Abstract
Obesity is of global health concern. There are well-described inverse relationships between female pubertal timing and obesity. Recent genome-wide association studies of age at menarche identified several obesity-related variants. Using data from the ReproGen Consortium, we employed meta-analytical techniques to estimate the associations of 95 a priori and recently identified obesity-related (body mass index (weight (kg)/height (m)2), waist circumference, and waist:hip ratio) single-nucleotide polymorphisms (SNPs) with age at menarche in 92,116 women of European descent from 38 studies (1970–2010), in order to estimate associations between genetic variants associated with central or overall adiposity and pubertal timing in girls. Investigators in each study performed a separate analysis of associations between the selected SNPs and age at menarche (ages 9–17 years) using linear regression models and adjusting for birth year, site (as appropriate), and population stratification. Heterogeneity of effect-measure estimates was investigated using meta-regression. Six novel associations of body mass index loci with age at menarche were identified, and 11 adiposity loci previously reported to be associated with age at menarche were confirmed, but none of the central adiposity variants individually showed significant associations. These findings suggest complex genetic relationships between menarche and overall obesity, and to a lesser extent central obesity, in normal processes of growth and development.
Keywords: adiposity, body mass index, genetic association studies, menarche, obesity, waist circumference, waist:hip ratio, women's health
There have been dramatic changes in the prevalence of obesity worldwide (1). Although higher-income countries have been at the forefront of the obesity epidemic, developing countries have recently shown striking trends in obesity (1, 2). Given that obesity may influence multiple physiological functions in children and adolescents (3, 4), recent trends may have lasting health impacts.
Age at menarche is considered a marker of hypothalamic-pituitary-driven pubertal development in girls. Secular trends towards earlier age at menarche began during the late 19th century in European countries (5). Early menarche is a risk factor for increased adult adiposity (5), type 2 diabetes (6, 7), breast cancer (8, 9), adolescent risk-taking behaviors (10) and all-cause mortality (11).
Global and local obesogenic changes in the environment and gene-environment interactions are hypothesized to account for recent shifts in the distributions of both age at menarche and adult obesity (5). A negative association between childhood obesity and timing of menarche has been described (12), and a recent Mendelian randomization study suggested a possible causal relationship for this association (13). Another study has shown that earlier menarche is associated with adult obesity and predicts faster growth tempo among a woman's children (14).
Although the biological mechanisms of the relationship(s) among increased childhood/adolescent adiposity, menarcheal timing, and adult obesity have not been elucidated (3, 5), there is evidence for genetic influences (15, 16). For instance, the fat mass and obesity-associated gene (FTO) has been associated with a decrease in age at menarche and increased body mass index (BMI; weight (kg)/height (m)2) in childhood and adulthood (17). Recent genome-wide association studies (GWAS) have identified several loci for age at menarche (18–22), several of which were also associated with measures of obesity over the life course (18, 23, 24). In a candidate single-nucleotide polymorphism (SNP) study, Elks et al. (18) recently described 11 adiposity-related (BMI and waist circumference) loci that were also associated with age at menarche (P < 0.05) among women of European ancestry.
Two recent large GWAS from the Genetic Investigation of Anthropometric Traits (GIANT) Consortium have substantially expanded the number of validated overall (BMI) and central adiposity (waist:hip ratio adjusted for BMI) loci by reporting divergent sets of novel loci (18 and 13, respectively) for these traits in persons of European ancestry (25, 26). Genetic and environmental factors may contribute to increased prepubertal adiposity, resulting in early menarche, shorter stature, and increase postpubertal weight gain in women (5). Thus, understanding the relationship between age at menarche and obesity may have important implications for public health. Therefore, we aimed to systematically investigate the association between newly identified BMI-, waist circumference-, and waist:hip ratio-related SNPs and age at menarche in 92,116 women of European descent.
MATERIALS AND METHODS
Population and outcome
Data from 38 studies in the ReproGen Consortium (see Web Table 1 and the Web Appendix, available at http://aje.oxfordjournals.org/) were included (18). Each study consisted of women with genome-wide association genetic information who identified themselves as being of European descent and who reported their age at menarche as being between 9 and 17 years of age (n = 92,116). Exclusion of women with extreme ages at menarche was intended to capture the normal range of pubertal variation by excluding precocious or late-onset puberty or amenorrhea, which might be due to rare genetic variants with large effect sizes (e.g., Mendelian disorders).
The institutional review board of the University of North Carolina at Chapel Hill reviewed and approved this research.
SNP selection
An a priori list of adiposity SNPs was compiled using the National Human Genome Research Institute GWAS catalog (27) for reported associations in the literature (P < 1 × 10−5) with adult BMI, waist circumference, and waist:hip ratio as of August 11, 2011 (25, 26, 28–37). Sixty-eight of the resulting 95 candidate SNPs were previously reported to be genome-wide significant (P < 5 × 10−8; Web Table 2). When information on direction of effect was absent in the literature, the direction of each SNP-adiposity association was supplemented by publicly available data from overall (BMI) or central (waist:hip ratio adjusted for BMI) adiposity analyses, as appropriate (25, 26). Further information on SNP selection is presented in Web Figure 1, parts A–D.
Linkage disequilibrium among variants
Because of our reliance on multiple sources to generate our candidate SNP list, some loci contained more than 1 SNP or were associated with more than 1 obesity-related phenotype. The 11 adiposity SNPs previously reported to be associated with menarche in the literature are presented in Web Table 3 (18). An additional 14 SNPs at these 11 loci and 70 SNPs at 60 loci not previously described as being associated with menarche were then assessed for dependence with respect to phenotype using publicly available information derived from HapMap CEU data (38). Variants that were within 500 kilobases of one another and in low linkage disequilibrium with each other (r2 < 0.2) were considered to not represent the same underlying signal. If 2 or more SNPs in linkage disequilibrium (r2 ≥ 0.2) were available, the most replicated SNP in the GWAS literature (27) was selected to represent the region (Table 1, Web Table 4). The other SNPs are shown in Web Table 5.
Table 1.
Associations Between Body Mass Indexa and Single-Nucleotide Polymorphisms at Novel Age-at-Menarche Loci and Loci Previously Reported to Be Associated With Age at Menarche (P < 0.05/95b) Among Women (n ≤ 92,105) From 38 Studies in the ReproGen Consortium, 1970–2010
| Reference SNP No. | Chromosome | Position,c base pairs | Nearest Gene | No. of Studies | No. of Women | Allele |
Fixed Effects |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coded | Other | Coded Frequency | Estimate (SE), daysd | P Value | P for Heterogeneity | ||||||
| Novel AAM Loci | |||||||||||
| rs1514175 | 1 | 74,764,232 | TNNI3K | 37 | 89,922 | A | G | 0.43 | −12.2 (2.4) | 4.0 × 10−7 | 0.07 |
| rs713586 | 2 | 25,011,512 | RBJ | 38 | 92,078 | T | C | 0.53 | 11.7 (2.4) | 8.4 × 10−7 | 0.82 |
| rs887912 | 2 | 59,156,381 | FANCL | 38 | 92,063 | T | C | 0.28 | −11.1 (2.7) | 3.8 × 10−5 | 0.91 |
| rs10769908d | 11 | 8,440,665 | STK33 | 36 | 86,344 | T | C | 0.48 | 9.6 (2.4) | 7.8 × 10−5 | 0.98 |
| rs2241423 | 15 | 65,873,892 | MAP2K5 | 38 | 92,085 | A | G | 0.22 | 13.1 (2.9) | 6.1 × 10−6 | 0.25 |
| rs12444979 | 16 | 19,841,101 | GPRC5B | 37 | 88,557 | T | C | 0.13 | 13.5 (3.7) | 2.6 × 10−4 | 0.20 |
| Previously Reported AAM Locie | |||||||||||
| rs2568958 | 1 | 72,537,704 | NEGR1 | 38 | 92,071 | A | G | 0.61 | −13.4 (2.4) | 3.3 × 10−8 | 0.41 |
| rs543874 | 1 | 176,156,103 | SEC16B | 38 | 92,053 | A | G | 0.81 | 19.2 (3.1) | 6.7 × 10−10 | 0.81 |
| rs7561317e | 2 | 634,953 | TMEM18 | 38 | 92,083 | A | G | 0.17 | 18.4 (3.2) | 8.5 × 10−9 | 0.70 |
| rs9816226 | 3 | 187,317,193 | ETV5 | 38 | 92,067 | A | T | 0.18 | 12.5 (3.1) | 7.0 × 10−5 | 0.74 |
| rs7481311e | 11 | 27,539,705 | BDNFf | 37 | 89,927 | T | C | 0.23 | −12.7 (2.9) | 1.3 × 10−5 | 0.14 |
| rs6265d,e | 11 | 27,636,492 | BDNFf | 37 | 89,934 | T | C | 0.18 | 13.6 (3.2) | 1.7 × 10−5 | 0.51 |
| rs8050136e | 16 | 52,373,776 | FTO | 37 | 89,844 | A | C | 0.40 | −16.7 (2.4) | 9.5 × 10−12 | 0.23 |
| rs29941 | 19 | 39,001,372 | KCTD15 | 38 | 92,065 | A | G | 0.32 | 11.4 (2.6) | 9.3 × 10−6 | 0.11 |
Abbreviations: AAM, age at menarche; BMI, body mass index; GIANT, Genetic Investigation of Anthropometric Traits; SE, standard error; SNP, single-nucleotide polymorphism.
a Weight (kg)/height (m)2.
b All associations shown had a P value less than a Bonferroni correction for 95 tests or P < 0.00053.
c Position from HapMap Build 36 (http://hapmap.ncbi.nlm.nih.gov/).
d The T allele for rs6265 at BDNF has been associated with increases in both menarche and BMI. All other SNP associations in the table have inverse associations for the same coded allele with menarche and BMI from the literature. However, information on direction of association with BMI for rs10769908 at STK33 was supplemented by publicly available data from the GIANT Consortium as well as information from another SNP (rs4929949 at RPL27AI, shown in Web Table 4) in linkage disequilibrium (r2 = 0.97).
e Unlike those listed in Web Table 3, these SNPs have not previously been reported to be associated with age at menarche. However, they were generally in linkage disequilibrium (r2 ≥ 0.2) with other SNPs previously reported to be associated with both adiposity traits and menarche (18) (see Web Table 3). Nonetheless, the associations presented here are larger by up to 5,389 additional samples and now include heterogeneity P values. Additional SNP associations at 3 of these previously reported menarche-adiposity loci are reported in Web Table 4 (TMEM18, BDNF, and FTO), because they were in linkage disequilibrium with the noted SNPs (r2 ≥ 0.2).
f The BDNF locus had evidence of 2 signals (represented here by rs7481311 and rs6265) based on linkage disequilibrium (r2 < 0.2).
Only 1 locus (defined as ±500 kilobases in size) had evidence of 2 signals based on our above methodology: the brain-derived neurotrophic factor gene (BDNF) (Table 1). Multiple SNPs at the melanocortin 4 receptor (MC4R) and neurexin-3 (NRXN3) loci have been reported in association with various adiposity phenotypes (25, 31). rs489693 and rs12970134 at the MC4R locus were considered to represent waist circumference and BMI, respectively (Web Table 4), whereas NRXN3 was considered to represent BMI only (31). Therefore, rs10150332 is presented in Web Table 4 and a second NRXN3 variant, rs10146997, in perfect linkage disequilibrium with rs10150332 (r2 = 1.0) is presented in Web Table 5.
Quality control
This study used existing genetic data from the ReproGen Consortium. Study-specific call rates, minor allele frequencies, Hardy-Weinberg equilibrium, and other quality control measures are described briefly in the Web Appendix and more in depth elsewhere (18). Imputed SNPs used in analyses were required to have a quality score greater than 0.3 (MACH software, variable rsq (www.sph.umich.edu/csg/abecasis/MACH/index.html)) or greater than 0.4 (IMPUTE software, proper_info (https://mathgen.stats.ox.ac.uk/impute/impute.html)).
Meta-analytical techniques
Investigators in each participating study performed a linear regression of age at menarche (in years) on the number of coded alleles per SNP, while adjusting for birth year, population stratification, and study center (as applicable). We tested between-study heterogeneity for all SNP associations and considered a given association potentially heterogeneous when the heterogeneity P value was less than 0.10.
Study-specific results were meta-analyzed using inverse variance weighting fixed-effect meta-analysis (METAL) (39). All estimated changes in age at menarche per allele (i.e., slopes) were then converted to represent changes in menarche onset in days per allele (365.25βyear = βdays) and are referred to as effect estimates.
In the presence of potential heterogeneity, we used random-effects meta-analysis and performed meta-regression using restricted maximum likelihood in Stata 11 (StataCorp LP, College Station, Texas). The following study-specific characteristics were used (Web Table 1): population-based cohort study of unrelated individuals (vs. non-population-based/family study), population isolate (vs. nonendogamous source population), European country (vs. United States/Australia) with an obesity epidemic (vs. without an obesity epidemic, defined as <20% of the adult female population being obese in the most recent publicly available survey (40)), coded allele frequency (0–1), imputed SNP (vs. genotyped SNP), imputation quality score (0–1 for imputed SNP, 1 for genotyped SNP), age at report (mean and standard deviation), and birth year (mean and standard deviation). We also performed a meta-regression of age at menarche on birth year to account for differences in the precision of study-level average menarcheal ages.
Multiple testing
We performed a Bonferroni correction for all estimated SNP associations (n = 95) without consideration of the adiposity phenotypes, genetic signal dependence, or previous reports of significance in the literature (18). Therefore, we considered SNP associations not previously described as involving additional associations with menarche as significant if their 2-sided P value was below 0.0005. For the 11 adiposity SNPs previously reported to be associated with menarche, we considered directionally consistent effects as confirmatory of the results of Elks et al. (18), because of the overlap of approximately 87,800 stage 1 samples in this study and the previous report.
Alternative multiple testing methods were considered comprising 1 additional family-wise error rate method and 2 false discovery rate methods (Web Appendix).
Cumulative genetic effects
Genetic risk scores for overall (BMI) and central (waist circumference/waist:hip ratio) adiposity were calculated using comparable methods (Web Appendix) to investigate the cumulative effects of these variants on age at menarche (41, 42).
Pathway and protein-protein interaction analyses
As described in the Web Appendix, we used 3 methods to further investigate biological processes, based on observed patterns of association (25, 43–45).
RESULTS
Menarche secular trends and birth cohort effects
The mean age at menarche among the 38 studies, which collected data between 1970 and 2010, ranged from 12.4 years in the Women's Genome Health Study to 13.7 years in the CROATIA-Korcula and KORA studies (Web Table 1). The average age at which women reported having undergone menarche ranged between 12.7 years (Western Australian Pregnancy Cohort) and 69.6 years (Rotterdam Study I). No study measured age at menarche prospectively in the entire sample. However, through parent-assisted reports, investigators in the Western Australian Pregnancy Cohort assessed menarche prospectively in the majority of its sample (596 of 614 girls) at an average age of 12.7 years. Similarly, in the 1958 British Birth Cohort, researchers assessed menarche during adolescence at an average of 16.1 years—however, they did so retrospectively.
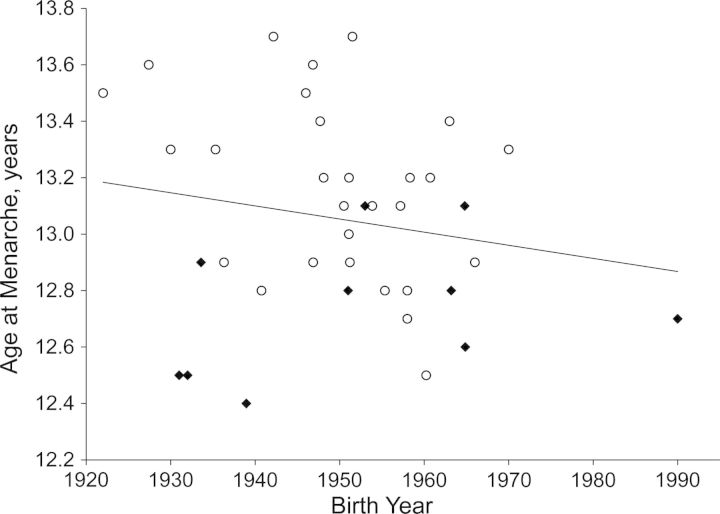
Secular trends at the population level exist for age at menarche (5). A similar trend is shown in the ReproGen Consortium (Figure 1), in which studies with more recent birth years also had lower average ages of menarche (a 14.0-day decline in average age at menarche per decade; estimated in a meta-regression not shown). The oldest birth cohort was the Nurses' Health Study, and the youngest was the Western Australian Pregnancy Cohort (Web Table 1).
Figure 1.
Secular trends in mean age at menarche among 38 studies of women of European descent in the ReproGen Consortium, by continent of origin (Europe (n = 28; white circles) or United States/Australia (n = 10; black diamonds)). The ReproGen Consortium studies were conducted in Europe, the United States, and Australia between 1970 and 2010 (see Web Table 1). The best-fit line includes all studies and corresponds to an 11.2-day decline in age at menarche per decade.
Genetic findings
Among the 60 adiposity loci not previously associated with age at menarche, 6 BMI loci contained at least 1 SNP that was significantly associated with age at menarche after adjustment for multiple testing (Table 1; Web Table 6), whereas 54 loci showed weaker evidence of association (P ≥ 0.0005; Web Figure 1B and Web Tables 4 and 5). No waist circumference or waist:hip ratio SNPs were significantly associated with age at menarche.
All 11 adiposity loci previously reported to be associated with age at menarche were also associated with menarche in this study (P < 0.05), of which 9 SNP associations were significant after Bonferroni correction (Web Table 3). Seven loci contained at least 1 additional statistically significant SNP (P < 7 × 10−5; Table 1) in high linkage disequilibrium with the index SNP.
Description of 6 BMI loci with novel menarche associations
Of the significant BMI variants (shown in Table 1 by chromosomal location and in Web Figure 2 by decreasing magnitude of effect), GPRC5B on chromosome 16 showed the strongest magnitude of effect on age at menarche (rs12444979-T; a 13.5-day increase (Web Figure 2A)). A signal at MAP2K5 on chromosome 15 had the next strongest effect (rs2241423-A; a 13.1-day increase (Web Figure 2B)). Both coded alleles confer an increase in age at menarche and a decrease in BMI (25).
rs1514175-A at TNNI3K, previously reported to increase adult BMI (25), was associated with menarche onset that was decreased by 12.2 days, but the locus showed evidence of between-study heterogeneity (P = 0.07). As compared with the fixed-effect estimate (Web Figure 2C), the random-effects estimate was larger and more imprecise (decreases in menarche per A allele were 12.2 days (95% confidence interval: –16.8, –7.7) and 14.6 days (95% confidence interval: –21.2, –8.0), respectively). In a meta-regression with study-specific characteristics, we estimated the variance between studies (τ2) to be zero across the 37 studies with information on rs1514175. Moreover, it indicated that population-based studies or those with genotyped data tended to have smaller effect estimates as compared with non-population-based designs or studies with imputed data (P ≤ 0.01). No other study characteristics appeared to significantly predict study-specific effect estimates (P ≥ 0.05).
The association of each rs713586-T allele at the RBJ locus was estimated to reflect an 11.7-day increase in menarche (Web Figure 2D) and a decrease in BMI (25). rs887912 at FANCL was associated with an 11.1-day decrease in menarche per T allele (Web Figure 2E) and has been reported to be associated with increased BMI (25). Lastly, rs10769908 (near STK33) was associated with an increased age at menarche of 9.6 days for each T allele (Web Figure 2F). Although the direction of effect of rs10769908 on BMI was not published (37), publicly available data from the GIANT Consortium indicated that each additional T allele was associated with decreased BMI (25). Moreover, rs10769908 was in high linkage disequilibrium (r2 = 0.97) with another SNP, rs4929949, which had a similar magnitude and precision (Web Table 5) as rs10769908 and has a documented negative effect on BMI (25). Each of these SNPs (or a proxy thereof) showed inverse associations with BMI and age at menarche.
Loci with no statistically significant evidence of association
Fifty-four adiposity loci containing 55 independent genetic signals and 8 dependent genetic signals (Web Tables 4 and 5, respectively) were not associated with age at menarche after the Bonferroni correction. Of these 54 loci, 12 BMI and waist circumference loci contained at least 1 nominally significant SNP association (P < 0.05; Web Tables 4 and 5)—10 loci having inverse associations and 2 loci having parallel effects on adiposity and menarche.
Previously reported menarche-adiposity loci
Of the 11 previously reported adiposity-menarche loci (18), all index SNPs were significant (P < 0.05), were directionally consistent, and had inverse or unknown (e.g., rs11084753) effects on adiposity and menarche (Web Table 3). Nine index SNPs also had evidence of association with age at menarche below a Bonferroni correction (P < 0.0005), and an additional index SNP at ETV5, rs7647305, was in tight linkage disequilibrium (r2 = 0.79) with another ETV5 SNP with strong evidence of association (rs9816226; Table 1).
In addition to the 11 index SNPs previously described (18), 7 loci (NEGR1, SEC16B, TMEM18, ETV5, BDNF, FTO, and KCTD15) contained 14 SNP associations (8 are presented in Table 1 (P < 7 × 10−5) and 6 in Web Table 5 (P < 0.03)). Notably, all additional BMI SNPs at BDNF were associated with similar magnitudes of effect (P < 1.7 × 10−5; Table 1 and Web Table 5), but linkage disequilibrium patterns indicated that rs10767664 and rs6265 (r2 = 0.77, P < 0.021) and rs925946 and rs7481311 (r2 = 0.57, P < 0.017) were in tighter linkage disequilibrium within themselves than with the other group (r2 < 0.15). Similar to previous work (18), 4 index SNPs at or near GNPDA2, TFAP2B, MSRA, and FAIM2 were confirmed to be associated with age at menarche (Web Table 3), and 4 BMI loci (MTCH2, MC4R, SH2B1, NRXN3) were not associated with age at menarche.
Multiple testing
Because of the highly nonuniform distribution of P values among the 95 candidate SNP associations (Web Figure 3, parts A and B), various approaches were contrasted. Holm P values were comparable to Bonferroni's (Web Tables 7 and 8). In contrast, the false discovery rate and positive false discovery rate, which yielded less conservative P values, increased the number of significant associations to 35 and 45, respectively.
Cumulative genetic effects
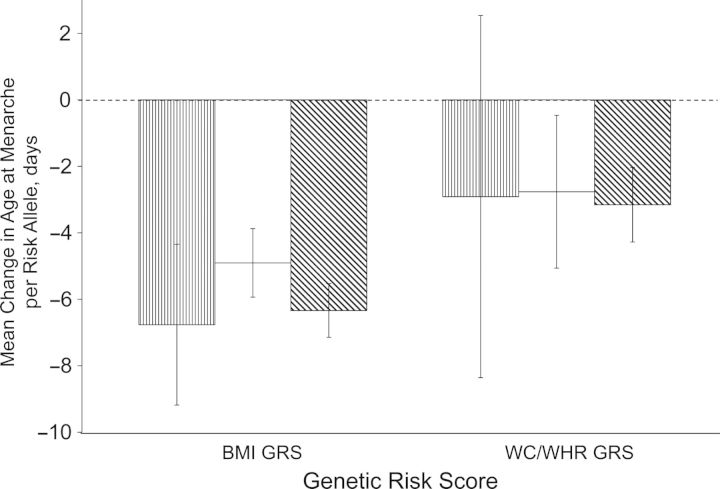
Both overall (BMI) and central (waist circumference/waist:hip ratio) genetic risk scores were normally distributed in the Atherosclerosis Risk in Communities and Women's Health Genome studies (Web Figure 4, parts A–F). The BMI genetic risk score was associated with a larger decline in menarche than the waist circumference/waist:hip ratio genetic risk score after adjustment for birth year, population stratification, and center (as appropriate) (Figure 2). Although the association between BMI genetic risk score and menarche was significant in each study and when we used SNP-level data to estimate the genetic risk score (P < 4 × 10−8), the waist circumference/waist:hip ratio genetic risk score was not significant in the Atherosclerosis Risk in Communities Study, probably because of imprecision (P = 0.3). Results did not change considerably in either of the 2 studies when no adjustments for population stratification were made (Web Figure 4, parts A and D).
Figure 2.
Effect estimates for the association between overall and central adiposity genetic risk scores (GRS) and age at menarche in the Atherosclerosis Risk in Communities (ARIC) Study (n = 4,775; bars with vertical thin stripes) (52), the Women's Genome Health Study (WGHS) (n = 22,863; white bars) (53), and 38 studies in the ReproGen Consortium, using SNP-level fixed-effect estimates from a sample of up to 92,105 women (bars with diagonal thick stripes) (for references, see Web Appendix). The ARIC and WGHS studies were conducted in the United States in 1987–1989 and 1992–1994, respectively; other ReproGen studies included in the SNP-level fixed-effect estimates were conducted in Europe, the United States, and Australia between 1970 and 2010 (see Web Table 1). Cumulative genetic risk was defined as the sum of the menarche risk alleles per individual. Genetic risk scores were calculated for overall (BMI) and central adiposity (WC/WHR) variants separately. Adjustments were made for birth year, population stratification, and center (appropriate for the ARIC Study only). Further details about the use of SNP-level fixed-effect estimates can be found in the Web Appendix. Bars, 95% confidence interval. BMI, body mass index; SNP, single-nucleotide polymorphism; WC, waist circumference; WHR, waist:hip ratio.
Pathway and protein-protein interaction analyses
MAGENTA pathway analyses (http://www.broadinstitute.org/mpg/magenta/) identified several genes in pathways near BMI loci that were associated with menarche in our study (Web Table 9), but genes near these loci did not significantly cluster in particular BMI pathways according to g:Profiler (http://biit.cs.ut.ee/gprofiler/) (Web Tables 10–13). Protein-protein interaction analysis revealed 6 direct interactions among the proteins coded by nearby genes of menarche-associated adiposity loci (P < 0.0005; Web Figure 5); none of these genes overlapped with any of the genes identified by the MAGENTA pathway analysis.
DISCUSSION
Recent studies of the genetics of menarche have agnostically focused on the discovery of novel loci (18–22). We implemented a candidate-SNP approach to test the hypothesis that adiposity loci are also associated with age at menarche. We aimed to characterize the SNP effects and evaluate potential heterogeneous effects that may be represented by loci influencing early growth and development and thus both adiposity and menarche.
We identified 6 adiposity loci (7 SNPs; Web Figure 1A) associated with age at menarche that have been described in a recent GWAS of adult BMI as part of the GIANT Consortium (25), which overlaps substantially with the cohorts of the ReproGen Consortium (approximately 82,000 samples). Eleven previously described adiposity-menarche loci (11 SNPs; Web Table 3) were confirmed (18), and an additional 14 SNPs were found to be associated with age at menarche at these loci (Web Figure 1C). Interestingly, all 17 loci exhibited inverse relationships with adiposity and menarche timing and were related to BMI (or waist circumference unadjusted for BMI). One such locus showed evidence of heterogeneity, which appeared to be primarily driven by data quality. Therefore, our study brings the known number of such loci to 17 (32 SNPs) and implies that there are extensive genetic influences on overall adiposity and pubertal development.
Because of the known relationship between central adiposity, elevated androgen exposure, and reduced ovulatory function, as in polycystic ovary syndrome (46), we had predicted that genetic variants associated with increased central adiposity would be associated with menarche timing. Genetic loci associated with the distribution of body fat in gluteal/femoral regions may possibly influence menarche, since fat in these regions is more readily mobilized for reproductive function than abdominal fat depots (47, 48). However, our study shows a lack of individual SNP associations of central adiposity loci (independent of BMI) with age at menarche but some evidence of a cumulative association using the genetic risk score (Figure 2). The 2 waist circumference loci (TFAP2B, MSRA) previously reported to be associated with age at menarche were both unadjusted for BMI and therefore may inadvertently represent BMI genetic signals (33). In fact, the TFAP2B locus has recently also been associated with adult BMI (25). These findings may suggest that the high energy cost of pregnancy and lactation (which becomes an issue as soon as a female reaches menarche and can conceive) drives a stronger shared genetic basis between overall adiposity and menarche than with fat distribution (49, 50).
Because of the candidate-SNP design of our study, we observed a skewed distribution towards lower P values. Therefore, we applied alternative multiple testing adjustments to our data, including the false discovery rate, which appeared to be the best alternative to Bonferroni correction. Using this method, we could expect 1–2 of our 35 positive findings to be false. Nonetheless, such less conservative multiple correction methods may be advantageous in prioritizing specific genetic variants for future studies of the genetics of early growth and development.
Searching known biological pathways and protein-protein interactions for loci influencing both adiposity and menarche was inconclusive, since there was no overlap between the outcomes of the various approaches. In part, this may be due to limited knowledge of the relevant biology. Temporal action earlier in the life course could also drive the association with menarche of some adiposity variants over others in the same pathways.
Our study had several strengths, including its large sample size, availability of detailed study characteristics, and evaluation of between-study heterogeneity. We used a conservative threshold for significance but also investigated the impact of alternative multiple testing adjustments.
However, our study was restricted to women of European descent, and thus the results may not be generalizable to pubertal timing in males or other ancestral populations. Our study may have been subject to the “winner's curse,” and the findings should be replicated in future studies. Even though we had greater than 80% power to detect 93 of 95 nominal associations (see Web Appendix and Web Figure 6, parts A and B), this number decreased substantially after Bonferroni correction—especially for variants of low frequency or weak effects. This study primarily relied on a retrospective measure of age at menarche. However, the reliability of self-reported menarche is generally considered to be good as a major milestone for girls (51). The resulting misclassification is probably independent of both genotype and childhood BMI. Given the effect of earlier menarche on all-cause mortality, there is potential for selection bias among older cohorts of women in this meta-analysis. Nonetheless, we observe that these common genetic variants have modest effects on age at menarche ( <20-day decline in menarche per risk allele) generally with little heterogeneity across studies. Moreover, at TNNI3K, age at report was not a significant predictor of effect heterogeneity. Lastly, we were unable to assess whether a variant's effects on menarche and adiposity resulted from pleiotropic or mediated effects, since data on adiposity measures were not available for most studies at the time of menarche.
Our study expands current knowledge on menarche-associated SNPs and may help to increase understanding of how adiposity variants play a role in growth and development during puberty. Its findings do not strongly support the existence of associations between menarche and central obesity, a risk factor for metabolic disease. Instead, they indicate that overall adiposity is a phenotype more closely related to normal growth and development. Because loci that increase adiposity and decrease age at menarche may have particularly strong effects on adult-onset outcomes related to cumulative estradiol and fatness exposures, this study and others like it may contribute to the identification of genetic variants that are important for outcomes such as breast cancer (8) or cardiovascular diseases.
Future researchers can investigate the biological mechanisms of these variants using observational and animal studies. In addition, longitudinal study designs may help to identify temporal relationships of pre- and postpubertal BMI with age of menarche as well as optimal intervention windows. Understanding the complex interface between childhood/adolescent adiposity, early menarche, and adult obesity may help with the investigation of growth and developmental trajectories, which influence disease throughout the life course.
Supplementary Material
ACKNOWLEGMENTS
Author affiliations: Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Kari E. North); Center for Statistical Genetics, University of Michigan, Ann Arbor, Michigan (John R. B. Perry); Centre for Population Health Sciences, College of Medicine and Veterinary Medicine, University of Edinburgh, Edinburgh, United Kingdom (Igor Rudan, James F. Wilson); Department of Public Health, School of Medicine, University of Split, Split, Croatia (Ozren Polasek); Department of Biological Psychology, Faculty of Psychology and Education, Vrije Universiteit Amsterdam, Amsterdam, the Netherlands (Jouke-Jan Hottenga, Gonneke Willemsen, Dorret I. Boomsma); Department of Biostatistics, School of Public Health, Boston University, Boston, Massachusetts (Kathryn L. Lunetta); Department of Biostatistics, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Ran Tao); Department of Biotechnology, Institute of Molecular and Cell Biology, University of Tartu, Tartu, Estonia (Andres Metspalu, Tõnu Esko, Evelin Mihailov); Department of Clinical Genetics, Erasmus Medical Center, Rotterdam, the Netherlands (Ben Oostra); Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark (Bjarke Feenstra, Heather A. Boyd); Department of Epidemiology, Erasmus Medical Center, Rotterdam, the Netherlands (Cornelia M. van Duijn, Linda Broer, Albert Hofman, André G. Uitterlinden); Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Lindsay Fernández-Rhodes, Charles Poole, Kari E. North, Nora Franceschini); Department of Genetics, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Kari E. North); Department of Genetics, Harvard Medical School, Boston, Massachusetts (Joel N. Hirschhorn); Department of Internal Medicine, Erasmus Medical Center, Rotterdam, the Netherlands (M. Carola Zillikens, Lisette Stolk, Fernando Rivadeneira, Jenny A. Visser, André G. Uitterlinden); Department of Medical and Molecular Genetics, School of Medicine, Indiana University, Indianapolis, Indiana (Daniel L. Koller, Dongbing Lai); Department of Medical Genetics, Faculty of Biology and Medicine, University of Lausanne, Lausanne, Switzerland (Zoltán Kutalik); Department of Medicine, Division of Endocrinology, Diabetes and Nutrition, School of Medicine, University of Maryland at Baltimore, Baltimore, Maryland (Laura M. Yerges-Armstrong, Patrick F. McArdle); Department of Medicine, Internal Medicine Unit, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland (Peter Vollenweider); Department of Nutrition and Epidemiology, Harvard School of Public Health, Boston, Massachusetts (David J. Hunter, Peter Kraft, Frank B. Hu); Department of Obstetrics and Gynaecology, Campus Grosshadern, Ludwig-Maximilians-University, Munich, Germany (Doris Stöckl); Department of Psychiatry, School of Medicine, Washington University, St. Louis, Missouri (Laura J. Bierut, Peng Lin, Sherri L. Fisher); Department of Public Health, School of Medicine, Indiana University, Indianapolis, Indiana (Chunyan He); Department of Twin Research and Genetic Epidemiology, Division of Genetics and Molecular Medicine, School of Medicine, King's College London, London, United Kingdom (John R. B. Perry, Tim D. Spector, Massimo Mangino, Guangju Zhai); Department of Biomedical Sciences, Faculty of Medicine and Surgery, University of Sassari, Sassari, Italy (Eleonora Porcu); Discipline of Genetics, Faculty of Medicine, Memorial University of Newfoundland, St. John's, Canada (Guangju Zhai); Division of Cell Biology, San Raffaele Scientific Institute, Milan, Italy (Tanguy Corre, Daniela Toniolo); Division of Epidemiology and Community Health, School of Public Health, University of Minnesota, Minneapolis, Minnesota (Ellen W. Demerath, Jill G. Dreyfus); Division of Genetics, San Raffaele Scientific Institute, Milan, Italy (Tanguy Corre, Daniela Toniolo); Division of Population Health Sciences and Education, St. George's Hospital Medical School, University of London, London, United Kingdom (David P. Strachan); Division of Preventive Medicine, Brigham and Women's Hospital, Boston, Massachusetts (Paul M. Ridker, Daniel I. Chasman, Julie E. Buring, Lynda M. Rose); Estonian Genome Center, University of Tartu, Tartu, Estonia (Andres Metspalu, Tõnu Esko, Evelin Mihailov); Gen Info Ltd., Zagreb, Croatia (Ozren Polasek); Genetic Epidemiology, Molecular Epidemiology and Neurogenetics Laboratories, Queensland Institute of Medical Research, Brisbane, Australia (Enda M. Byrne, Nicholas G. Martin, Grant W. Montgomery); Genetics of Complex Traits Research Group, Peninsula Medical School, University of Exeter, Exeter, United Kingdom (Anna Murray, John R. B. Perry); Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts (Paul M. Ridker, Daniel I. Chasman, Julie E. Buring); Icelandic Heart Association, Reykjavik, Iceland (Albert V. Smith, Vilmundur Gudnason); Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (Doris Stöckl); Institute of Genetic Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (Christian Gieger, Eva Albrecht); Institute for Molecular Medicine Finland, University of Helsinki, Helsinki, Finland (Elisabeth Widen, Diana L. Cousminer); Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche, Cagliari, Italy (Laura Crisponi, Eleonora Porcu); Laboratory of Epidemiology, Demography, and Biometry, Intramural Research Program, National Institute on Aging, Bethesda, Maryland (Tamara B. Harris, Lenore Launer); MRC Epidemiology Unit, University of Cambridge, Cambridge, United Kingdom (Cathy E. Elks, Ken K. Ong); MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, Western General Hospital, Edinburgh, United Kingdom (Caroline Hayward); Netherlands Consortium for Healthy Aging, Netherlands Genomics Initiative, Leiden, the Netherlands (M. Carola Zillikens, Cornelia M. van Duijn, Linda Broer, Lisette Stolk, André G. Uitterlinden, Fernando Rivadeneira); Netherlands Twin Register, Amsterdam, the Netherlands (Jouke-Jan Hottenga, Gonneke Willemsen, Dorret I. Boomsma); Program in Personalized and Genomic Medicine, School of Medicine, University of Maryland at Baltimore, Baltimore, Maryland (Laura M. Yerges-Armstrong, Patrick F. McArdle); Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany (Harald Grallert); School of Women's and Infants’ Health, University of Western Australia, Perth, Australia (Nicole M. Warrington, Craig E. Pennell); Section of General Internal Medicine, Department of Medicine, School of Medicine, Boston University, Boston, Massachusetts (Joanne M. Murabito); Swiss Institute of Bioinformatics, Lausanne, Switzerland (Zoltán Kutalik); Complex Traits Genomics Group, Queensland Brain Institute, University of Queensland, Brisbane, Australia (Enda M. Byrne); and Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom (John R. B. Perry).
All of the authors contributed equally to this work.
This work was supported by the US National Heart, Lung, and Blood Institute (grants 5T32HL007055 to L.F.R., T32HL007779 to J.G.D., and K02 DA021237 to L.J.B.) and the Wellcome Trust (Sir Henry Wellcome Fellowship 092447/Z/10/Z to J.R.B.P.).
We acknowledge the contributions made to this work by the participating studies and investigators of the ReproGen Consortium.
P.V. received an unrestricted grant from GlaxoSmithKline to build the CoLaus Study, and L.J.B. is listed as an inventor on Issued United States Patent 8,080,371, “Markers for Addiction,” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
REFERENCES
- 1.Popkin BM. Recent dynamics suggest selected countries catching up to US obesity. Am J Clin Nutr. 2009;91(1):284S–288S. doi: 10.3945/ajcn.2009.28473C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377(9765):557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jasik CB, Lustig RH. Adolescent obesity and puberty: the “perfect storm.”. Ann N Y Acad Sci. 2008;1135:265–279. doi: 10.1196/annals.1429.009. [DOI] [PubMed] [Google Scholar]
- 4.Pietrobelli A, Boner AL, Tato L. Adipose tissue and metabolic effects: new insight into measurements. Int J Obes (Lond) 2005;29(suppl 2):S97–S100. doi: 10.1038/sj.ijo.0803079. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed ML, Ong KK, Dunger DB. Childhood obesity and the timing of puberty. Trends Endocrinol Metab. 2009;20(5):237–242. doi: 10.1016/j.tem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Stockl D, Doring A, Peters A, et al. Age at menarche is associated with prediabetes and diabetes in women (aged 32–81 years) from the general population: the KORA F4 Study. Diabetologia. 2012;55(3):681–688. doi: 10.1007/s00125-011-2410-3. [DOI] [PubMed] [Google Scholar]
- 7.He C, Zhang C, Hunter DJ, et al. Age at menarche and risk of type 2 diabetes: results from 2 large prospective cohort studies. Am J Epidemiol. 2010;171(3):334–344. doi: 10.1093/aje/kwp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, Chasman DI, Dreyfus J, et al. Reproductive aging-associated common genetic variants and the risk of breast cancer. Breast Cancer Res. 2012;14(2):R54. doi: 10.1186/bcr3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velie EM, Nechuta S, Osuch JR. Lifetime reproductive and anthropometric risk factors for breast cancer in postmenopausal women. Breast Dis. 2005-2006;24(1):17–35. doi: 10.3233/bd-2006-24103. [DOI] [PubMed] [Google Scholar]
- 10.Michaud PA, Suris JC, Deppen A. Gender-related psychological and behavioural correlates of pubertal timing in a national sample of Swiss adolescents. Mol Cell Endocrinol. 2006;254-255:172–178. doi: 10.1016/j.mce.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen BK, Heuch I, Kvale G. Association of low age at menarche with increased all-cause mortality: a 37-year follow-up of 61,319 Norwegian women. Am J Epidemiol. 2007;166(12):1431–1437. doi: 10.1093/aje/kwm237. [DOI] [PubMed] [Google Scholar]
- 12.Freedman DS, Khan LK, Serdula MK, et al. Relation of age at menarche to race, time period, and anthropometric dimensions: the Bogalusa Heart Study. Pediatrics. 2002;110(4):e43. doi: 10.1542/peds.110.4.e43. [DOI] [PubMed] [Google Scholar]
- 13.Mumby HS, Elks CE, Li S, et al. Mendelian randomisation study of childhood BMI and early menarche. J Obes. 2011;2011:180729. doi: 10.1155/2011/180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong KK, Northstone K, Wells JC, et al. Earlier mother's age at menarche predicts rapid infancy growth and childhood obesity. PLoS Med. 2007;4(4):e132. doi: 10.1371/journal.pmed.0040132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaprio J, Rimpela A, Winter T, et al. Common genetic influences on BMI and age at menarche. Hum Biol. 1995;67(5):739–753. [PubMed] [Google Scholar]
- 16.Wang W, Zhao LJ, Liu YZ, et al. Genetic and environmental correlations between obesity phenotypes and age at menarche. Int J Obes (Lond) 2006;30(11):1595–1600. doi: 10.1038/sj.ijo.0803322. [DOI] [PubMed] [Google Scholar]
- 17.Frayling TM, Ong K. Piecing together the FTO jigsaw. Genome Biol. 2011;12(2):104. doi: 10.1186/gb-2011-12-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C, Kraft P, Chen C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41(6):724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YZ, Guo YF, Wang L, et al. Genome-wide association analyses identify SPOCK as a key novel gene underlying age at menarche. PLoS Genet. 2009;5(3):e1000420. doi: 10.1371/journal.pgen.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry JR, Stolk L, Franceschini N, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41(6):648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulem P, Gudbjartsson DF, Rafnar T, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41(6):734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 23.den Hoed M, Ekelund U, Brage S, et al. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59(11):2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Bradfield JP, Li M, et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring) 2009;17(12):2254–2257. doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Divission of Genomic Medicine, National Human Genome Research Institute. A Catalog of Published Genome-Wide Association Studies. Bethesda, MD: National Human Genome Research Institute; 2011. http://www.genome.gov/gwastudies/#1. (Accessed August 11, 2011) [Google Scholar]
- 28.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 29.Fox CS, Heard-Costa N, Cupples LA, et al. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8(suppl 1):S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heard-Costa NL, Zillikens MC, Monda KL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5(6):e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson A, Marroni F, Hayward C, et al. Linkage and genome-wide association analysis of obesity-related phenotypes: association of weight with the MGAT1 gene. Obesity (Silver Spring) 2010;18(4):803–808. doi: 10.1038/oby.2009.359. [DOI] [PubMed] [Google Scholar]
- 33.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JZ, Medland SE, Wright MJ, et al. Genome-wide association study of height and body mass index in Australian twin families. Twin Res Hum Genet. 2010;13(2):179–193. doi: 10.1375/twin.13.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 37.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Global Database on Body Mass Index: An Interactive Surveillance Tool for Monitoring Nutrition Transition. Geneva, Switzerland: World Health Organization; 2012. http://apps.who.int/bmi/index.jsp. (Accessed June 8, 2012) [Google Scholar]
- 41.Dastani Z, Hivert MF, Timpson N, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson T Comprehensive R Archive Network. gtx: Genetics ToolboX. Vienna, Australia: Comprehensive R Archive Network; 2012. http://cran.r-project.org/web/packages/gtx/index.html. (Accessed November 5, 2012) [Google Scholar]
- 43.Rossin EJ, Lage K, Raychaudhuri S, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reimand J, Arak T, Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39(Web Server issue):W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimand J, Kull M, Peterson H, et al. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goudas VT, Dumesic DA. Polycystic ovary syndrome. Endocrinol Metab Clin North Am. 1997;26(4):893–912. doi: 10.1016/s0889-8529(05)70286-3. [DOI] [PubMed] [Google Scholar]
- 47.Rebuffe-Scrive M, Lonnroth P, Marin P, et al. Regional adipose tissue metabolism in men and postmenopausal women. Int J Obes. 1987;11(4):347–355. [PubMed] [Google Scholar]
- 48.Power ML, Schulkin J. The Evolution of Obesity. Baltimore, MD: Johns Hopkins University Press; 2009. [Google Scholar]
- 49.Frisch RE. Menarche and fatness: reexamination of the critical body composition hypothesis. Science. 1978;200(4349):1509–1513. doi: 10.1126/science.200.4349.1509. [DOI] [PubMed] [Google Scholar]
- 50.Frisch RE, Revelle R. Height and weight at menarche and a hypothesis of critical body weights and adolescent events. Science. 1970;169(943):397–399. doi: 10.1126/science.169.3943.397. [DOI] [PubMed] [Google Scholar]
- 51.Must A, Phillips SM, Naumova EN, et al. Recall of early menstrual history and menarcheal body size: after 30 years, how well do women remember? Am J Epidemiol. 2002;155(7):672–679. doi: 10.1093/aje/155.7.672. [DOI] [PubMed] [Google Scholar]
- 52.The ARIC Investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 53.Ridker PM, Chasman DI, Zee RY, et al. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy American women. Clin Chem. 2008;54(2):249–255. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.