Abstract
Deregulation of microRNA (miRNA) expression has been documented in diffuse large B-cell lymphoma (DLBCL). However, the impact of miRNAs and their machinery in DLBCL is not fully determined. Here, we assessed the role of miRNA expression and their processing genes in DLBCL development. Using microarray and RT-qPCR approaches, we quantified global miRNAs and core components of miRNA-processing genes expression in 75 DLBCLs (56 de novo and 19 transformed) and 10 lymph nodes (LN). Differential miRNA signatures were identified between DLBCLs and LNs, or between the de novo and transformed DLBCLs. We also identified subsets of miRNAs associated with germinal center B-cell phenotype, BCL6 and IRF4 expression, and clinical staging. In addition, we showed a significant over-expression of TARBP2 in de novo DLBCLs as compared with LNs, and decreased expression of DROSHA, DICER, TARBP2 and PACT in transformed as compared with de novo cases. Interestingly, cases with high TARBP2 and DROSHA expression had a poorer chemotherapy response. We further showed that TARBP2 can regulate miRNA-processing efficiency in DLBCLs, and its expression inhibition decreases cell growth and increases apoptosis in DLBCL cell lines. Our findings provide new insights for the understanding of miRNAs and its machinery in DLBCL.
Keywords: microRNA, TARBP2, gene expression, diffuse large B-cell lymphoma
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a highly aggressive B-cell lymphoma. The majority of cases are diagnosed de novo (primary DLBCL), whereas a subset is transformed from a previous indolent lymphoma. This disease is very heterogeneous with significantly different clinical outcomes,1, 2, 3 and advanced knowledge of the biology of DLBCL is crucial for future advances in treatment management. Considerable clinical heterogeneity and broad survival variation among DLBCL patients and investigations for identifying new prognostic markers that predict the clinical behavior of such tumors with greater accuracy than is currently possible are warranted. While there has been progress in understanding the molecular genetics of these tumors,1, 2, 4 the detailed network of events leading to DLBCL development and progression has yet to be elucidated. Here, we aimed to determine the role of microRNAs (miRNAs) and their processing factors in DLBCL development, as well as their clinical implications.
miRNAs are a class of short non-coding RNAs involved in gene regulation through binding to 3′-untranslated regions of target genes.5 miRNAs are transcribed as long primary transcripts (pri-miRNAs), which are subsequently processed by the miRNA machinery. Pri-miRNAs are converted in the nucleus to ∼70 nucleotides (nt) stem-loop structures called precursor miRNAs (pre-miRNAs) by the RNase-III enzyme DROSHA (or RNASEN) and its binding partner DGCR8.6 Pre-miRNAs are further processed in the cytoplasm to ∼22 nt mature miRNAs by a second RNase-III enzyme known as DICER. DICER associates with dsRNA-binding proteins such as TARBP2 and PACT (or PRKRA).7, 8
miRNAs have key roles in many cellular processes including differentiation of hematopoietic lineages.5, 9 Thus, miRNAs are thought to have important roles in hematological malignancies. In line with such notions, several miRNAs such as miR-15a, miR-16-1, miR-155 and miR-17-92 cluster have been implicated in leukemogenesis and lymphomagenesis.10, 11, 12 miRNA expressions are altered in DLBCL.11, 13, 14 Interestingly, DLBCL shows the most heterogeneous miRNA profiles among various lymphoma types15 and, concordantly with mRNA profilings, distinct miRNA signatures are associated with molecular subtypes of DLBCL.13, 16
It has also been shown that certain miRNAs have prognostic and diagnostic value in some cancer types including lymphomas and leukemia.17, 18 Despite miRNA profiling being reported in DLBCLs, their clinical impact is not fully understood. Besides miRNAs, the miRNA machinery genes have also been associated with cancer development, progression and prognosis in several cancer types including ovarian,19 colorectal,20 melanoma21 and T-cell lymphoma.22 However, the role of miRNA machinery genes in DLBCL has yet to be determined.
In this study, we investigated the expression of miRNAs and their machinery genes in DLBCLs, their expression association with clinical and histopathological features, and the functional role of TARBP2 in DLBCL.
Materials and methods
Clinical samples
Cohort 1 includes a total of 75 frozen tumor biopsies from primary DLBCL (56 de novo and 19 transformed) collected at Uppsala University Hospital, Sweden. Cohort 2 includes total RNA extracted from 47 DLBCL FFPE-specimens (38 de novo and 9 transformed) collected from the John Radcliffe Hospital, Oxford, as previously described.14 In addition, 15 anonymized samples of histopathologically verified reactive lymph nodes (LNs) (10 frozen and 5 FFPE) collected at the Karolinska University Hospital or Uppsala University Hospital were included as references of non-neoplastic lymphatic tissue.
The clinical, histopathological and follow-up details of all DLBCL cases from both cohorts are given in Supplementary Tables S1 and S2 and summarized in Table 1.
Table 1. Summary of clinical and molecular features of the DLBCL casesa.
| Parameter |
Cohort 1 |
Cohort 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| De novo |
Transformed |
De novo |
Transformed |
|||||
| No. of informative | No. of cases | No. of informative | No. of cases | No. of informative | No. of cases | No. of informative | No. of cases | |
| Gender | 56 | 19 | 37 | 9 | ||||
| Male | 31 | 10 | 13 | 3 | ||||
| Female | 25 | 9 | 24 | 6 | ||||
| Age at diagnosis (median) | 56 (68 years) | 19 (66 years) | 37 (63 years) | 9 (69 years) | ||||
| ⩽ median | 30 | 11 | 19 | 5 | ||||
| > median | 26 | 8 | 18 | 4 | ||||
| Stage | 53 | 19 | 36 | 7 | ||||
| I | 13 | 1 | 17 | 3 | ||||
| II | 16 | 5 | 7 | 0 | ||||
| III | 10 | 6 | 9 | 2 | ||||
| IV | 14 | 7 | 3 | 2 | ||||
| Regime treatment | 48 | 18 | 36 | 9 | ||||
| CHOP | 22 | 7 | 5 | 3 | ||||
| R-CHOP | 17 | 7 | 17 | 4 | ||||
| No treatment | 1 | 0 | 5 | 0 | ||||
| Others | 8 | 4 | 9 | 2 | ||||
| IPI score | 51 | 19 | 36 | 6 | ||||
| Low (0-1-2) | 36 | 13 | 27 | 4 | ||||
| High (3-4-5) | 15 | 6 | 9 | 2 | ||||
| GCB phenotype | 54 | 16 | 35 | 9 | ||||
| GCB | 20 | 13 | 18 | 5 | ||||
| non-GCB | 34 | 3 | 17 | 4 | ||||
| BCL2 expression | 55 | 0 | 20 | 3 | ||||
| Negative | 17 | — | 9 | 1 | ||||
| Positive | 38 | — | 11 | 2 | ||||
| BCL6 expression | 54 | 16 | 35 | 9 | ||||
| Negative | 30 | 5 | 15 | 3 | ||||
| Positive | 24 | 11 | 20 | 6 | ||||
| IRF4 expression | 54 | 16 | 35 | 9 | ||||
| Negative | 22 | 11 | 16 | 4 | ||||
| Positive | 32 | 5 | 19 | 5 | ||||
| CD10 expression | 54 | 16 | 35 | 9 | ||||
| Negative | 35 | 3 | 23 | 5 | ||||
| Positive | 19 | 13 | 12 | 4 | ||||
| Follow-up | 56 | 0 | 35 | 8 | ||||
| Alive | 24 | — | 26 | 4 | ||||
| Dead | 10 | — | 1 | 0 | ||||
| DOD | 22 | — | 8 | 4 | ||||
Ethical permission for the use of all samples was obtained from the local ethics committee.
DLBCL cell lines
Three de novo DLBCL cell lines OCI-Ly-1, OCI-Ly-7 and OCI-Ly-3 were kindly provided by Dr Mark Minden (University Health Network, Toronto, ON, Canada). OCI-Ly-1 and OCI-Ly-7 are germinal center B-cell (GCB)-subtype and OCI-Ly-3 is activated B-cell (ABC)-subtype.1 OCI-Ly-1 and OCI-Ly-3 were cultured in Iscove's modified Dulbecco's medium (IMDM) with 20% fetal bovine serum. OCI-Ly-7 was grown in IMDM with 2-Mercaptoethanol (1:10 000) and 10% fetal bovine serum. All cells were supplemented with 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) and cultured at 37 °C and 5% CO2 in a humidified incubator.
Immunohistochemistry
Twenty-five de novo DLBCL cases without immunophenotype classification from cohort 1 were analyzed for expression of CD10, BCL6, IRF4 and BCL2 by immunohistochemistry as previously described.3, 4 In brief, antigen retrieval was performed using heat treatment with Target Retrieval Solution (pH 9.0) (S2367, Dako, Glostrup, Denmark) at 95–99 °C for 20 min. Sections were incubated with anti-CD10 (1:25; M7308, Dako), anti-BCL6 (1:10; M7211, Dako), anti-IRF4 (1:25; M7259, Dako) or anti-BCL2 (1:50; M0887, Dako) at 4 °C overnight. An anti-mouse HRP-conjugated secondary antibody was applied and signal detection was performed using DAB chromogen (K5007, EnVision Detection System, Dako). The expression was scored as positive if >30% of the tumor cells were immunoreactive for CD10, BCL6 or IRF4 and >50% for BCL2. The immunostaining was evaluated by a hematopathologist (R-M A).
RNA extraction
Total RNA was isolated from tissues and cell lines using the mirVana miRNA Isolation Kit (Applied Biosystems/Ambion, Austin, TX, USA) and quantified with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
miRNA microarray
Agilent human miRNA microarray system (miRBase release 9.1; Agilent, Santa Clara, CA, USA) was adopted to evaluate miRNA profiles in 45 DLBCLs (32 de novo DLBCL and 13 DLBCL-t) from cohort 1 and 10 LNs. Microarray experiments were performed as previously described.23 Cluster 3 software was used for normalization and arrays median centering.24 Normalized miRNA probes with <50% missing values across the samples were used for clustering and statistical analyses. Hierarchical clustering was performed using Cluster 3.0 and visualized with Treeview v1.60 (http://rana.lbl.gov/EisenSoftware.htm). Significance analysis of microarrays (SAM) and prediction analysis of microarrays (PAM) were used to identify differentially expressed miRNAs between sample groups (http://www-stat.stanford.edu/Btibs/). The microarray data have been deposited at NCBI Gene Expression Omnibus (GEO accession number, GSE42906).
Reverse transcription quantitative PCR for mature miRNAs and mRNAs
Reverse transcription quantitative PCR (RT-qPCR) was performed to evaluate expression of mature miRNAs and mRNAs using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). For miRNA quantification, 25 ng of total RNA was converted into cDNA using Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). Predesigned TaqMan miRNA assays were purchased from Applied Biosystems for quantification of miR-155 (ID 002623), miR-21 (ID 000397), miR-150 (ID 000473), miR-494 (ID 002365), miR-638 (ID 001582), miR-145 (ID 002278), miR-143 (ID 002249), miR-103 (ID 000439), miR-107 (ID 000443), miR-142-3p (ID 000464), miR-142-5p (ID 002248), miR-29a (ID 002112), miR-29b (ID 000413), miR-146a (ID 000468), miR-140 (ID 002234), let-7i (ID 002221), miR-16 (ID 000391), miR-195 (ID 000494) and miR-497 (ID 001043). RNU6B (ID 001093) was used for the normalization of miRNA quantification. For mRNA expression analysis, 100 ng of total RNA (250 ng for FFPE samples) were used to synthesize cDNA with High Capacity cDNA Reverse Transcription kit (Applied Biosystems), and RT-qPCR was performed using specific TaqMan assays (Applied Biosystems) for DICER (ID 00998578), DROSHA (ID 01095029), TARBP2 (ID 00998379), DGCR8 (ID 00987089) and PACT (ID 00269379). Normalization of mRNA expression was performed against 18S (ID 99999901). For both mRNA and miRNA analyses, relative expression was determined using the comparative CT method and reported as 2−ΔCT. All reactions were performed in triplicate.
SYBR-Green RT-qPCR assay for primary and precursor miRNAs
SYBR-Green RT-qPCR was performed to evaluate the relative expression of primary and precursor forms of miR-155 and miR-146a. In brief, cDNA was reverse transcribed from 100 ng of total RNA using 10 μM of a specific reverse primer (miR-155-reverse: 5′-GCTAATATGTAGGAGTCAGTTGGA-3′ or miR-146a-reverse: 5′-GCTGAAGAACTGAATTTCAGAG-3′) and subsequently quantified by SYBR-Green RT-qPCR, as previously described.25 RT-qPCR was performed using the following primers for miR-155: 5′-GTTAATGCTAATCGTGATAGGG-3′ (pri+pre-forward) or 5′-GAAGGGGAAATCTGTGGTTTA-3′ (pri-forward) and the miR-155-reverse primer; for miR-146a: 5′-TTGAGAACTGAATTCCATGG-3′ (pri+pre-forward) or 5′-TCTTTGCACCATCTCTGA-3′ (pri-forward) and the miR-146a-reverse primer. RNU6B was used as a reference gene. The expression level of pre-miRNA was calculated as follows:
Pre-miR=2−ΔCT (pri-miRNA+pre-miRNA)−2−ΔCT (pri-miRNA)
The miRNA processing efficiency is presented as ‘relative processing efficiency', which was calculated as the ratio of mature to precursor miRNA.
Transfection experiments
TARBP2 siRNA or non-targeting control siRNA (sc-106846 and sc-36869, respectively; Santa Cruz Biotechnology Inc.) was transiently transfected into DLBCL cell lines (OCI-Ly-1 and OCI-Ly-3) using Nucleofector technology (Lonza, Basel, Switzerland). A total of 1 × 106 cells were resuspended in Nucleofector solution T mixed with 200 nM of siRNA and electroporated using program G-16. Cells were then seeded in T-25 flasks with IMDM media and cultured for 48 h. The expression levels of TARBP2 were assessed by western blot. All transfection experiments were repeated at least three times and used for cell growth and apoptosis assays.
Western blot analysis
Western blots were performed as previously described.26 Primary antibodies included anti-DROSHA (1:400; ab12286; Abcam), anti-DICER (1:200; ab14601), anti-TARBP2 (1:1000; ab72547), anti-DGCR8 (1:2000; ab90579) and anti-PACT (1:400; ab75749). Secondary antibodies were either anti-mouse IgG-HRP (1:10 000; sc-2005; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or anti-rabbit IgG-HRP (1:3000; 170-6515; Bio-Rad Laboratories, Hercules, CA, USA). Subsequent incubation with a GAPDH antibody (1:5000; sc-47724; Santa Cruz Biotechnology Inc.) was performed for normalization. Protein levels were quantified on X-ray films from immunoblots using ImageJ software (http://rsb.info.nih.gov/ij/).
WST-1 colorimetric assay
Cell viability was assessed in OCI-Ly-1 and OCI-Ly-3 cells using WST-1 colorimetric assay (Roche Applied Science, Mannheim, Germany), as previously described.26 All transfections and WST-1 assays were repeated independently three times and six replicates were performed during each experiment. Cell viability was calculated by comparing the absorbance values of the samples after background subtraction. The fraction of viable cells was normalized to siRNA control-treated cells.
Apoptosis caspase-3/CPP32 colorimetric assay
Apoptosis was evaluated 48 h post-transfection using the Caspase-3/CPP32 Colorimetric Assay Kit (K106-25; BioVision, Mountain View, CA, USA).26 Apoptosis was calculated by comparing the absorbance values of the siTARBP2-treated cells with the respective negative control-treated cells. All experiments were replicated three times.
Statistical analyses
All analyses were conducted using Statistica 8.0 (StatSoft, Inc., Tulsa, OK, USA) or MS Office Excel. The patient population was divided in two groups based on high or low expression of DICER, DROSHA, TARBP2, DGCR8 and PACT according to the median expression levels. The comparison between miRNA/mRNA expressions among or between different groups was conducted using one-way ANOVA or unpaired student's t-test. Paired student's t-test was performed to analyze transfection experiments. The association between miRNA machinery genes expression levels and clinical parameters was analyzed using χ2 test. Kaplan–Meier analysis was carried out to analyze different factors potentially associated with overall or disease-free survival. For overall survival, patients who were still alive at the end of the follow-up were considered as censored. In disease-free survival, censored group included patients who did not show any sign of disease or died of unknown or DLBCL-unrelated reasons. Survival curves for each group of patients were compared using log-rank test. All the analyses were two-tailed and P-values <0.05 were regarded as significant.
Results
miRNA profilings in DLBCLs and LNs
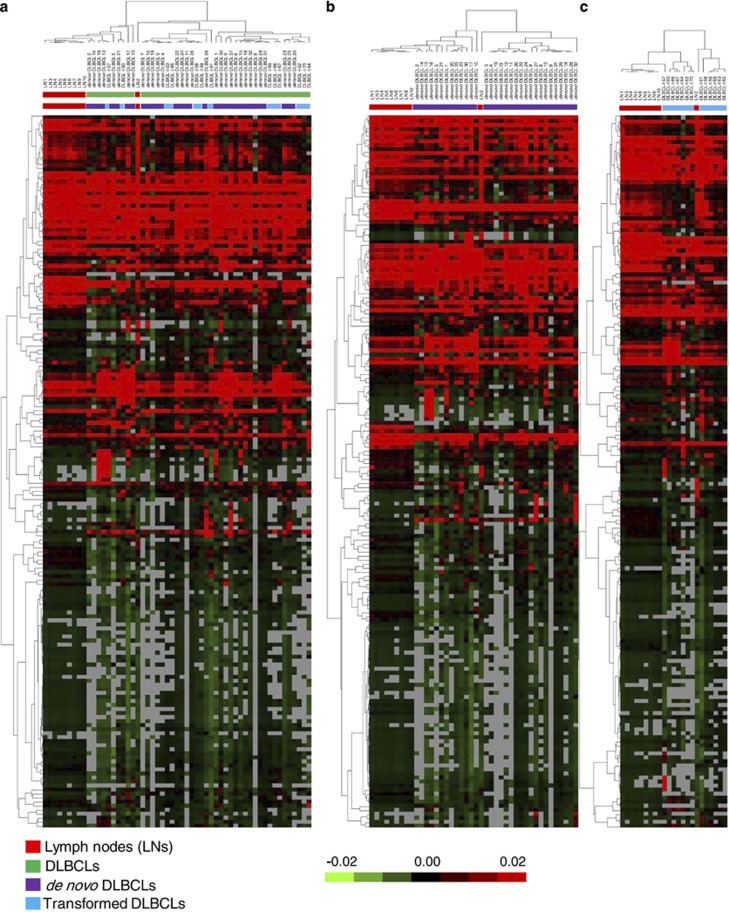
We determined global miRNA expression profiles in 45 DLBCLs and 10 LNs using a microarray approach. After data normalization and filtering, we performed unsupervised clustering of 177 filtered miRNAs among all the DLBCLs and LNs. The analysis revealed several distinct clusters (Figure 1). Nine of the 10 LNs were grouped together in a distinct cluster and the remaining LN (that is, LN 2) was loosely clustered with the majority of DLBCLs in a separate sub-tree. Among the DLBCLs, we observed several distinct clusters, without distinction between de novo and transformed DLBCLs.
Figure 1.
Unsupervised clustering of miRNA expression by microarray analysis among reactive lymph nodes (LNs) and DLBCLs (cohort 1). The heat maps illustrate unsupervised clustering of miRNA expression profiling in: (a) 10 LN and 45 DLBCLs (including 32 de novo and 13 transformed cases), (b) 10 LNs and 32 de novo DLBCLs, and (c) 10 LNs and 13 transformed DLBCLs. Samples were clustered using the uncentered correlation and complete linkage. Median centered values for each miRNA are represented. The green and red colors indicate relatively low and high expression, respectively. Missing values are indicated in gray.
We performed SAM analysis to identify the most significant miRNAs that could distinguish DLBCLs from LNs. The analysis identified 55 deregulated miRNAs with a false-discovery rate of <20% (Supplementary Table S3). We further used the PAM to determine the minimal number of miRNAs that could distinguish all DLBCLs from LNs. The analysis identified a signature of 23 deregulated miRNAs that best discriminate the two groups with a prediction accuracy of 96% as determined by 10-fold cross-validation (Supplementary Figure S1). Similarly, a minimal number of 25 miRNAs could correctly classify the de novo DLBCLs from LNs with an accuracy of 98%, and a set of 32 miRNAs for distinguishing transformed DLBCLs from LNs with 93% accuracy (Supplementary Figure S1).
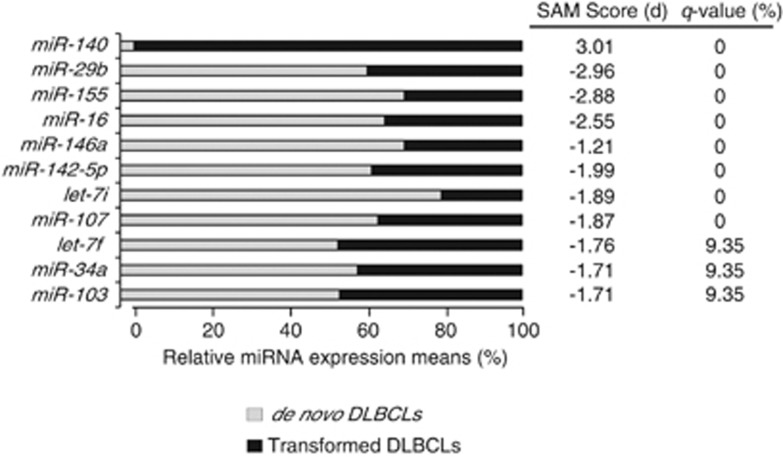
By comparing the miRNA classifiers of the de novo DLBCLs vs LNs and transformed DLBCLs vs LNs, we found 22 common miRNAs between the two subgroups (Supplementary Figure S2). We also found several deregulated miRNAs that could be specific for each tumor subgroups. To identify a miRNA classifier to distinguish the two tumor subgroups, we compared directly the de novo and transformed DLBCLs using SAM analysis. The analysis identified 10 miRNAs (miR-29b, miR-155, miR-16, miR-146a, miR-142-5p, let-7i, miR-107, let-7f, miR-34a and miR-103) with higher expression and one (miR-140) with lower expression in de novo as compared with transformed DLBCLs (false-discovery rate<20% Figure 2).
Figure 2.
Array-based SAM results for differentially expressed miRNAs between de novo and transformed DLBCLs (cohort 1). The bar charts show the relative expression of 11 deregulated miRNAs between de novo (n=32) and transformed (n=13) DLBCL tumors analyzed by microarray. The expression levels are presented as means. Positive or negative SAM scores indicate over-expressed or under-expressed miRNAs in transformed cases, respectively. Note that only miRNAs with false-discovery rate (q-value) <20% are shown.
Validation of the miRNA expression signatures by RT-qPCR
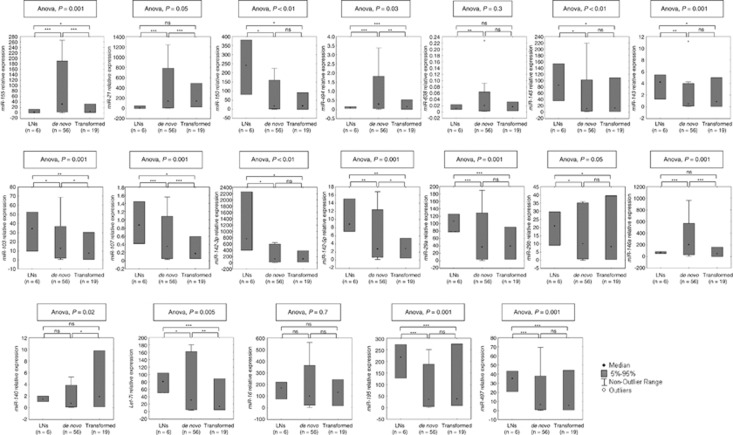
To validate the microarray results, we measured 19 miRNAs by RT-qPCR in an extended cohort of 75 tumors from cohort 1 and 6 LNs. The miRNAs were selected based on their high scores in the SAM or PAM analyses for different comparisons or because their involvement in DLBCL or other tumor types. Thirteen miRNAs (miR-150, miR-29b, miR-29a, miR-142-3p, miR-142-5p, miR-145, miR-143, miR-494, miR-638, miR-21, miR-195, miR-497 and miR-155) were significantly deregulated between DLBCLs and LNs (Supplementary Figure S1; Supplementary Table S3), whereas the remaining six miRNAs (miR-103, miR-107, miR-146a, miR-140, let-7i and miR-16) were differentially expressed between the de novo and transformed DLBCLs (Figure 2). Of note, the three deregulated miRNAs (miR-29b, miR-155 and miR-142-5p) found in the comparison between all DLBCLs and LNs were also differentially expressed between the two tumor subgroups (Figure 2).
Concordantly with the microarray findings, all 13 candidate DLBCL-associated miRNAs were significantly deregulated in the tumor samples and seven of the subgroup-associated miRNAs were also significant as determined by RT-qPCR (Figure 3; Supplementary Tables S4 and S5). The expression levels of miR-16 and miR-29b were not significantly different between the two subgroups by RT-qPCR (Figure 3; Supplementary Table S5). On the other hand, miR-494 was significantly over-expressed in de novo as compared with transformed DLBCLs by RT-qPCR (Figure 3).
Figure 3.
Validation of differentially expressed miRNAs by RT-qPCR (cohort 1). The box plots illustrate relative expression of the respective miRNA in six LNs, 56 de novo and 19 transformed DLBCLs. One-way ANOVA and unpaired Student's t-test were used to compare expression levels among or between groups and P<0.05 was considered significant. *P<0.05; **P<0.01; ***P<0.001; ns, not significant.
miRNAs associated with clinico-histopathological features of de novo DLBCLs
We investigated miRNA expression in relation to clinico-histopathological parameters such as GCB/non-GCB phenotype, staging, IPI score (International Prognostic Index) and immunohistochemical expressions of CD10, BCL6, IRF4 or BCL2 in de novo DLBCLs.
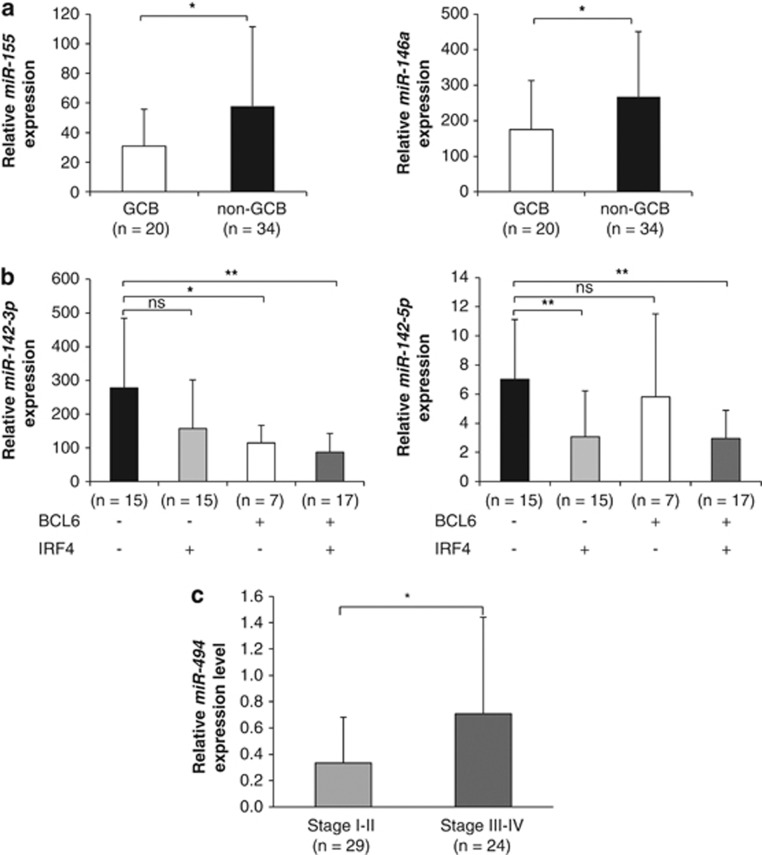
Based on the microarray data, we identified a subset of six miRNAs (let-7g, miR-155, miR-29c, miR-146a, miR-451 and miR-16) that could discriminate GCB from non-GCB cases using SAM (Supplementary Table S6). Three of the six GCB/non-GCB-associated miRNAs (miR-155, miR-146a and miR-16) were chosen for validation using RT-qPCR. The results support significant over-expression of miR-155 and miR-146a in the non-GCB group (Figure 4a), whereas miR-16 did not show any significant difference between the two groups (data not shown).
Figure 4.
miRNA signatures associated with pathological and molecular features of de novo DLBCLs (cohort 1) validated by RT-qPCR. (a) Histograms show expression of miR-155 and miR-146a in GCB and non-GCB cases. (b) Histograms show the expression levels of miR-142-3p and miR-142-5p in de novo DLBCLs in relation to BCL6 and IRF4 immunoreactivities. (c) Histogram show relative expression levels of miR-494 in stage I–II vs stage III–IV de novo DLBCLs. Unpaired t-test was used to compare miRNA expressions between groups and P<0.05 was considered significant. *P<0.05; **P<0.01; ns, not significant.
Given that CD10, BCL6, IRF4 or BCL2 are commonly deregulated in DLBCLs and have prognostic impact of this tumor type,27 we sought to determine whether miRNA expression profiles correlated with expression levels of these factors. SAM analyses identified four and seven miRNAs associated with BCL6-positive and IRF4-positive DLBCLs, respectively (Supplementary Table S6). Interestingly, decreased expression of miR-142-3p and miR-142-5p were associated with both BCL6-positive and IRF4-positive DLBCLs (Figure 4b; Supplementary Table S6). However, no significant association between miRNA profile and CD10 or BCL2 expression was found (data not shown).
Next, we investigated whether miRNAs were associated with disease stage of de novo DLBCLs. Comparing miRNA profiles of stage I–II vs III–IV tumors, we identified a single miRNA (miR-494) that was upregulated in stage III–IV (Supplementary Table S6). The result was further confirmed by RT-qPCR in 53 de novo cases (P=0.03, unpaired t-test; Figure 4c).
We also examined the relationship between miRNAs and IPI score, which is widely used for risk stratification of lymphoma patients with significantly different responses to therapy and survival. The DLBCL cases were divided into two groups with low (0–2) or high IPI score (3–5). However, we could not identify any miRNA that could distinguish these two groups of patients (data not shown).
Expression of miRNA processing factors in DLBCLs
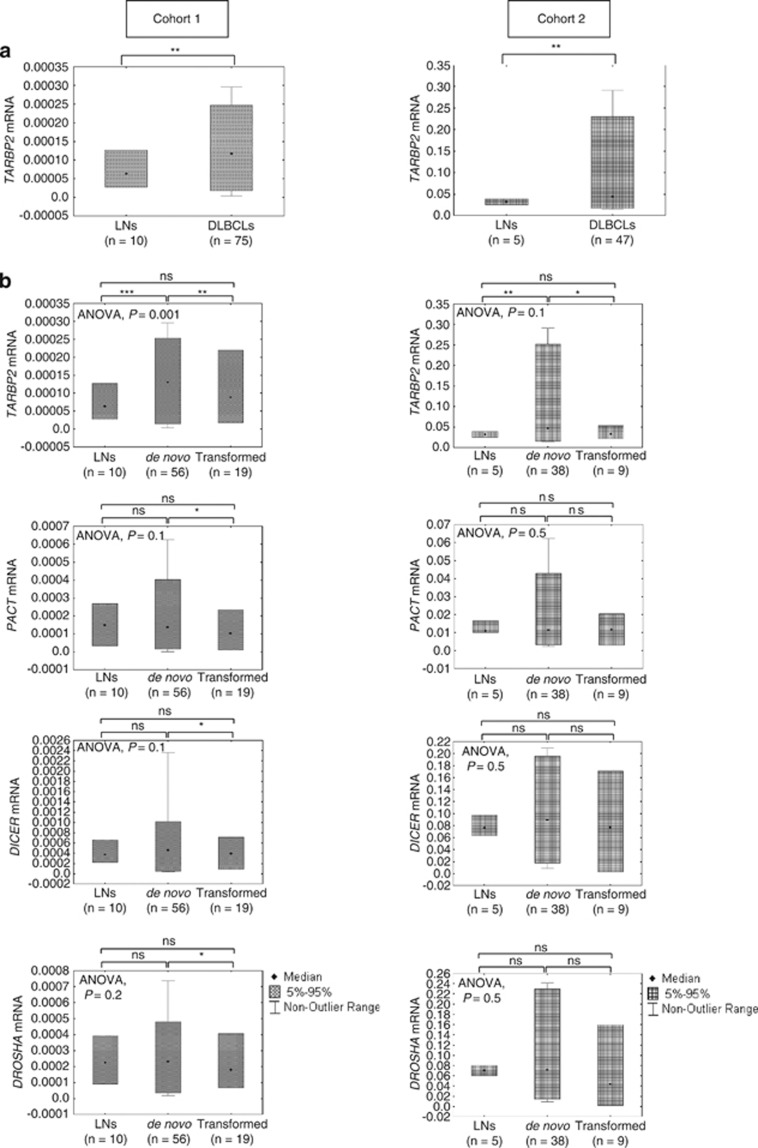
The deregulation of miRNA expression observed in DLBCLs could be caused by abnormal regulation of the miRNA-processing factors. Thus, we evaluated the mRNA expression levels of the main components of miRNA machinery, including DROSHA, DICER, DGCR8, PACT and TARBP2, in 75 DLBCLs (cohort 1) and 10 LNs using RT-qPCR (Supplementary Figure S3). Our results show a significant over-expression of TARBP2 in all DLBCLs or de novo DLBCLs in comparison to LNs (P<0.001, unpaired t-test) but not for the other four genes (Figures 5a and b). However, comparison between de novo and transformed DLBCLs revealed a decreased expression of TARBP2, DICER, PACT and DROSHA in transformed DLBCLs (P=0.004, P=0.021, P=0.016, P=0.033, respectively; unpaired t-test) (Figure 5b), whereas DGCR8 did not show any significant difference between the two groups (data not shown).
Figure 5.
Differential expression of miRNA machinery genes in de novo and transformed DLBCLs and in comparison to LNs, as determined by RT-qPCR. (a) Box plot showing differences in mRNA expression levels for TARBP2 in LNs and DLBCLs for both cohorts. (b) Box plots show relative mRNA expression levels for TARBP2, DICER, PACT and DROSHA in LNs, de novo DLBCLs and transformed DLBCLs for both cohorts. The mRNAs expression among and between groups were compared using one-way ANOVA or unpaired t-test, respectively. P-values <0.05 were considered significant. *P<0.05; **P<0.01; ***P<0.001; ns, not significant.
To validate our results, we analyzed the expression of miRNA machinery genes in an independent cohort (cohort 2) using RT-qPCR. We confirmed a statistically significant over-expression of TARBP2 in all DLBCLs and de novo DLBCLs in comparison to LNs (P<0.01and P<0.001, respectively; unpaired t-test) and no changes in expression for the other genes (Figures 5a and b). Moreover, we did not observe any significant deregulation of miRNA machinery between the two tumor subtypes (Figure 5b).
We also determined the mRNA expression of DICER, DROSHA, TARBP2, DGCR8 and PACT in three DLBCL cell lines. The analysis showed varying levels of expression among the cell lines but we did not observe any obvious difference for the five genes analyzed (Supplementary Figure S4a). Furthermore, protein levels analyzed by western blot were concordant with mRNA expression levels (Supplementary Figure S4b).
miRNA machinery genes expression associated with clinical data of de novo DLBCLs
In cohort 1, we found that higher expression levels of DGCR8 were significantly correlated with high IPI score (3–5) (P=0.012, OR=5.6 (CI95% 1.343–23.358)). Furthermore, patients with high TARBP2 and DROSHA expression had a poorer response to the chemotherapy treatment (P=0.02, OR=9.2 (CI95% 1.005–84.259) and P=0.04, OR=7.3 (CI95% 0.803–65.713), respectively), suggesting that these two genes could be potential predictors of chemotherapy response in de novo DLBCLs. No significant associations were observed for age, gender, disease stage, GCB phenotype, CD10, BCL6, IRF4 or BCL2 expression (Supplementary Table S7).
In cohort 2, we confirmed that high expression of DROSHA (P=0.03, OR=10 (CI95% 0.944–105.922)), but not TARBP2, is associated with a poor response to the treatment. In addition, we observed a significant association between high levels of TARBP2 and non-GCB phenotype (P=0.004, OR=8.45 (CI95% 1.843–38.753)) (Supplementary Table S8).
We then performed a univariate analysis to investigate whether any of the clinical and molecular parameters could have a prognostic impact among patients with de novo DLBCLs. We identified high IPI score (3–5), advanced tumor stage (III–IV) and non-GCB phenotype (P=0.01, P=0.01, P=0.02, respectively) as predictive factors for poorer overall survival (Supplementary Figure S5a). High IPI score (P=0.03) and advance tumor stage (P=0.04) were also associated with shorter disease-free survival (Supplementary Figure S5b). However, none of the miRNA-processing factors showed a significant association with survival of DLBCL patients in both cohorts (data not shown).
Deregulation of miRNA machinery genes affects miRNA processing in DLBCLs
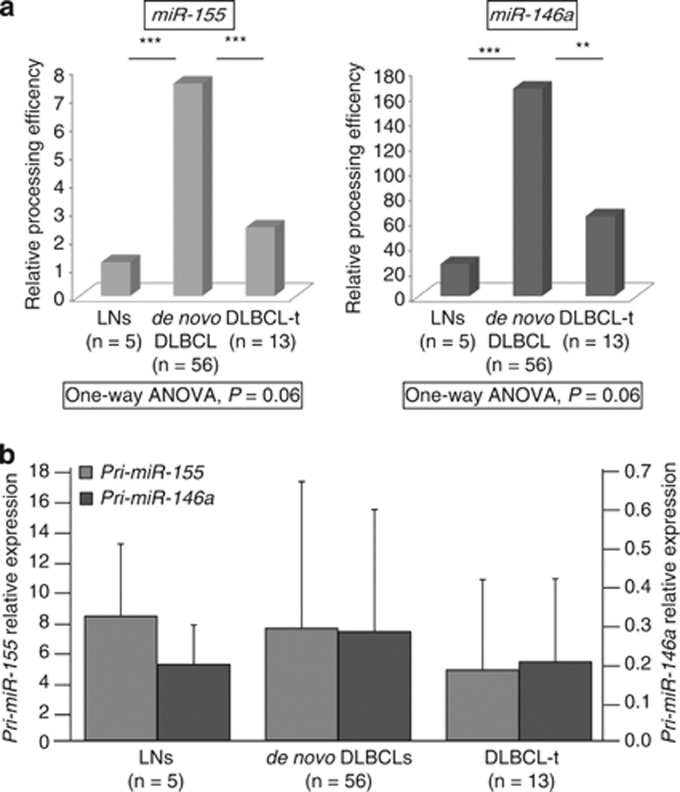
To investigate whether the deregulation of miRNA-processing genes affects miRNA biogenesis, we evaluated the processing efficiency of pre-miR-155 and pre-miR-146a in 5 LNs, 56 de novo DLBCLs and 13 transformed DLBCLs. These two miRNAs were selected because of their differential expression between de novo and transformed DLBCLs. We found a significant increase in the precursor miRNA processing efficiency in de novo DLBCLs as compared with LNs and transformed DLBCLs. However, we did not observe any significant change in pri-miRNA levels among the three groups (Figure 6).
Figure 6.
Deregulation of miRNA machinery affects miRNA processing in DLBCLs (cohort 1). (a) Histograms show a significantly higher miRNA processing efficiency for both miR-155 and miR-146a in de novo DLBCLs as compared with transformed DLBCLs and LNs. (b) The levels of pri-miRNA transcripts do not show any significant difference among the three groups. One-way ANOVA and unpaired t-test were used to determine differences among and between groups and P<0.05 was considered significant. **P<0.01; ***P<0.001.
Inhibition of TARBP2 reduces cell proliferation and increases apoptosis in DLBCL cells
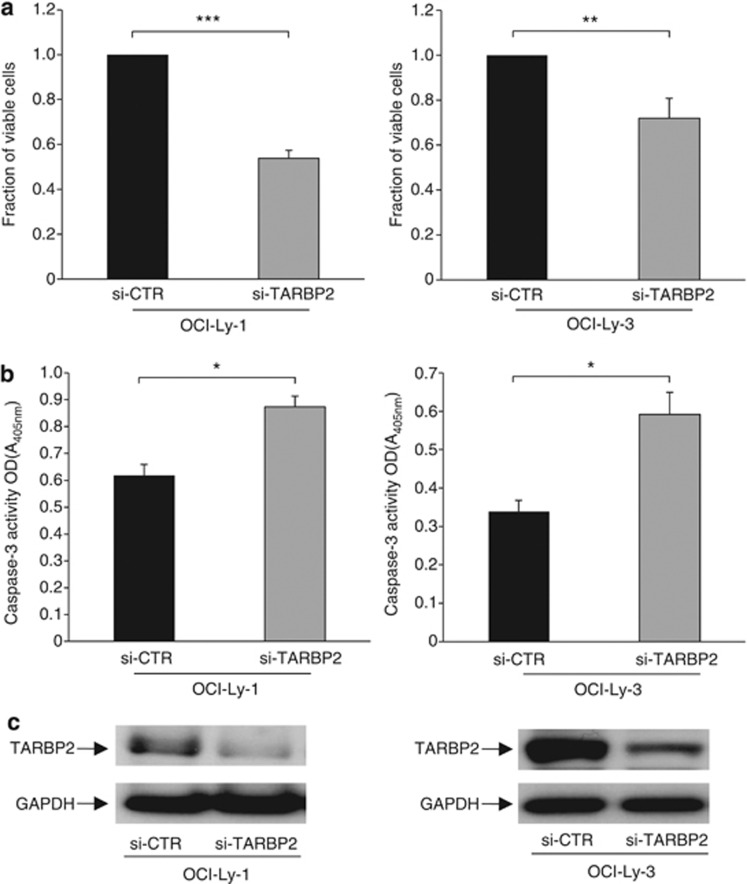
The observed over-expression of TARBP2 in de novo DLBCLs suggests its tumor-promoting activity in this disease. To investigate the functional role of TARBP2, we silenced TARBP2 expression in two DLBCL cell lines (OCI-Ly-1 and OCI-Ly-3) and determined its effect on cell growth and apoptosis. Inhibition of TARBP2 expression significantly decreased cell viability and increased apoptosis in both cell lines as compared with that in siRNA control-treated cells (Figure 7).
Figure 7.
Silencing of TARBP2 reduces cell proliferation and increases apoptosis in DLBCL cell lines. Histograms show the effect of (a) cell viability measured by WST-1 colorimetric assay and (b) cell death evaluated by Caspase-3/CPP32 colorimetric assay in OCI-Ly-1 and OCI-Ly-3 cells transfected with TARBP2 siRNA as compared with si-CTR cells. Data presented represent mean of three independent experiments. Error bars represent standard deviations from the mean. All comparisons were evaluated by paired t-test. P-values <0.05 were considered significant. *P<0.05; **P<0.01; ***P<0.001. (c) Representative western blots showing TARBP2 levels after transfection with TARBP2 siRNA or control siRNA. GAPDH was used as loading control.
Discussion
miRNA expression profiles of DLBCLs
miRNAs have an important role in hematopoiesis and lymphomagenesis. However, the role of miRNAs and their clinical impact in DLBCL are still not fully understood. Here, we show that miRNA expression patterns could clearly distinguish between DLBCLs and LNs, and between de novo and transformed DLBCLs. Despite the identification of DLBCL subtype-specific miRNAs, we found a substantial number of commonly deregulated miRNAs in the two tumor subtypes, suggesting that both de novo and transformed DLBCLs may involve overlapping genetic pathways in DLBCL pathogenesis. Among others, we identified over-expression of miR-21 and under-expression of miR-150, miR-145, miR-143, miR-142-3p/-5p, miR-195 and miR-497 in DLBCLs. Notably, deregulation of most of these miRNAs has been previously reported in DLBCL and/or other malignancies.
In accordance with our findings, over-expression of miR-21 has been found in de novo and transformed DLBCLs in comparison to normal B-cells,13, 14 as well as in many other tumor types.28, 29 High level of miR-21 was also found in the serum of DLBCL patients associated with relapse-free survival,30 suggesting its potential as a non-invasive prognostic marker for DLBCL. miR-150 has a key role in hematopoiesis, including the development of B-cell,31 T-cell32 and NK-cell33 lineages. Downregulation of miR-150 has been described in leukemias and lymphomas, including DLBCL.13, 34 Functionally, increased expression of miR-150 decreases cell proliferation and induces apoptosis in NK/T-cell lymphoma lines.35 Given its crucial role in hematopoiesis, it is tempting to speculate that miR-150 may function as a general tumor suppressor in hematopoietic malignancies. Decreased expression of miR-143 or miR-145 has been shown in many tumor types, including breast cancer, DLBCL, Burkitt's lymphoma and EBV-transformed cell lines.36, 37 Ectopic expression of miR-143 and miR-145 reduces cell growth in lymphoma cell lines.37, 38 Furthermore, miR-142-3p/-5p were among the most downregulated miRNAs in mantle cell lymphoma,39 whereas low levels of miR-195 and miR-497 were recently described in adrenocortical carcinoma26 and breast cancer.40 We previously showed that miR-195/-497 cluster could directly regulate TARBP2 expression in adrenocortical carcinoma.41 Noteworthy, miR-195/-497 and TARBP2 showed an inverse expression pattern in the DLBCL cases (miR-195: Corr=−0,25, P=0.03; miR-497: Corr=−0.3, P=0.008), suggesting that TARBP2 could also be regulated by miR-195 and miR-497 in DLBCL.
In addition, we identified a miRNA classifier that distinguishes de novo and transformed DLBCLs. Notably, most of the differentially expressed miRNAs in our classifier are different from the findings reported previously.14 However, in both studies, transformed DLBCLs were mostly characterized by under-expressed miRNAs as compared with de novo DLBCLs, suggesting that miRNA processing efficiency is different between the two subtypes.
miRNAs associated with pathological and molecular features of de novo DLBCL
We identified a subset of six miRNAs that could classify GCB and non-GCB immunophenotypes among the de novo DLBCLs. In agreement with previous studies, we show higher expression of miR-155 and miR-146a in non-GCB subtype.11, 13, 16 Recently, Thompson et al.42 proposed that high NF-kB activity in the ABC DLBCLs downregulates CD10 expression partly by miR-155; CD10 is highly expressed in the GCB subtype and is low or absent in the ABC subtype.1 In addition, Dagan et al.43 demonstrated that miR-155 targets the germinal center-specific gene HGAL (Human Germinal center-Associated Lymphoma) and increases lymphoma cell motility. Taken together, these observations support a significant oncogenic role for miR-155 over-expression in non-GCB/ABC tumor cells.11, 13, 14, 16 Besides these two miRNAs, we also identified four other miRNAs (that is, let-7g, miR-29c, miR-451 and miR-16) with higher expression in the non-GCB subtype that have not been observed in other studies. The discrepancies may be due to interpretations of immunohistochemical staining, different microarray platforms or the use of cell lines instead of clinical samples to identify miRNA signature.
Moreover, the levels of some miRNAs were inversely correlated with the expression of BCL6 and IRF4. BCL6 is a proto-oncogene that encodes a transcription repressor; it is frequently translocated or mutated in DLBCL and is implicated in DLBCL pathogenesis.44, 45 BCL6 was shown to promote follicular helper T-cell differentiation through repression of multiple miRNAs.46 Interestingly, three of the BCL6-responsive miRNAs (miR-142-3p, miR-142-5p and miR-101) showed lower expression levels in our BCL6-positive DLBCLs, indicating that these miRNAs could be directly suppressed by BCL6 in DLBCL.
Furthermore, we found that DLBCLs in advanced stage of the disease (stage III and IV) had higher expression of miR-494. Over-expression of miR-494 has been observed in follicular lymphoma,47 retinoblastoma48 and Waldenström's macroglobulinemia.49 On the other hand, reduced expression of miR-494 was observed in head and neck squamous cell carcinoma,50 cholangiocarcinoma51 and gastrointestinal stromal tumor.52 miR-494 has been shown to target the tumor suppressor PTEN in myeloid-derived suppressor cells53 and KIT oncogene in gastrointestinal stromal tumor.52 Decreased expression of miR-494 inhibits tumor growth and metastasis in murine breast cancer model,53 whereas its increased expression suppresses cell proliferation in several cancer cell types including head and neck squamous cell carcinoma, cholangiocarcinoma and gastrointestinal stromal tumor.50, 51, 52 Despite that miR-494 over-expression did not affect cell proliferation in the Namalwa Burkitt's lymphoma cell line,54 its functional role in DLBCL remains to be investigated.
Deregulation of miRNA processing factors in DLBCL
Different studies have shown the involvement of miRNA machinery genes in tumor development.19, 20, 55, 56 Here, we found a significant over-expression of TARBP2 in de novo DLBCLs as compared with LNs, and decreased expression of DROSHA, DICER, TARBP2 and PACT in transformed than de novo DLBCLs.
TARBP2 is known to interact with DICER and is involved in the processing of precursor miRNAs to mature miRNAs.7 We therefore investigated whether deregulation of TARPB2 expression affects miRNA biogenesis in DLBCL. Using miR-155 and miR-146a as examples, we showed higher processing efficiency of pre-miRNAs in de novo DLBCLs in comparison to LNs and transformed DLBCLs. However, the expression levels of the pri-miRNA transcripts were similar between the DLBCLs (de novo and transformed cases) and LNs. In addition, previous studies have shown that altered TARBP2 expression in colorectal cancer cell lines could affect, among others, the maturation of miR-29b and members of let-7 family.20, 57 Our findings suggest that over-expression of TARBP2 in de novo DLBCLs may contribute to miRNA deregulation.
Furthermore, we showed that inhibition of TARBP2 expression decreases cell viability and increases cell death in DLBCL cell lines, implying a tumorigenic role of TARBP2 in this tumor type. In line with our findings, over-expression of TARBP2 enhances cell proliferation and transformation in both cell culture systems and mouse models.58, 59 On the contrary, restoration of TARBP2 expression in colorectal cancer cells reduces cell viability and malignancy in vitro and in vivo, supporting its role as a tumor suppressor in colorectal cancer.20 Together, these findings may suggest a tumor specificity of TARBP2 deregulation and function in different cancer types.
In summary, we identified miRNA signatures that are specific for DLBCLs, histological and molecular subtypes. We also show that deregulation of miRNA-processing factors may be involved in the development of DLBCL. In particular, we demonstrate a potential oncogenic role of TARBP2 in DLBCL development. Our study highlights the clinical impact of miRNAs and the involvement of miRNA machinery factors in the pathogenesis of DLBCL.
Acknowledgments
We thank the members of the Medical Genetics research group for their help and suggestions, and Dr Mark Minden (University Health Network, Toronto, Canada) for providing the DLBCL cell lines used in the study. This work was supported by grants from Åke Olsson's Foundation for Haematological Research, Swedish Research Council (523-2009-3517 and 521-2010-3518), Swedish Cancer Foundation, Åke Wiberg's Foundation, Cancer Research Funds of Radiumhemmet, Karolinska Institutet and Stockholm County Council.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
SC, MB and WOL designed the study. SC, LL, DMÖ, PA and HX performed the experiments. SC, LL, PGH, RMA and WOL analyzed the data. MB, GE and CHL provided the clinical materials and information. SC, WOL and CL contributed to writing the manuscript. All authors read and approved the paper.
Supplementary Material
References
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Berglund M, Thunberg U, Amini RM, Book M, Roos G, Erlanson M, et al. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod Pathol. 2005;18:1113–1120. doi: 10.1038/modpathol.3800396. [DOI] [PubMed] [Google Scholar]
- Berglund M, Enblad G, Flordal E, Lui WO, Backlin C, Thunberg U, et al. Chromosomal imbalances in diffuse large B-cell lymphoma detected by comparative genomic hybridization. Mod Pathol. 2002;15:807–816. doi: 10.1097/01.MP.0000024375.04135.2B. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Chi J, Taylor S, Tramonti D, Ballabio E, Palazzo S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13:1248–1260. doi: 10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lisio L, Sanchez-Beato M, Gomez-Lopez G, Rodriguez ME, Montes-Moreno S, Mollejo M, et al. MicroRNA signatures in B-cell lymphomas. Blood Cancer J. 2012;2:e57. doi: 10.1038/bcj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma Z, Swede H, Cassarino D, Fleming E, Fire A, Dadras SS. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencak J, Schmid K, Trautinger F, Wallnofer W, Muellauer L, Soleiman A, et al. High expression of Dicer reveals a negative prognostic influence in certain subtypes of primary cutaneous T cell lymphomas. J Dermatol Sci. 2011;64:185–190. doi: 10.1016/j.jdermsci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Caramuta S, Egyhazi S, Rodolfo M, Witten D, Hansson J, Larsson C, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J Invest Dermatol Aug. 130:2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Caramuta S, Velazquez-Fernandez D, Akcakaya P, Xie H, Hoog A, et al. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18:643–655. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 2011;117:7053–7062. doi: 10.1182/blood-2010-12-326629. [DOI] [PubMed] [Google Scholar]
- Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011;208:2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Saunders NJ, Soneji S, Palazzo S, Dunlop HM, Cooper CD, et al. MicroRNA expression in lymphocyte development and malignancy. Leukemia. 2008;22:1440–1446. doi: 10.1038/sj.leu.2405083. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N, Kuroda J, Nagoshi H, Yamamoto M, Kobayashi S, Tsutsumi Y, et al. Bcl-2 is a better therapeutic target than c-Myc, but attacking both could be a more effective treatment strategy for B-cell lymphoma with concurrent Bcl-2 and c-Myc overexpression Exp Hematol 201139817–828.e811. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- Caramuta S, Lee L, Ozata DM, Akcakaya P, Xie H, Hoog A, et al. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocr Relat Cancer. 2013;20:551–564. doi: 10.1530/ERC-13-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Herscovitch M, Zhao I, Ford TJ, Gilmore TD. NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem. 2011;286:1675–1682. doi: 10.1074/jbc.M110.177063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan LN, Jiang X, Bhatt S, Cubedo E, Rajewsky K, Lossos IS. miR-155 regulates HGAL expression and increases lymphoma cell motility. Blood. 2012;119:513–520. doi: 10.1182/blood-2011-08-370536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, et al. Frequent somatic hypermutation of the 5' noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Arribas AJ, Campos-Martin Y, Gomez-Abad C, Algara P, Sanchez-Beato M, Rodriguez-Pinilla MS, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119:e9–e21. doi: 10.1182/blood-2011-02-339556. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, et al. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- Roccaro AM, Sacco A, Chen C, Runnels J, Leleu X, Azab F, et al. microRNA expression in the biology, prognosis, and therapy of Waldenstrom macroglobulinemia. Blood. 2009;113:4391–4402. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaru AV, Ghiaur G, Yamanaka S, Luvsanjav D, An F, Popescu I, et al. MicroRNA down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology. 2011;54:2089–2098. doi: 10.1002/hep.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Park M, Kim YK, Tae YK, Yang HK, Lee JM, et al. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17:7584–7594. doi: 10.1158/1078-0432.CCR-11-0166. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F, et al. MicroRNA-494 is required for the accumulation and functions of tumor-expanded myeloid-derived suppressor cells via targeting of PTEN. J Immunol. 2012;188:5500–5510. doi: 10.4049/jimmunol.1103505. [DOI] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- Melo S, Villanueva A, Moutinho C, Davalos V, Spizzo R, Ivan C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci USA. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, et al. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim H, Ryu CH, Kim JY, Choi BH, Lim Y, et al. Merlin, a tumor suppressor, interacts with transactivation-responsive RNA-binding protein and inhibits its oncogenic activity. J Biol Chem. 2004;279:30265–30273. doi: 10.1074/jbc.M312083200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.