AML1-binding sites/1 kb; mean±1 s.d.; P-value=0.144). As expected, those 31 genes, whose promoters and gene products were identified by all three methods showed highest enrichment in AML1-binding sites (2.3±0.9 AML1-binding sites/1 kb; P-value<0.001). We therefore concluded that indeed we could distinguish direct and indirect TEL-AML1 targets by our approach, whereas using solely information on mRNA expression would have obscured this result.
Functional annotation of TEL-AML1-regulated genes
To assess the global downstream functional consequence of TEL-AML1 transcriptional regulation, we investigated the functional annotation of TEL-AML1-regulated genes with concomitant change in protein output. Upregulated genes were enriched mainly in unspecific intracellular and cytoplasm structural components. In contrast, we could identify 48 GO terms and 9 functional protein terms enriched specifically for downregulated genes (Supplementary Table 3). Of those, 46 GO terms were also represented by genes identified as direct targets in this study. Translation-related GO terms were the most enriched (GO:0006412, P<0.001) represented by 20% of downregulated genes. GO term clustering (Supplementary Table 4) revealed downregulation of mRNAs with simultaneous protein changes of genes involved in a cascade of translational processes (cluster 1, 5 and 6), noncoding RNA biogenesis and metabolism (clusters 3 and 9) and methylation-related processes (cluster 8) aside from structural clusters. In all processes, direct targets of TEL-AML1 expression were also identified by TEL-AML1 immunoprecipitation, leading to a complex phenotype with a focus on translational processes as possible early events in the selective advantage of a pre-leukemic clone carrying the TEL-AML1 fusion protein.
Comparison with gene expression data of TEL-AML1-positive patients
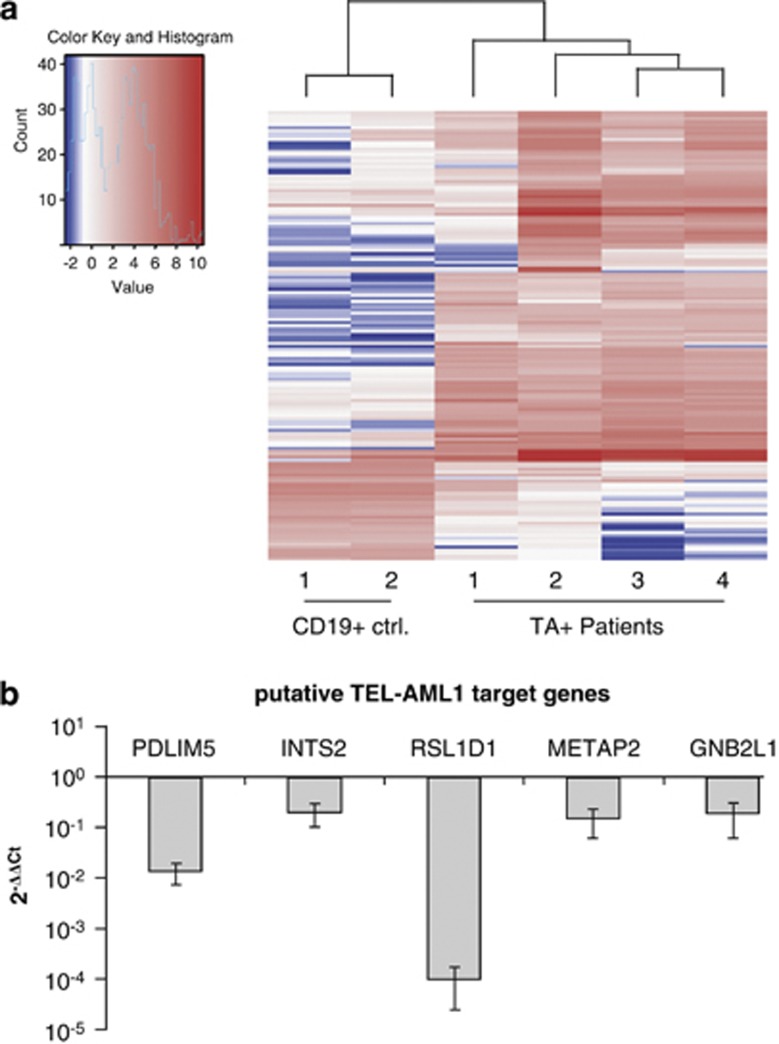
We next investigated whether the regulation of early direct TEL-AML1 targets identified in our mouse precursor B-cell model persist up to leukaemia-onset in paediatric patients. Therefore, we recorded the differential expression of mRNAs from children with precursor-B-ALL with TEL-AML1 fusion protein in comparison to normal CD19+ cells (Figure 5a, Supplementary Table 5). In addition, we used data from a previously published paediatric ALL patient cohort with a total of 132 primary specimen including 20 patients with TEL-AML1.35 Twenty-three of the downregulated and thirty of the upregulated mRNAs, which were directly targeted by TEL-AML1 in our model, were also concomitantly regulated in at least one of the two patient cohorts at significance level (Supplementary Table 6), again most of them not found in TEL or AML1 immunoprecipitation experiments. Four mRNAs were not uni-directional regulated in at least one of the patient cohorts and our cell line model. Quantitative reverse transcription-PCR of directly targeted mRNAs of five selected genes (PDLIM5, INTS2, RSL1D1, METAP2 and GNB2L1) covering the range of downregulation as determined in our cell line model confirmed those as downregulated in our patient cohort (Figure 5b).
Figure 5.
Differential gene expression in TEL-AML1-positive patients. (a) The 150 most variant differentially expressed genes between four TEL-AML1-positive pre-B-ALL patients and two CD19-positive healthy donor controls (Ctrls) are shown. Unsupervised hierarchical clustering of samples using the Ward method revealed a separation of patients and normal Ctrls. (b) mRNAs of five direct TEL-AML1 targets covering the range of downregulation as determined in our cell line model were validated by quantitative reverse transcription-PCR in our patient cohort and compared with normal Ctrls.
GO and functional annotation of directly downregulated mRNAs in our cell line model and patients revealed terms related to cell proliferation and homeostasis, splicing, noncoding RNA processing, cellular transport mechanisms and receptor signalling pathways (Supplementary Table 7). Interestingly, the genes encoding for Strap and Smurf2, both negative regulators of transforming growth factor-β (TGF-β) signalling36, 37 were among the identified downregulated genes. Genes identified to be directly upregulated could be specifically sorted into GO terms mainly related to cellular adhesion and migration, cellular stress response, regulation of transcription, cell cycle and, interestingly, immune response (Supplementary Table 8). Hence, TEL-AML1 exerts an early repressive effect on translation as well as epigenetic processes like methylation as exemplified by our cell line model and a persisting inhibitory effect on cellular processes involving cell proliferation. A persisting positive effect could be identified on cellular stress responses including immune responses. This is leading to a complex early homeostatic function of TEL-AML1 in our pre-disposition cell line model as well as its prevalence in TEL-AML1-positive leukemic patients.
Discussion
Several mRNA profiling studies using microarray technology have been used to identify genes differentially regulated in TEL-AML1-positive patients in comparison to other subgroups.19, 35, 38, 39, 40 However, the direct targets of the fusion transcript in early pre-leukemic cells have only been identified for single candidates. We therefore aimed to first globally identify and distinguish direct and indirect TEL-AML1 targets using a cell line model and screen a TEL-AML1-positive patient cohort to validate those. In order to avoid detection of effects because of secondary aberrations as present in patient samples and the patient-derived cell lines carrying a TEL-AML1 fusion, we used an established in vitro pro-B-cell progenitor cell line with inducible TEL-AML1 fusion protein.22 This cell model closely resembles the cell stage previously shown to harbour the translocation in contrast to hematopoietic stem- and precursor cells.4 Nonetheless, we cannot rule out that other cell stages may contribute to the pre-leukemic phenotype in patients. Our study revealed promoter regions corresponding to 217 genes binding to TEL-AML1 and being differentially expressed on mRNA level after TEL-AML1 induction. This is by far less than the over 400 mRNAs downregulated after TEL-AML1 knockdown using an small hairpin RNA approach20 or the over 2500 downregulated genes in TEL-AML1-positive patients samples compared with TEL-AML1-negative patient samples.26 This difference might be attributed to the lack of secondary aberrations because of the short induction time and the possibility to distinguish between direct and indirect targets in our study. Other species-specific and cell line-specific B-cell maturation effects might as well account for the difference between the mouse and the human data sets. For examples, the BA/F3 cells, described to mature to Ig-producing cells in vivo,41 do not express Pax5,42 a factor shown to be indispensable for B-cell lineage maintenance and maturation.43 However, TEL-AML1-bound regions of the human precursor B-ALL cell line NALM-6 stably expressing the fusion construct showed overlapping regions with the regions identified in BA/F3. In addition, our approach let us to identify 118 mRNAs and proteins indirectly regulated by TEL-AML1 expression, most of them unidirectional. The mRNA of Rab8b was the only mRNA upregulated with concomitant downregulation of the protein level. The 3′-untranslated region of the corresponding transcript indeed harbours a predicted miRNA binding site for miR-19a/b that were induced upon TEL-AML1 expression. Rab8b, a member of the Ras oncogene family, has been implicated in vesicular transport44 and has been recently discovered as sensitizer to bortezomib in multiple myeloma.45
The binding and thus function of the TEL-AML1 fusion protein has been proposed to be mediated through the Runt DNA-binding domain of the AML1 fusion part.46 Indeed, we identified a statistical significant enrichment over background of the canonical AML1-binding motif in direct TEL-AML1 target promoters in this study in contrast to the use of merely mRNA expression data in a knockdown study of TEL-AML1.20 As 88% of identified direct target promoters were specific for TEL-AML1 as compared with TEL and AML1 ChIP in this study and very recent AML1 ChIP-seq results,34 other determinants of binding seem to be involved. These might be differences in co-factors or higher affinity binding to DNA of the fusion protein itself as has been shown for the AML1-ETO and inv-16 triggered AML1 fusion proteins47, 48 and proposed for the TEL-AML1 fusion protein as well.46
Mainly repressive GO terms are enriched in regulated genes of TEL-AML1 as identified here. This is in line with repressive function previously exemplified for single TEL-AML1 targets (reviewed in Zelent et al.46) as well as globally in a small hairpin RNA-mediated cell culture knockdown study.20 The downregulated translational processes might resemble the differentiation arrest described as early event of TEL-AML1 expression.7 Furthermore, genes involved in acetylation and methylation processes were also downregulated as direct consequence of TEL-AML1 induction with binding of TEL-AML1 fusion protein to the respective promoter regions, stressing the importance of epigenetic mechanisms for modulation of transcription factor activity. These direct targets included granzyme B, previously shown to be repressed by the fusion protein through recruitment of histone deacetylases.26
To investigate if identified early direct targets of TEL-AML1 in a progenitor B-cell model persist in the leukemic blasts at diagnosis, we evaluated gene expression in children with TEL-AML1-positive ALL compared with CD19+ cells of healthy donors or compared with TEL-AML1-negative ALL patients.35 A total of 53 genes were still significantly differentially regulated in either study after years of TEL-AML1 expression and multiple other factors absent in our cell culture model.49 The persistence of direct TEL-AML1 effects in two independent patient cohorts is in line with an in vivo transplantation mouse study, which showed an indispensable role of the fusion protein for cell survival and replenishment.21 Greaves et al. described a persistent growth of TEL-AML1-induced BA/F3 cells after TGF-β treatment, whereas without TGF-β TEL-AML1-induced cells had a growth disadvantage over control cells.22 We identified a binding of TEL-AML1 to promoter regions and a concomitant downregulation of the mRNA of inhibitors of the ‘classical' SMAD-dependent TGF-β pathway, STRAP and SMURF2, which were also found in TEL-AML1-positive patients at significant levels. It has been demonstrated that the TEL-AML1 fusion protein binds to Smad3 downstream of its activation, thus impairing signal transduction of the ‘classical' TGF-β pathway. However, the possible hyperactivation of TGF-β receptors through downregulation of negative effectors might also result in activation of Smad-independent TGF-β receptor-mediated pathways like the JNK, p38MAPK or Erk/MAPK pathways (reviewed in Derynck and Zhan50). We find the mRNA of DUSP7, an inactivator of ERKs, directly upregulated after TEL-AML1 induction; however, it was not identified as significantly regulated in patients. Aside from the TGF-β pathway, Smurf2 deficiency has been shown to attenuate p16 expression thus impairing senescence response and increasing the susceptibility of mice for spontaneous tumorigenesis of mainly B-cell lymphoma.51 Furthermore, ablation of Smurf2 leads to dysregulation of the epigenetic landscape through histone modifications and thus loss of chromosomal stability in a mouse model leading to a wide range of malignancies including lymphomas.52
In summary, we globally identified early direct and indirect targets of the TEL-AML1 fusion protein by integration of chromatin immunoprecipitation, mRNA expression and differential stable isotope labelling of proteins from the same cell system for the first time. Unexpectedly, TEL-AML1 binds to specific promoter regions mostly distinct from AML1-bound promoter regions in murine cell line models, although binding seems to be, at least in part, mediated by the Runt DNA-binding domain as the consensus signal is enriched in direct targets over background and also over indirect TEL-AML1 targets defined here. We could also demonstrate an immediate suppressive effect on mRNA transcription upon TEL-AML1 promoter binding resulting in enrichment of respective biological processes and pathways only for downregulated genes. Our work represents a resource for the future in vitro and more importantly in vivo study of the underlying cause of premalignant transformation of early lymphoid precursors in the development of TEL-AML1-positive ALL and is useful for interpretation of patient-derived data as exemplified on two patient cohorts in this work.
Acknowledgments
We are very grateful to Anthony Ford and Mel Greaves for providing the inducible murine cell system and the corresponding control cell line. We thank the Biologisch-Medizinisches Forschungszentrum team and especially René Deenen for Array Hybridization and Scanning. Furthermore, we are grateful to Wolfgang Kaisers for bioinformatic support. This work was aided by grants of the Research Commission of the Medical Faculty of the University of Dusseldorf (YL, AN), the German-Israeli-Foundation for Scientific Research and Development (GIF; Jerusalem, Israel, to AB), Federal Ministry of Education and Research (BMBF; to AB, PL), the German Cancer Aid (Krebshilfe, to AB, PL) and the German research foundation (DFG, grant no. LA2983/2-1 to PL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
AB and AN perceived the study; YL, MK, AN, PL and AB designed research; YL, MK, MG and AN performed research; EK and JH contributed new reagents/patients samples; YL, SG, RT, MK and PL analyzed data; YL, PL and AB wrote the paper.
Supplementary Material
References
- Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, et al. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff SA, Buijs A, Behm FG, Rubnitz JE, Raimondi SC, Hancock ML, et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotfilder M, Rottgers S, Rosemann A, Jurgens H, Harbott J, Vormoor J. Immature CD34+CD19- progenitor/stem cells in TEL/AML1-positive acute lymphoblastic leukemia are genetically and functionally normal. Blood. 2002;100:640–646. doi: 10.1182/blood.v100.2.640. [DOI] [PubMed] [Google Scholar]
- Fischer M, Schwieger M, Horn S, Niebuhr B, Ford A, Roscher S, et al. Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. Oncogene. 2005;24:7579–7591. doi: 10.1038/sj.onc.1208931. [DOI] [PubMed] [Google Scholar]
- Morrow M, Horton S, Kioussis D, Brady HJ, Williams O. TEL-AML1 promotes development of specific hematopoietic lineages consistent with preleukemic activity. Blood. 2004;103:3890–3896. doi: 10.1182/blood-2003-10-3695. [DOI] [PubMed] [Google Scholar]
- Tsuzuki S, Seto M, Greaves M, Enver T. Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice. Proc Natl Acad Sci USA. 2004;101:8443–8448. doi: 10.1073/pnas.0402063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood. 2011;118:4910–4918. doi: 10.1182/blood-2011-05-354266. [DOI] [PubMed] [Google Scholar]
- Ford AM, Bennett CA, Price CM, Bruin MC, Van Wering ER, Greaves M. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc Natl Acad Sci USA. 1998;95:4584–4588. doi: 10.1073/pnas.95.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- Lausten-Thomsen U, Madsen HO, Vestergaard TR, Hjalgrim H, Nersting J, Schmiegelow K. Prevalence of t(12;21)[ETV6-RUNX1]-positive cells in healthy neonates. Blood. 2011;117:186–189. doi: 10.1182/blood-2010-05-282764. [DOI] [PubMed] [Google Scholar]
- Morrow M, Samanta A, Kioussis D, Brady HJ, Williams O. TEL-AML1 preleukemic activity requires the DNA binding domain of AML1 and the dimerization and corepressor binding domains of TEL. Oncogene. 2007;26:4404–4414. doi: 10.1038/sj.onc.1210227. [DOI] [PubMed] [Google Scholar]
- Fears S, Gavin M, Zhang DE, Hetherington C, Ben-David Y, Rowley JD, et al. Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc Natl Acad Sci USA. 1997;94:1949–1954. doi: 10.1073/pnas.94.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenrick R, Amann JM, Lutterbach B, Wang L, Westendorf JJ, Downing JR, et al. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert SW, Sun W, Davis JN, Golub T, Shurtleff S, Buijs A, et al. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho JK, Kim JH, Yu J, Choe SY. Correlation between cellular localization of TEL/AML1 fusion protein and repression of AML1-mediated transactivation of CR1 gene. Biochem Biophys Res Commun. 2002;297:91–95. doi: 10.1016/s0006-291x(02)02075-2. [DOI] [PubMed] [Google Scholar]
- Gunji H, Waga K, Nakamura F, Maki K, Sasaki K, Nakamura Y, et al. TEL/AML1 shows dominant-negative effects over TEL as well as AML1. Biochem Biophys Res Commun. 2004;322:623–630. doi: 10.1016/j.bbrc.2004.07.169. [DOI] [PubMed] [Google Scholar]
- McLean TW, Ringold S, Neuberg D, Stegmaier K, Tantravahi R, Ritz J, et al. TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood. 1996;88:4252–4258. [PubMed] [Google Scholar]
- Gandemer V, Rio AG, de Tayrac M, Sibut V, Mottier S, Ly Sunnaram B, et al. Five distinct biological processes and 14 differentially expressed genes characterize TEL/AML1-positive leukemia. BMC Genomics. 2007;8:385. doi: 10.1186/1471-2164-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuka G, Kauer M, Kofler R, Haas OA, Panzer-Grumayer R. The leukemia-specific fusion gene ETV6/RUNX1 perturbs distinct key biological functions primarily by gene repression. PLoS One. 2011;6:e26348. doi: 10.1371/journal.pone.0026348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuka G, Kantner HP, Grausenburger R, Inthal A, Bauer E, Krapf G, et al. Silencing of ETV6/RUNX1 abrogates PI3K/AKT/mTOR signaling and impairs reconstitution of leukemia in xenografts. Leukemia. 2012;26:927–933. doi: 10.1038/leu.2011.322. [DOI] [PubMed] [Google Scholar]
- Ford AM, Palmi C, Bueno C, Hong D, Cardus P, Knight D, et al. The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells. J Clin Invest. 2009;119:826–836. doi: 10.1172/JCI36428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Linka RM, Risse SL, Bienemann K, Werner M, Linka Y, Krux F, et al. Loss-of-function mutations within the IL-2 inducible kinase ITK in patients with EBV-associated lymphoproliferative diseases. Leukemia. 2012;26:963–971. doi: 10.1038/leu.2011.371. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkova J, Madzo J, Cario G, Kalina T, Ford A, Zaliova M, et al. The identification of (ETV6)/RUNX1-regulated genes in lymphopoiesis using histone deacetylase inhibitors in ETV6/RUNX1-positive lymphoid leukemic cells. Clin Cancer Res. 2007;13:1726–1735. doi: 10.1158/1078-0432.CCR-06-2569. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Niebuhr B, Kriebitzsch N, Fischer M, Behrens K, Gunther T, Alawi M, et al. Runx1 is essential at two stages of early murine B-cell development. Blood. 2013;122:413–423. doi: 10.1182/blood-2013-01-480244. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Datta PK, Moses HL. STRAP and Smad7 synergize in the inhibition of transforming growth factor beta signaling. Mol Cell Biol. 2000;20:3157–3167. doi: 10.1128/mcb.20.9.3157-3167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, et al. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem J. 2005;386 (Pt 3:461–470. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine BM, Stanulla M, Schrappe M, Ho M, Viehmann S, Harbott J, et al. Gene expression patterns associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood. 2004;103:1043–1049. doi: 10.1182/blood-2003-05-1518. [DOI] [PubMed] [Google Scholar]
- Moos PJ, Raetz EA, Carlson MA, Szabo A, Smith FE, Willman C, et al. Identification of gene expression profiles that segregate patients with childhood leukemia. Clin Cancer Res. 2002;8:3118–3130. [PubMed] [Google Scholar]
- Holleman A, den Boer ML, de Menezes RX, Cheok MH, Cheng C, Kazemier KM, et al. The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood. 2006;107:769–776. doi: 10.1182/blood-2005-07-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- Sigvardsson M, Clark DR, Fitzsimmons D, Doyle M, Akerblad P, Breslin T, et al. Early B-cell factor, E2A, and Pax-5 cooperate to activate the early B cell-specific mb-1 promoter. Mol Cell Biol. 2002;22:8539–8551. doi: 10.1128/MCB.22.24.8539-8551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- Chen S, Liang MC, Chia JN, Ngsee JK, Ting AE. Rab8b and its interacting partner TRIP8b are involved in regulated secretion in AtT20 cells. J Biol Chem. 2001;276:13209–13216. doi: 10.1074/jbc.M010798200. [DOI] [PubMed] [Google Scholar]
- Zhu YX, Tiedemann R, Shi CX, Yin H, Schmidt JE, Bruins LA, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117:3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
- Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112:1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Wee HJ, Liu PP, Osato M, Ito Y. Molecular basis for a dominant inactivation of RUNX1/AML1 by the leukemogenic inversion 16 chimera. Blood. 2004;103:3200–3207. doi: 10.1182/blood-2003-07-2188. [DOI] [PubMed] [Google Scholar]
- Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Ramkumar C, Kong Y, Cui H, Hao S, Jones SN, Gerstein RM, et al. Smurf2 regulates the senescence response and suppresses tumorigenesis in mice. Cancer Res. 2012;72:2714–2719. doi: 10.1158/0008-5472.CAN-11-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med. 2012;18:227–234. doi: 10.1038/nm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.