Abstract
Drug resistance remains a major clinical obstacle to successful treatment in ovarian cancer patients, and the evidence of microRNAs involvement in drug resistance has been emerging recently. In this report, we investigated the role of let-7e in the development of cisplatin-resistant ovarian cancer. On the cellular level, let-7e expression was significantly reduced in cisplatin-resistant human epithelial ovarian cancer (EOC) cell line A2780/CP compared with parental A2780 cell and decreased in a concentration-dependent manner in A2780, SKOV3 and ES2 cells treated with cisplatin. Overexpression of let-7e by transfection of agomir could resensitize A2780/CP and reduce the expression of cisplatin-resistant-related proteins enhancer of zeste 2 (EZH2) and cyclin D1 (CCND1), whereas let-7e inhibitors increased resistance to cisplatin in parental A2780 cells. Quantitative methylation-specific PCR analysis showed hypermethylation of the CpG island adjacent to let-7e in A2780/CP cells, and demethylation treatment with 5-aza-CdR or transfection of pYr-let-7e-shRNA plasmid containing unmethylated let-7e DNA sequence could restore let-7e expression and partly reduce the chemoresistance. In addition, cisplatin combined with let-7e agomirs inhibited the growth of A2780/CP xenograft more effectively than cisplatin alone. Diminished expression of EZH2 and CCND1 and higher cisplatin concentrations in tumor tissue of mice subjected to administration of let-7e agomirs in addition to cisplatin were revealed by immunohistochemistry and atomic absorption spectroscopy, respectively. Taken together, our findings suggest that let-7e may act as a promising therapeutic target for improvement of the sensibility to cisplatin in EOC.
Keywords: let-7e, cisplatin, epithelial ovarian cancer, methylation, agomir
Introduction
Epithelial ovarian cancer is one of the leading causes of gynecologic malignancy-related mortality in women.1 Platinum-based chemotherapeutic strategies are commonly required for patients with advanced ovarian cancer, and clinical drug resistance remains a major barrier to the successful ovarian cancer treatment.2 Resistance to platinum has been reported to be associated with several mechanisms, including increased drug efflux, drug inactivation, alterations in drug target, processing of drug-induced damage and evasion of apoptosis.3, 4, 5 However, the evidence for these mechanisms has not been clearly established.
MicroRNAs (miRNAs) represent a class of small non-coding RNAs that regulate gene expression by targeting mRNAs through sequence-specific base pairing with the 3′-untranslated regions of mRNAs, resulting in RNA degradation and/or translational repression.6, 7 The human let-7 family contains 13 members located on nine different chromosomes and they are downregulated in many different types of cancer, such as breast cancer, lung cancer, ovarian cancer and prostate cancer.8, 9, 10, 11 More importantly, recent evidence linked several miRNAs to chemoresistance in cancers.12 Downregulation of miR-21, miR-342 and miR-489 in MCF-7 cells are responsible for the resistant to tamoxifen.13 The expression of miR-200b, miR-200c, let-7b, let-7c, let-7d and let-7e was significantly downregulated in gemcitabine-resistant cells.14 miRNA microarray showed that miR-30c, miR-130a and let-7e were downregulated in paclitaxel- and cisplatin-resistant ovarian cells.15 In the present study, we focused on the role of let-7e in the development of cisplatin-resistant ovarian cancer.
Results
Repression of let-7e is associated with development of resistance to cisplatin
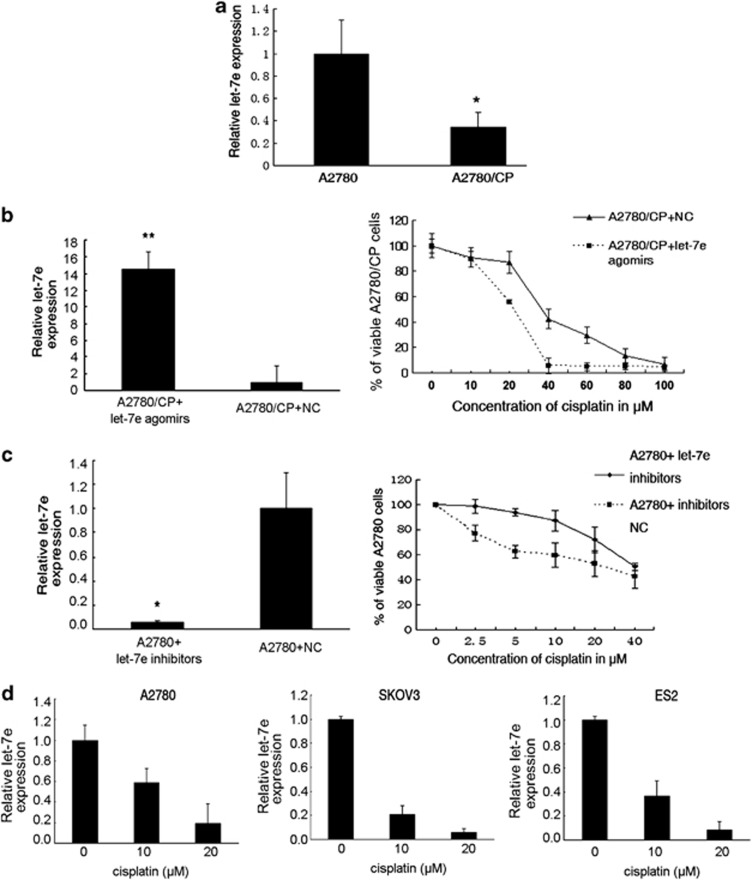
Using quantitative reverse transcription-PCR we found that let-7e level was remarkably decreased (by 66%) in the cisplatin-resistant A2780 (A2780/CP) cells when compared with its parent line A2780 (P=0.016, Figure 1a). We then manipulated let-7e expression in A2780/CP and A2780 cells using specific agomirs (cholesterol-conjugated 2'-O-methyl-modified let-7e mimics) and inhibitors, respectively, and MTT assays were used to determine the resulting changes in drug sensitivity. Let-7e expression in A2780/CP was increased over 14-fold after agomirs treatment, accompanied by significant decrease in IC50 of cisplatin compared with negative control (NC) (IC50: 19.42 versus 46.55 μM, Figure1b). Inversely, inhibitors decreased let-7e expression effectively and, simultaneously, increased the IC50 of cisplatin in A2780 cells compared with NC (IC50: 37.57 versus 21.79 μM, Figure1c). Furthermore, 72 h after treatment with various concentrations of cisplatin (0, 10 and 20 μM), the expression of let-7e decreased in a concentration-dependent manner in A2780, SKOV3 and ES2 cells (P<0.05, Figure 1d). Above results suggest that decreased let-7e expression level is tightly associated with platinum resistance in ovarian cancer.
Figure 1.
Deregulation of let-7e in epithelial ovarian cancer cells. (a) Quantitative reverse transcription-PCR (qRT-PCR) analysis of let-7e expression in ovarian cancer cells. The values correspond to the let-7e/U6 RNA ratio (2−ΔΔCT). The let-7e level in A2780 cells was set as 1. Student's t-test, *P<0.05, compared with parental cells. Bars, s.d. (b) Left, qRT-PCR analysis of let-7e expression in cisplatin-resistant ovarian cancer cell line A2780/CP after transfection with agomirs of let-7e. Bars, s.d. Student's t-test, *P<0.05, *P<0.01, compared with NC. Right, MTT analysis of cell viability after transfection with agomirs of let-7e. Data are represented as mean±s.d., each one done in triplicate. (c) Left, qRT-PCR analysis of let-7e expression in cisplatin-resistant ovarian cancer cell line A2780/CP after transfection with inhibitors of let-7e. Bars, s.d. Student's t-test, *P<0.05, *P<0.01, compared with NC. Right, MTT analysis of cell viability after transfection with inhibitors of let-7e. Data are represented as mean±s.d., each one done in triplicate. (d) qRT-PCR analysis of let-7e expression in epithelial ovarian cancer cell line A2780, SKOV3 and ES2 after treated with cisplatin (0, 10 and 20 μM) for 72 h. Bars, s.d. One-way ANOVA.
DNA hypermethylation contributes to let-7e silencing in ovarian cancer
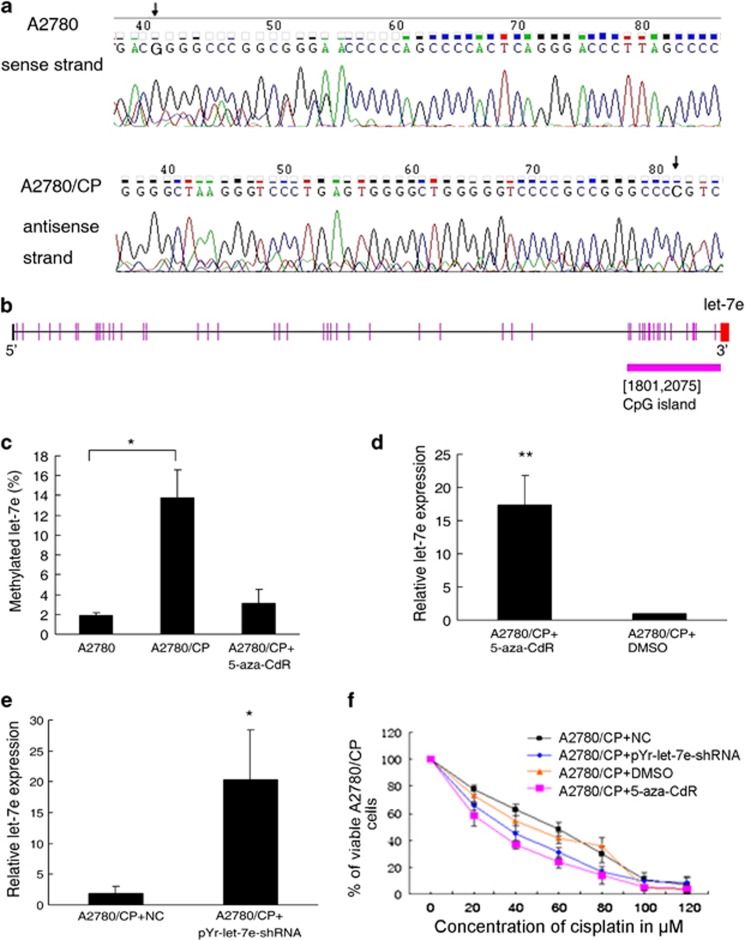
It has been reported that a G→A mutation located at 19 nt downstream of the let-7e precursor led to a significant reduction of mature let-7e.16 However, we could observe such a mutation neither in ovarian cancers (n=20) nor in A2780 and A2780/CP cells using DNA sequencing (Figure 2a). So we hypothesized that epigenetic mechanism may have a role in the regulation of let-7e expression in ovarian cancer. Using a CpG Island Searcher web tool (http://www.cpgislands.com/), a CpG island was identified upstream (−250 bp) of the region encoding let-7e (Figure 2b). Quantitative methylation-specific PCR revealed a 7.38-fold increase in let-7e methylation level in A2780/CP cells when compared with A2780 (P<0.05, Figure 2c). Treatment with 5 μM of DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR) could impair the methylation and restore let-7e expression (Figures 2c and d).
Figure 2.
Epigenetic silencing of let-7e by CpG hypermethylation in cisplatin-resistant ovarian cancer cells. (a) Upper, DNA sequencing result for downstream of the let-7e precursor in A2780 cells (sense strand, 5′-3′), the arrow indicates the G located at 19 nt downstream of the let-7e precursor. Lower, DNA sequencing result for downstream of the let-7e precursor in A2780/CP cells (antisense strand, 3′-5′), the arrow indicates the G located at 19 nt downstream of the let-7e precursor. (b) Schematic presentation of the 274-bp length CpG island upstream (−250 bp) of the region encoding let-7e. The red segment represents let-7e. (c) Results of quantitative methylation-specific PCR (qMSP) analysis of let-7e methylation level in A2780 and A2780/CP cells after demethylating with 5-aza-CdR. Student's t-test, *P<0.05, compared with A2780 cells. Bars, s.d. (d) qRT-PCR analysis of let-7e expression in A2780/CP cells transfected with plasmid and NC. Student's t-test, *P<0.05, compared with NC. Bars, s.d. (e) qRT-PCR analysis of let-7e expression in A2780/CP cells after effective demethylation. Data are showed as fold change. Bars, s.d. Student's t-test, **P<0.01, compared with dimethyl sulfoxide (DMSO) treatment. (f) The viability of cells was measured using MTT assay after effective demethylation, each one done in triplicate. Data are represented as mean±s.d.
After transfection with pYr-let-7e-shRNA plasmid that contains DNA fragment of unmethylated-CpG island and pri-let-7e, the let-7e expression level in A2780/CP cells was significantly increased (Figure 2e). These results verified our hypothesis that let-7e expression was regulated by DNA methylation in adjacent CpG island. Moreover, both treatment with 5-aza-CdR and tansfection with pYr-let-7e-shRNA plasmid could sensitize A2780/CP cells to cisplatin. The IC50 for A2780/CP cells treated with 5-aza-CdR and dimethyl sulfoxide was 24.69 μM and 56.44 μM, respectively, and the IC50 for A2780/CP cells transfected with pYr-let-7e-shRNA plasmid and NC were 35.12 μM and 49.63 μM, respectively (Figure 2f).
EZH2 and CCND1 are potentially regulated by let-7e
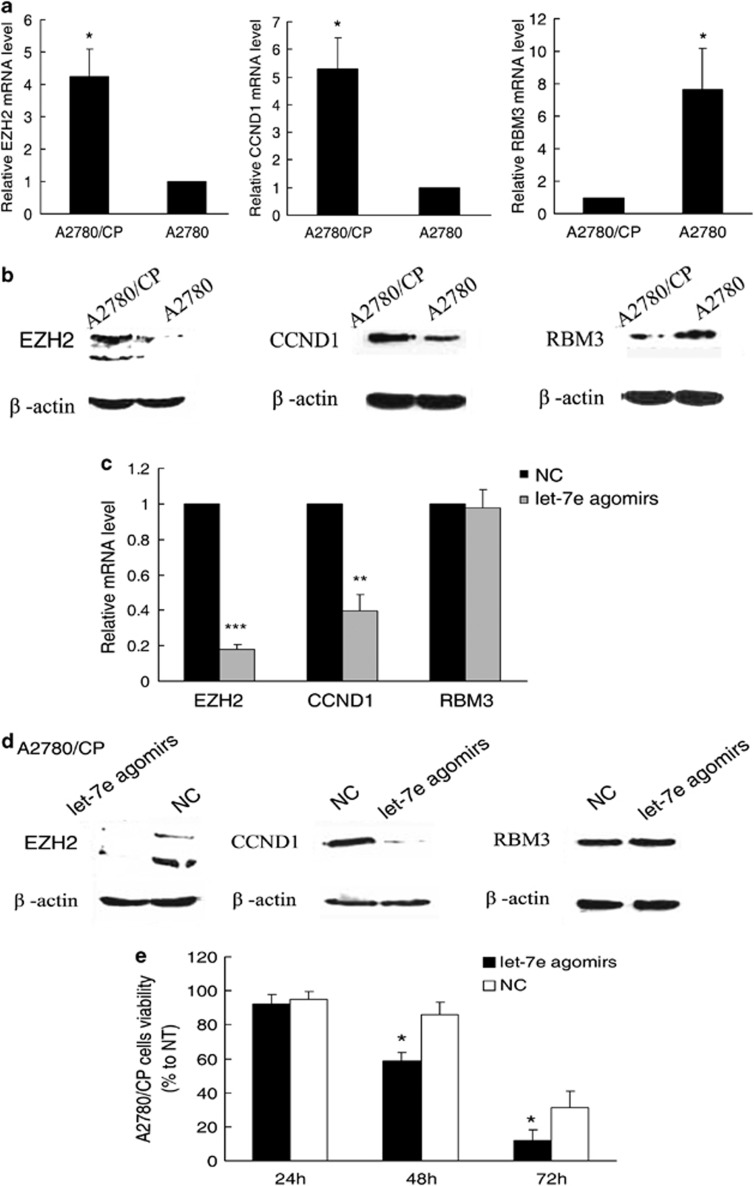
To further investigate the mechanisms of how let-7e impacts drug resistance, we used TargetScan (http://www.targetscan.org/)17 and PicTar (http://pictar.mdc-berlin.de/)18 to predict its target genes. Among the search results, enhancer of zeste 2 (EZH2) and cyclin D1 (CCND1) captured our attention because of their involvement in drug resistance.19, 20 In addition, RNA-binding motif protein 3 (RBM3) was also studied, which has been reported to be a predictor of response to platinum-based chemotherapy in epithelial ovarian cancer. 21 Their expression levels in cell lines were detected by real-time PCR and western blots. We found that A2780/CP cells expressed more EZH2 and CCND1 but less RBM3 compared with A2780 (Figures 3a and b). After treatment with let-7e agomirs, EZH2 and CCND1 expression was markedly decreased in A2780/CP cells. However, no significant alteration in RBM3 expression was observed (Figures 3c and d).
Figure 3.
EZH2 and CCND1 are potential regulated by let-7e. (a) Results of qRT-PCR analysis of EZH2, CCND1 and RBM3 mRNA expression in A2780/CP cells. Student's t-test, *P<0.05, compared with parental cells. Bars, s.d. (b) Results of immunoblotting analysis of EZH2, CCND1 and RBM3 protein expression in A2780/CP and A2780 cells. The two bands of EZH2 are at 100 and 125 kDa, which may represent different isoforms of the protein. (c) qRT-PCR analysis of EZH2, CCND1 and RBM3 mRNA expression in A2780/CP cells treated with let-7e agomirs. Student's t-test, **P<0.01, ***P<0.001, compared with NCC. Bars, s.d. (d) Immunoblotting analysis of EZH2, CCND1 and RBM3 protein expression in A2780/CP cells treated with let-7e agomirs or NC. (e) Trypan blue exclusion viable cell analysis of A2780/CP cells at 24, 48 and 72 h after treatment with let-7e agomirs or NC. Student's t-test, *P<0.05, compared with NC. Bars, s.d.
In addition, viable A2780/CP cells were counted after treatment with cisplatin for 24, 48 and 72 h using typan blue staining. Cell viability of A2780/CP transfected with let-7e agomirs was significantly suppressed compared with cells transfected with NC (Figure 3e). Given that induction of cell cycle progression and promotion of cell growth are the major functions of CCND1, we speculated that let-7e may affect cell cycle regulation in ovarian cancer. Nevertheless, altered cell cycle distribution induced by transfection with agomirs in A2780/CP cells was not detected by flow-cytometry (data not shown).
Re-expression of let-7e could reverse resistance to cisplatin in ovarian cancer in vivo
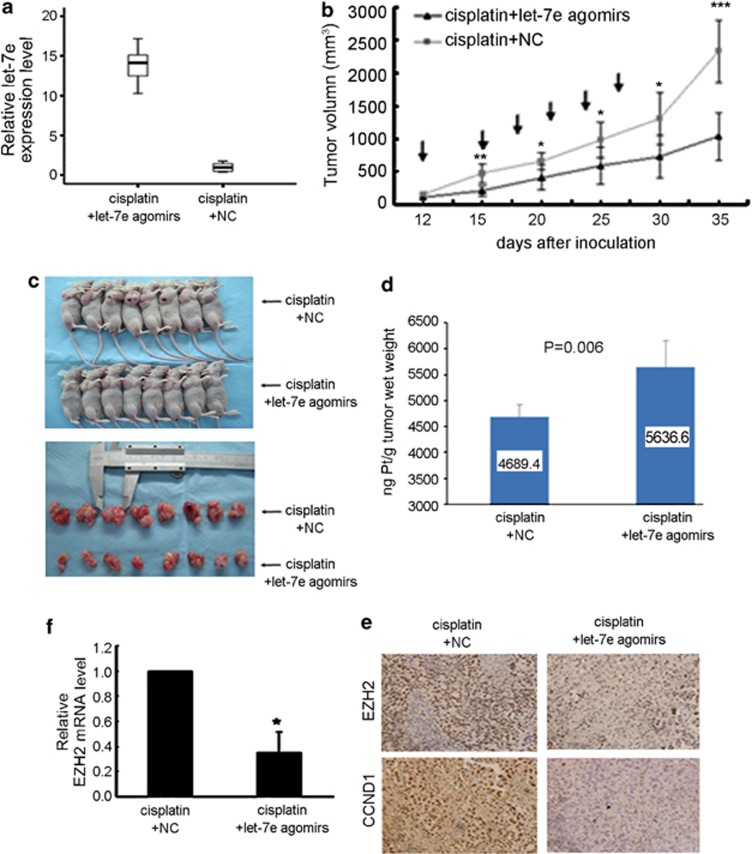
To investigate whether re-expression of let-7e is useful in inhibiting the growth of drug-resistant ovarian cancer in vivo or not, we established A2780/CP subcutaneous xenograft tumor model in nude mice. Twelve days after inoculation, the mice were treated with cisplatin alone or in combination with let-7e agomirs. In the combination group, as gauged by real-time quantitative reverse transcription, let-7e levels were markedly increased in tumor when compared with cisplatin alone group (P<0.001; Figure 4a), suggesting that let-7e agomirs effectively increased let-7e expression in tumor in vivo. The growth curves of xenografts showed that the tumors in the combination group tended to grow more slowly than those in cisplatin alone group. At the start of treatment (12 days after inoculation), there was no difference in tumor size between these two groups. Three days after the first administration of cisplatin, the difference in tumor size was significant (P<0.001) and it increased as the treatment continued. The tumor size was 1031.94±364.90 mm3 on day 35 after inoculation in mice who received combination therapy, whereas tumor size was 2339.61±475.75 mm3 in those given cisplatin alone (Figures 4b, c and P<0.001). In addition, atomic absorption spectroscopy assays showed the cisplatin concentration in tumor tissue was significantly higher in combination group (5636.6 versus 4689.4ng Pt/g tumor wet weight, P=0.006, Figure 4d). In line with results of our experiments in vitro, immunohistochemistry staining of tumor sections revealed a dramatic decrease in the expression of EZH2 and CCND1 in tumors received let-7e agomirs injection (Figure 4e). The mRNA level of EZH2 in combination group was decreased 65% when compared with the control group (Figure 4f).
Figure 4.
Overexpression of let-7e could reverse the chemoresistance in vivo. (a) Box plots show results of qRT-PCR analysis of let-7e expression in nude mice treated with let-7e agomirs and NC. The vertical ordinate is the let-7e expression level normalized to U6 RNA (2−ΔΔCT). Each box shows median, 75th and 25th percentile value. (b) Tumor volumes of nude mice treated with let-7e agomirs and cisplatin. Student's t-test, *P<0.05, **P<0.01, ***P<0.001, compared with treatment of NC and cisplatin. The arrows indicated treatment. (c) Upper, the nude mouse tumor xenograft model treated with cisplatin combined with let-7e agomirs or NC. Lower, the comparation of tumor sizes in the two groups after the xenografted tumors were excised. (d) Determination of cisplatin in tumor tissues by Atomic absorption spectroscopy. Bars, s.d. Student's t-test, P=0.006, compared with NC. (e) Representative results of immunohistochemical staining of EZH2 and CCND1 in the tumor sections of nude mice (magnification × 400). Left, control group; right, let-7e agomirs group. (f) Results of qRT-PCR analysis of EZH2 mRNA expression in xenografted tumors treated with let-7e agomirs. Student's t-test, *P<0.05, compared with NC. Bars, s.d.
Discussion
Indeed, recent studies have shown that miRNAs have a critical role in tumor cells by offering resistance to cytotoxic anticancer therapy.22, 23, 24 Although miRNA genetic alterations, such as deletions, mutations and amplifications, have been increasingly found in human cancers and shown to alter expression levels of mature and/or precursor miRNA transcripts (reviewed in Rossi et al.25), less is known about epigenetic mechanisms contributing to mRNAs expression regulation. Recent studies have reported that miRNA-129-2, miRNA-34b/4 and miRNA-31 undergo 5'-CpG island hypermethylation-associated silencing.26, 27, 28 In our study, after exclusion of the existence of the mutation affecting let-7e expression, we demonstrated that let-7e downregulation in A2780/CP was due to the hypermethylation in its neighboring CpG island. Besides 5-aza-CdR, which results in genome-wide demethylation, site-specific plasmid was used. In A2780/CP cells, when the CpG island adjacent to let-7e was converted to unmethylated form by transfection with pYr-let-7e-shRNA, the transcription activity of pri-let-7e was significantly reduced, suggesting that the CpG island is probably located in a crucial regulatory element of let-7e promoter region. Most importantly, such methylation is associated with chemo-response, and thus provides novel promising therapeutic targets for drug-resistant tumor. That is further verified by using the nude mice model in this study. In general, reports about methylation of miRNA related to drug resistance are very few. More recently, Ma et al.29 reported that DNA methylation-regulated miR-193a-3p dictated the 5-fluorouracil resistance of hepatocellular cancer cells via the repression of the expression of serine/arginine-rich splicing factor 2.
EZH2, the catalytic subunit of the polycomb repressive complex 2 with histone methyltransferase activity, has an essential role in the epigenetic maintenance of the trimethylation on lysine-27 of histone H3 (H3K27me3) repressive chromatin mark. Silencing of EZH2 can suppress cell proliferation and cause G2/M cell cycle arrest in A2780/CP cells as well as in resensitize drug-resistant ovarian cancer cells to cisplatin.22 Although we found re-expression of let-7e resulted in downregulation of EZH2, the underlying mechanisms is still unclear. A recent study provided evidence that Let-7 family of miRNAs is responsible for the regulation of EZH2 in human prostate cancer and the let-7a, let-7b, let-7c and let-7b could strongly inhibit EZH2 3′UTR luciferase activity.30 However, it remains an open question whether EZH2 is a direct target of let-7e.
CCND1 is a member of the highly conserved cyclin family and its overexpression has been frequently observed in a variety of tumors and considered to be associated with tumorigenesis through affecting cell cycle progression.31 Numerous studies have implicated CCND1 overabundance in cisplatin resistance.20, 32, 33 We found transfection with let-7e agomir attenuated CCND1 expression in A2780/CP cells but did not influence cell cycle significantly. That may be because let-7e inversely regulated cell cycle through other mechanism.
Our results showed that cisplatin concentration was increased in let-7e agomirs-treated tumor tissue, suggesting let-7e can promote cellular uptake of cisplatin and/or inhibit its efflux. The traditional view is that cisplatin enters cells mainly by passive diffusion. Gradually, as research continues, it is recognized that several positive transport systems are also involved. Among them, Cu transporters has been most extensively investigated, including the influx transporter 1 (Cu transporter 1, CTR1),34, 35 transporter 2 (Cu transporter 2, CTR2)36 and the efflux transporter ATP7B.37, 38 Studies attempting to identify the impact of let-7e on cell permeability and drug-transport should be performed to more exactly explain our results. However, it must be pointed out that cisplatin triggers cell apoptosis just when it binds to DNA. Thus, we also analyzed cisplatin concentration in DNA isolated from xenografts using atomic absorption spectroscopy, but the results were considered inaccurate because the concentration in DNA was too low (in pg category, data not shown) exceeding the maximum measurement range of our instrument. More accurate methods, for example, high performance liquid chromatography-inductively coupled plasma mass spectrometry (HPLC-ICP-MS), should be used in this case.
Our data indicated that treatment with let-7e agomirs combined with cisplatin could increase the intratumoral cisplatin concentration by about 20.2% and significantly reduce but not stop the growth of tumors in vivo, suggesting higher let-7e level might be required for more effective tumor growth inhibition through further elevated cisplatin concentration in tumor cell. On the other hand, other mechanisms involved in cisplatin resistance should be considered. Cisplatin binds to and causes crosslinking of DNA, which ultimately triggers cell apoptosis. Although let-7e agomirs increased intracellular cisplatin, the cytotoxicity of cisplatin could still be limited by the inhibition of apoptosis and activation of DNA repair.39 Thus, simultaneous intervention on multiple mechanisms of drug resistance may provide a better chance of being able to overcome this challenge.
In summary, our findings link epigenetic-silencing of let-7e to the development of cisplatin resistance, which inspires potential therapeutic strategies targeting let-7e to improve the treatment of cisplatin-resistant ovarian cancer.
Materials and methods
Cell lines and cell culture
The human ovarian epithelial carcinoma cell lines A2780, ES2 and SKOV3 (ATCC) were cultured in RPMI1640 medium (Gibco BRL, New York, NY, USA) supplemented with 50 IU/ml of penicillin, 50 μg/ml streptomycin and 10% fetal bovine serum. The cisplatin-resistant A2780 (A2780/CP) cells were generated by exposure to increasing concentration of cisplatin.40 The A2780/CP cells used in the present study were cultured in media without cisplatin for 1 month to exclude the acute stress reactions mediated by the administration of cisplatin and the subline was approximately three to five fold more resistant to cisplatin than was the parent line (A2780).
Real-time PCR for let-7e
Total RNA of cell lines and tissues were isolated using Trizol (Invitrogen, Carlsbad, CA, USA). Expression level of let-7e was analyzed using the Bulge-Loop miRNA real-time PCR Primer Set (Cat no. MQP-0101, RiboBio, Guangzhou, China), following the manufacturer's instructions. The universally expressed U6 small nuclear RNA was used for let-7e template normalization. The cycle parameters for the reverse transcription reaction were 70 °C for 10 min, followed by 42 °C for 60 min and 70 °C for 10 min. SYBR green real-time PCR was carried out on an ABI PRISM 7300 instrument (Applied Biosystems, Foster City, CA, USA). The PCR conditions included initial incubation step of 20s at 95 °C, 40 cycles of denaturing at 94 °C for 10s, annealing at 55 °C for 30s and extension at 70 °C for 30s. Comparative real-time PCR was done in triplicate, including no-template controls. The fold change for miRNA expression level was calculated using 2−ΔΔCT method.41
Transfections
The let-7e agomirs were synthesized by RiboBio Corporation and inhibitors were purchased from Genepharma Corporation (Shanghai, China). Cells were plated in six-well plates (2 × 105/well) and they were transfected with either 100 nmol let-7e agomirs or inhibitors at 60% confluence.
Real-time PCR analysis for mRNA expression of EZH2, RBM3 and CCND1
Real-time PCR was performed to determine the level of EZH2, RBM3 and CCND1 mRNA expression. Briefly, total RNA of cell lines and tissues were isolated using Trizol (Invitrogen). Reverse transcription was run using RevertAid First Strand complementary DNA Synthesis Kit (MBI Fermentas, Flamborough, ON, Canada) according to the supplied protocol. The primers were as follows: EZH2: 5′-TTGTTGGCGGAAGCGTGTAAAATC-3′ and 5′-TCCCTAGTCCCGCGCAATGAGC-3′. CCND1: 5′-TGCCCTCTGTGCCACAGATG-3′ and 5′-TCTGGAGAGGAAGCGTGTGA-3′. RBM3: 5′-CTTCAGCAGTTTCGGACCTA-3′ and 5′-ACCATCCAGAGACTCTCCGT-3′. Reactions were performed in 25 μl PCR mixture included 2 × SYBR Green Real-time PCR mix (Toyobo, Japan), 2 μg of complementary DNA template and 0.3 μM of each primer. The GAPDH was used as internal control. The PCR profile was 95°C for 60s, and then followed by 40 cycles of 95 °C for 15s, 60 °C for 15 s and 72 °C for 45s. The fold change for EZH2, RBM3 and CCND1 mRNA expression level was calculated using 2−ΔΔCT method.41
Drug sensitivity assay
The 50% inhibitory drug concentration (IC50) was determined using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazoliumbromide) assay. Cells were plated in 96-well plates (5000 cells/well), and after cellular adherence, the cells were exposed to various concentrations of cisplatin. After incubation for 48 h, the cells were incubated with 20 μl MTT (at a final concentration of 0.5 mg/ml) at 37°C for 4 h. The medium was removed and the precipitated formazan was dissolved in 100 μl dimethyl sulfoxide. The absorbance at 490 nm was detected using microplate reader. The IC50 was estimated by the relative survival curve. Each assay was performed in triplicate.
Sequencing analysis of let-7e precursor in ovarian cancer cell lines
The sequencing analysis of let-7e precursor in ovarian cancer cell lines and tissues was performed by the Sangon Biotech Co. Ltd (Shanghai, China) using an ABI PRISM 3730 DNA sequencer.
5-aza-CdR treatment and quantitative methylation-specific PCR analysis
Using the CpG Island Searcher web tool (http://www.cpgislands.com/), a CpG island was identified upstream (−250 bp) of the region encoding let-7e. A2780/CP cells were treated with 5-aza-CdR at 5 μM or dimethyl sulfoxide for 72 h. For methylation analysis, genomic DNA was extracted from the tumor samples or the cells by using Genomic DNA extraction kit (Tiangen Biotech, Beijing, China). One microgram of genomic DNA was subjected to treatment with the CpGenome DNA Modification Kit (Chemicon, Billerica, MA, USA) according to the manufacturer's instructions. The primers for methylated sequence are 5′ -CGTGTCGGGGGTTTATTATC-3′ (forward) and 5′-CTACCTCAACCCGAAAAACG-3′ (reverse), and the primers for unmethylated sequence are 5′-GTTTGTGTTGGGGGTTTATTATT-3′ (forward) and 5′-CTACCTCAACCCAAAAAACACAA-3′ (reverse). The PCR conditions were as follows: 95 °C for 15 min followed by 40 cycles of 94 °C for 30s, 51 °C for 45 s and 72 °C for 1 min. The percentage of CpG methylation in a sample was estimated according to the method described by Lu L et al.42
Plasmids preparation
An unmethylated DNA fragment spanning positions −250 to +250 containing the CpG island (−250 bp) and pri-let-7e was synthesized. This DNA fragment was cloned into the vector pYr-miR30-shRNA-2 (Genechem, Shanghai, China) and the plasmid was named pYr-let-7e-shRNA.
Immunohistochemistry for the detection of EZH2, RBM3 and CCND1
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded tissues. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Antigen retrieval was carried out in citrate buffer (10 mM, pH-6) for 20 min at 100 °C. Tissue sections were incubated first with the primary antibodies of EZH2 (1:100, Invitrogen), RBM3 (1:25, PTG, Chicago, IL, USA) and CCND1 (1:25, CST, Danvers, MA, USA) for 16 h at 4 °C and then subsequently with a secondary biotinylated antibody (1:200, Boster, China) for 2 h at 37 °C followed by incubation with streptavidin–peroxidase complex for 20 min at room temperature.
Western blot assay
Proteins from cell lysates were separated and detected using western blotting as previously described.19 The primary antibodies used in the present study were as follow: anti-mouse EZH2 antibody (1:500; Cell Signal, Beverly, MA, USA), anti-rabbit RBM3 antibody (1:400; PTG) and anti-rabbit CCND1 antibody (1:200; CST).
Cell cycle assay
Seventy-two hours after transfection, cells were harvested by trypsinization, washed three times with ice-cold phosphate-buffered saline and then fixed with 70% ethanol overnight at 4 °C. The fixed cells were rehydrated in phosphate-buffered saline and subjected to propidium iodide/RNase staining followed by FACS analysis (Fluorescence Activated Cell Sorter Scan FACS Calibur; Becton Dickinson, Mountain View, CA, USA). The percentage of cells in each phase of the cell cycle was estimated using ELITE software (Gainesville, FL, USA).
Trypan blue exclusion viable cell assay
The viability of cells treated with cisplatin after transfection with let-7e agomirs was assessed by Trypan blue exclusion method. Cells were harvested and resuspended in equal volumes of medium and Trypan blue (0.05% solution) and viable cells were counted in triplicate using a haemocytometer.
In vivo assay
All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Centre, Tongji Medical College, Huazhong University of Science and Technology, Wuhan. Female nude mice (BALB/c, 4-6wk) were purchased from Hunan SLAC laboratory Animal Co. Ltd. (SLAC, Hunan, China). For preparation of subcutaneous xenograft model, 0.2 ml A2780/CP ovarian cancer cells (2.0 × 106 in phosphate-buffered saline/100 μl) were injected subcutaneously into the right flank of the nude mice. Twelve days after tumor cell inoculation with confirmation of successful maturation of tumors, mice were divided randomly into two groups (eight mice per group). They were treated with cisplatin in combination with let-7e agomirs (combination group) or with NC of agomirs (cisplatin alone group) every 3 days for 2 weeks. Cisplatin was administrated by intraperitoneal injection at a dose of 4 mg/kg body weight and let-7e agomirs (2 nmol; RiboBio, Guangzhou, China) were given locally by direct injection into the xenografts. The tumors were monitored with a caliper every day and tumor volumes were determined (in cubic millimeter) by measuring in two directions and was calculated as tumor volume=length × (width)2/2. One week after the last administration of cisplatin and agomirs, all mice were killed according to the animal experimental guidelines. The xenografted tumors were excised and paraffin-embedded or cryopreservation at −80 °C.
Atomic absorption spectroscopy for detection of cisplatin in tumor tissue
Pt concentrations in xenografts were measured by flameless atomic absorption spectrometry (Varian SpectrAA 240FS, Palo Alto, CA, USA) using a standard curve covering the range of 0-450 ppb. Before analyses, the tumor samples were digested in concentrated nitric acid with heating to 140 °C for 60 min, followed by evaporation to near dryness, and the digests were dissolved in 2 ml deionized water for Pt detection. All samples were analyzed in duplicate, and the duplicate values were within 10% relative s.d. of each other in all cases.
Statistical analysis
Student's t-test, one-way analysis of variance with LSD (Fisher's least significant difference) post hoc tests were used for statistical analyses using SPSS v. 13.0.0 (Chicago, IL, USA). All P-values are two-tailed; P-values<0.05 were considered significant.
Acknowledgments
This study is supported by National Natural Science Foundation of China (81101961) and China Postdoctoral Science Foundation (Grand No.20100480904).
The authors declare no conflict of interest.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat. 2009;12:114–126. doi: 10.1016/j.drup.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Brabec V, Kasparkova J. Molecular aspects of resistance to antitumor platinum drugs. Drug Resist Updat. 2002;5:147–161. doi: 10.1016/s1368-7646(02)00047-x. [DOI] [PubMed] [Google Scholar]
- Morin PJ. Drug resistance and the microenvironment: nature and nurture. Drug Resist Updat. 2003;6:169–172. doi: 10.1016/s1368-7646(03)00059-1. [DOI] [PubMed] [Google Scholar]
- Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, IeM Shih, Zhang Y, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, L'Abbe D, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710–1716. doi: 10.1093/carcin/bgn073. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Hu S, Yu L, Li Z, Shen Y, Wang J, Cai J, et al. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;10:788–795. doi: 10.4161/cbt.10.8.12913. [DOI] [PubMed] [Google Scholar]
- Noel EE, Yeste-Velasco M, Mao X, Perry J, Kudahetti SC, Li NF, et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am J Pathol. 2010;176:2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlén A, Brennan DJ, Nodin B, O'Connor DP, Eberhard J, Alvarado-Kristensson M, et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- Rossi S, Sevignani C, Nnadi SC, Siracusa LD, Calin GA. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm Genome. 2008;19:526–540. doi: 10.1007/s00335-008-9119-8. [DOI] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, He Y, Zhang H, Fei Q, Niu D, Wang D, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287:5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, et al. Loss of let-7 upregulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 2012;7:e33729. doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen ME, Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med. 2004;10:158–162. doi: 10.1016/j.molmed.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Biliran H, Jr, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, et al. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–6086. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- Warenius HM, Seabra LA, Maw P. Sensitivity to cis-diamminedichloroplatinum in human cancer cells is related to expression of cyclin D1 but not c-raf-1 protein. Int J Cancer. 1996;67:224–231. doi: 10.1002/(SICI)1097-0215(19960717)67:2<224::AID-IJC13>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Ishida S, McCormick F, Smith-McCune K, Hanahan D. Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator. Cancer Cell. 2010;17:574–583. doi: 10.1016/j.ccr.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer AK, Manorek GH, Howell SB. Contribution of the major copper influx transporter CTR1 to the cellular accumulation of cisplatin, carboplatin, and oxaliplatin. Mol Pharmacol. 2006;70:1390–1394. doi: 10.1124/mol.106.022624. [DOI] [PubMed] [Google Scholar]
- Blair BG, Larson CA, Safaei R, Howell SB. Copper transporter 2 regulates the cellular accumulation and cytotoxicity of cisplatin and carboplatin. Clin Cancer Res. 2009;15:4312–4321. doi: 10.1158/1078-0432.CCR-09-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katano K, Kondo A, Safaei R, Holzer A, Samimi G, Mishima M, et al. Acquisition of resistance to cisplatin is accompanied by changes in the cellular pharmacology of copper. Cancer Res. 2002;62:6559–6565. [PubMed] [Google Scholar]
- Safaei R, Otani S, Larson BJ, Rasmussen ML, Howell SB. Transport of cisplatin by the copper efflux transporter ATP7B. Mol Pharmacol. 2008;73:461–468. doi: 10.1124/mol.107.040980. [DOI] [PubMed] [Google Scholar]
- Shahzad MM, Lopez-Berestein G, Sood AK. Novel strategies for reversing platinum resistance. Drug Resist Updat. 2009;12:148–152. doi: 10.1016/j.drup.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens BC, Hamilton TC, Masuda H, Grotzinger KR, Whang-Peng J, Louie KG, et al. Characterization of a cis-diamminedichloroplatinum(II)-resistant human ovarian cancer cell line and its use in evaluation of platinum analogues. Cancer Res. 1987;47:414–418. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]